# These authors contribute equally.
1. Introduction
The Solute carrier (SLC) transporters comprise a superfamily of hundreds of trans-membrane proteins that facilitate the transport of a variety of molecules including sugars, amino acids, and external chemicals such as drugs across membranes by using the ionic gradient [
1,
2]. Although the relevance for drug development is increasingly being recognized, the SLC transporters are highly understudied, and at least 30% are still considered entirely orphans in terms of knowledge of their substrate and biological function [
3].
bloated tubules (
blot) was identified as an orphan SLC involved in cortical F-actin organization of Malpighian tubules and early embryos of
Drosophila melanogaster [
4]. The phylogenetic analysis revealed that blot belonged to SLC6, which was characterized as a family of Na
+ and Cl
- dependent neurotransmitter transporters [
4,
5]. It has been proposed that the substrate transported by Blot is required for F-actin polymerization[
4]. Nevertheless, such substrate has not been determined yet.
In the first 2 hours,
Drosophila embryo develops as a syncytial blastoderm. After 13 continuous nuclear division, cellularization occurs at the interphase of cycle 14, converting the one-cell syncytium to a multicellular blastoderm. The process of cellularization is well documented [
6]. Briefly, the plasma membranes between adjacent nuclei invaginate, and the membranes at the invaginating front form furrow canals, where F-actin and its associated proteins such as myosin, anillin are enriched. At the end of cellularization, furrow canals widen, and fuse with the neighbors, enclosing the individual nuclei.
The successful initiation and progression of cellularization depend on the tight regulation of F-actin. Maternally depleting F-actin regulator, such as Dia [
7,
8,
9], Arp2/3 [
10], Scar [
11], and Slam [
12,
13], results in the failure of cellularization. In this study, we uncovered Blot was required during cellularization, and localized at the membrane with the enrichment at furrow canals. Further, we identified a Blot-binding partner, the unconventional RhoGEF ELMO/DOCK, which is required for Blot-mediated F-actin organization and cellularization. We observed that Malpighian tubules were widened in the absence of ELMO, resembling the reported
blot phenotype [
4]. Blot and ELMO interact physically, and colocalize at the furrow canal in cellularizing embryo. Moreover, we found that localization of ELMO depended on Blot. Our study thus reveals that instead of being a neurotransmitter transporter, Blot regulates F-actin organization by recruiting ELMO to the cell membrane during development.
2. Results
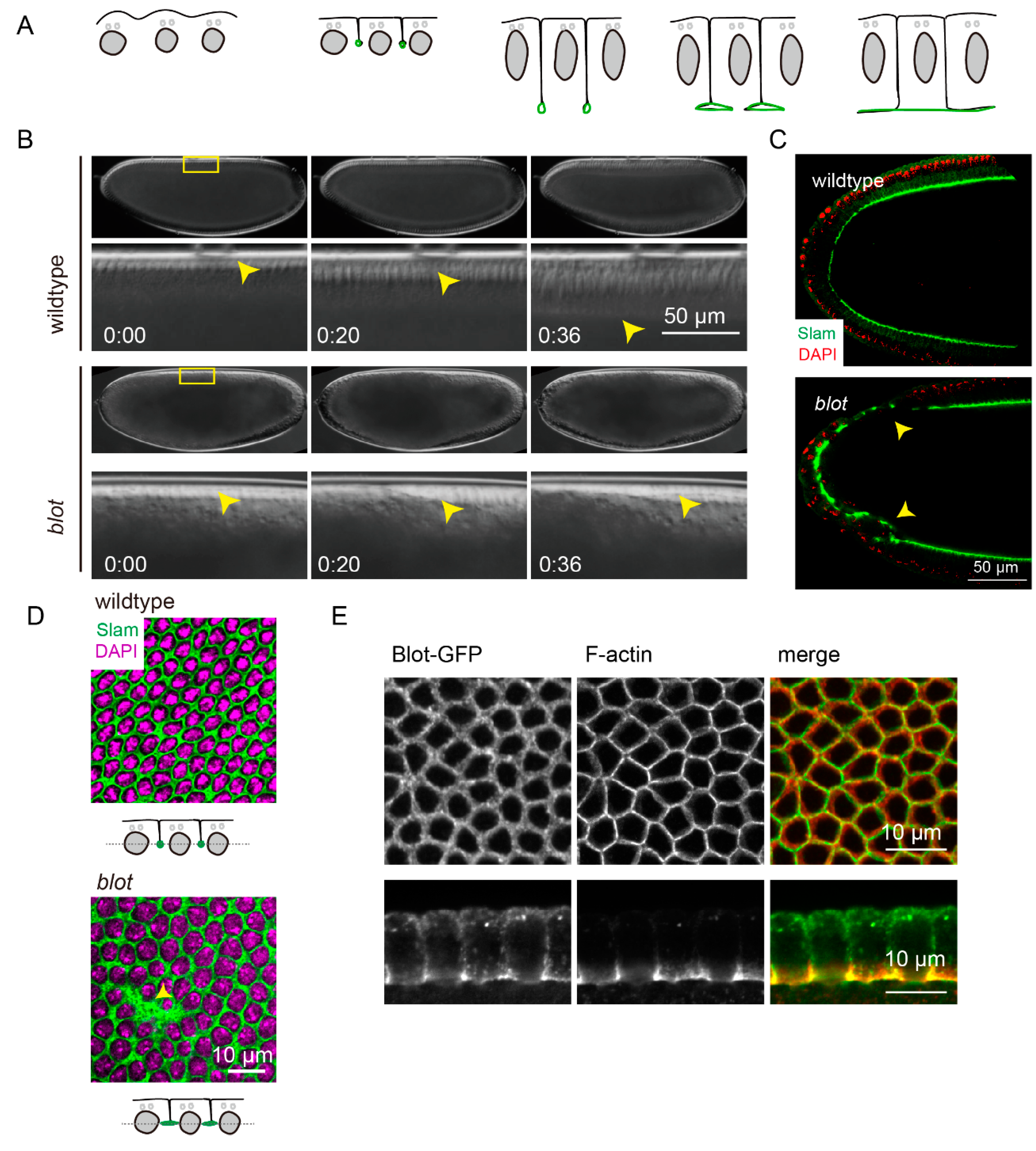
Cellularization in Drosophila embryogenesis is an ideal model to investigate the interaction between cell membrane and F-actin [
6]. The invaginating membranes between adjacent nuclei and the cortical F-actin collectively enclose the nuclei and form the monolayer epithelial cells (
Figure 1A).
To determine whether Blot is involved in cellularization, we recorded the embryonic development of the embryos from females with germline clones of
blot (hereafter called
blot embryo) using differential interference contrast (DIC) microscope (
Figure 1B). The front of membrane invagination is visible in DIC as indicated by yellow arrowhead. The membranes invaginated to 45 µm in 36 min in wildtype embryo, whereas the cellularization process was delayed dramatically, albeit the furrow canals were visible in
blot embryo. Next, we fixed and immuno-stained for Slam, which localizes at furrow canal exclusively. A large population of the embryos showed the incomplete cellularization (
Figure 1C, arrowhead). We also noticed that the furrow canals in
blot embryo were wider (
Figure 1D, arrowhead). Further, we utilized two independent RNAi lines against
blot, crossing with 67,15-Gal4, a maternal Gal4 line, to reduce the
blot gene expression level. We observed a consistent phenotype in RNAi embryos as in
blot germline clone embryos using DIC and immuno-staining (Supp. Figure), confirming that Blot is necessary for cellularization.
To further study the function of Blot, we generated transgenic flies expressing Blot-GFP under the control of UAS. Surprisingly, overexpression of Blot maternally induced embryonic lethality. DIC recordings revealed that the cellularization process was disrupted, similar with the one in
blot germline clone embryos. A small fraction of the embryos overexpressing Blot-GFP remained normal structure. In such embryos, we found Blot-GFP localized along the plasma membrane with an enrichment at furrow canal during cellularization, overlapped with F-actin distribution (
Figure 1E).
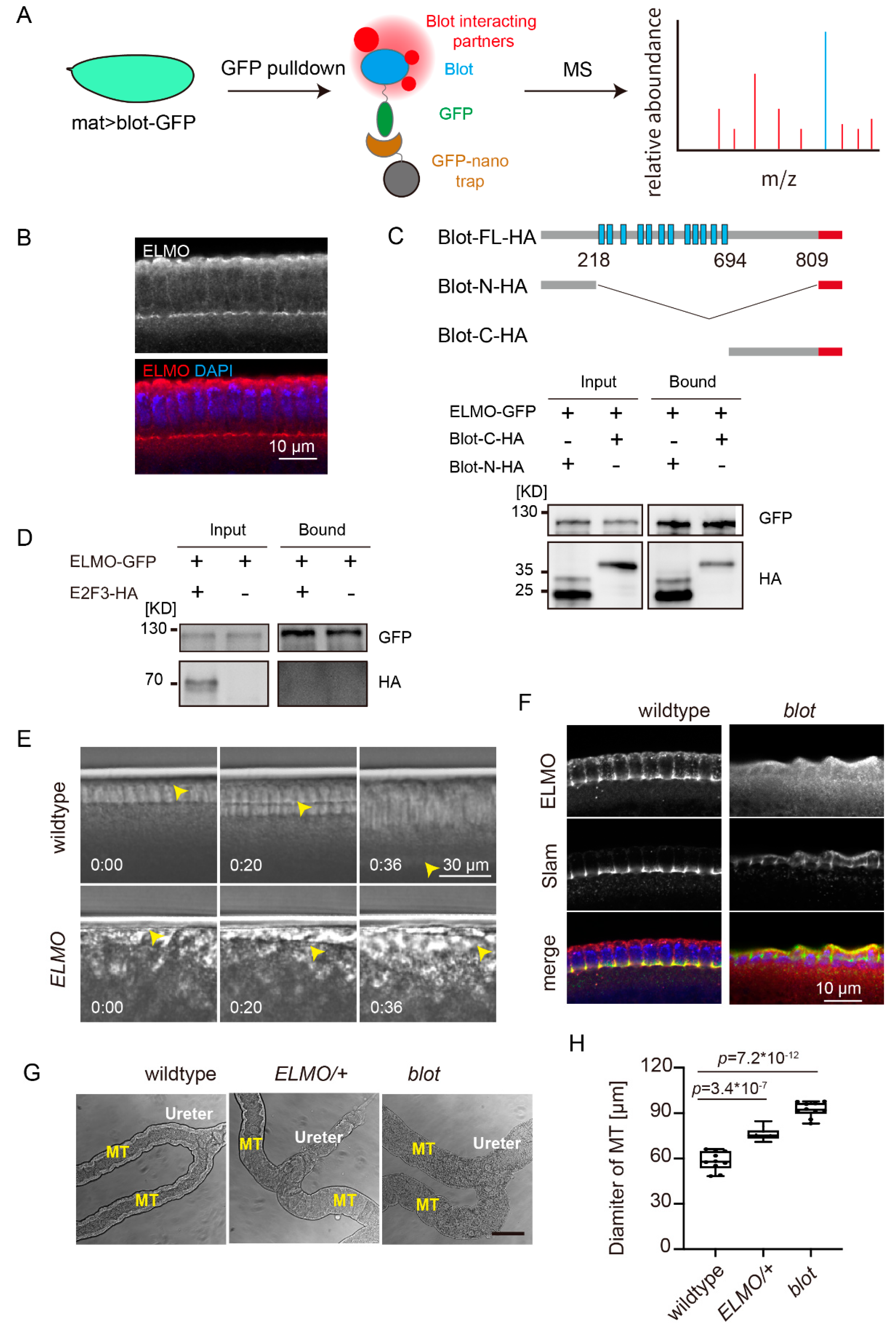
Our data, together with previous study[
4], imply that Blot is involved in F-actin organization. Blot belongs to SLC6 family, which transports neurotransmitter and amino acids. How is a transporter protein required for F-actin organization? To address this question, we performed GFP pulldown– mass spectrometry experiments using the lysates from embryos overexpressing Blot-GFP as a bait (
Figure 2A). On the list of the binding partners, ELMO is of particular interest since it is a subunit of an unconventional RhoGEF ELMO/DOCK180 complex [
14,
15]. We purified a fragment of ELMO (aa240–aa480) and immunized mouse to generate the polyclonal antibodies. Interestingly, ELMO was found enriched at the furrow canals by immunostaining, sharing a similar subcellular localization to Blot (
Figure 2B).
To test whether ELMO and Blot binds directly, we preformed GFP-pulldown assay using HEK293 cells co-transfected Blot-HA and ELMO-GFP. However, only a very small fraction of cells was able to express Blot after transfection, making it difficult to detect by western blot. To circumvent this problem, we transfected the soluble fractions of Blot, i.e., N- and C-terminal regions rather than the full-length which contains 12 transmembrane regions to the HEK293 cells. Following expression of ELMO-GFP and Blot-N/C-HA in HEK293 cells and immunoprecipitation with GFP antibodies, Blot-N and -C terminal cytoplasmic domains were detected in the bound fraction (
Figure 2C). To confirm the interaction of Blot and ELMO is specific, we co-transfected the transcriptional factor E2F3-HA and ELMO-GFP and precipitated by GFP-nanobodies. In the bound fraction, we could not detect E2F3 (
Figure 2D), implying a specific interaction of Blot and ELMO.
ELMO is required for proper cortical F-actin formation [
16] and adherens junction assembly [
15]. We asked whether ELMO was also required for cellularization. DIC time lapse images from
ELMO germline clone embryos showed that the membrane invagination was impaired dramatically, which is reminiscent of
blot embryos (
Figure 2E).
Next, we checked the localization of ELMO in
blot embryo. Strikingly, furrow canal localization of ELMO was disappeared in the absence of Blot, albeit the furrow canal structure was present in
blot embryos as Slam signal indicated (
Figure 2F).
Based on our data, we propose that Blot functions as an F-actin regulator by recruiting RhoGEF ELMO/DOCK180 complex to the membrane, rather than transporting unknown substrates as a neurotransmitter transporter. We then asked whether this was true in other contexts. It has been reported that the Malpighian tubules display bloated appearance in blot mutant larvae [
4]. We dissected the Malpighian tubules from
ELMO heterozygous third instar larvae, since the homozygote cannot develop to larvae stage. The morphology of the
ELMO Malpighian tubules was indistinguishable from the one of
blot, and both were wider than wildtype (
Figure 2G and H), implying that Blot regulates F-actin via recruiting ELMO is not restricted to the cellularization process.
3. Discussion
SLC proteins transport sugars, proteins, metabolic waste, and neurotransmitters, playing an important role in homeostatic control, making them being often considered to be therapeutic targets in the field of drug development. However, SLCs are relatively understudied compared to other gene families with similar status[
3,
17].
In this study, we investigated the biological function of an orphan SLC, Blot, during
Drosophila embryo cellularization process.
blot was identified as a novel gene expressing in the Malpighian tubules by P-element enhancer trap [
4]. The Malpighian tubules are wider in the absence of
blot (hence the name
bloated tubules) and the cortical F-actin is ill-structured in
blot embryo [
4]. Blot is clustered in SLC6 family based on its sequence [
4,
5]. It was proposed that the potential molecules transported by Blot was vital for F-actin polymerization [
4]. Nevertheless, no substrate of Blot has been identified yet. Blot contains relative larger cytoplasmic domains compared to other members in SLC6 family [
5], which provides potential sites for post-translational modification and interaction with other proteins.
By GFP-pulldown and mass spectrometry we identified that ELMO bound Blot in
Drosophila embryo lysate. ELMO was first identified in
C.elegans as an essential gene for engulfment of the apoptotic cells with the name of
ced-12 [
18]. Molecular cloning and characterization showed Ced-12 functioned as an upstream factor of Ced-10, Rac in C.elegans [
19]. At the same time, the homolog of
ced-12 in human was identified, and renamed as
ELMO (engulfment and cell motility) [
20]. ELMO has no intrinsic catalytic activity and instead binds to DOCK180, forming a functional complex with RhoGEF activity [
21,
22]. It has been reported that ELMO and Sponge (the ortholog of DOCK180 in
Drosophila) are essential for cortical F-actin organization in syncytial blastoderm [
16,
23]. Although PH domain of ELMO might be involved in the interaction with phosphatidylinositol lipids, the experiments in
C.elengas have shown that PH domain is not sufficient for its membrane localization [
19]. How ELMO/Sponge complex is activated or recruited remained unknown.
Unexpectedly, we uncovered the transmembrane protein Blot recruited ELMO to the membrane. Blot binds to ELMO physically via its N- and C-terminal cytoplasmic regions. ELMO is not able to localize at the furrow canal when Blot is depleted.
We propose Blot plays a role in F-actin organization as an upstream factor of ELMO by recruiting ELMO to the target membrane compartments. Yet we were not able to exclude the contribution of transport activities from Blot, which needs to be further investigations.
4. Materials and Methods
4.1. Drosophila Genetics
Flies were kept at 25 °C under standard conditions. The following fly lines were used:
| Drosophila genotype
|
Resource or Reference
|
| y w |
maintained in the lab |
|
blot[MI05046]/TM3 |
BDSC: 38188 |
|
w; ELMO[367]/CyO |
[15] |
| 67,15-Gal4 |
maintained in the lab |
| hs-flp; ovoD[3L] Frt3L/TM3 |
maintained in the lab |
| hs-flp; ovoD[2L] Frt2L/CyO |
maintained in the lab |
| vas-int; attPZH-86Fb |
BDSC: 24749 |
| UASp-blot-GFP/TM3, Sb |
made in this study |
To get the blot germline clone females, blot[MI05046]/TM3 was recombined with Frt3L, crossed with hs-flp; ovoD[3L] Frt3L/TM3, then the F1 larvae were heat shocked at 37℃ for 2 h in two consecutive days. The eggs from the blot[MI05046] Frt3L/ovoD[3L] Frt3L females were regarded as the eggs without Blot protein or its mRNA supplied maternally. ELMO germline clone females were obtained in the same way.
Genotypes by figure:
Figure 1B (upper panel), C (upper panel) and D (upper panel): y w
Figure 1B (lower panel), C (lower panel) and D (lower panel): embryos from
blot[MI05046] Frt3L/ovoD[3L] Frt3L females
Figure 1E: embryos from 67-Gal4/+; 15-Gal4/UASp- blot-GFP females
Figure 2A: embryos from 67-Gal4/+; 15-Gal4/UASp- blot-GFP females
Figure 2E (upper panel), F (left panel) and G (left panel): y w
Figure 2E (lower panel): embryo from
ELMO[367] Frt2L/ovoD[2L] Frt2L females
Figure 2F (left panel): embryos from
blot[MI05046] Frt3L/ovoD[3L] Frt3L females
Figure 2G (middle panel):
ELMO[367] /CyO
Figure 2G (right panel):
blot[MI05046]/TM3
Supplementary figure: the embryos from 67-Gal4/+; 15-Gal4/ UAS-blot-RNAi
4.2. Antibody Generation
The ELMO antibody was generated in mice after immunization with a purified recombinant ELMO fragment (aa240–aa480) with 6xHis tag. The serum from each mouse was collected and tested by immnostaining.
4.3. DIC Live Imaging
The staged embryos were collected and dechorionated with 50% hypochlorite bleach for 90 s, washed thoroughly, aligned on coverslips, covered with halocarbon oil. DIC images were acquired on an inverted microscopy (ZEISS Axiovert 5) with a time interval of 1 min/frame. The Malpighian tubules were dissected from the third instar larvae. The debris were removed by rinsing the Malpighian tubules in PBS and the images were taken on an inverted microscopy (ZEISS Axiovert 5) as soon as possible. Images were edited using Fiji [
24].
4.4. Histology and Microscopy
The dechorionated embryos were fixed by 4% formaldehyde in PBS/heptane for 20 min at room temperature. The vitelline membranes were manually removed. Fixed embryos were stained in PBST (PBS with 0.1% Tween-20) consecutively with primary antibodies, fluorescent secondary antibodies/ Phalloidin, DNA dyes and mounted in Aqua polymount (Polysciences). Antibodies against the following antigens were used: Slam (rabbit, 1:5000, gift from Grosshans lab), ELMO (mouse, 1:1000, made in this study). Secondary antibodies were labeled with Alexa dyes (Invitrogen, 5 μg/mL). Phalloidin-Alexa 555 (Invitrogen) stained F-actin. GFP was stained by a nanobody labeled with ATTO 488 dye (GFP-Booster, chromotek). DNA was stained by DAPI (Sigma).
Confocal images were acquired on ZEISS LSM 900, 63 oil objective (Carl Zeiss, ×63/oil, NA1.4). Images were edited using Fiji [
24].
4.5. GFP-pulldown and mass spectrometry
2-4 hours aged embryos (0.1 gram, roughly 10,000 embryos) for each sample were collected. The genotypes of the fly females were yw (control) and 67-Gal4/+; 15-Gal4/UAS-Blot-GFP (Blot-GFP maternal overexpression). The embryos were dechorionated, weighted, snap frozen in liquid nitrogen, transferred in lysis buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 0.5% NP-40, supplemented with protease inhibitor (Roche, 11697498001), 0.1 gram embryos in 1 mL lysis buffer) and crushed by Dounce homogenizer 5~10 times on ice. The lysates were centrifuged at 12,000 rpm for 10 min at 4°C to remove the debris. The clear supernatants were incubated with rinsed GFP-trap magnetic agarose resin (Chromotek, gtma) at 4°C for 2 hours, rinsed 3x and resuspended in 2x Laemmli buffer without bromophenol blue.
Protein samples were further purified on 12% SDS-PAGE and treated with in-gel trypsin digestion. Peptides were harvested analyzed by liquid chromatography (LC)-tandem mass spectrometry (MS/MS) with a Q-Exactive Mass Spectrometer.
The spectra were searched against UniProt Drosophila melanogaster database by Mascot2.2 with the following parameter: Dynamical modifications: Oxidation (M); Fixed modifications: Carbamidomethyl (C); Max Missed Cleavages: 2; ProteomicsTools: 3.1.6; Filter by score ≥ 20
4.6. Cell culture, immunoprecipitation and Western blotting
HEK293T cells were cultured followed standard protocol. Lipo3000 liposome transfection system was used for transfection. The transfected cells were collected after 2 days transfection, and lysed in lysis buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 0.5% NP-40, supplemented with protease inhibitor (Roche, 11697498001). Co-immunoprecipitation was performed using GFP-trap magnetic beads (ChromoTek, gtma), following the manufacturer's protocol. Briefly, the lysates were incubated with the rinsed GFP-trap beads at 4°C for 2 hours, rinsed 3x and resuspended in 2x Laemmli buffer. Protein sample were separated by 10% SDS-PAGE, blotted to PVDF membranes by wet transfer (100 mA per mini gel, 3 hours). The PVDF membranes were blocked with 5% fat-free milk in PBST (PBS with 0.1% Tween-20), incubated with the primary antibodies [rabbit-anti-GFP (1:2000, 2956T, CST), rabbit-anti-HA (1:4000, 3724T, CST)] for 2 hours at room temperature or overnight at 4°C. After rinsed 3x15 min, the PVDF were incubated with the secondary antibody [Goat Anti-Rabbit IgG (1:2000, HS101-01, TRANS)] for 1 hour, and detected by enhanced chemiluminescence (Advansta, Menlo Park, CA, USA).
Author Contributions
Conceptualization, C.Y. and Z.L.; methodology, G.W., Z.Z, C.Y.; writing—original draft preparation, C.Y.; writing—review and editing, Z.L.; supervision, Z.L.; funding acquisition, Z.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Natural Science Foundation of China, grant number 32070786, the Young Taishan Scholars Program of Shandong Province, grant number qnts20191090155.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article. Raw data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank Jörg Großhans for Slam antibodies; Bloomington Drosophila Stock Center for fly stocks; Bo Dong for supporting the HEK293 cell culture.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lin, L., et al., SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov, 2015. 14(8): p. 543-60. [CrossRef]
- Colas, C., P.M. Ung, and A. Schlessinger, SLC Transporters: Structure, Function, and Drug Discovery. Medchemcomm, 2016. 7(6): p. 1069-1081. [CrossRef]
- Cesar-Razquin, A., et al., A Call for Systematic Research on Solute Carriers. Cell, 2015. 162(3): p. 478-87. [CrossRef]
- Johnson, K., E. Knust, and H. Skaer, bloated tubules (blot) encodes a Drosophila member of the neurotransmitter transporter family required for organisation of the apical cytocortex. Dev Biol, 1999. 212(2): p. 440-54.
- Thimgan, M.S., J.S. Berg, and A.E. Stuart, Comparative sequence analysis and tissue localization of members of the SLC6 family of transporters in adult Drosophila melanogaster. J Exp Biol, 2006. 209(Pt 17): p. 3383-404. [CrossRef]
- Sokac, A.M., N. Biel, and S. De Renzis, Membrane-actin interactions in morphogenesis: Lessons learned from Drosophila cellularization. Semin Cell Dev Biol, 2023. 133: p. 107-122. [CrossRef]
- Grosshans, J., et al., RhoGEF2 and the formin Dia control the formation of the furrow canal by directed actin assembly during Drosophila cellularisation. Development, 2005. 132(5): p. 1009-20.
- Afshar, K., B. Stuart, and S.A. Wasserman, Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development, 2000. 127(9): p. 1887-97. [CrossRef]
- Yan, S., et al., The F-BAR protein Cip4/Toca-1 antagonizes the formin Diaphanous in membrane stabilization and compartmentalization. J Cell Sci, 2013. 126(Pt 8): p. 1796-805.
- Stevenson, V., et al., Arp2/3-dependent pseudocleavage [correction of psuedocleavage] furrow assembly in syncytial Drosophila embryos. Curr Biol, 2002. 12(9): p. 705-11.
- Zallen, J.A., et al., SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol, 2002. 156(4): p. 689-701.
- Acharya, S., et al., Function and dynamics of slam in furrow formation in early Drosophila embryo. Dev Biol, 2014. 386(2): p. 371-84. [CrossRef]
- Lecuit, T., R. Samanta, and E. Wieschaus, slam encodes a developmental regulator of polarized membrane growth during cleavage of the Drosophila embryo. Dev Cell, 2002. 2(4): p. 425-36. [CrossRef]
- Geisbrecht, E.R., et al., Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev Biol, 2008. 314(1): p. 137-49. [CrossRef]
- Schmidt, A., Z. Lv, and J. Grosshans, ELMO and Sponge specify subapical restriction of Canoe and formation of the subapical domain in early Drosophila embryos. Development, 2018. 145(2). [CrossRef]
- Winkler, F., et al., Fluctuation Analysis of Centrosomes Reveals a Cortical Function of Kinesin-1. Biophys J, 2015. 109(5): p. 856-68. [CrossRef]
- Hediger, M.A., et al., The ABCs of membrane transporters in health and disease (SLC series): introduction. Mol Aspects Med, 2013. 34(2-3): p. 95-107. [CrossRef]
- Ellis, R.E., D.M. Jacobson, and H.R. Horvitz, Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics, 1991. 129(1): p. 79-94. [CrossRef]
- Zhou, Z., et al., The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev Cell, 2001. 1(4): p. 477-89. [CrossRef]
- Gumienny, T.L., et al., CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell, 2001. 107(1): p. 27-41.
- Cote, J.F. and K. Vuori, GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol, 2007. 17(8): p. 383-93. [CrossRef]
- Biersmith, B., et al., The DOCK protein sponge binds to ELMO and functions in Drosophila embryonic CNS development. PLoS One, 2011. 6(1): p. e16120. [CrossRef]
- Postner, M.A., K.G. Miller, and E.F. Wieschaus, Maternal effect mutations of the sponge locus affect actin cytoskeletal rearrangements in Drosophila melanogaster embryos. J Cell Biol, 1992. 119(5): p. 1205-18. [CrossRef]
- Schindelin, J., et al., Fiji: an open-source platform for biological-image analysis. Nat Methods, 2012. 9(7): p. 676-82. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).