Submitted:
23 May 2023
Posted:
23 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Copper uses in hospital settings
3. Copper bioinorganic chemistry
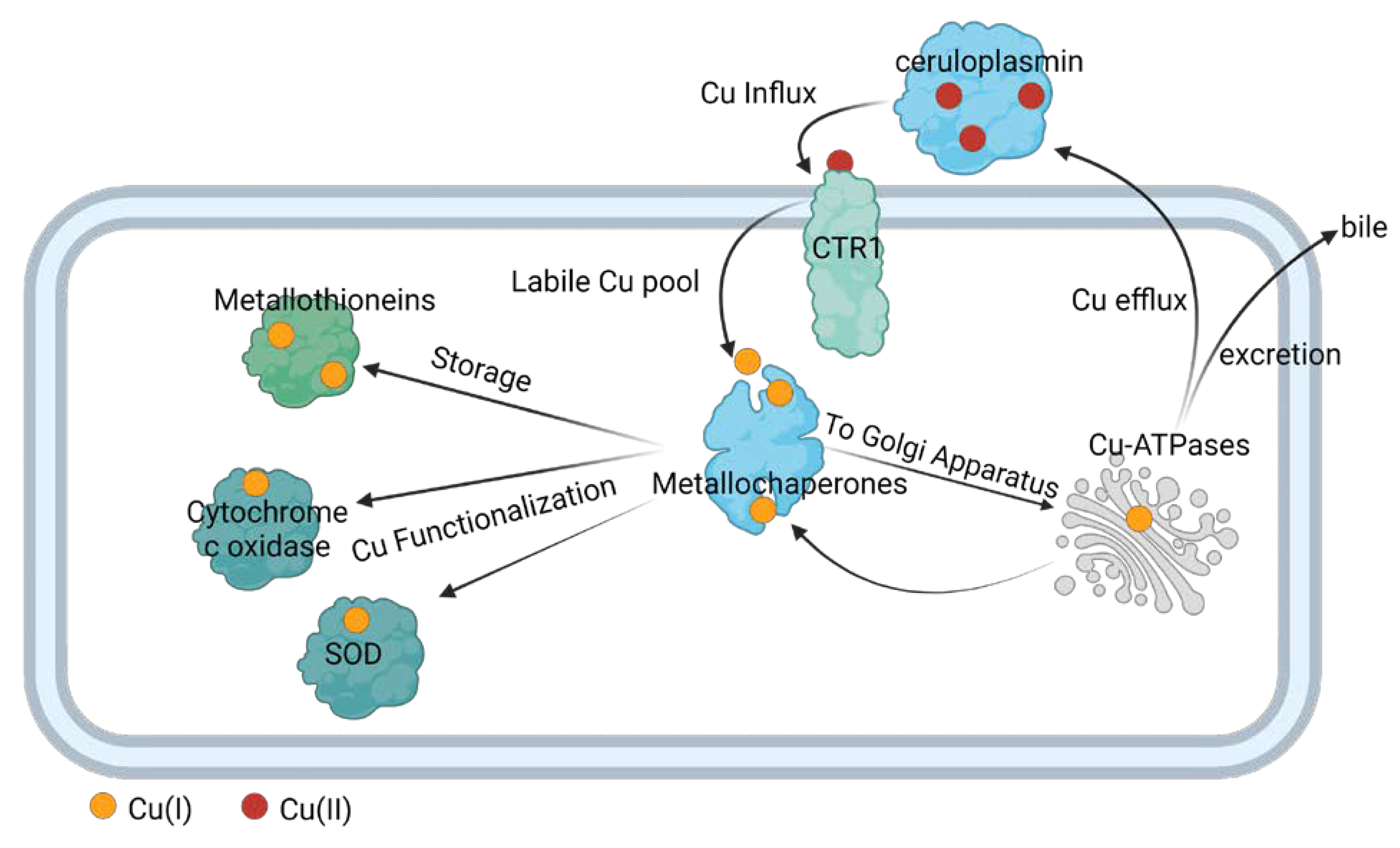
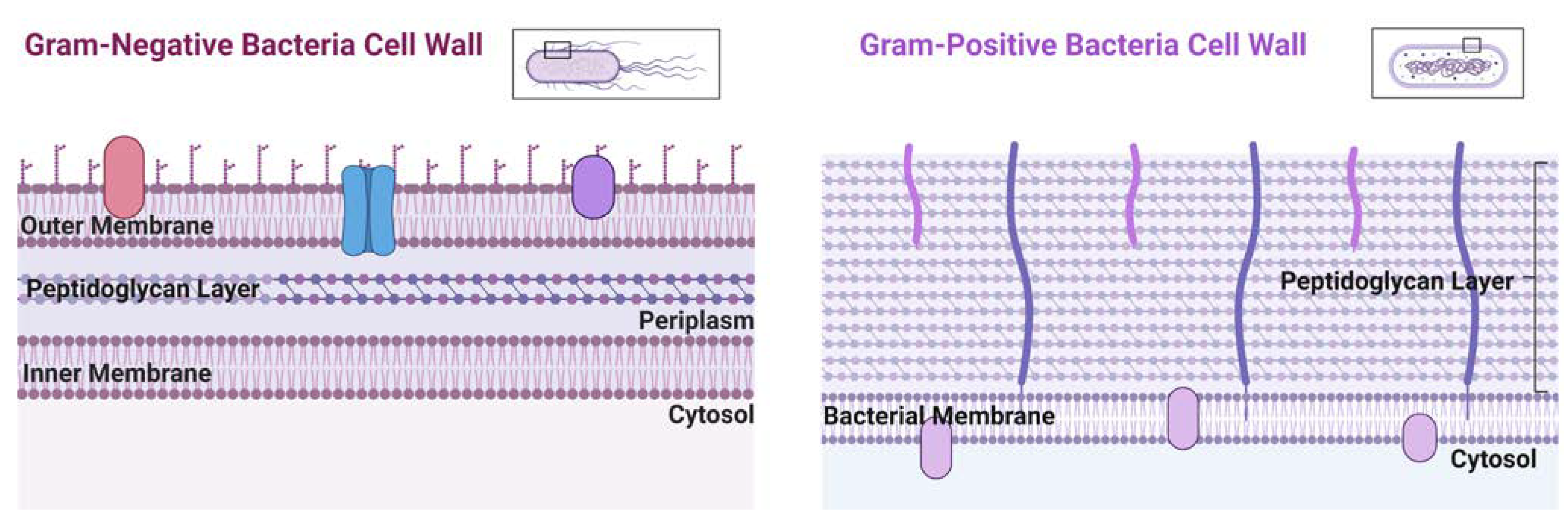
4. Copper transport in eukaryote and prokaryote cells
5. Copper utility against bacterial infections
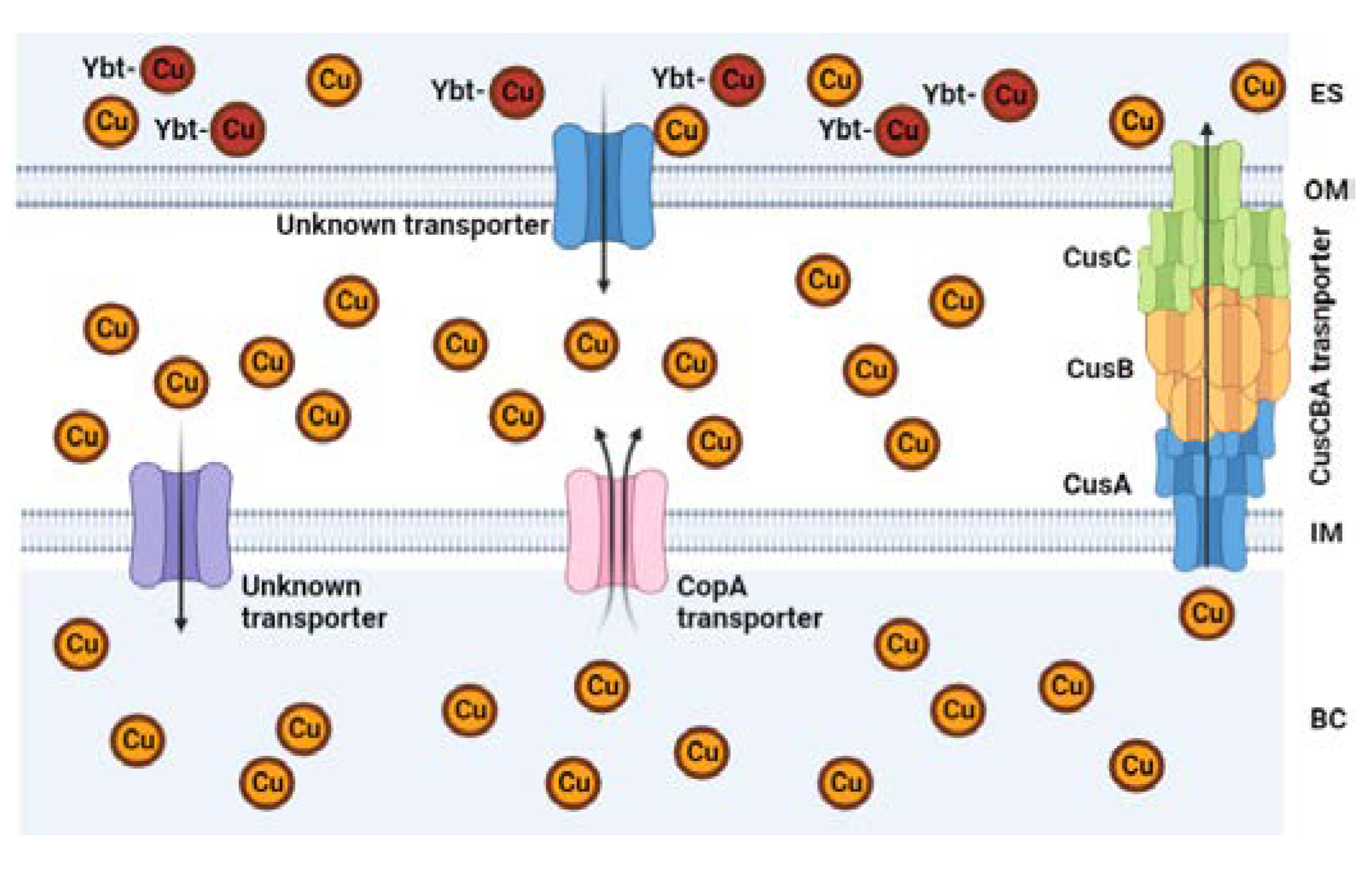
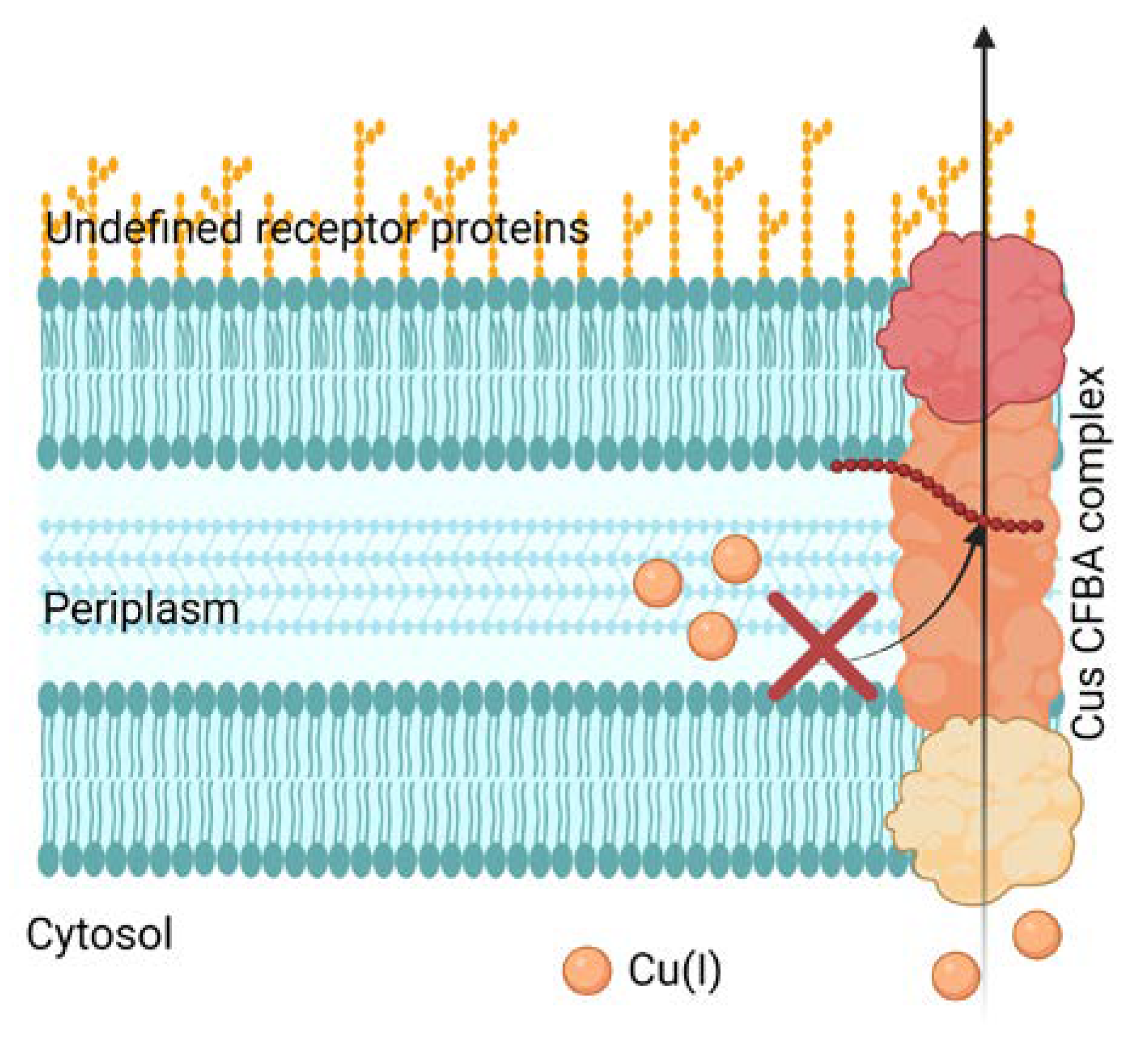
5.1. Inhibition of Cu efflux pathways
5.2. Targeting bacterial Cu storage may restore Cu sensitivity in bacteria
5.3. Cu toxicity by displacing iron and iron-sulfur clusters
6. Exploring Cu(II) chelation for potential drug designs
6.1. Cu(II) coordination complexes
6.2. Cu dependent inhibitors as potential synergistic treatment with traditional antibiotics
6.3. Peptide based Cu(II) chelators
6.4. Antibacterial Cu(II) compound isolated from bacteria
6.5. Cu(II) prochelators as potential multimodal antibiotic drugs
7. Copper nanoparticles against pathogens
7.1. Cu NP surface modification
7.2. Synergizing Cu NPs with antibiotics
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C.W. Editorial: Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Front. Microbiol. 2019, 10, 1933. [Google Scholar] [CrossRef]
- Murray, C.; Ikuta, K.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. , et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022. Centers for Disease, C., Prevention, National Center for, E., Zoonotic Infectious, D., Division of Healthcare Quality, P., Eds. Hyattsville, MD, 2022. [CrossRef]
- Millanao, A.R.; Mora, A.Y.; Villagra, N.A.; Bucarey, S.A.; Hidalgo, A.A. Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents. Molecules 2021, 26. [Google Scholar] [CrossRef]
- Sarkar, P.; Yarlagadda, V.; Ghosh, C.; Haldar, J. A review on cell wall synthesis inhibitors with an emphasis on glycopeptide antibiotics. Med. Chem. Comm. 2017, 8, 516–533. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Sköld, O. Sulfonamide resistance: mechanisms and trends. Drug. Resist. Updat. 2000, 3, 155–160. [Google Scholar] [CrossRef]
- Frei, A.; Zuegg, J.; Elliott, A.G.; Baker, M.; Braese, S.; Brown, C.; Chen, F.; C, G.D.; Dujardin, G.; Jung, N. , et al. Metal complexes as a promising source for new antibiotics. Chem. Sci. 2020, 11, 2627–2639. [Google Scholar] [CrossRef]
- Arendsen, L.P.; Thakar, R.; Sultan, A.H. The Use of Copper as an Antimicrobial Agent in Health Care, Including Obstetrics and Gynecology. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Arendsen, L.P.; Thakar, R.; Bassett, P.; Sultan, A.H. A double blind randomized controlled trial using copper impregnated maternity sanitary towels to reduce perineal wound infection. Midwifery 2021, 92, 102858. [Google Scholar] [CrossRef]
- Hunsaker, E.W.; Franz, K.J. Emerging Opportunities To Manipulate Metal Trafficking for Therapeutic Benefit. Inorg. Chem. 2019, 58, 13528–13545. [Google Scholar] [CrossRef]
- Institute of Medicine Standing Committee on the Scientific Evaluation of Dietary Reference, I. ; its Panel on Folate, O.B.V.; Choline. The National Academies Collection: Reports funded by National Institutes of Health. In Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B(6), Folate, Vitamin B(12), Pantothenic Acid, Biotin, and Choline, National Academies Press (US)Copyright © 1998, National Academy of Sciences.: Washington (DC), 1998. [Google Scholar]
- Montero, D.A.; Arellano, C.; Pardo, M.; Vera, R.; Gálvez, R.; Cifuentes, M.; Berasain, M.A.; Gómez, M.; Ramírez, C.; Vidal, R.M. Antimicrobial properties of a novel copper-based composite coating with potential for use in healthcare facilities. Antimicrob. Resis. Infect. Control 2019, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.G.; Von Dessauer, B.; Benavente, C.; Benadof, D.; Cifuentes, P.; Elgueta, A.; Duran, C.; Navarrete, M.S. Copper surfaces are associated with significantly lower concentrations of bacteria on selected surfaces within a pediatric intensive care unit. Am. J. Infect. Control 2016, 44, 203–209. [Google Scholar] [CrossRef]
- Butler, J.P. Effect of copper-impregnated composite bed linens and patient gowns on healthcare-associated infection rates in six hospitals. J. Hosp. Infect. 2018, 100, e130–e134. [Google Scholar] [CrossRef]
- Monegro, A.F.; Muppidi, V.; Regunath, H. Hospital Acquired Infections. In StatPearls, StatPearls Publishing: Treasure Island (FL), 2022.
- CDC. Could you or your loved one have C. diff? Availabe online: https://www.cdc.gov/cdiff/what-is.html (accessed on April 26, 2023).
- Schmidt, M.G.; Attaway, H.H.; Fairey, S.E.; Howard, J.; Mohr, D.; Craig, S.; Schaffner, D.W. Self-Disinfecting Copper Beds Sustain Terminal Cleaning and Disinfection Effects throughout Patient Care. Appl. Environ. Microbiol. 2019, 86, e01886–e01819. [Google Scholar] [CrossRef] [PubMed]
- Hinsa-Leasure, S.M.; Nartey, Q.; Vaverka, J.; Schmidt, M.G. Copper alloy surfaces sustain terminal cleaning levels in a rural hospital. Am. J. Infect. Control 2016, 44, e195–e203. [Google Scholar] [CrossRef]
- Parma, M.; Elli, E.; Terruzzi, E.; Fedele, M.; Doni, E.; Stasia, A.; Pogliani, E.M.; Pioltelli, P. Low dose of Deferasirox treatment in patients actually free of transfusion who present iron overload after bone marrow transplantation. Bone Marrow Transplantation 2015, 50, S421–S421. [Google Scholar]
- Green, J.J. US EPA, Pesticide product label, Antimicrobial copper alloys- Group III. US EPA, Office of Chemical Safety and Pollution Prevention: 2014.
- Bryce, E.A.; Velapatino, B.; Khorami, H.A.; Donnelly-Pierce, T.; Wong, T.; Dixon, R.; Asselin, E. In vitro evaluation of antimicrobial efficacy and durability of three copper surfaces used in healthcare. Biointerphases 2020, 15, 011005. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Douglas, S.P.; Wu, G.; MacRobert, A.J.; Allan, E.; Knapp, C.E.; Parkin, I.P. A rugged, self-sterilizing antimicrobial copper coating on ultra-high molecular weight polyethylene: a preliminary study on the feasibility of an antimicrobial prosthetic joint material. J. Mater. Chem. B 2019, 7, 3310–3318. [Google Scholar] [CrossRef]
- Rowe, L.A.; Degtyareva, N.; Doetsch, P.W. DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Free Radic. Biol. Med. 2008, 45, 1167–1177. [Google Scholar] [CrossRef]
- Robinett, N.G.; Peterson, R.L.; Culotta, V.C. Eukaryotic copper-only superoxide dismutases (SODs): A new class of SOD enzymes and SOD-like protein domains. J. Biol. Chem. 2018, 293, 4636–4643. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.D. Copper as a Cofactor and Regulator of Copper,Zinc Superoxide Dismutase. J. Nutr. 1992, 122, 636–640. [Google Scholar] [CrossRef]
- Barber-Zucker, S.; Shaanan, B.; Zarivach, R. Transition metal binding selectivity in proteins and its correlation with the phylogenomic classification of the cation diffusion facilitator protein family. Sci. Rep. 2017, 7, 16381–16381. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Gray, H.B.; Stiefel, E.I.; Selverstone-Valentine, J. Biological Inorganic Chemistry Structure and Reactivity; University Science Books: Herndon, 2007; pp. 1–3. [Google Scholar]
- Fu, Y.; Chang, F.-M.J.; Giedroc, D.P. Copper Transport and Trafficking at the Host–Bacterial Pathogen Interface. Acc. Chem. Res. 2014, 47, 3605–3613. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.N.; Xing, G.; Miller, C.J.; Waite, T.D. Fenton-like copper redox chemistry revisited: Hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production. J. Catal. 2013, 301, 54–64. [Google Scholar] [CrossRef]
- Dabrowiak, J.C. , 2nd ed.John Wiley & Sons, Ltd: Hoboken, NJ, 2017.
- Soldatović, T. Correlation between HSAB Principle and Substitution Reactions in Bioinorganic Reactions. In Photophysics, Photochemical and Substitution Reactions - Recent Advances, IntechOpen: 2021; [CrossRef]
- MacPherson, I.S.; Murphy, M.E. Type-2 copper-containing enzymes. Cell. Mol. Life Sci. 2007, 64, 2887–2899. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, F.; McDougall, C.; Degnan, B.M. Origin, evolution and classification of type-3 copper proteins: lineage-specific gene expansions and losses across the Metazoa. BMC Evol. Biol. 2013, 13, 96. [Google Scholar] [CrossRef]
- Ge, E.J.; Bush, A.I.; Casini, A.; Cobine, P.A.; Cross, J.R.; DeNicola, G.M.; Dou, Q.P.; Franz, K.J.; Gohil, V.M.; Gupta, S. , et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat. Rev. Cancer 2022, 22, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Rubino, J.T.; Franz, K.J. Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J Inorg. Biochem. 2012, 107, 129–143. [Google Scholar] [CrossRef]
- Hellman, N.E.; Gitlin, J.D. Ceruloplasmin Metabolism and Function. Annu. Rev. Nutr. 2002, 22, 439–458. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.Y.-S.; Posimo, J.M.; Lee, S.; Tsang, T.; Davis, J.M.; Brady, D.C.; Chang, C.J. Activity-based ratiometric FRET probe reveals oncogene-driven changes in labile copper pools induced by altered glutathione metabolism. Proceedings of the National Academy of Sciences 2019, 116, 18285–18294. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Kozyreva, T.; Zovo, K.; Palumaa, P. Affinity gradients drive copper to cellular destinations. Nature 2010, 465, 645–648. [Google Scholar] [CrossRef]
- Shen, C.; New, E.J. What has fluorescent sensing told us about copper and brain malfunction? Metallomics : integrated biometal science 2015, 7, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, C.M.; Lee, S.; Chang, C.J. Analytical Methods for Imaging Metals in Biology: From Transition Metal Metabolism to Transition Metal Signaling. Analytical Chemistry 2017, 89, 22–41. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Chang, C.J. Copper signaling in the brain and beyond. The Journal of biological chemistry 2018, 293, 4628–4635. [Google Scholar] [CrossRef] [PubMed]
- Pezacki, A.T.; Matier, C.D.; Gu, X.; Kummelstedt, E.; Bond, S.E.; Torrente, L.; Jordan-Sciutto, K.L.; DeNicola, G.M.; Su, T.A.; Brady, D.C. , et al. Oxidation state-specific fluorescent copper sensors reveal oncogene-driven redox changes that regulate labile copper(II) pools. Proceedings of the National Academy of Sciences of the United States of America 2022, 119, e2202736119. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, Jack H. ; Maryon, Edward B. How Mammalian Cells Acquire Copper: An Essential but Potentially Toxic Metal. Biophys J 2016, 110, 7–13. [Google Scholar] [CrossRef]
- Pang, W.L.; Kaur, A.; Ratushny, A.V.; Cvetkovic, A.; Kumar, S.; Pan, M.; Arkin, A.P.; Aitchison, J.D.; Adams, M.W.W.; Baliga, N.S. Metallochaperones Regulate Intracellular Copper Levels. PLOS Comput Biol 2013, 9, e1002880. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.J.; Winge, D.R. Copper metallochaperones. Annu Rev Biochem 2010, 79, 537–562. [Google Scholar] [CrossRef]
- Kaplan, J.H.; Lutsenko, S. Copper Transport in Mammalian Cells: Special Care for a Metal with Special Needs. J. Bio. Chem. 2009, 284, 25461–25465. [Google Scholar] [CrossRef]
- Andrei, A.; Öztürk, Y.; Khalfaoui-Hassani, B.; Rauch, J.; Marckmann, D.; Trasnea, P.-I.; Daldal, F.; Koch, H.-G. Cu Homeostasis in Bacteria: The Ins and Outs. Membranes 2020, 10, 242. [Google Scholar] [CrossRef]
- Jiang, W.; Saxena, A.; Song, B.; Ward, B.B.; Beveridge, T.J.; Myneni, S.C.B. Elucidation of Functional Groups on Gram-Positive and Gram-Negative Bacterial Surfaces Using Infrared Spectroscopy. Langmuir 2004, 20, 11433–11442. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, E.R.; Martin, R.J. Asthma and Atypical Bacterial Infection. Chest 2007, 132, 1962–1966. [Google Scholar] [CrossRef]
- Ladomersky, E.; Petris, M.J. Copper tolerance and virulence in bacteria. Metallomics : integrated biometal science 2015, 7, 957–964. [Google Scholar] [CrossRef]
- Argüello, J.M.; Raimunda, D.; Padilla-Benavides, T. Mechanisms of copper homeostasis in bacteria. Front. Cell Infect. Microbiol. 2013, 3. [Google Scholar] [CrossRef]
- Ward, S.K.; Abomoelak, B.; Hoye, E.A.; Steinberg, H.; Talaat, A.M. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2010, 77, 1096–1110. [Google Scholar] [CrossRef] [PubMed]
- Wolschendorf, F.; Ackart, D.; Shrestha, T.B.; Hascall-Dove, L.; Nolan, S.; Lamichhane, G.; Wang, Y.; Bossmann, S.H.; Basaraba, R.J.; Niederweis, M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. PNAS 2011, 108, 1621–1626. [Google Scholar] [CrossRef]
- Dennison, C.; David, S.; Lee, J. Bacterial copper storage proteins. J. Biol. Chem. 2018, 293, 4616–4627. [Google Scholar] [CrossRef] [PubMed]
- Calvo, J.; Jung, H.; Meloni, G. Copper metallothioneins. IUBMB Life 2017, 69, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J. Memory in the innate and adaptive immune systems. Microbes Infect. 2004, 6, 1410–1417. [Google Scholar] [CrossRef]
- Healy, C.; Munoz-Wolf, N.; Strydom, J.; Faherty, L.; Williams, N.C.; Kenny, S.; Donnelly, S.C.; Cloonan, S.M. Nutritional immunity: the impact of metals on lung immune cells and the airway microbiome during chronic respiratory disease. In Respiratory Research, BioMed Central Ltd: 2021; Vol. 22.
- Hood, M.I.; Skaar, E.P. Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef]
- He, R.; Zuo, Y.; Zhao, L.; Ma, Y.; Yan, Q.; Huang, L. Copper stress by nutritional immunity activates the CusS-CusR two-component system that contributes to Vibrio alginolyticus anti-host response but affects virulence-related properties. Aquaculture 2021, 532, 736012. [Google Scholar] [CrossRef]
- Yadav, R.; Noinaj, N.; Ostan, N.; Moraes, T.; Stoudenmire, J.; Maurakis, S.; Cornelissen, C.N. Structural Basis for Evasion of Nutritional Immunity by the Pathogenic Neisseriae. Front. Microbiol. 2019, 10, 2981. [Google Scholar] [CrossRef] [PubMed]
- Cassat, J.E.; Skaar, E.P. Metal ion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Semin. Immunopathol. 2012, 34, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Benhalima, L.; Amri, S.; Bensouilah, M.; Ouzrout, R. Antibacterial effect of copper sulfate against multi-drug resistant nosocomial pathogens isolated from clinical samples: Effect of copper sulfate on clinical isolates. Pak J Med Sci 2019, 35. [Google Scholar] [CrossRef]
- Febré, N.; Silva, V.; Báez, A.; Palza, H.; Delgado, K.; Aburto, I.; Silva, V. Comportamiento antibacteriano de partículas de cobre frente a microorganismos obtenidos de úlceras crónicas infectadas y su relación con la resistencia a antimicrobianos de uso común. Rev. Med. Chile 2016, 144, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Meir, A.; Lepechkin-Zilbermintz, V.; Kahremany, S.; Schwerdtfeger, F.; Gevorkyan-Airapetov, L.; Munder, A.; Viskind, O.; Gruzman, A.; Ruthstein, S. Inhibiting the copper efflux system in microbes as a novel approach for developing antibiotics. PLOS ONE 2019, 14, e0227070. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Dennison, C. Cytosolic Copper Binding by a Bacterial Storage Protein and Interplay with Copper Efflux. Int. J. Mol. Sci. 2019, 20, 4144. [Google Scholar] [CrossRef] [PubMed]
- Vita, N.; Landolfi, G.; Baslé, A.; Platsaki, S.; Lee, J.; Waldron, K.J.; Dennison, C. Bacterial cytosolic proteins with a high capacity for Cu(I) that protect against copper toxicity. Sci. Rep. 2016, 6, 39065. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.L. Microbial Growth. In Encyclopedia of Infection and Immunity, Elsevier: 2022; 10.1016/B978-0-12-818731-9.00190-7pp. 324-335.
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D. , et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Steunou, A.S.; Bourbon, M.L.; Babot, M.; Durand, A.; Liotenberg, S.; Yamaichi, Y.; Ouchane, S. Increasing the copper sensitivity of microorganisms by restricting iron supply, a strategy for bio-management practices. Microb. Biotech. 2020, 13, 1530–1545. [Google Scholar] [CrossRef]
- Macomber, L.; Imlay, J.A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. PNAS 2009, 106, 8344–8349. [Google Scholar] [CrossRef] [PubMed]
- Fung, D.K.; Lau, W.Y.; Chan, W.T.; Yan, A. Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J. Bacteriol. 2013, 195, 4556–4568. [Google Scholar] [CrossRef]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. In Pathogens, 2021; Vol. 10.
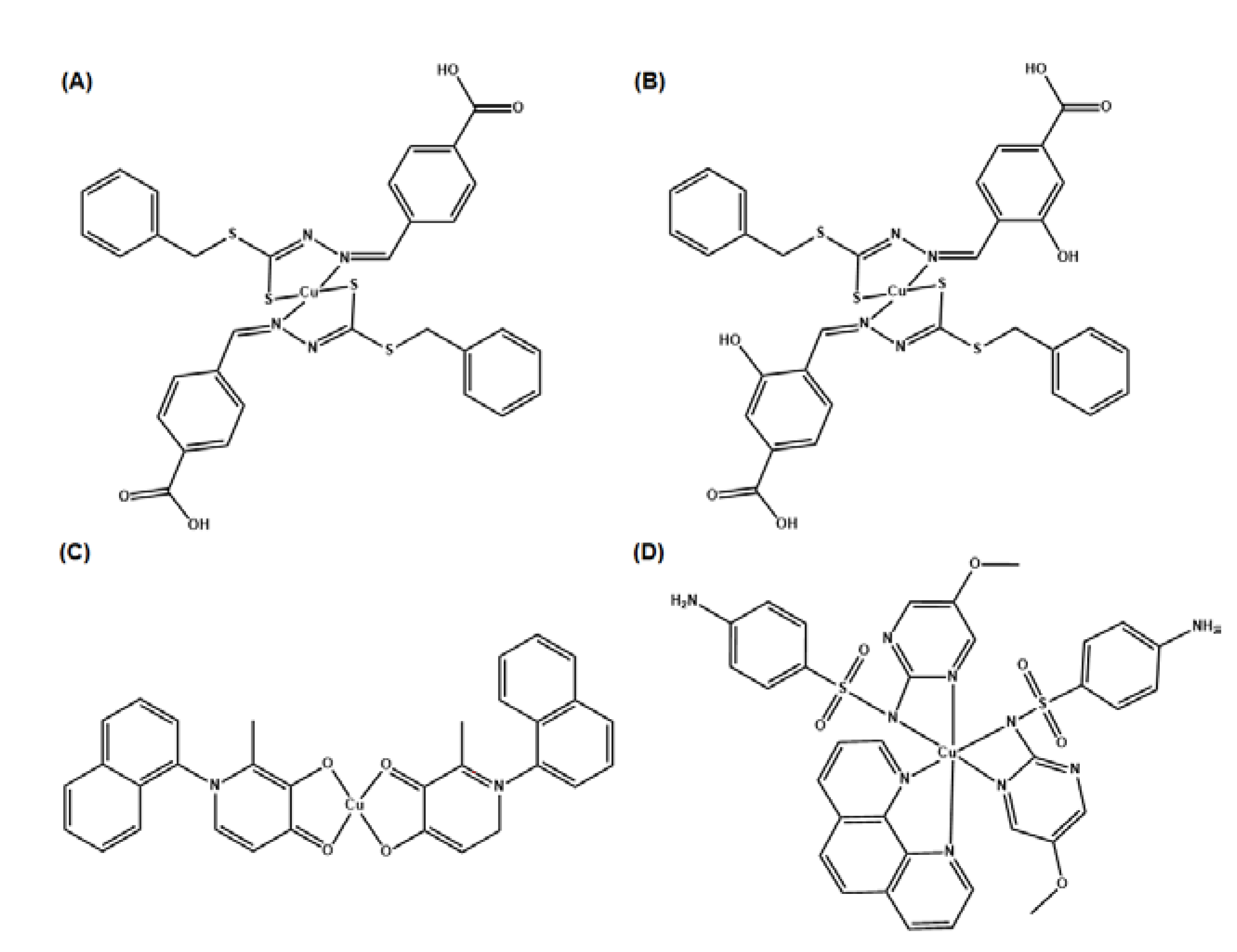
- Ejidike, I.P. Cu(II) Complexes of 4-[(1E)-N-{2-[(Z)-Benzylidene-amino]ethyl}ethanimidoyl]benzene-1,3-diol Schiff Base: Synthesis, Spectroscopic, In-Vitro Antioxidant, Antifungal and Antibacterial Studies. Molecules 2018, 23, 1581. [Google Scholar] [CrossRef] [PubMed]
- Ceramella, J.; Iacopetta, D.; Catalano, A.; Cirillo, F.; Lappano, R.; Sinicropi, M.S. A Review on the Antimicrobial Activity of Schiff Bases: Data Collection and Recent Studies. Antibiotics (Basel) 2022, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.L. The penicillins: A review and update. J. Midwifery Womens Health 2002, 47, 426–434. [Google Scholar] [CrossRef]
- Bruniera, F.R.; Ferreira, F.M.; Saviolli, L.R.; Bacci, M.R.; Feder, D.; da Luz Gonçalves Pedreira, M.; Sorgini Peterlini, M.A.; Azzalis, L.A.; Campos Junqueira, V.B.; Fonseca, F.L. The use of vancomycin with its therapeutic and adverse effects: a review. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 694–700. [Google Scholar] [PubMed]
- Chung, P.Y.; Khoo, R.E.Y.; Liew, H.S.; Low, M.L. Antimicrobial and antibiofilm activities of Cu(II) Schiff base complexes against methicillin-susceptible and resistant Staphylococcus aureus. Ann Clin Microbiol Antimicrob 2021, 20, 67. [Google Scholar] [CrossRef]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Meletiadis, J.; Pournaras, S.; Roilides, E.; Walsh Thomas, J. Defining Fractional Inhibitory Concentration Index Cutoffs for Additive Interactions Based on Self-Drug Additive Combinations, Monte Carlo Simulation Analysis, and In Vitro-In Vivo Correlation Data for Antifungal Drug Combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2010, 54, 602–609. [Google Scholar] [CrossRef]
- Irto, A.; Cardiano, P.; Chand, K.; Cigala, R.M.; Crea, F.; De Stefano, C.; Gano, L.; Gattuso, G.; Sammartano, S.; Santos, M.A. New bis-(3-hydroxy-4-pyridinone)-NTA-derivative: Synthesis, binding ability towards Ca2+, Cu2+, Zn2+, Al3+, Fe3+ and biological assays. Journal of Molecular Liquids 2018, 272, 609–624. [Google Scholar] [CrossRef]
- Leite, A.; Bessa, L.J.; Silva, A.M.G.; Gameiro, P.; de Castro, B.; Rangel, M. Antibacterial activity of naphthyl derived bis-(3-hydroxy-4-pyridinonate) copper(II) complexes against multidrug-resistant bacteria. J. Inorg. Biochem. 2019, 197, 110704. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, D.H.; de Paiva, R.E.F.; Lustri, W.R.; Corbi, P.P. Sulfonamide-containing copper(ii) complexes: new insights on biophysical interactions and antibacterial activities. New J. Chem. 2020, 44, 17236–17244. [Google Scholar] [CrossRef]
- Nakahata, D.H.; de Paiva, R.E.F.; Lustri, W.R.; Ribeiro, C.M.; Pavan, F.R.; da Silva, G.G.; Ruiz, A.; de Carvalho, J.E.; Corbi, P.P. Sulfonamide-containing copper(II) metallonucleases: Correlations with in vitro antimycobacterial and antiproliferative activities. J. Inorg. Biochem. 2018, 187, 85–96. [Google Scholar] [CrossRef]
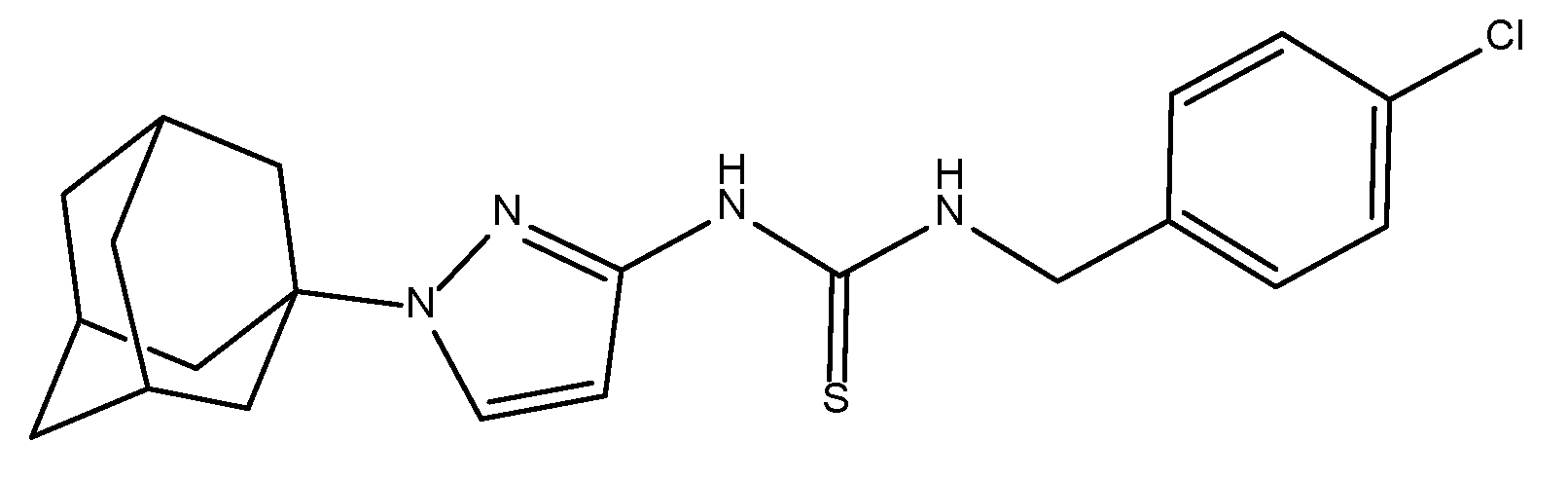
- Dalecki, A.G.; Haeili, M.; Shah, S.; Speer, A.; Niederweis, M.; Kutsch, O.; Wolschendorf, F. Disulfiram and Copper Ions Kill Mycobacterium tuberculosis in a Synergistic Manner. Antimicrob. Agents Chemother. 2015, 59, 4835–4844. [Google Scholar] [CrossRef]
- Dalecki, A.G.; Crawford, C.L.; Wolschendorf, F. Chapter Six - Copper and Antibiotics: Discovery, Modes of Action, and Opportunities for Medicinal Applications. In Advances in Microbial Physiology, Poole, R.K., Ed. Academic Press: 2017; Vol. 70, pp. 193–260.
- Crawford, C.L.; Dalecki, A.G.; Perez, M.D.; Schaaf, K.; Wolschendorf, F.; Kutsch, O. A copper-dependent compound restores ampicillin sensitivity in multidrug-resistant Staphylococcus aureus. Sci. Rep. 2020, 10, 8955. [Google Scholar] [CrossRef]
- Libardo, M.D.; Nagella, S.; Lugo, A.; Pierce, S.; Angeles-Boza, A.M. Copper-binding tripeptide motif increases potency of the antimicrobial peptide Anoplin via Reactive Oxygen Species generation. Biochem. Biophys. Res. Comm. 2015, 456, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Libardo, M.D.; Gorbatyuk, V.Y.; Angeles-Boza, A.M. Central Role of the Copper-Binding Motif in the Complex Mechanism of Action of Ixosin: Enhancing Oxidative Damage and Promoting Synergy with Ixosin B. ACS Infect. Dis. 2016, 2, 71–81. [Google Scholar] [CrossRef]
- Libardo, M.D.J.; Bahar, A.A.; Ma, B.; Fu, R.; McCormick, L.E.; Zhao, J.; McCallum, S.A.; Nussinov, R.; Ren, D.; Angeles-Boza, A.M. , et al. Nuclease activity gives an edge to host-defense peptide piscidin 3 over piscidin 1, rendering it more effective against persisters and biofilms. Febs. J. 2017, 284, 3662–3683. [Google Scholar] [CrossRef]
- Hayden, R.M.; Goldberg, G.K.; Ferguson, B.M.; Schoeneck, M.W.; Libardo, M.D.; Mayeux, S.E.; Shrestha, A.; Bogardus, K.A.; Hammer, J.; Pryshchep, S. , et al. Complementary Effects of Host Defense Peptides Piscidin 1 and Piscidin 3 on DNA and Lipid Membranes: Biophysical Insights into Contrasting Biological Activities. J. Phys. Chem. B 2015, 119, 15235–15246. [Google Scholar] [CrossRef]
- Chen, W.; Cotten, M.L. Expression, purification, and micelle reconstitution of antimicrobial piscidin 1 and piscidin 3 for NMR studies. Protein. Expr. Purif. 2014, 102, 63–68. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.G.; Spago, F.R.; Simionato, A.S.; Navarro, M.O.P.; da Silva, C.S.; Barazetti, A.R.; Cely, M.V.T.; Tischer, C.A.; San Martin, J.A.B.; de Jesus Andrade, C.G.T. , et al. Bioactive Organocopper Compound from Pseudomonas aeruginosa Inhibits the Growth of Xanthomonas citri subsp. citri. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Kumar, C.S.K.V.; Rao, A.S. Inoculum potential, disease development and penetration of host byAlternaria triticina. Incitant of leaf blight of wheat. Proc. Indian Acad. Sci. 1979, 88, 359–365. [Google Scholar] [CrossRef]
- Afonso, L.; Andreata, M.F.; Chryssafidis, A.L.; Alarcon, S.F.; das Neves, A.P.; da Silva, J.V.; Gonçalves, G.D.; Abussafi, L.D.; Simionato, A.S.; Cely, M.V. , et al. Fluopsin C: A Review of the Antimicrobial Activity against Phytopathogens. In Agronomy, 2022; Vol. 12.
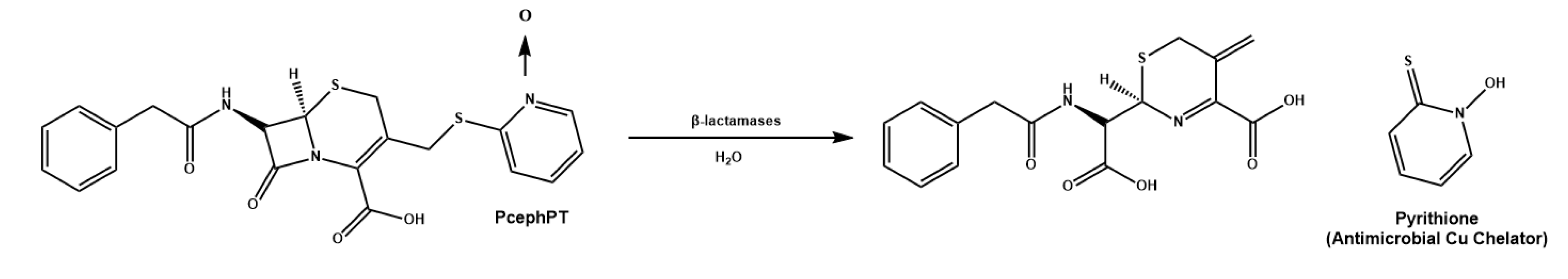
- Hyman, L.M.; Stephenson, C.J.; Dickens, M.G.; Shimizu, K.D.; Franz, K.J. Toward the development of prochelators as fluorescent probes of copper-mediated oxidative stress. Dalton Trans. 2010, 39, 568–576. [Google Scholar] [CrossRef]
- Folk, D.S.; Franz, K.J. A Prochelator Activated by β-Secretase Inhibits Aβ Aggregation and Suppresses Copper-Induced Reactive Oxygen Species Formation. J. Amer. Chem. Soc. 2010, 132, 4994–4995. [Google Scholar] [CrossRef] [PubMed]
- Zaengle-Barone, J.M.; Jackson, A.C.; Besse, D.M.; Becken, B.; Arshad, M.; Seed, P.C.; Franz, K.J. Copper Influences the Antibacterial Outcomes of a β-Lactamase-Activated Prochelator against Drug-Resistant Bacteria. ACS Infect. Dis. 2018, 4, 1019–1029. [Google Scholar] [CrossRef]
- Valodkar, M.; Rathore, P.S.; Jadeja, R.N.; Thounaojam, M.; Devkar, R.V.; Thakore, S. Cytotoxicity evaluation and antimicrobial studies of starch capped water soluble copper nanoparticles. J. Hazard. Mater. 2012, 201-202, 244–249. [Google Scholar] [CrossRef]
- Tao, B.; Lin, C.; Deng, Y.; Yuan, Z.; Shen, X.; Chen, M.; He, Y.; Peng, Z.; Hu, Y.; Cai, K. Copper-nanoparticle-embedded hydrogel for killing bacteria and promoting wound healing with photothermal therapy. J. Mat. Chem. B 2019, 7, 2534–2548. [Google Scholar] [CrossRef] [PubMed]
- Mehdizadeh, T.; Zamani, A.; Abtahi Froushani, S.M. Preparation of Cu nanoparticles fixed on cellulosic walnut shell material and investigation of its antibacterial, antioxidant and anticancer effects. Heliyon 2020, 6, e03528. [Google Scholar] [CrossRef]
- Bastos, C.A.P.; Faria, N.; Wills, J.; Malmberg, P.; Scheers, N.; Rees, P.; Powell, J.J. Copper nanoparticles have negligible direct antibacterial impact. NanoImpact 2020, 17, 100192. [Google Scholar] [CrossRef]
- Chatterjee, A.K.; Chakraborty, R.; Basu, T. Mechanism of antibacterial activity of copper nanoparticles. Nanotech. 2014, 25, 135101. [Google Scholar] [CrossRef]
- Datta Majumdar, T.; Ghosh, C.K.; Mukherjee, A. Dual Role of Copper Nanoparticles in Bacterial Leaf Blight-Infected Rice: A Therapeutic and Metabolic Approach. ACS Agric. Sci. Technol. 2021, 1, 160–172. [Google Scholar] [CrossRef]
- Naqvi, S.S.; Anwer, H.; Siddiqui, A.; Zohra, R.R.; Ali, S.A.; Shah, M.R.; Hashim, S. Novel Synthesis of Maltol Capped Copper Nanoparticles and Their Synergistic Antibacterial Activity with Antibiotics. Plasmonics 2021, 16, 1915–1928. [Google Scholar] [CrossRef]
- Zou, Z.; Sun, J.; Li, Q.; Pu, Y.; Liu, J.; Sun, R.; Wang, L.; Jiang, T. Vancomycin modified copper sulfide nanoparticles for photokilling of vancomycin-resistant enterococci bacteria. Colloids Surf. B 2020, 189, 110875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, Z.; Ma, Y.; Zhang, N.; Wei, D.; Zhang, H.; Zhang, H. Response of partial nitrification sludge to the single and combined stress of CuO nanoparticles and sulfamethoxazole antibiotic on microbial activity, community and resistance genes. Sci. Total Environ. 2020, 712, 135759. [Google Scholar] [CrossRef]
- Antimicrobial resistance. Availabe online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 02-15-2023).
- Qian, J.; Dong, Q.; Chun, K.; Zhu, D.; Zhang, X.; Mao, Y.; Culver, J.N.; Tai, S.; German, J.R.; Dean, D.P. , et al. Highly stable, antiviral, antibacterial cotton textiles via molecular engineering. Nature Nanotech. 2022, 18, 168–176. [Google Scholar] [CrossRef]
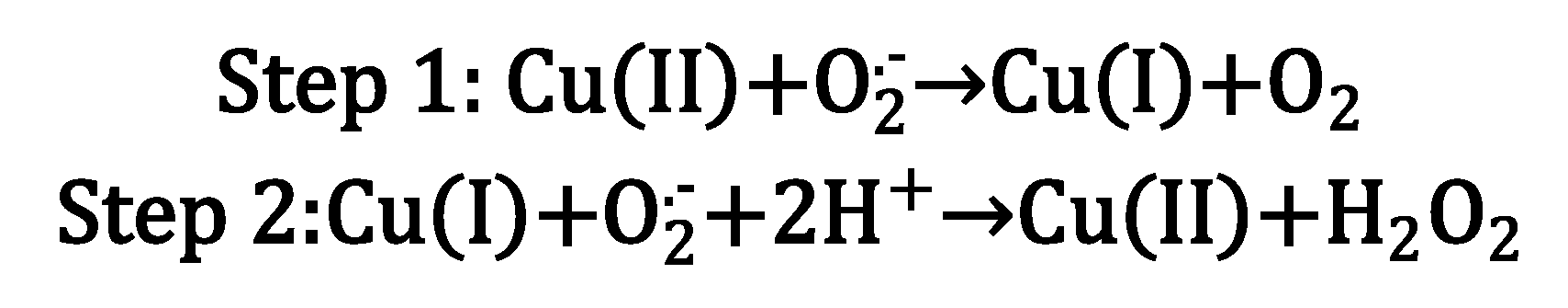
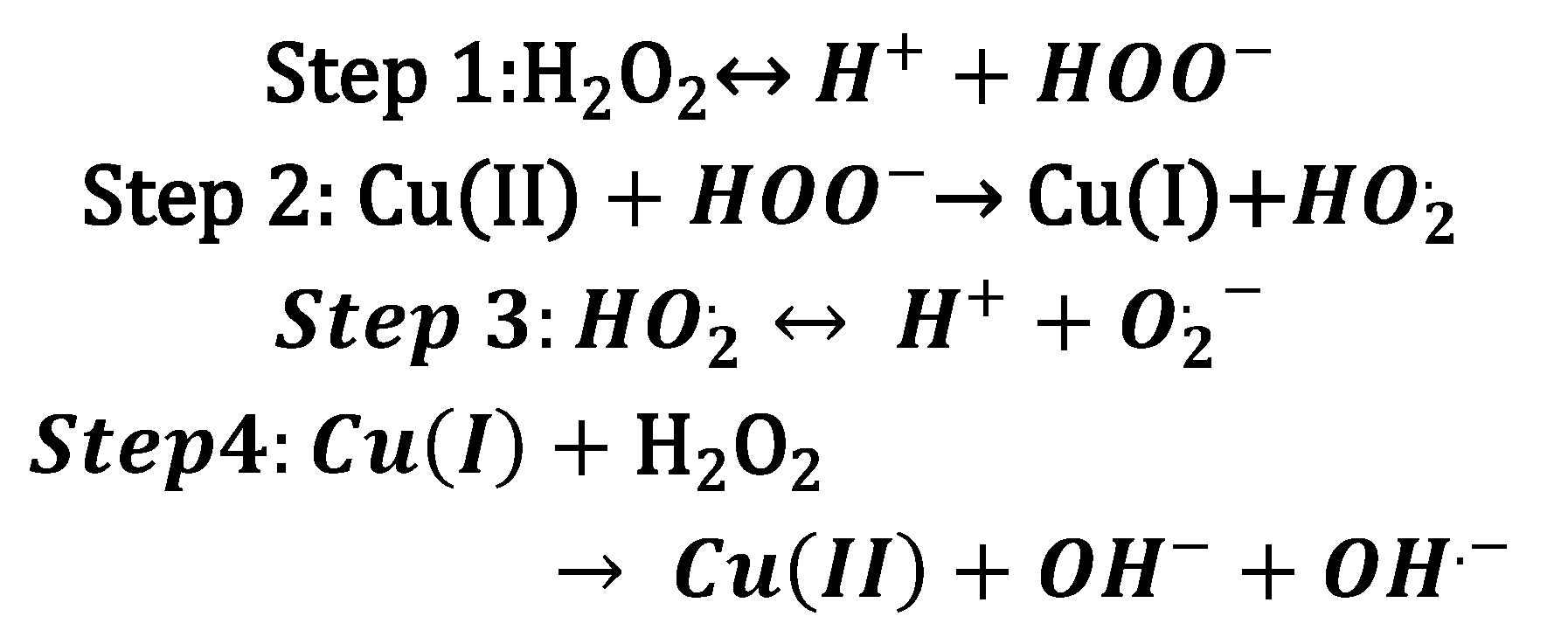
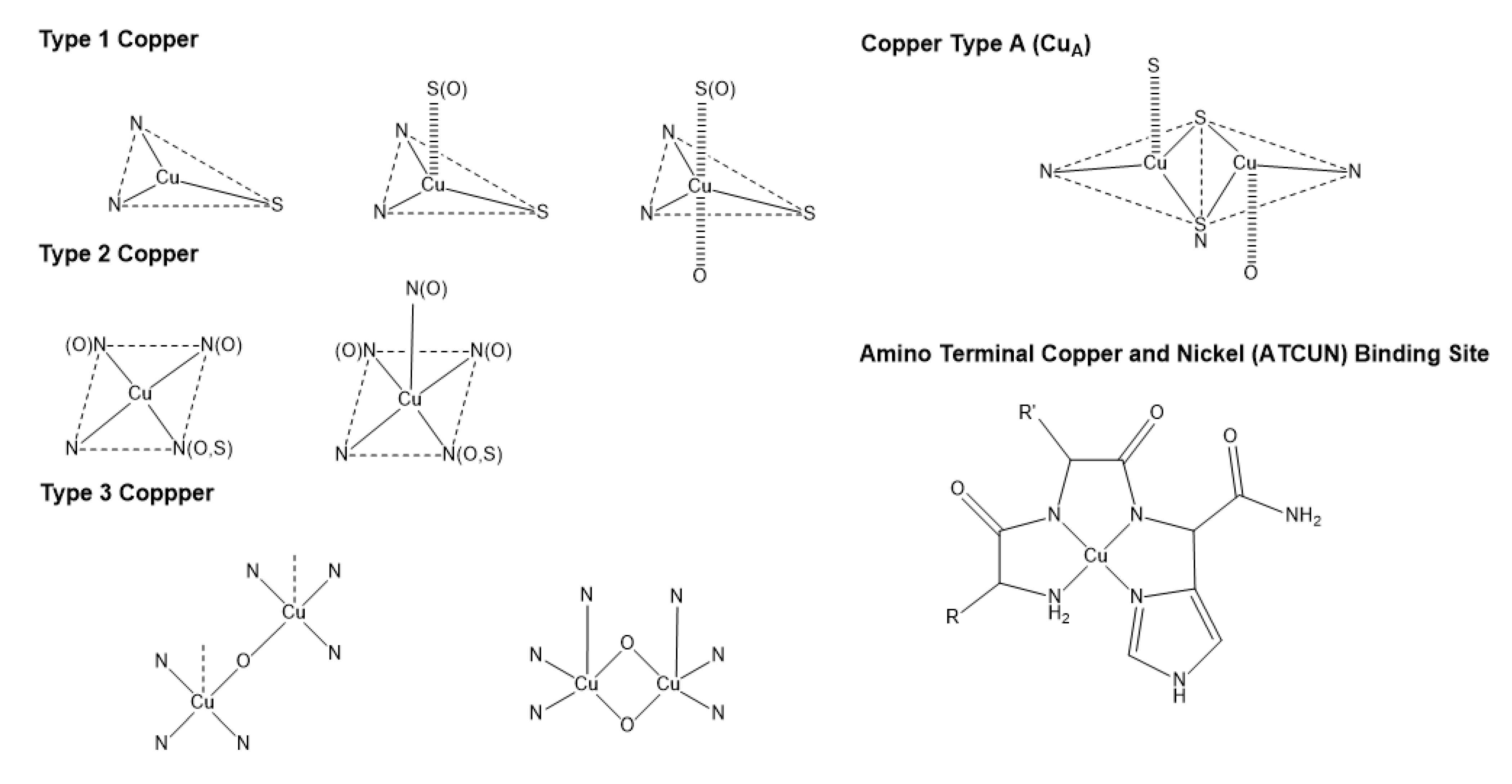
| Metal ion | Coordination number, Geometry | Preferred ligands | Preferred amino acid ligands |
|---|---|---|---|
| Copper, Cu(I) (d10) | 4, Tetrahedral | S-donor, thiolate, N-donors, imidazole | Cysteine (Cys/C), Lysine (Lys/K), Arginine (Arg/R), Histidine (His/H) |
| 3, Trigonal planar | N-donors, imidazole | (His/H) | |
| Copper, Cu(II) (d9) | 4, Tetrahedral | S-donor, thiolate, N-donors, imidazole | Cys/C, Lys/K, Arg/R, His/H |
| 4, Square planar | O-donor, Carboxylate, N-donors, imidazole | Glutamate (Glu/E), Aspartate (Asp/D), Lys/K, Arg/R, His/H | |
| 6, Tetragonal | O-donor, Carboxylate, N-donors, imidazole | Glu/E, Asp/D, Lys/K, Arg/R, His/H | |
| Iron, Fe (II) (d6) | 4, Tetrahedral | S-donor, thiolate | Cys/C |
| 6, Octahedral | O-donor, carboxylate, alkoxide, oxide, phenolate, N-donor, imidazole, porphyrins (heme groups) | Glu/E, Asp/D, Lys/K, Arg/R, His/H, Serine (Ser/S) | |
| Iron, Fe (III) (d5) | 6, Octahedral | O-donor, Carboxylate, carbonyl | Glu/E, Asp/D, His/H, Tyrosine (Tyr/Y) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





