Submitted:
27 April 2023
Posted:
28 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. DMD Equations
3. Methodology
3.1. Solver Settings
3.2. Geometry, Domain, and Boundary Conditions
3.3. Discretization Scheme
3.4. DMD Workflow
- Step 1: Collect multiple time snapshots of the system of interest,
- Step 2: Create a low-dimensional subspace using the SVD or Truncated SVD (TSVD) methods,
- Step 3: Obtain an eigendecomposition of the low-dimensional subspace,
- Step 4: Using the eigendecomposed low-dimensional subspace, assemble the mode shapes and their associated oscillation frequencies, called the ’Time Dynamics’ or TD for short,
- Step 5: Use the mode shapes and TD to assemble the DMD output equations,
- Step 6: Use the DMD solution to predict (or reconstruct) the flow field.
3.5. Data Collection Strategy
4. Results
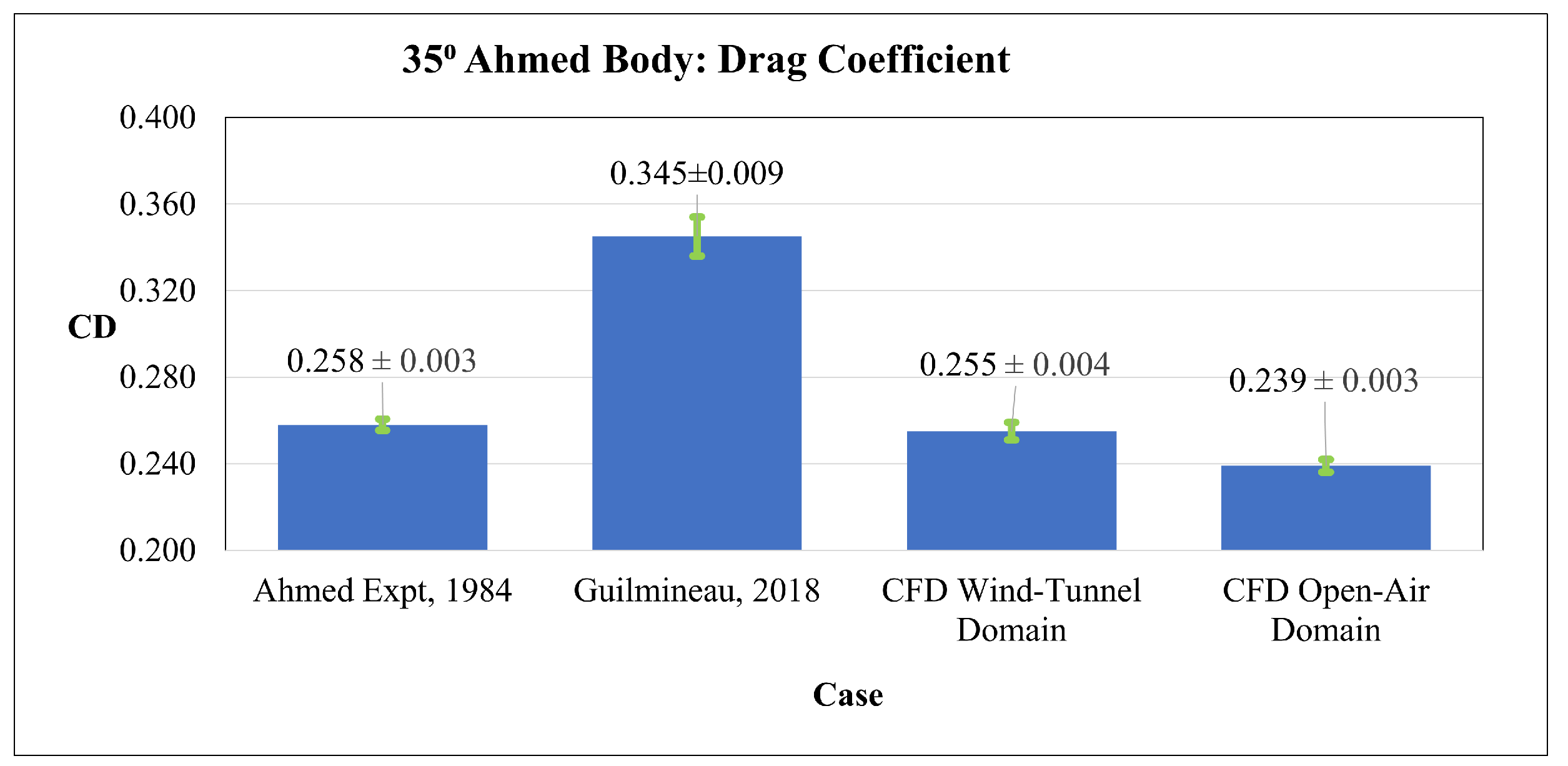
4.1. CFD Validation
4.2. Application of DMD to a Canonical Flow Case
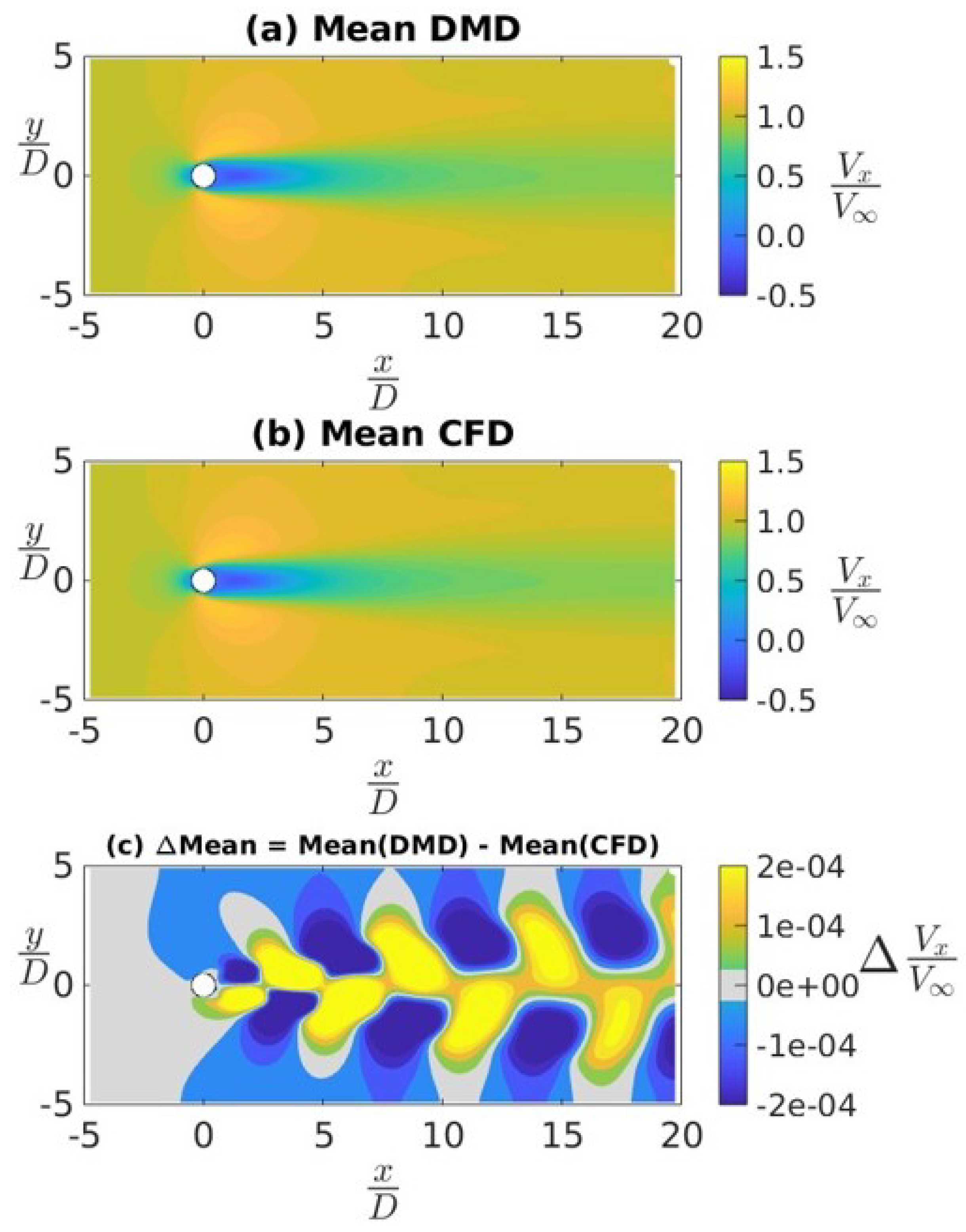
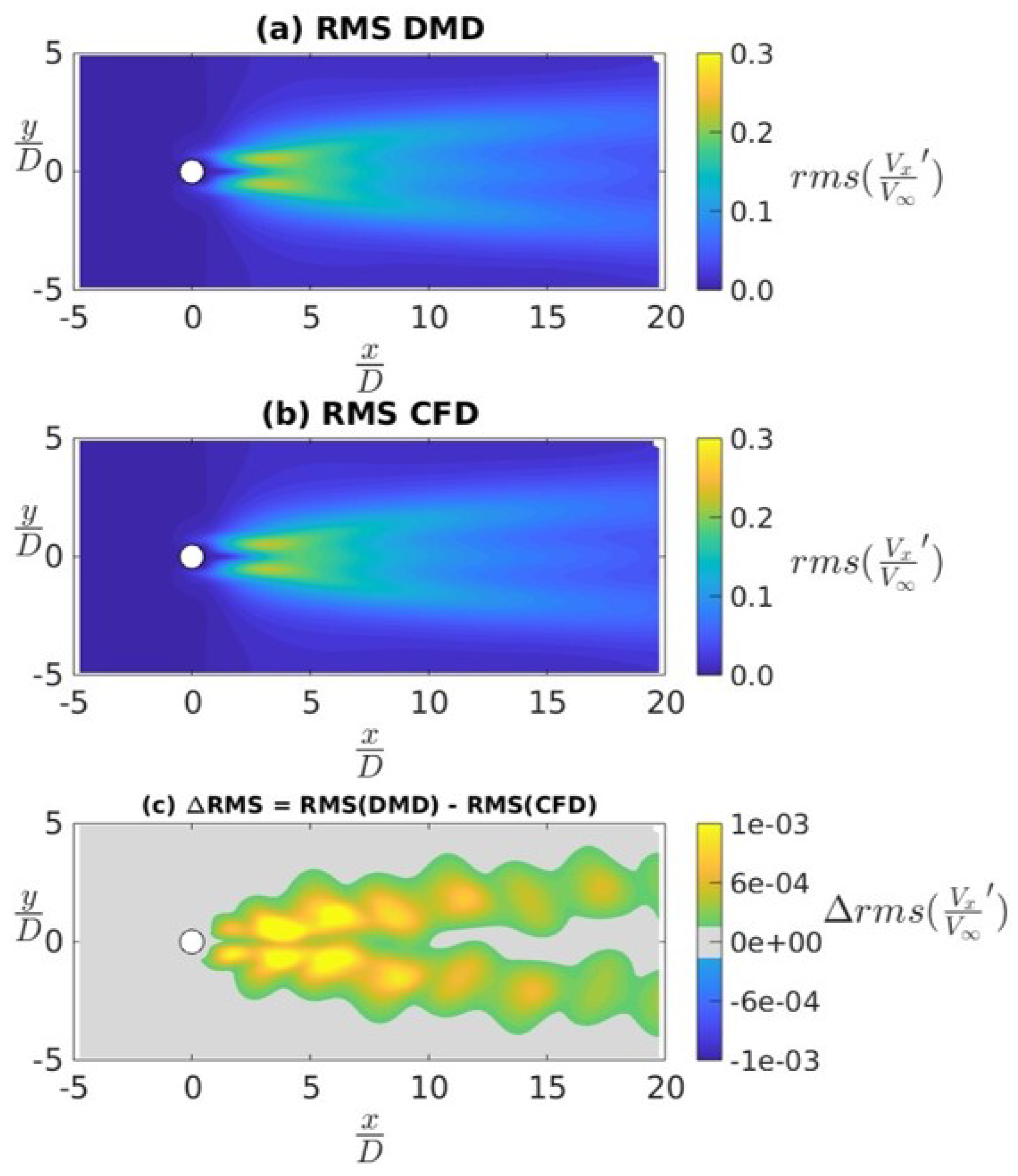
4.3. Ahmed Body Simulations
4.3.1. Data Sampled at 4 kHz
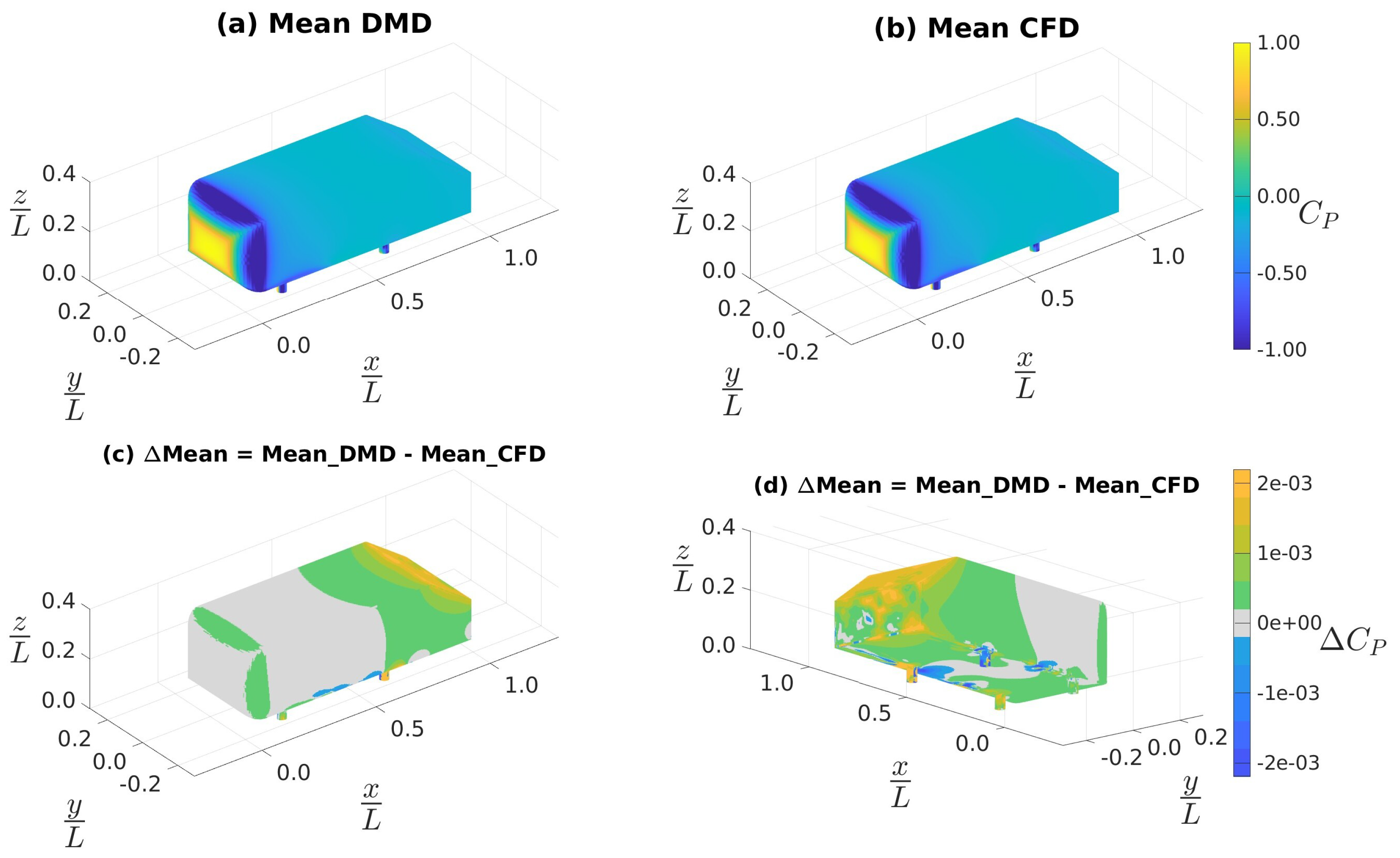
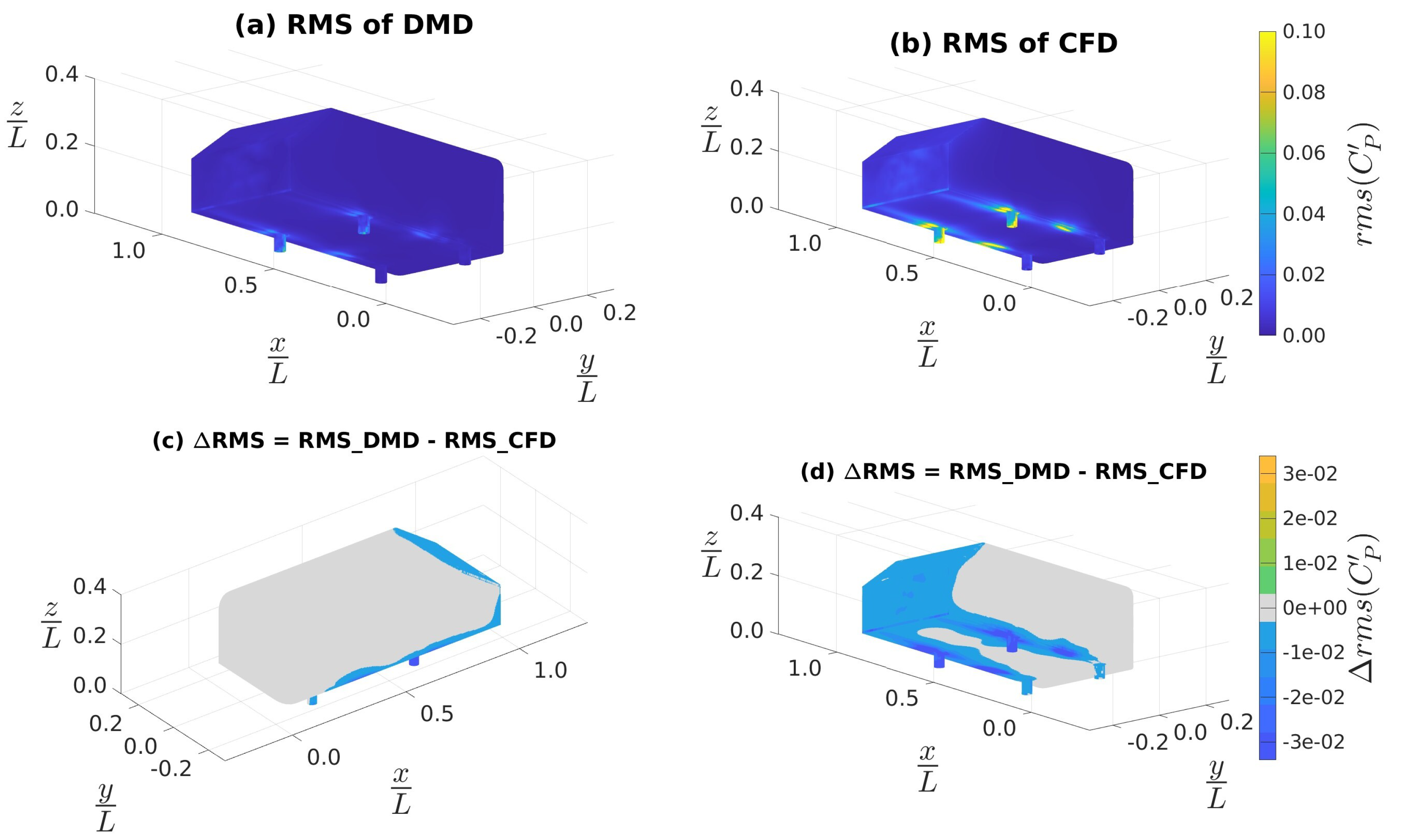
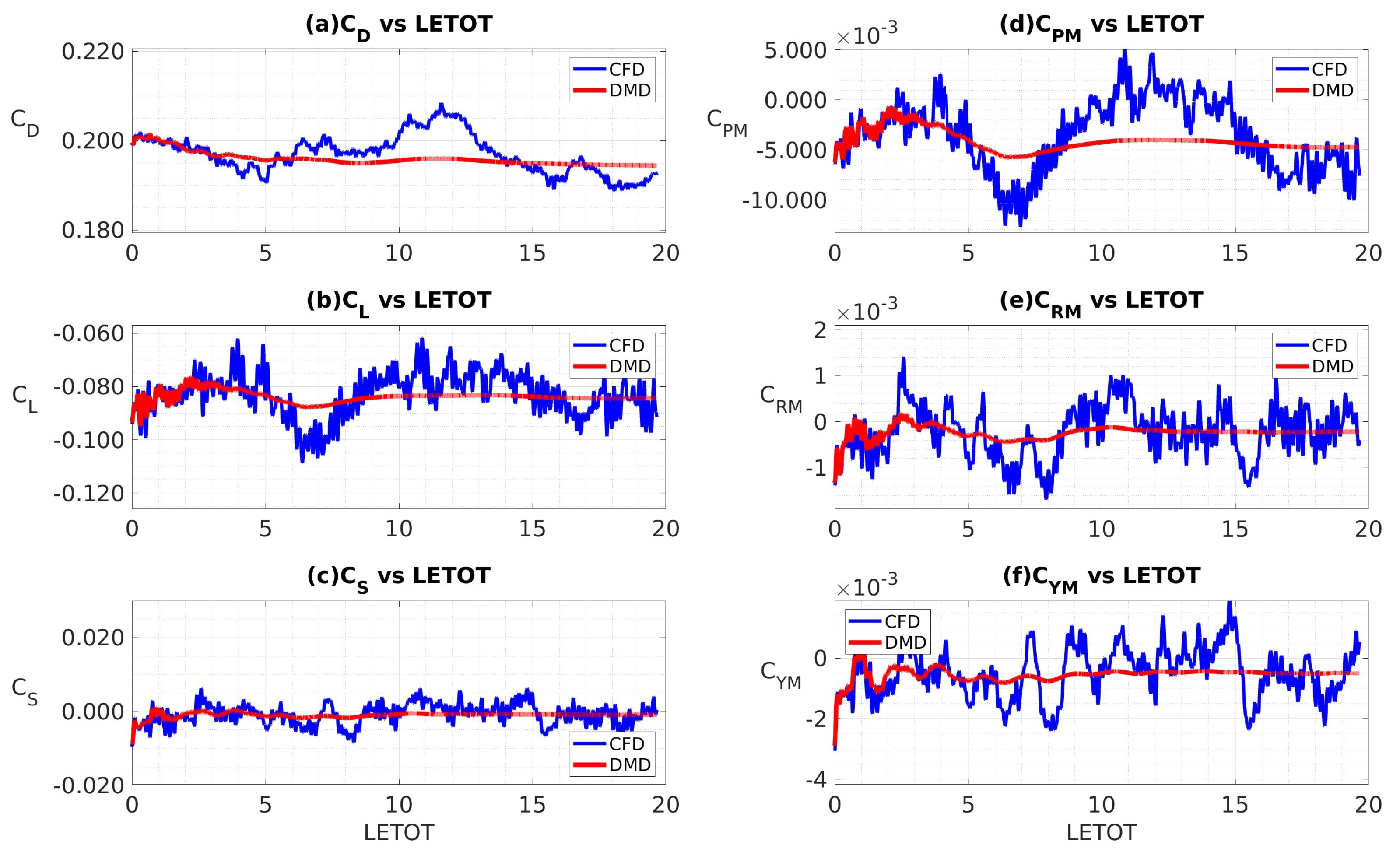
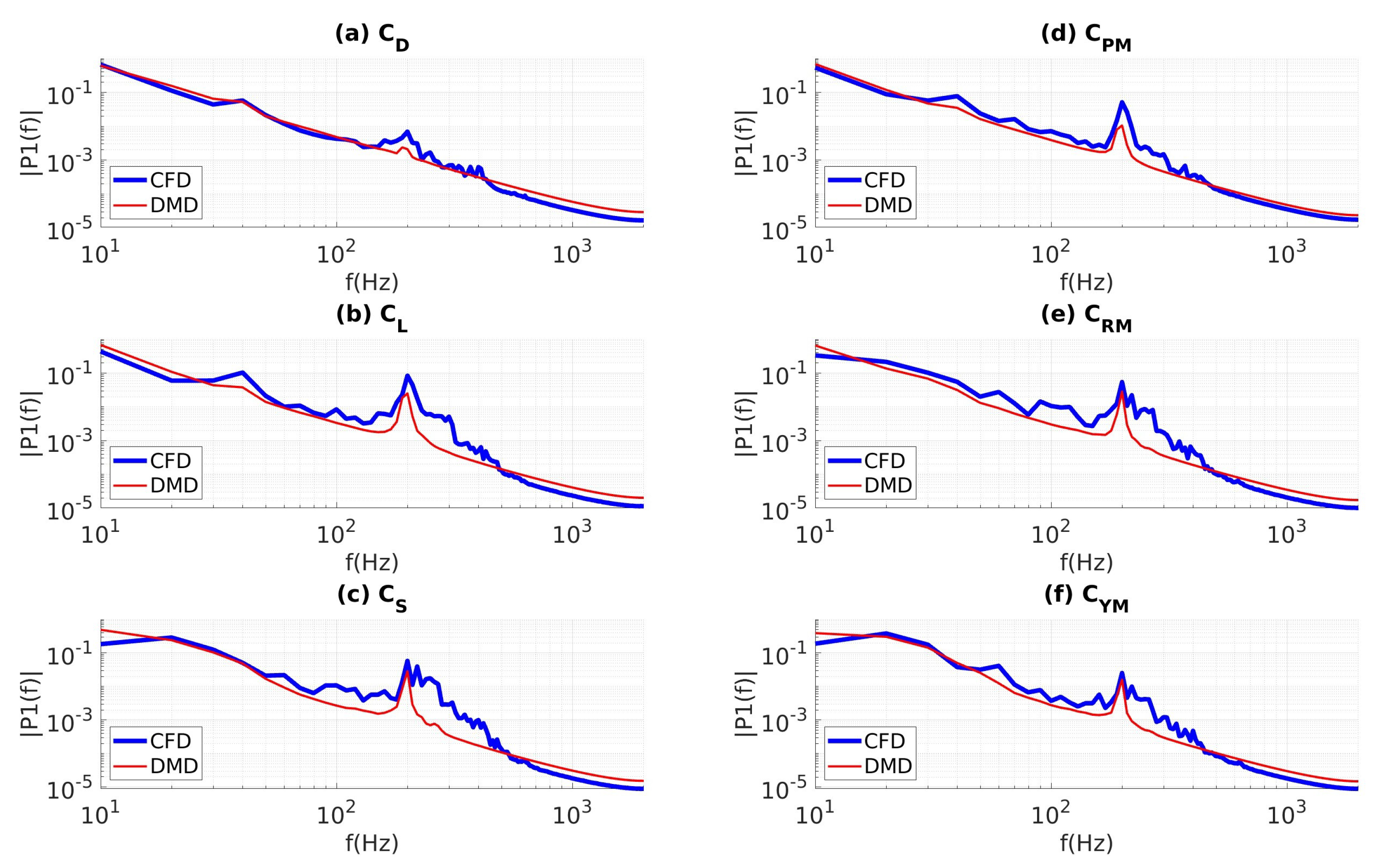
4.3.2. Data Sampled at 10 kHz
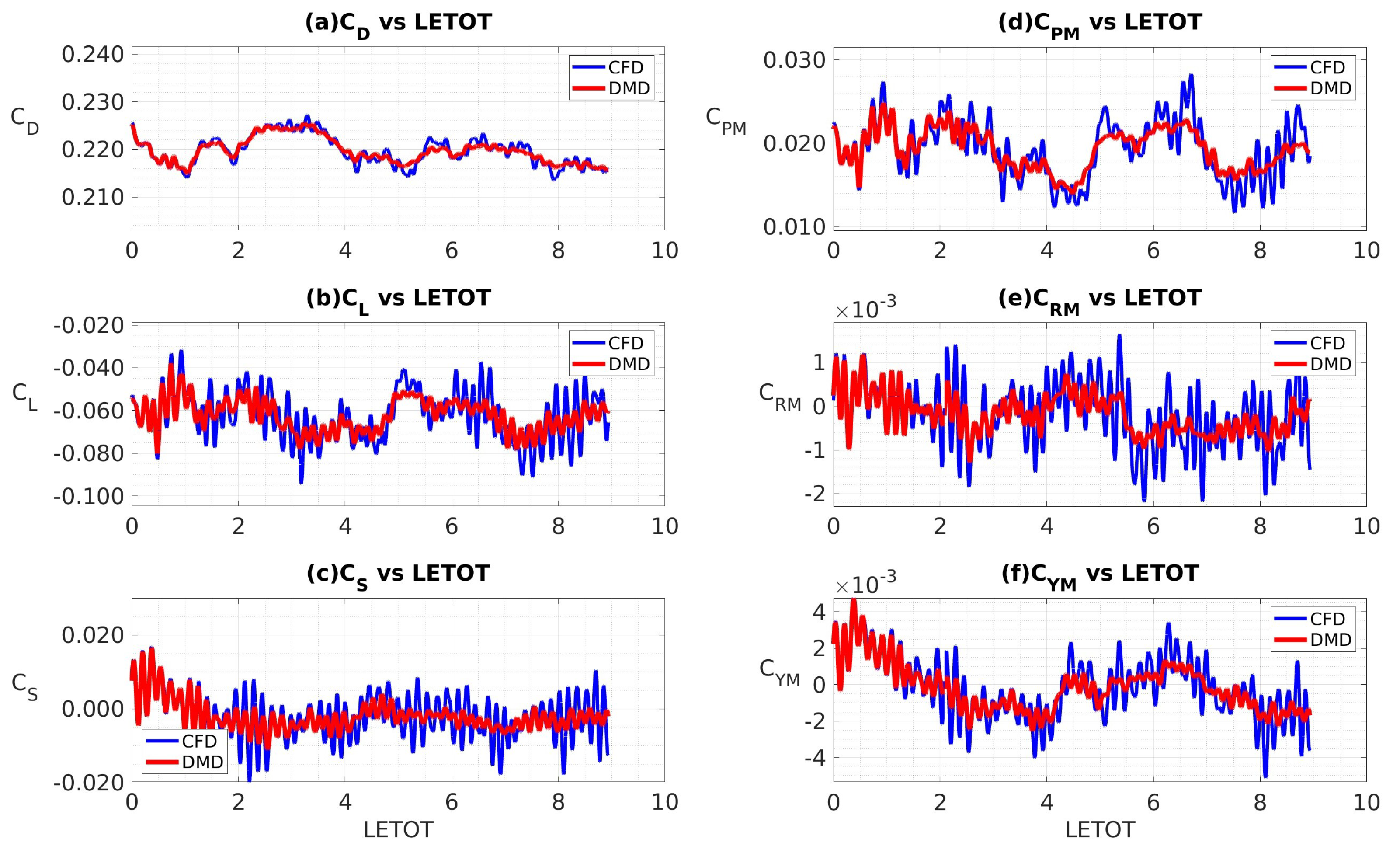
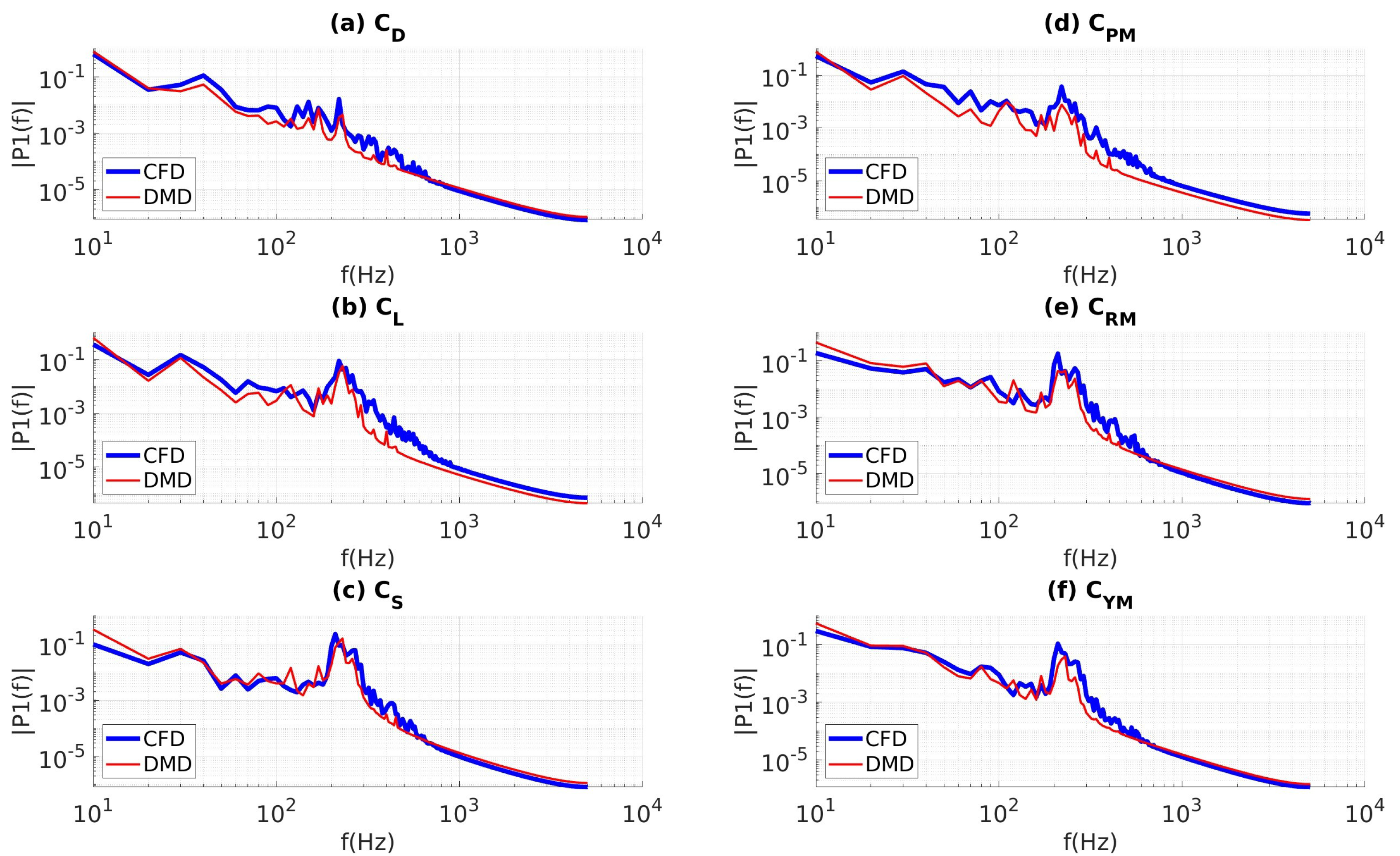
4.3.3. Custom Filtering with Data Sampled at 10 kHz
- The first filter was a low-pass filter applied to the modes identified based upon their maximum instantaneous amplitude in the time dynamics term from Equation (5). The modes having a maximum instantaneous amplitude greater than of the zero-frequency mode were removed.
- The second filter was applied to the modes based upon their frequency and their amplitude, given by the RMS version Equation (14). The second filter was designed to remove high-frequency modes having non-physically excessive energy. To accomplish this, the modes were plotted in frequency space against the amplitudes; among the high frequency-modes ( Hz), the spurious modes were identified using a clustering-based anomaly detection algorithm. Outliers were defined as modes having an amplitude greater than a moving mean of 10 samples by more than a single local standard deviation. The outliers thus identified had their associated modes removed.
- The third filter was designed to remove modes which contribute negligible energy to the system. The remaining modes were sorted based upon their contribution towards the total cumulative energy in the system. In this example, modes contributing collectively less than to total energy were removed; we suspect that these mode may arise from the numerical noise. However, this aspect and the effects of the mode-cut-off energy limit need to be further investigated.
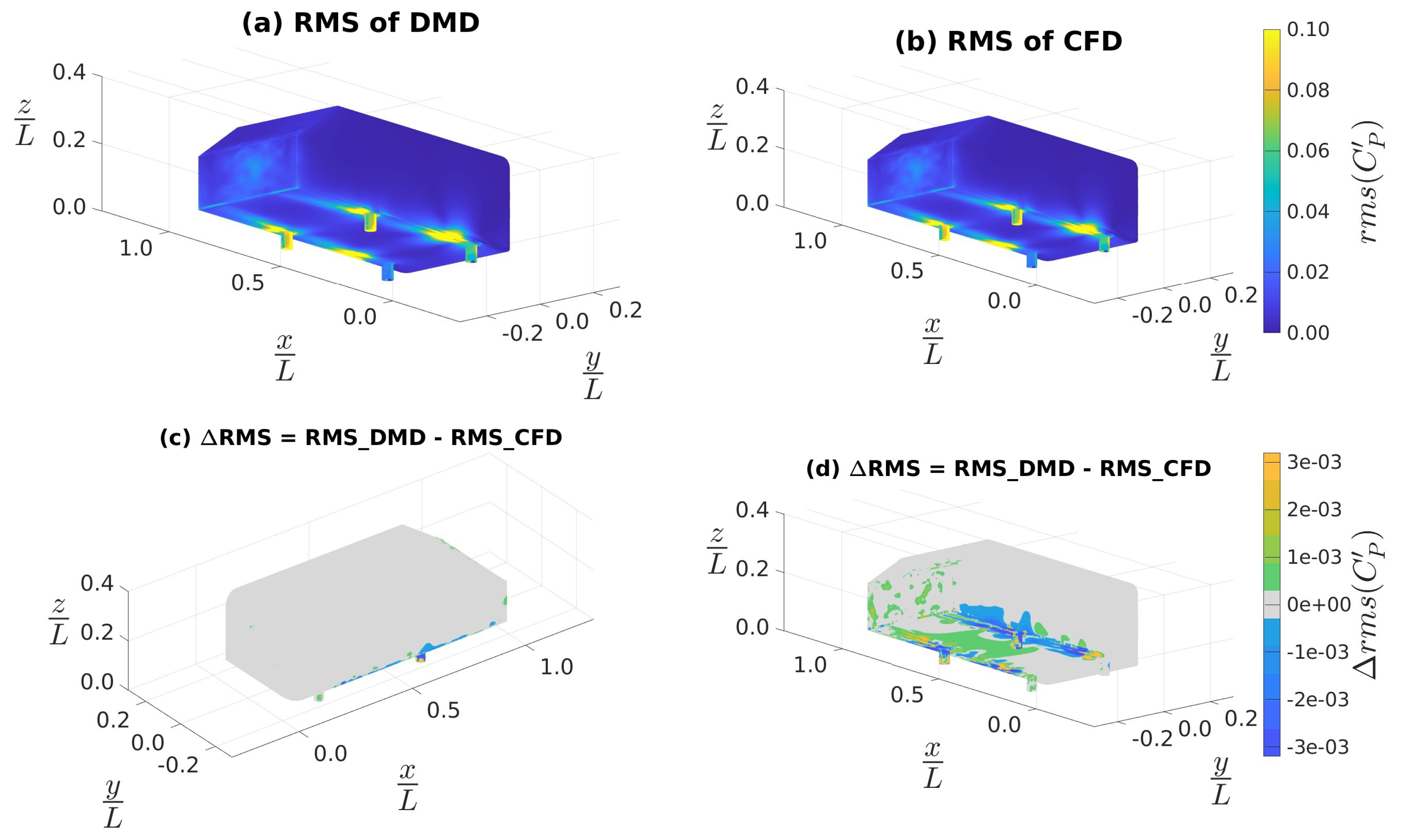
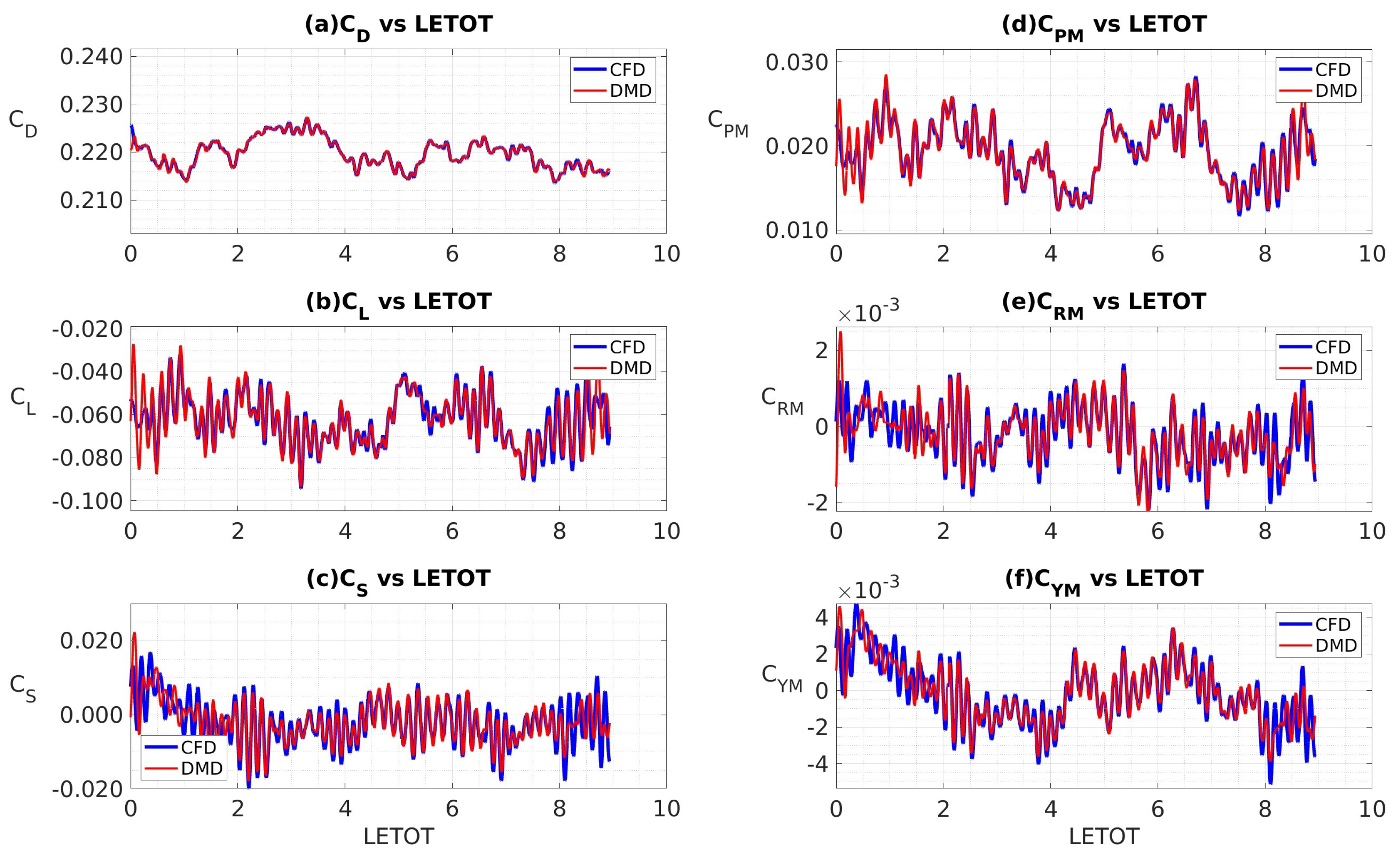
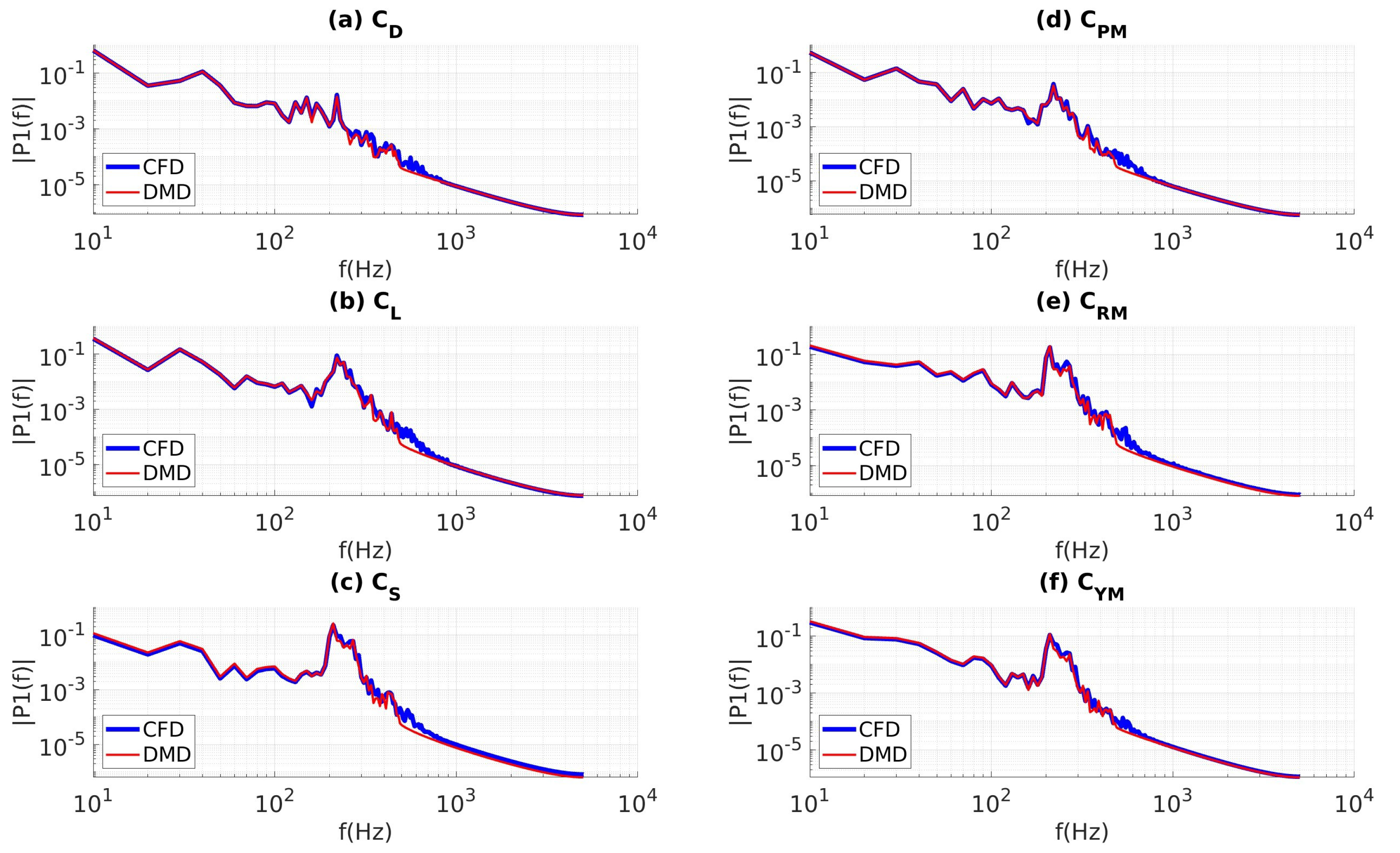
4.3.4. Coefficients of Aerodynamic Forces and Moments
4.4. Future State Predictions Using DMD
4.5. Computational Resources
5. Conclusion
| Parameter | CFD | DMD |
|---|---|---|
| Processors | 144 | 1 |
| CPU time for the entire timeseries | 100 hrs | s |
| CPU time for a single time snapshot | 5 s | s |
| Storage needed | 20 GB | GB |
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CFD | Computational Fluid Dynamics |
| DDES | Delayed Detached Eddy Simulation |
| DES | Detached Eddy Simulation |
| DMD | Dynamic Mode Decomposition |
| DNS | Direct Numerical Simulation |
| GV | Ground Vehicle |
| GVSC | Ground Vehicles Systems Center |
| IDDES | Improved Delayed Detached Eddy Simulation |
| LES | Large Eddy Simulation |
| PSD | Power Spectral Density |
| RANS | Reynolds-Averaged Navier-Stokes |
| SGS | Sub Grid Scale |
| SRS | Scale Resolved Simulation |
| SST | Shear Stress Transport |
| SVD | Singular Value Decomposition |
| TD | Time Dynamics |
| VWT | Virtual Wind Tunnel |
| WT | Wind Tunnel |
References
- Ahani, H.; Nielsen, J.; Uddin, M. The Proper Orthogonal and Dynamic Mode Decomposition of Wake Behind a Fastback DrivAer Model. Technical Report 2022-01-0888; SAE Technical Paper; United States, 2022. [Google Scholar]
- Ikeda, J.; Matsumoto, D.; Tsubokura, M.; Uchida, M.; Hasegawa, T.; Kobayashi, R. Dynamic mode decomposition of flow around a full-scale road vehicle using unsteady CFD. In Proceedings of the 34th AIAA Applied Aerodynamics Conference; 2016; p. 3727. [Google Scholar]
- Jacuzzi, E.; Granlund, K. Passive flow control for drag reduction in vehicle platoons. Journal of Wind Engineering and Industrial Aerodynamics 2019, 189, 104–117. [Google Scholar] [CrossRef]
- Mohammadikalakoo, B.; Schito, P.; Mani, M. Passive flow control on Ahmed body by rear linking tunnels. Journal of Wind Engineering and Industrial Aerodynamics 2020, 205, 104330. [Google Scholar] [CrossRef]
- Hanfeng, W.; Yu, Z.; Chao, Z.; Xuhui, H. Aerodynamic drag reduction of an Ahmed body based on deflectors. Journal of Wind Engineering and Industrial Aerodynamics 2016, 148, 34–44. [Google Scholar] [CrossRef]
- Siddiqui, N.; Chaab, M. A simple passive device for the drag reduction of an ahmed body. Journal of Applied Fluid Mechanics 2020, 14, 147–164. [Google Scholar]
- Tian, J.; Zhang, Y.; Zhu, H.; Xiao, H. Aerodynamic drag reduction and flow control of Ahmed body with flaps. Advances in Mechanical Engineering 2017, 9, 1687814017711390. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, K.; Zhou, Y.; To, S.; Tu, J. Active drag reduction of a high-drag Ahmed body based on steady blowing. Journal of Fluid Mechanics 2018, 856, 351–396. [Google Scholar] [CrossRef]
- Joseph, P.; Amandolese, X.; Aider, J.L. Drag reduction on the 25° slant angle Ahmed reference body using pulsed jets. Experiments in fluids 2012, 52, 1169–1185. [Google Scholar] [CrossRef]
- Joseph, P.; Amandolese, X.; Edouard, C.; Aider, J.L. Flow control using MEMS pulsed micro-jets on the Ahmed body. Experiments in fluids 2013, 54, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Cui, W.; Jia, Q.; Li, Q.; Yang, Z.; Morzyński, M.; Noack, B.R. Explorative gradient method for active drag reduction of the fluidic pinball and slanted Ahmed body. Journal of Fluid Mechanics 2022, 932, A7. [Google Scholar] [CrossRef]
- Bruneau, C.H.; Creusé, E.; Depeyras, D.; Gilliéron, P.; Mortazavi, I. Coupling active and passive techniques to control the flow past the square back Ahmed body. Computers & Fluids 2010, 39, 1875–1892. [Google Scholar]
- Sudin, M.N.; Abdullah, M.A.; Shamsuddin, S.A.; Ramli, F.R.; Tahir, M.M. Review of research on vehicles aerodynamic drag reduction methods. International Journal of Mechanical and Mechatronics Engineering 2014, 14, 37–47. [Google Scholar]
- Mukut, A.M.I.; Abedin, M.Z. Review on aerodynamic drag reduction of vehicles. International Journal of Engineering Materials and Manufacture 2019, 4, 1–14. [Google Scholar] [CrossRef]
- Bounds, C.P.; Rajasekar, S.; Uddin, M. Development of a Numerical Investigation Framework for Ground Vehicle Platooning. Fluids 2021, 6, 404. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, T.; Yan, F.; Qiao, S.; Wang, X. Analysis and design on intervehicle distance control of autonomous vehicle platoons. ISA transactions 2020, 100, 446–453. [Google Scholar] [CrossRef]
- Sivanandham, S.; Gajanand, M. Platooning for sustainable freight transportation: an adoptable practice in the near future? Transport Reviews 2020, 40, 581–606. [Google Scholar] [CrossRef]
- Proctor, J.L.; Brunton, S.L.; Kutz, J.N. Dynamic mode decomposition with control. SIAM Journal on Applied Dynamical Systems 2016, 15, 142–161. [Google Scholar] [CrossRef]
- Tennekes, H.; Lumley, J.L.; Lumley, J.L.; et al. A first course in turbulence; MIT press, 1972. [Google Scholar]
- Liu, K.; Zhang, B.; Zhang, Y.; Zhou, Y. Flow structure around a low-drag Ahmed body. Journal of Fluid Mechanics 2021, 913, A21. [Google Scholar] [CrossRef]
- Misar, A.S.; Uddin, M.; Pandaleon, T.; Wilson, J. Scale-Resolved and Time-Averaged Simulations of the Flow over a NASCAR Cup Series Racecar. Technical Report 2023-01-0735; SAE Technical Paper; United States, 2023. [Google Scholar]
- Guilmineau, E.; Deng, G.; Leroyer, A.; Queutey, P.; Visonneau, M.; Wackers, J. Assessment of hybrid RANS-LES formulations for flow simulation around the Ahmed body. Computers & Fluids 2018, 176, 302–319. [Google Scholar]
- Ashton, N.; West, A.; Lardeau, S.; Revell, A. Assessment of RANS and DES methods for realistic automotive models. Computers & fluids 2016, 128, 1–15. [Google Scholar]
- Kutz, J.N.; Brunton, S.L.; Brunton, B.W.; Proctor, J.L. Dynamic mode decomposition: data-driven modeling of complex systems; SIAM, 2016. [Google Scholar]
- Schmid, P.J. Dynamic mode decomposition and its variants. Annual Review of Fluid Mechanics 2022, 54, 225–254. [Google Scholar] [CrossRef]
- Schmidt, O.T.; Colonius, T. Guide to spectral proper orthogonal decomposition. Aiaa journal 2020, 58, 1023–1033. [Google Scholar] [CrossRef]
- Muld, T.W.; Efraimsson, G.; Henningson, D.S. Flow structures around a high-speed train extracted using proper orthogonal decomposition and dynamic mode decomposition. Computers & Fluids 2012, 57, 87–97. [Google Scholar]
- Sieber, M.; Paschereit, C.O.; Oberleithner, K. Spectral proper orthogonal decomposition. Journal of Fluid Mechanics 2016, 792, 798–828. [Google Scholar] [CrossRef]
- Berkooz, G.; Holmes, P.; Lumley, J.L. The proper orthogonal decomposition in the analysis of turbulent flows. Annual review of fluid mechanics 1993, 25, 539–575. [Google Scholar] [CrossRef]
- Taira, K.; Brunton, S.L.; Dawson, S.T.; Rowley, C.W.; Colonius, T.; McKeon, B.J.; Schmidt, O.T.; Gordeyev, S.; Theofilis, V.; Ukeiley, L.S. Modal analysis of fluid flows: An overview. Aiaa Journal 2017, 55, 4013–4041. [Google Scholar] [CrossRef]
- Schmid, P.J. Dynamic mode decomposition of numerical and experimental data. Journal of fluid mechanics 2010, 656, 5–28. [Google Scholar] [CrossRef]
- Schmid, P.J.; Li, L.; Juniper, M.P.; Pust, O. Applications of the dynamic mode decomposition. Theoretical and Computational Fluid Dynamics 2011, 25, 249–259. [Google Scholar] [CrossRef]
- Schmid, P.J.; Violato, D.; Scarano, F. Decomposition of time-resolved tomographic PIV. Experiments in fluids 2012, 52, 1567–1579. [Google Scholar] [CrossRef]
- Wynn, A.; Pearson, D.; Ganapathisubramani, B.; Goulart, P.J. Optimal mode decomposition for unsteady flows. Journal of Fluid Mechanics 2013, 733, 473–503. [Google Scholar] [CrossRef]
- Sakai, M.; Sunada, Y.; Imamura, T.; Rinoie, K. Experimental and numerical flow analysis around circular cylinders using POD and DMD. In Proceedings of the 44th AIAA Fluid Dynamics Conference; 2014; p. 3325. [Google Scholar]
- Hemati, M.S.; Rowley, C.W.; Deem, E.A.; Cattafesta, L.N. De-biasing the dynamic mode decomposition for applied Koopman spectral analysis of noisy datasets. Theoretical and Computational Fluid Dynamics 2017, 31, 349–368. [Google Scholar] [CrossRef]
- Erichson, N.B.; Mathelin, L.; Kutz, J.N.; Brunton, S.L. Randomized dynamic mode decomposition. SIAM Journal on Applied Dynamical Systems 2019, 18, 1867–1891. [Google Scholar] [CrossRef]
- Jovanović, M.R.; Schmid, P.J.; Nichols, J.W. Sparsity-promoting dynamic mode decomposition. Physics of Fluids 2014, 26, 024103. [Google Scholar] [CrossRef]
- Guéniat, F.; Mathelin, L.; Pastur, L.R. A dynamic mode decomposition approach for large and arbitrarily sampled systems. Physics of Fluids 2015, 27, 025113. [Google Scholar] [CrossRef]
- Mengmeng, W.; Zhonghua, H.; Han, N.; Wenping, S.; Le Clainche, S.; Ferrer, E. A transition prediction method for flow over airfoils based on high-order dynamic mode decomposition. Chinese Journal of Aeronautics 2019, 32, 2408–2421. [Google Scholar]
- Qiu, R.; Huang, R.; Wang, Y.; Huang, C. Dynamic mode decomposition and reconstruction of transient cavitating flows around a Clark-Y hydrofoil. Theoretical and Applied Mechanics Letters 2020, 10, 327–332. [Google Scholar] [CrossRef]
- Heft, A.I.; Indinger, T.; Adams, N.A. Experimental and numerical investigation of the DrivAer model. In Proceedings of the Fluids Engineering Division Summer Meeting; American Society of Mechanical Engineers, 2012; Volume 44755, pp. 41–51. [Google Scholar]
- Matsumoto, D.; Haag, L.; Indinger, T. Investigation of the unsteady external and underhood airflow of the DrivAer model by dynamic mode decomposition methods. International Journal of Automotive Engineering 2017, 8, 55–62. [Google Scholar] [CrossRef]
- Ahmed, S.R.; Ramm, G.; Faltin, G. Some salient features of the time-averaged ground vehicle wake. SAE Transactions 1984, 473–503. [Google Scholar]
- Uddin, M.; Nichols, S.; Hahn, C.; Misar, A.; Desai, S.; Tison, N.; Korivi, V. Aerodynamics of Landing Maneuvering of an Unmanned Aerial Vehicle in Close Proximity to a Ground Vehicle. Technical Report 2023-01-0118; SAE Technical Paper; United States, 2023. [Google Scholar]
- Lienhart, H.; Becker, S. Flow and turbulence structure in the wake of a simplified car model. SAE transactions 2003, 785–796. [Google Scholar]
- Misar, A.S. Insight into the Aerodynamics of Race and Idealized Road Vehicles Using Scale-Resolved and Scale-Averaged CFD Simulations. PhD thesis, University of North Carolina Charlotte, 2023, unpublished. [Google Scholar]
- Dylewsky, D.; Tao, M.; Kutz, J.N. Dynamic mode decomposition for multiscale nonlinear physics. Physical Review E 2019, 99, 063311. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.H. Dynamic mode decomposition: Theory and applications. PhD thesis, Princeton University, 2013. [Google Scholar]
- Kou, J.; Zhang, W. An improved criterion to select dominant modes from dynamic mode decomposition. European Journal of Mechanics-B/Fluids 2017, 62, 109–129. [Google Scholar] [CrossRef]
- Misar, A.S.; Uddin, M. Effects of Solver Parameters and Boundary Conditions on RANS CFD Flow Predictions over a Gen-6 NASCAR Racecar. Technical Report 2022-01-0372; SAE WCX Technical Paper; United States, 2022. [Google Scholar]
- Shur, M.L.; Spalart, P.R.; Strelets, M.K.; Travin, A.K. A hybrid RANS-LES approach with delayed-DES and wall-modelled LES capabilities. International journal of heat and fluid flow 2008, 29, 1638–1649. [Google Scholar] [CrossRef]
- Gritskevich, M.S.; Garbaruk, A.V.; Schütze, J.; Menter, F.R.; et al. Development of DDES and IDDES formulations for the k-ω shear stress transport model. Flow Turbulence and Combustion 2012, 88, 431. [Google Scholar] [CrossRef]
- Spalart, P.R. Comments on the Feasibility of LES for Wings and on the Hybrid RANS/LES Approach. In Proceedings of the First AFOSR International Conference on DNS/LES; 1997; pp. 137–147. [Google Scholar]
- Spalart, P.R.; Streett, C. Young-person’s guide to detached-eddy simulation grids. Technical report, NASA, NASA STI Help Desk NASA Center for AeroSpace Information 7121 Standard Drive Hanover, MD 21076-1320, 2001.
- Spalart, P.R.; Deck, S.; Shur, M.L.; Squires, K.D.; Strelets, M.K.; Travin, A. A new version of detached-eddy simulation, resistant to ambiguous grid densities. Theoretical and computational fluid dynamics 2006, 20, 181–195. [Google Scholar] [CrossRef]
- Piomelli, U.; Balaras, E. Wall-layer models for large-eddy simulations. Annual review of fluid mechanics 2002, 34, 349–374. [Google Scholar] [CrossRef]
- Menter, F.R. Two-equation eddy-viscosity turbulence models for engineering applications. AIAA journal 1994, 32, 1598–1605. [Google Scholar] [CrossRef]
- Menter, F. Zonal two equation kω turbulence models for aerodynamic flows. In Proceedings of the 23rd fluid dynamics, 778 plasmadynamics, and lasers conference; 1993; p. 2906. [Google Scholar]
- Menter, F.R.; Kuntz, M.; Langtry, R. Ten years of industrial experience with the SST turbulence model. Turbulence, heat and mass transfer 2003, 4, 625–632. [Google Scholar]
- Zhang, C.; Bounds, C.P.; Foster, L.; Uddin, M. Turbulence modeling effects on the CFD predictions of flow over a detailed full-scale sedan vehicle. Fluids 2019, 4, 148. [Google Scholar] [CrossRef]
- Aultman, M.; Wang, Z.; Duan, L. Effect of time-step size on flow around generic car models. Journal of Wind Engineering and Industrial Aerodynamics 2021, 219, 104764. [Google Scholar] [CrossRef]
- Misar, A.S.; Bounds, C.; Ahani, H.; Zafar, M.U.; Uddin, M. On the Effects of Parallelization on the Flow Prediction around a Fastback DrivAer Model at Different Attitudes. Technical Report 2021-01-0965; SAE WCX Technical Paper; United States, 2021. [Google Scholar]
- Daily, J.W.; James, W.; Harleman, D.R.; et al. Fluid dynamics; Addison-Wesley, 1966. [Google Scholar]
- Fu, C.; Bounds, C.P.; Selent, C.; Uddin, M. Turbulence modeling effects on the aerodynamic characterizations of a NASCAR Generation 6 racecar subject to yaw and pitch changes. Proceedings of the Institution of Mechanical Engineers, Part D: Journal of Automobile Engineering 2019, 233. [Google Scholar] [CrossRef]
- Hunt, J.C.; Wray, A.A.; Moin, P. Eddies, streams, and convergence zones in turbulent flows. In Studying turbulence using numerical simulation databases, 2. Proceedings of the 1988 summer program; 1988. [Google Scholar]
- Fu, C.; Uddin, M.; Zhang, C. Computational Analyses of the Effects of Wind Tunnel Ground Simulation and Blockage Ratio on the Aerodynamic Prediction of Flow over a Passenger Vehicle. Vehicles 2020, 2, 318–341. [Google Scholar] [CrossRef]
- Altinisik, A.; Kutukceken, E.; Umur, H. Experimental and numerical aerodynamic analysis of a passenger car: Influence of the blockage ratio on drag coefficient. Journal of Fluids Engineering 2015, 137. [Google Scholar] [CrossRef]
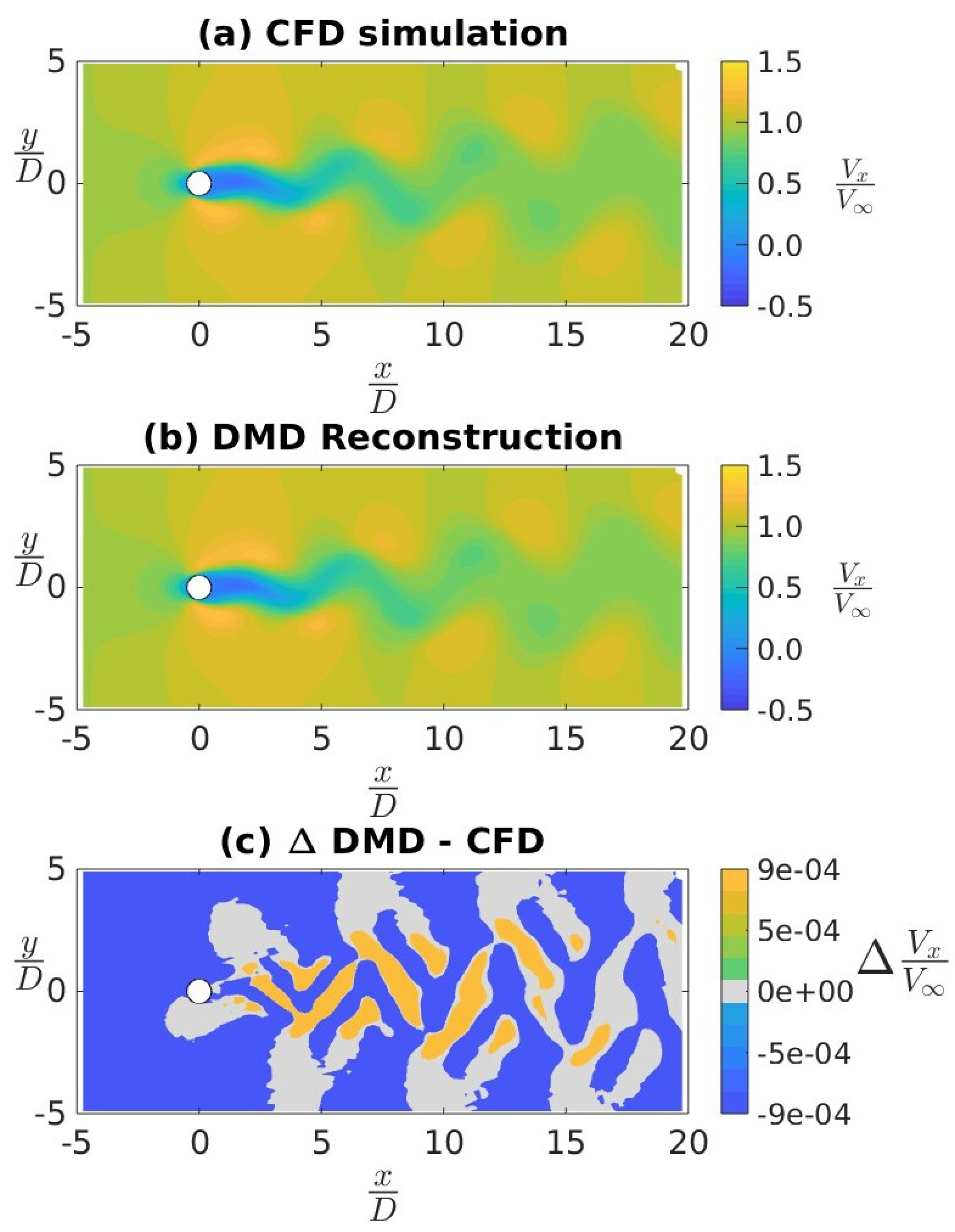
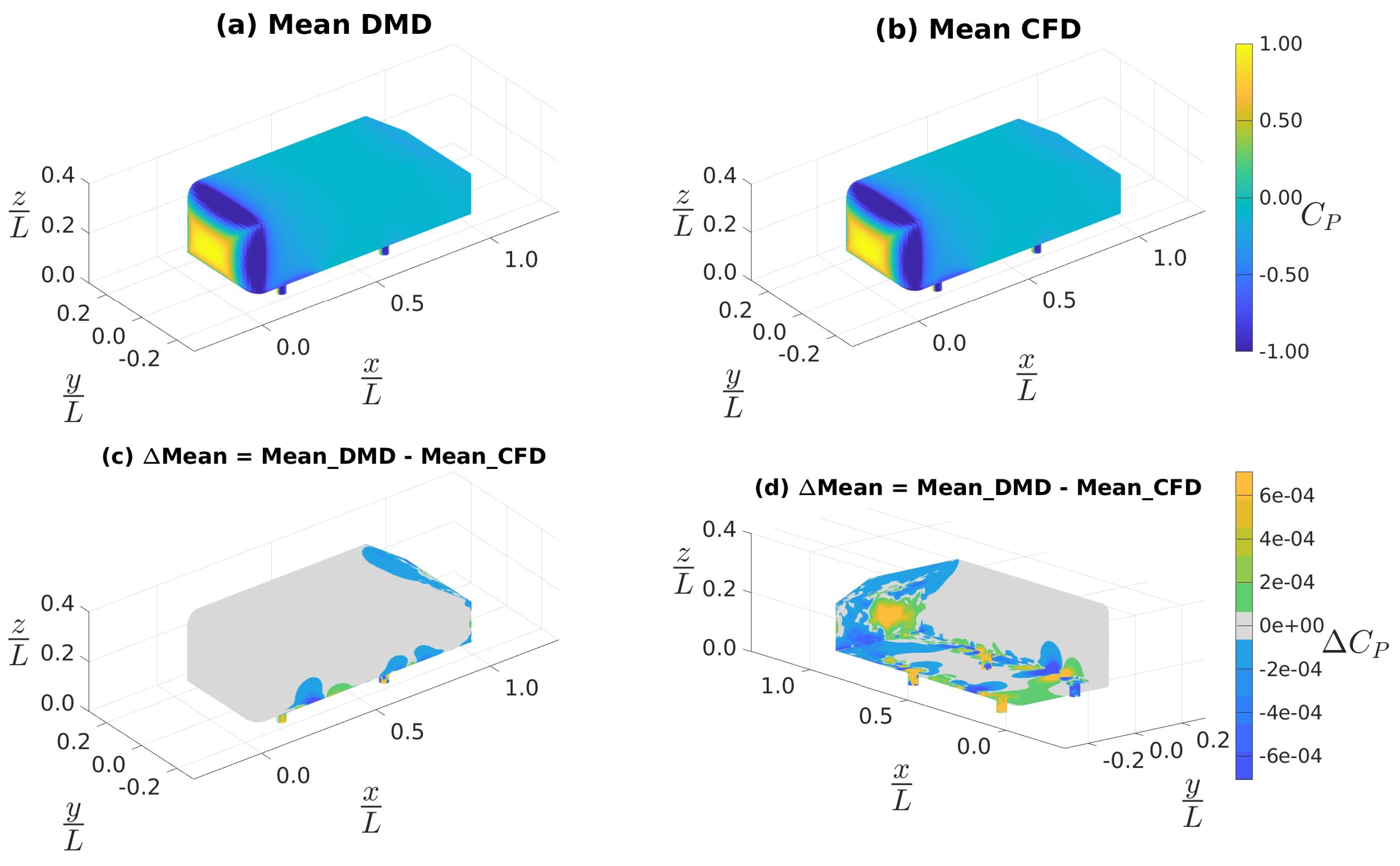
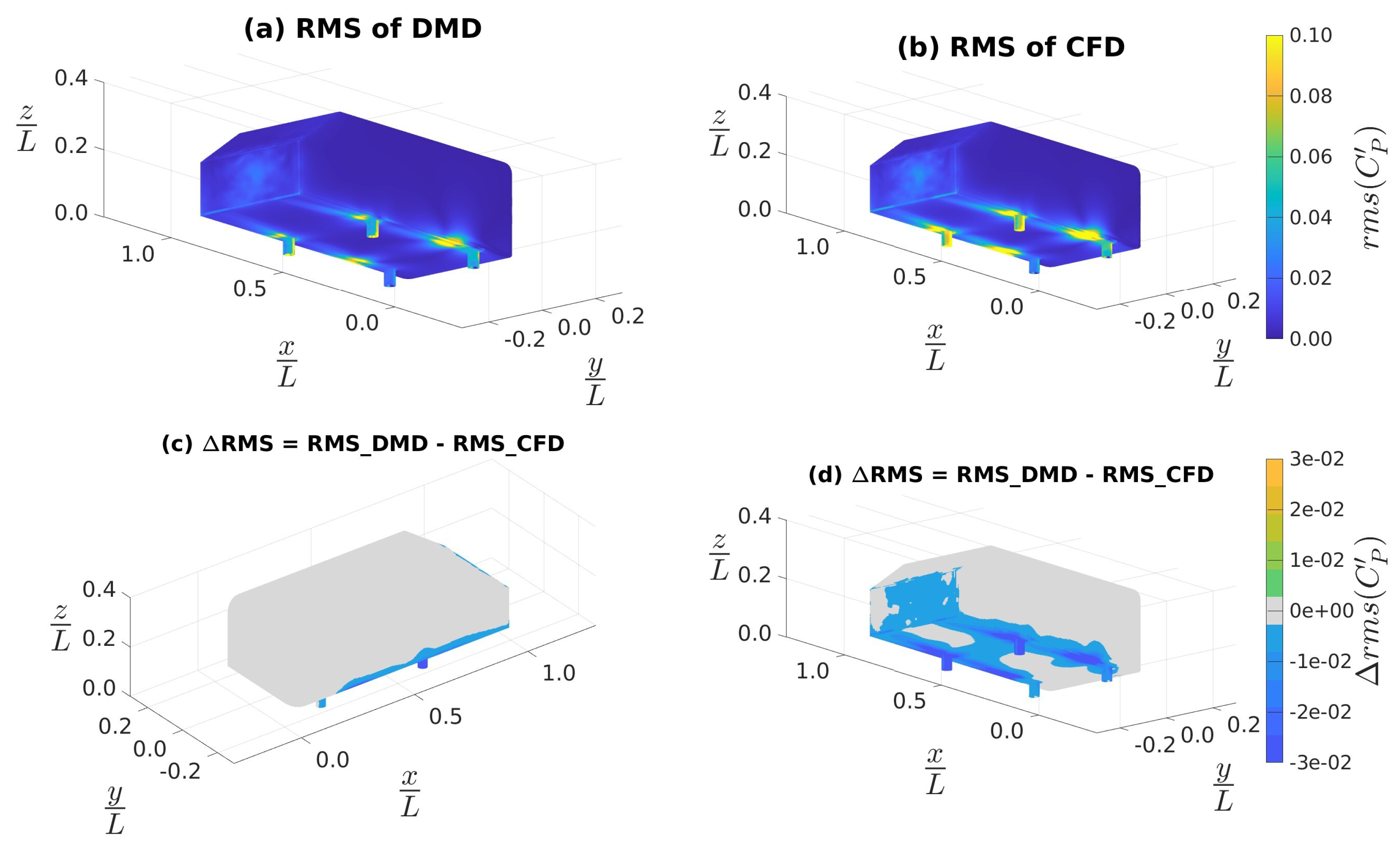
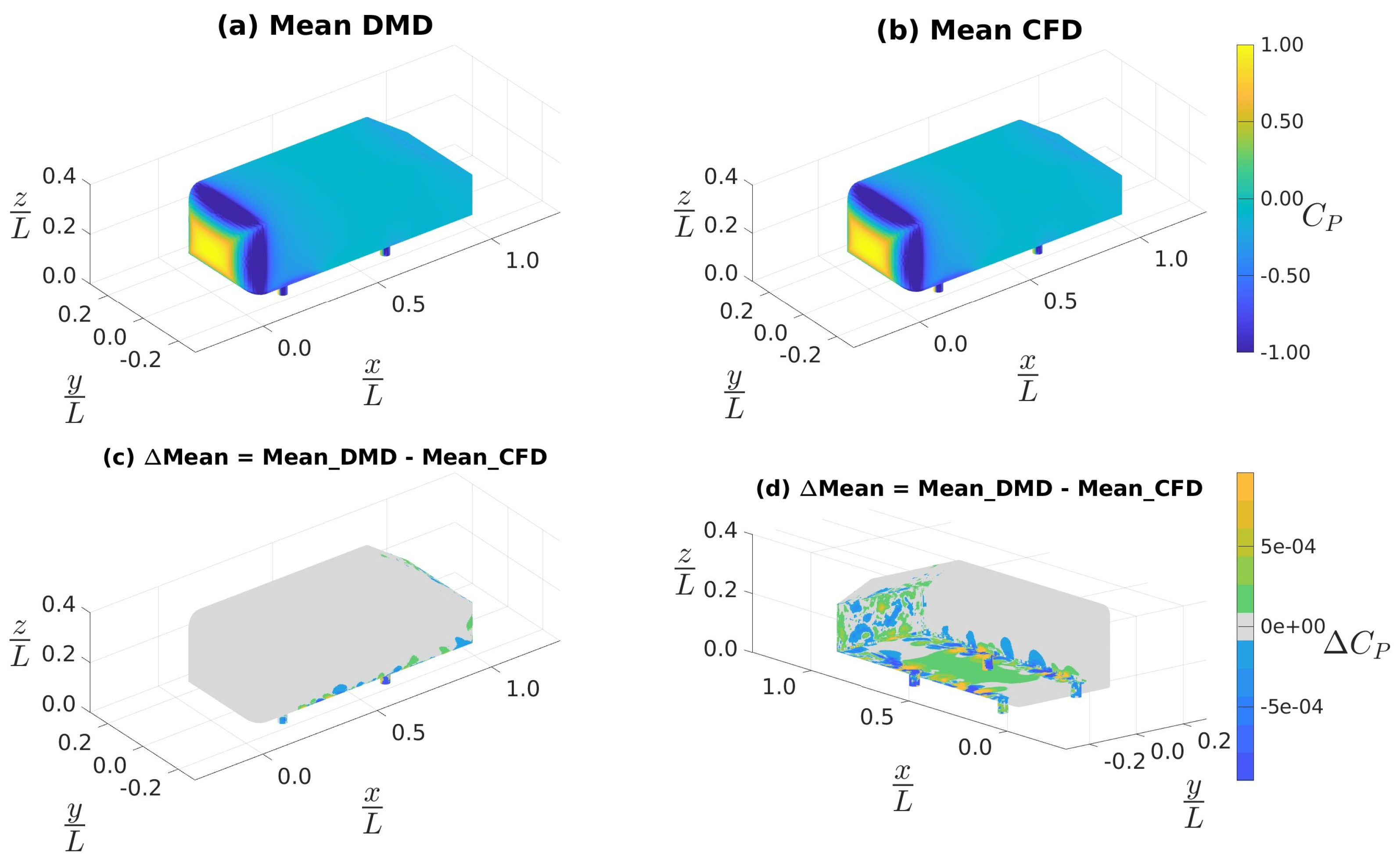
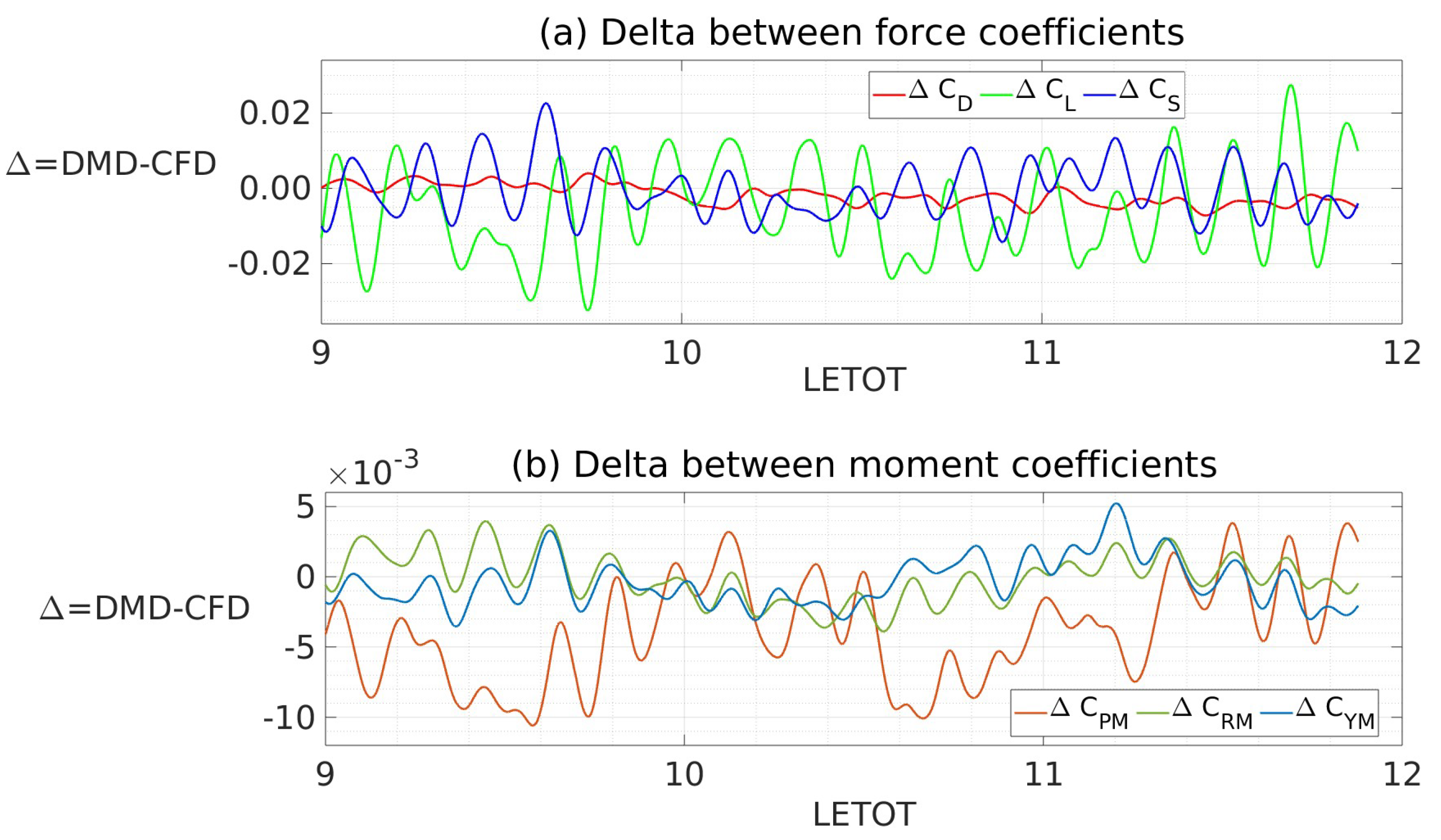
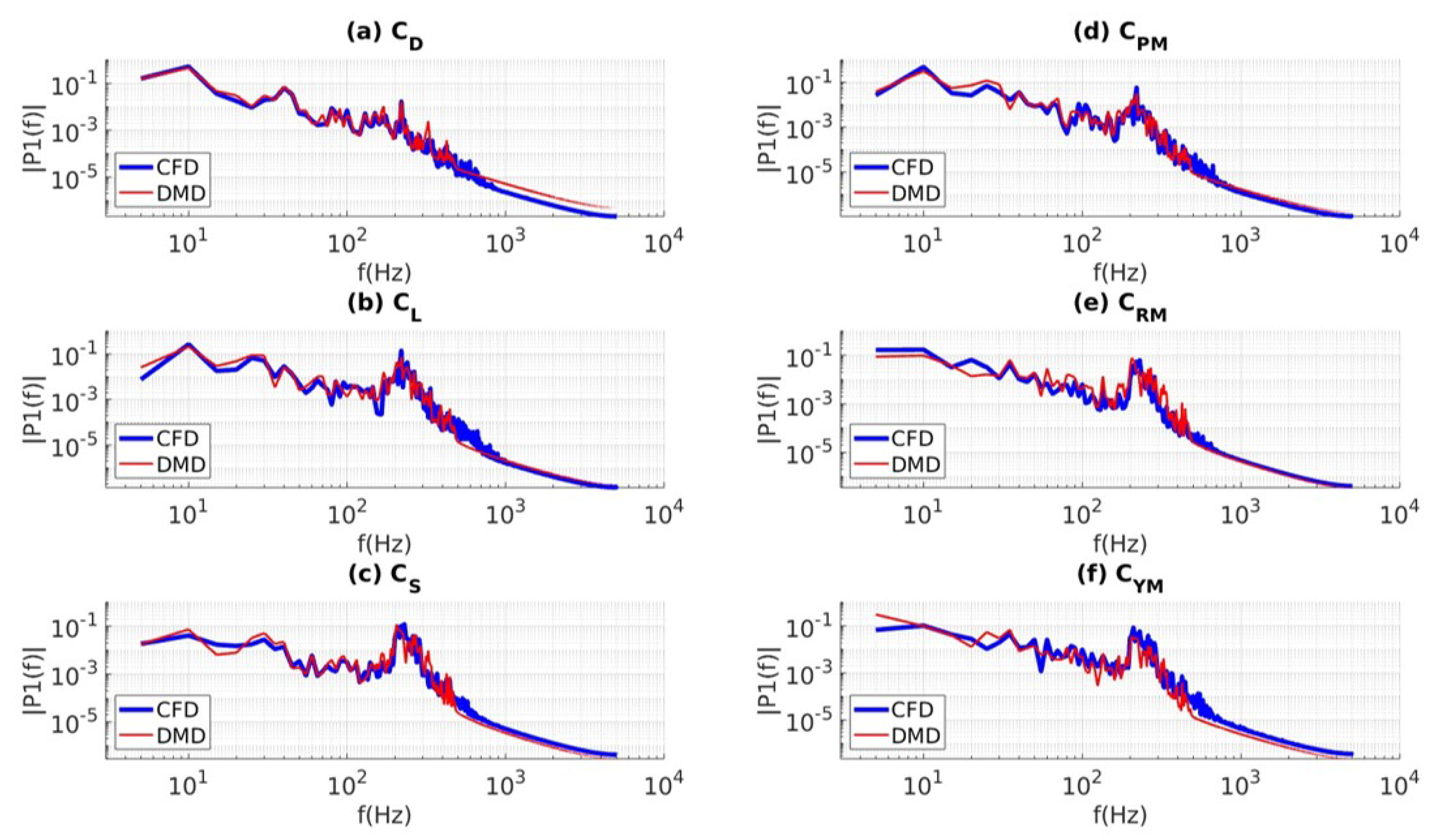
| Mean (CFD) | 0.220 | -0.062 | -0.002 | 0.019 | 0.000 | 0.000 |
| Mean (DMD) | 0.220 | -0.062 | -0.002 | 0.019 | 0.000 | 0.000 |
| RMS (CFD) | 0.003 | 0.012 | 0.006 | 0.004 | 0.002 | 0.001 |
| RMS (DMD) | 0.003 | 0.012 | 0.006 | 0.004 | 0.001 | 0.002 |
| Mean (CFD) | 0.220 | -0.059 | -0.001 | 0.022 | 0.000 | -0.001 |
| Mean (DMD) | 0.218 | -0.065 | -0.001 | 0.018 | 0.000 | -0.001 |
| RMS (CFD) | 0.002 | 0.011 | 0.005 | 0.003 | 0.001 | 0.001 |
| RMS (DMD) | 0.001 | 0.011 | 0.005 | 0.003 | 0.001 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





