1. Introduction
Malaria is an infectious disease, potentially fatal, caused by Plasmodium protozoan parasites and transmitted by Anopheles mosquitoes. According to the WHO, malaria is one of the leading causes of death worldwide. Globally, there were an estimated 247 million malaria cases in 2021 in 84 malaria-endemic countries, increasing from 245 million in 2020, with most of this increase coming from countries in the African Region. Furthermore, in 2020, malaria deaths increased by 10% compared with 2019, and between 2019 and 2021, there were 63 000 deaths due to disruptions to essential malaria services during the COVID-19 pandemic (WHO). Thus, malaria remains a major public health problem, especially in Africa [
1].
Data shows that individual risk for malaria infection and disease is multifactorial and modulated by host genetic factors [
2]. However, the mechanisms underlying the differences in malaria susceptibility between individuals have yet to be fully understood. Quantitative genetics have estimated that human genetic factors could explain 25% of individual variation in susceptibility to clinical malaria in Africa [
3]. Numerous studies have demonstrated a prominent role of red blood cell (RBC) Polymorphisms (SNP), such as haemoglobin-inherited disorders (thalassaemia, sickle cell disease), erythrocyte membrane protein polymorphisms (Duffy antigen) and erythrocyte enzymatic disorders (glucose-6-phosphate dehydrogenase (G6PD)) in malaria susceptibility [
4,
5,
6]. On the other hand, increasing evidence identifies polymorphisms in genes related to the immune system as essential determinant in susceptibility to malaria infection and disease. Immuno-genetic variants associated with diverse degrees of malaria susceptibility include polymorphisms in cytokine-related genes, which may affect protein levels and downstream functions, such as the production of C-reactive protein and immunoglobulin (Ig) isotype switching [
7,
8,
9,
10,
11].
The pathogenesis of malaria is complex and needs to be elucidated. During blood-stage infection, the host’s immune system produces proinflammatory cytokines to eliminate the parasite, including IL-6, IFN-γ, and TNF, which are pivotal in controlling the parasite’s growth and elimination. In many studies, the high levels of some pro-inflammatory cytokines have been protective in malaria [
12,
13]. Pro-inflammatory biomarkers were more elevated in cerebral malaria than in non-cerebral malaria patients [
14]. Regulatory cytokines such as transforming growth factor-β (TGF-β) and IL-10 balance the pro-inflammatory and anti-inflammatory responses. However, in many cases, cytokines have a double role. On the one hand, they contribute to parasitic clearance; on the other, they are responsible for pathological changes encountered in malaria. Cytokine-modulating strategies may represent a promising modern approach to disease management [
12].
Recently, the IL-17 cytokine has gained attention among malaria researchers because of its protective role in immunity against extracellular pathogens [
15,
16,
17] and for the clearance of intracellular pathogens [
18,
19,
20]. In addition to its essential role in protective immunity, IL-17 is critical in the pathogenesis of various autoimmune inflammatory diseases. IL-17 is a cytokine family that plays a vital role in innate and adaptive immune systems [
21,
22,
23,
24]. The IL-17 gene is located on chromosome 6p12, comprises three exons and two introns and is coded with six protein members (IL-17A-F). IL-17A is the most essential member of the IL-17 family. The IL-17 receptor family now comprises 5 members (IL-17RA, RB, RC, RD and RE) [
25,
26,
27].
In mice, it has been demonstrated that elevated IL-17 levels and high IL-4, IL-12α and IFN-γ levels may be a marker of protection against Plasmodium berghei [
28]. However, the role of IL-17 in human malarial infection outcomes is poorly described [
28], even if increased IL-17 levels in vivax and falciparum malaria and disease severity have been reported [
14,
29]. Further studies are needed to evaluate the implication of IL-17 cytokines levels and polymorphisms in malaria protection and/or pathogenesis.
This work analyzed IL-17A levels and gene polymorphisms in a Senegalese cohort. IL-17A gene and its flanking regions were sequenced in samples from a cohort of individuals, including healthy controls (CTR), uncomplicated malaria (UM) and Severe Malaria (SM) subjects. The Genetic variations, including single nucleotide polymorphisms (SNPs), were analyzed among individuals concerning malaria disease status to detect their influence on the IL-17A serum levels and associations with malaria severity.
2. Materials and Methods
2.1. Study participants
Our cohort included black Senegalese-born individuals whose parents and grandparents were born in Senegal, a malaria-endemic country in the Sahelian zone of West Africa. Malaria patients were enrolled from participating hospitals and corresponded to subjects with Plasmodium-positive Quantitative Buffy Coat (QBC). This test is more sensitive than Giemsa-stained thick films [
6,
30]. The malaria patients were classified into two groups: Mild Malaria (MM) and Severe Malaria (SM), according to the criteria defined by Saissy. et al. 2003, and previously described [
31]. To ensure homogeneity of data, inclusion criteria were: (1) Only black Senegalese individuals with P. falciparum infection confirmed in diagnosis; (2) persons born in Senegal; whose parents and grandparents were born in Senegal (3) individuals who have not travelled in the last three months of their hospital admission. Exclusion criteria were as follows: (1) other racial or ethnic group living in Senegal; (2) subjects with clinical signs of severity or any state that may interfere with the study, such as a recent pregnancy and childbirth; (3) previous use of antimalarial treatment or a possible stay out of town not older than three months;(4) people who travelled outside the areas few months before infection. The healthy control subjects corresponded to the exposed and uninfected subjects group in the same areas and belonging to Wolof ethnic group. A signed informed consent form was obtained from adult participants and parents or guardians of children involved in the study before blood sampling.
A total of 48 CTR, 54 UM and 71 SM subjects were included in the study. The study’s objectives have been explained clearly using the local dialect before including patients in hospital centers. The protocol has been reviewed according to the rules issued by the National Committee for Ethics for Health Research (CNERS) of Senegal and according to the procedures established by the Cheikh Anta Diop University of Dakar (UCAD) for the ethical approval of any research involving human participants. Written informed consent was obtained from adult participants and parents or legal representatives of children. In addition, based on the information provided, UCAD’s Committee on Research and Ethics (CER) considers that the research proposed respects the appropriate ethical standard and, as a result, approves its execution under “Protocole 0344/2018/CER-UCAD”. Furthermore, all patients enrolled in the cohort/or legal representative gave signed and informal written consent to provide a blood sample for further studies.
2.2. Serum collection and IL-17A quantification
Blood samples were collected from malaria patients and controls and drawn into EDTA vacutainer tubes. Samples were centrifuged, plasma aliquoted, and stored at −20°C until testing. Serum levels of IL-17A were quantified by enzyme-linked immunosorbent assays (ELISA) using the pre-designed kit (Bioassays Technology, China) as per the manufacturer. The sensitivity of the assay protocol was 2.38 pg/mL, and the coefficient of variation (CV) for intra-assays and inter-assays was <8% and <10%, respectively.
2.3. Genotyping
DNA was extracted from the peripheral blood of each subject by using standard Qiagen Kits according to the manufacturer’s recommendations. The concentration and quality of the extracted DNA were measured by using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The IL-17A polymorphisms were genotyped using the polymerase chain reaction method. The Oligonucleotide primers used to amplify promoters and exon regions are listed in
Table 1. The PCR reactions were performed using a Gotaq
®Green Master Mix (Promega, Germany) in a total volume of 25 µl containing 25 ng of genomic DNA (5 ng/µl) and 2.5 µL of each primer (10 μM). The PCR conditions were initial denaturation at 95 °C for 5 min, 35 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 5 min, with a final extension at 72 °C for 10 min. The amplicons were purified using BioGel P100 gels (Bio-Rad). Sequencing reactions (2 µL of PCR product) were performed using the dye terminator v3.1 method in an ABI PRISMs 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Sequencing conditions were: 96 °C for 5 min, 25 cycles of 96 °C for 10 s, 60 °C for 4 min and 15 °C forever, and PCR products were purified with Sephadex G50 superfine columns (GE Healthcare). Alignment of acquired sequences and SNP discovery were performed using NC_000014.9 as a reference. Analysis was performed with Genalys version 2.0b software [
32].
2.4. Statistical analysis
GraphPad Prism v9.5.1 was employed for all statistical analyses. Serum levels of IL-17A in malaria groups and controls were compared by one-way variance analysis (ANOVA). First, allelic frequencies and Hardy-Weinberg equilibrium were calculated, as described [
33]. Then, as reported previously, the differences in allelic frequencies between the three groups (SM, UM, CTR) were determined using the logistic regression analysis method [
34,
35]. Next, associations between SNPs and IL-17A serum levels and malaria outcomes were performed using the Mann–Whitney test, and then associations with P values < 0.05 were considered statistically significant.
3. Results
3.1. Characteristics of malaria patients and healthy controls
A total of 125 malaria patients and 48 controls were included in this retrospective study. The clinical characteristics of the malaria groups and the control group are summarized in
Table 2. A significant difference was observed in parameters while comparing malaria cases and controls, such as age, hemoglobin level, blood cell parameters and leukocyte cells. However, there were no statistically significant differences in monocytes, Basophils.
3.2. Severe malaria patients displayed higher serum IL-17A compared to uncomplicated malaria and controls
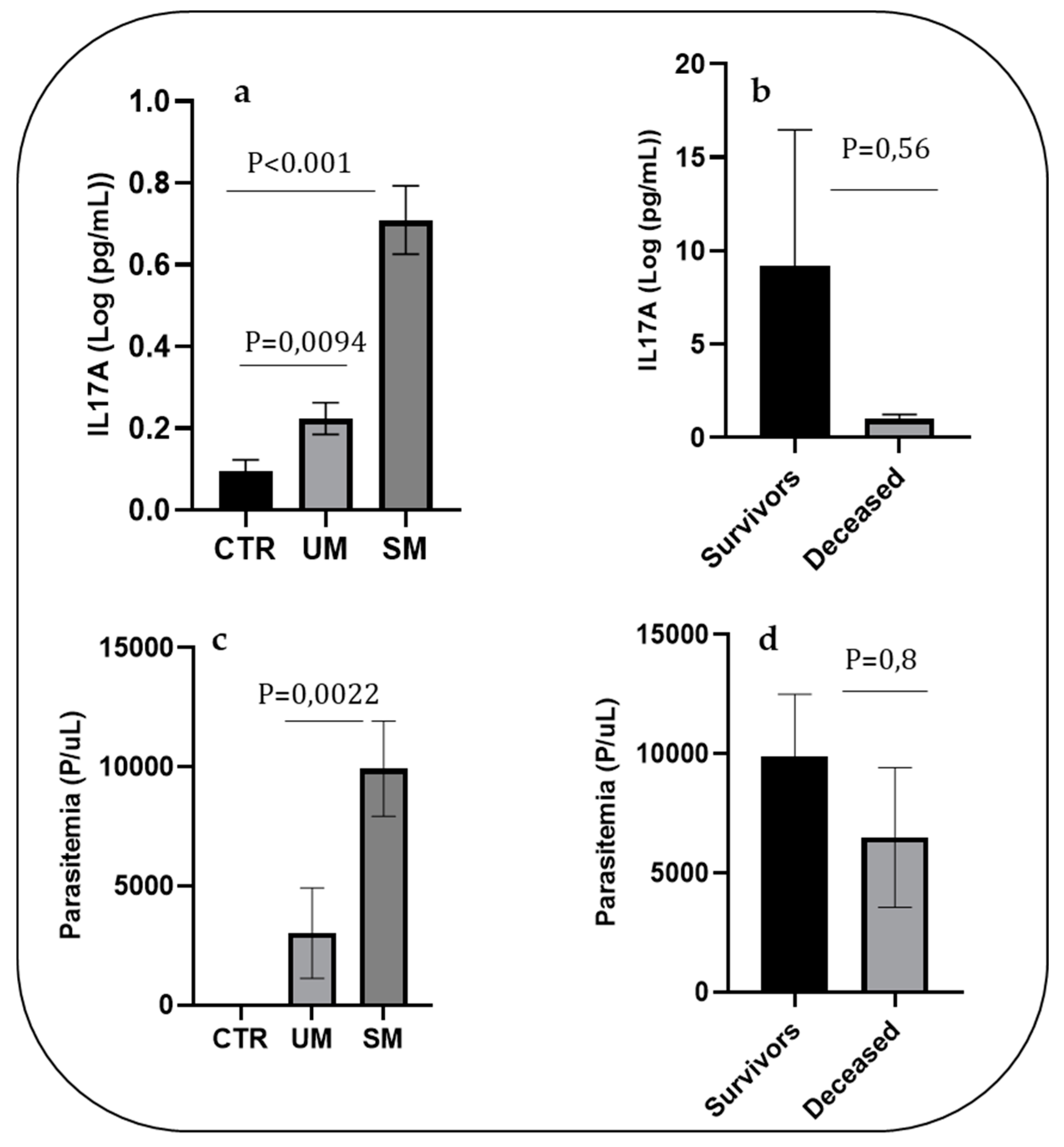
ELISA quantified the level of IL-17A. As shown in
Figure 1, the mean level of IL-17A was significantly higher in the SM group (mean±SE: 37.74±140 pg/mL) compared to that of the UM and control groups (mean±SD:1.69±2.4 pg/ml and 0.67±1.07 pg/mL, respectively; P<0.001) (
Figure 1a). In the SM group, we observed a lower level of IL-17A in patients who were deceased compared to those who survived (mean±SD: 25.92±46.48 and 40.78±150.67 pg/mL, respectively). Still, the difference was not significant (P=0.56); the lack of significance may be partly due to the small sample size (
Figure 1b). The parasitemia at the time of diagnosis in SM patients was higher than that of the UM patients (mean±SD: 9039.54±17891 and 3084±14092, respectively; P=0.0022). In addition, there was a decrease in parasitemia in the deceased compared to the survivors, but the difference was not significant (mean±SD: 9915.54±19185 and 6496±7161, respectively; P=0.8) (
Figure 1c,d).
3.3. Distribution of IL-17A polymorphisms and association with the risk of severe malaria outcome
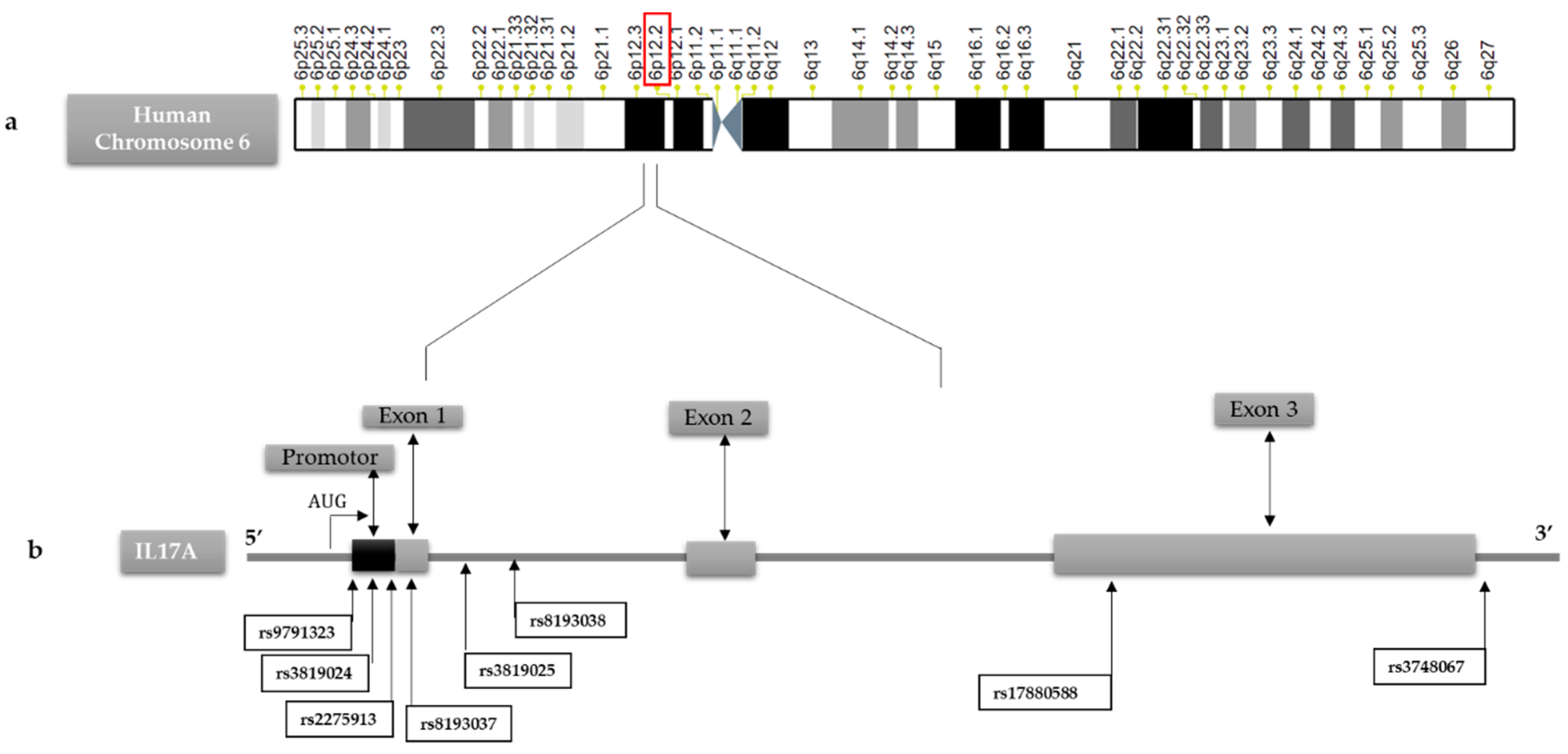
We analyzed genetic variations on the IL-17A gene in the promoter region and all the coding regions by sequencing and identified eight SNPs, including: 4 SNPs located in the 5′UTR: IL-17A +521A/C (rs9791323), IL-17A +606A/G (rs3819024), IL-17A +849G/A (rs2275913), IL-17A +973G/A (rs8193037); two SNP located within inton: IL-17A +1090G/A (rs3819025), IL-17A +1198A/G (rs8193038); one located within exon 3: IL-17A +3840G/A (rs17880588) and one located in the 3′UTR: IL-17A +5151C/T (rs3748067) (
Figure 2).
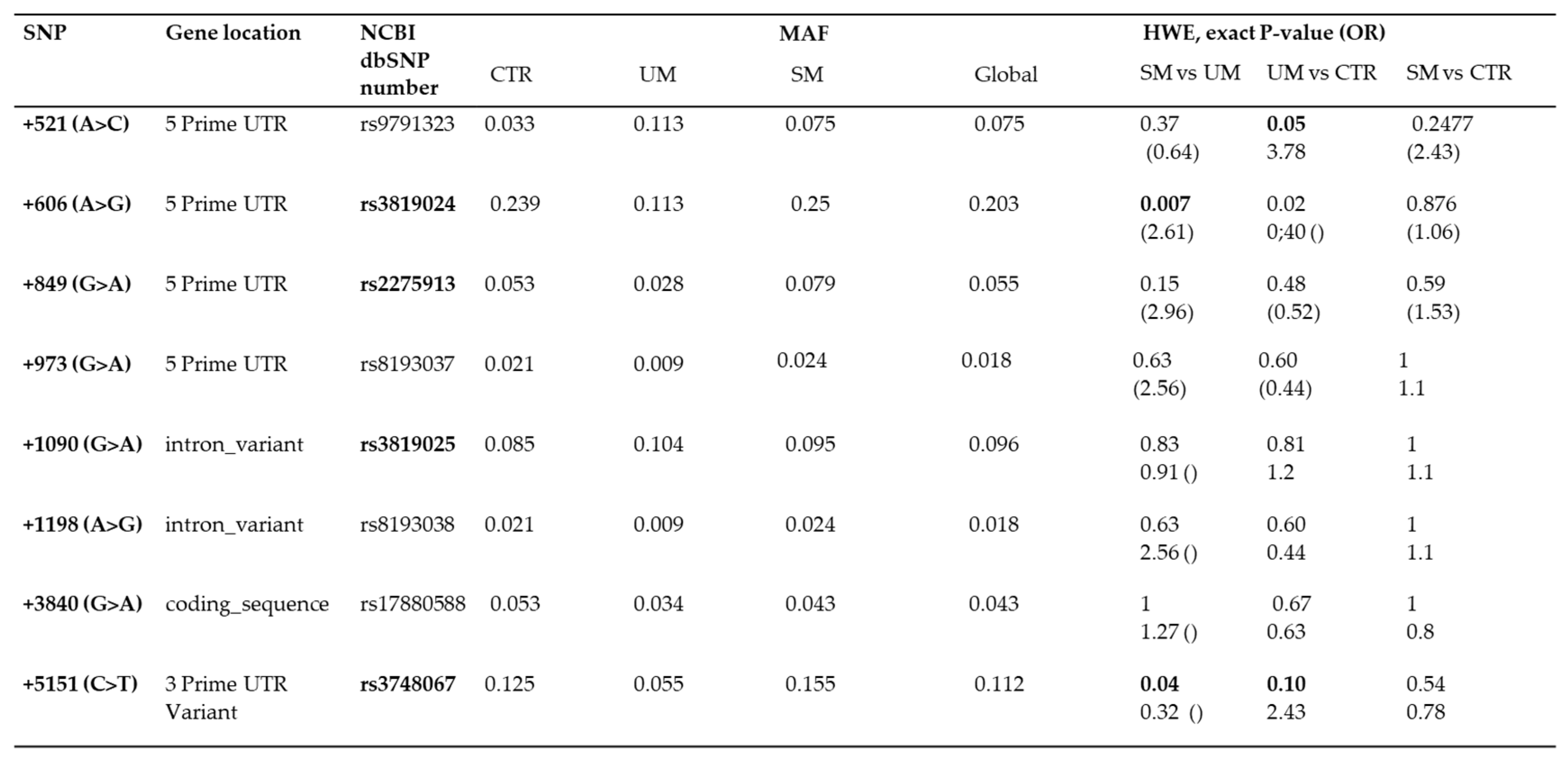
The Minor Allele frequencies (MAF) of the eight SNPs loci of the IL-17A gene were identified using the Hardy–Weinberg equilibrium test (
Table 3). Among them, 6 SNP (rs9791323, rs3819024, rs2275913, rs3819025, rs2233860 and rs3748067) were detected with high frequencies (with MAF > 3%), unlike 2 other SNPs (rs8193037 and rs8193038) with MAF < 3% were observed (
Table 3). First, comparisons were performed among the three groups to test whether polymorphisms were associated with malaria severity. Then, statistical IL-17A polymorphisms analysis were performed using logistic regression tests with an adjustment for potential confounders such as Hb polymorphisms. The SNPS rs3819024 and rs3748067 yielded a significant association with Severe Malaria. For SM vs UM, the p-value =0.007 (OR 2.61, 95% CI 0.35– 0.91), and p value= 0.04 (OR 0.32, 95% CI 1-2.1), respectively.
3.4. Association of SNP IL-17A+1198 A/G (rs8193038)) and serum IL-17A concentration
The relationship between the IL-17A polymorphisms and serum IL-17A concentration was analyzed regardless of the differences in the study groups. As shown in
Figure 3, the heterozygous rs8193038 AG genotype is significantly associated with higher levels of IL-17A amongst the whole study groups compared to the homozygous rs8193038 AA genotype (OR = 4.9, 95% CI = (2.01- 8.13), P<0.001).
4. Discussion
The analysis of the genetic effects of inflammatory response gene variants is a key step in malaria research to motivate experimental investigations of the underlying pathogenesis mechanisms. This knowledge will be critical to identify rational adjuvant therapies to prevent fatality or undesired malaria complications and subsequent long-term sequels that represent a high burden in endemic regions [
36]. Human populations display differences in susceptibility to infectious diseases such as malaria, and the basis for this differential susceptibility is due, at least in part, to genetic factors [
3,
37,
38]. It is understood that cytokine gene polymorphisms could affect the serum levels of cytokines by influencing transcriptional regulation. The role of single-nucleotide polymorphisms (SNPs) in some infectious diseases and immunological disorders has been previously reported [
39,
40,
41], and some exact genetic polymorphisms have been identified about malaria [
42,
43,
44]. Significant associations between cytokine polymorphisms and diseases support that cytokine gene polymorphisms have an unquestionable role in the orchestration of the immune response, leading to the different functional scenarios, which in turn influence the outcome of disease establishment and evolution [
36,
45,
46].
Thus, the present study explored IL-17A cytokine levels and polymorphisms in the Senegalese cohort and their association with malaria outcomes. In recent years, evaluating SNPs has been considered a common approach for testing human genetic variation [
47]. The IL-17 cytokine family is a relatively new family linked to adaptive and innate immune systems. IL-17A are members of the IL-17 cytokine family, essential for the pathogenic activity of IL-17 cells and the production of various proinflammatory mediators in the body [
18,
48].
We have determined the serum IL-17A levels and genotyped IL-17A variants in Senegalese severe and uncomplicated malaria patients and controls. We observed elevated IL-17A levels in SM patients compared to the UM and healthy cases. The high parasitemia in the SM group accompanied the IL-17A increase. This indicates that IL-17A has an essential regulatory role in malaria infection, controlling the intensity of the immune response, as described in the experimental model and human malaria and several other infectious diseases [
14,
28,
49,
50]. IL-17 production is associated with a very high occurrence of chronic inflammation and immunopathological conditions [
48]. Recent data suggest that IL-17 contributes to host protection against diverse infectious organisms during sepsis while inducing hyperinflammation with detrimental outcomes for the host under certain conditions [
51]. Earlier investigations in the experimental model have deciphered the essential role of IL-17. In P. vivax infection, authors suggest that increasing serum IL-17 levels in malaria patients could be considered a host adaptation mechanism to control changes in blood viscosity, and IL-17 could thus be used as an immunomodulatory agent [
52]. IL-17 appears to act on erythrocytes by remodeling their cell membrane; it is well-known that erythrocytes in malaria are very sensitive to osmotic shock [
52].
We found an elevated level of IL-17A in severe malaria patients whom survivors compare to those who were deceased. Our results seem to confirm the results of Helegbe et al., which showed elevated IL-17 levels together with high IL-4, IL-12α, and IFN-γ levels may be a marker of protection, and the mechanism may be controlled by host factors [
28]. Thus, pro-inflammatory IL-17A cytokine seems to have been protective against fatal malaria. Furthermore, the data agree with the observations of Oyegue-Liabagui et al. [
29], who noted a correlation between Th17 cell count and overall survival in patients with malaria in children.
Immuno-genetic variants are associated with diverse degrees of malaria susceptibility, including cytokine gene polymorphisms that modify their expression and their circulating protein levels to reflect inflammatory or anti-inflammatory responses [
53,
54,
55]. Polymorphisms in the IL-17A cytokine can impact the activity and expression of inflammatory mediators, which can affect interleukin-17 activity [
56,
57]. IL-17A polymorphisms have been linked to several malignancies, including gastric and breast cancer [
58,
59]. But little is known about the association between IL-17 gene variation and malaria.
One previous study has reported that IL-17F (rs6913472 and rs4715291) and IL-17RA (rs12159217 and rs41396547) polymorphisms independently modulate susceptibility to Cerebral Malaria and provide evidence that IL-17F protects against CM [
8]. In this study, we performed a genetic analysis of the variations of the IL-17A gene. For the first time, we identified two variants, rs3819024 and rs3748067, that are significantly associated with SM risk in the Senegalese population. For SM vs UM, the p-value =0.007 (OR 2.61, 95% CI 0.35– 0.91) and p value= 0.04 (OR 0.32, 95% CI 1-2.1), respectively. We also found that the heterozygous rs8193038 AG genotype is significantly associated with higher levels of IL-17A amongst the whole study groups compared to the homozygous rs8193038 AA genotype (OR = 4.9, 95% CI = (2.01- 8.13), P<0.001). These data suggest that the IL-17A gene rs8193038 polymorphism significantly affects IL-17A gene expression. Genetic variants controlling inflammatory responsiveness are proposed determinants of malaria clinical outcomes dependent on a history of exposure to infection [
2,
36].
A limitation of our study could be the need for a statistical correlation between IL-17A level, genotypes and malaria outcomes (SM and UM); this is due to a few numbers of patients. Therefore, further recruitment is needed to increase SM and UM patients.
5. Conclusions
The current report revealed an essential role of IL-17A in the pathogenesis of SM in Senegalese patients. Furthermore, heterozygous mutant and minor alleles of IL- rs3819024 and rs3748067 polymorphisms predisposed subjects for the development of SM. Interestingly, the current report further validated the functional relevance of IL-17A (rs8193038) variants and demonstrated the association of mutants with elevated IL-17A levels. However, further studies, including more significant sample-sized in the different populations, are required to validate the observations of the present study. In addition, further investigation on the role of IL-17 and its interplay with other immune factors needs to be conducted in clinical settings.
Author Contributions
FT conceived the study and the methodology and drafted the manuscript. GD conceived the study and the methodology. JFZ and CC conducted the data analysis, revised the manuscript and approved the final version. CD conducted the methodology, performed Sanger Sequencing and molecular biology experiments and approved the final version. BM conducted the malaria cohort recruitment, revised the manuscript from the hospital center, and approved the final version. AAMD contributed to the correction and revision of the manuscript and approved the final version. AT contributed to the correction and revision of the manuscript and approved the final version. MDM contributed to the correction and revision of the manuscript and approved the final version. MD contributed to the correction and revision of the manuscript and approved the final version. CMN contributed to the correction and revision of the manuscript and approved the final version. YD contributed to the correction, revising the manuscript and approving the final version. JFD & AD coordinated this study, revised the manuscript, and approved the final version. All authors read and approved the final manuscript.
Funding
This research received no external funding
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of UCAD’s Committee on Research and Ethics (CER) considers that the research proposed respects the appropriate ethical standard and, as a result, approves its execution under “Protocole 0344/2018/CER-UCAD”.
Informed Consent Statement
“Informed consent was obtained from all subjects involved in the study” and “Written informed consent has been obtained from the patient(s) to publish this paper”.
Data Availability Statement
Acknowledgments
The authors are grateful to all the patients and the medical staff who have generously collaborated in the Malaria Genomic Project Clayton Dedonder 2014 (Institut Pasteur de Dakar). In addition, the authors thank the team of the Centre National de Recherche en Génétique humaine (CNRGH) for assistance in performing the genotyping. The French Ministry of Education and Research supported the CNRGH-CEA.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. 2021. World malaria report 2020. Geneva: WHO, 1–178.
- Natama HM, Rovira-Vallbona E, Krit M, Guetens P, Sorgho H, Some MA, Traore-Coulibaly M, Valea I, Mens PF, Schallig H et al.: Genetic variation in the immune system and malaria susceptibility in infants: a nested case-control study in Nanoro, Burkina Faso. Malar J 2021, 20(1):94. [CrossRef]
- Mackinnon MJ, Mwangi TW, Snow RW, Marsh K, Williams TN: Heritability of malaria in Africa. PLoS Med 2005, 2(12):e340. [CrossRef]
- Driss A, Hibbert JM, Wilson NO, Iqbal SA, Adamkiewicz TV, Stiles JK: Genetic polymorphisms linked to susceptibility to malaria. Malar J 2011, 10:271. [CrossRef]
- Marquet S: Overview of human genetic susceptibility to malaria: From parasitemia control to severe disease. Infect Genet Evol 2018, 66:399-409. [CrossRef]
- Thiam F, Diop G, Coulonges C, Derbois C, Mbengue B, Thiam A, Nguer CM, Zagury JF, Deleuze JF, Dieye A: G6PD and HBB polymorphisms in the Senegalese population: prevalence, correlation with clinical malaria. PeerJ 2022, 10:e13487. [CrossRef]
- Prokunina-Olsson L, Morrison RD, Obajemu A, Mahamar A, Kim S, Attaher O, Florez-Vargas O, Sidibe Y, Onabajo OO, Hutchinson AA et al.: IFN-lambda4 is associated with increased risk and earlier occurrence of several common infections in African children. Genes Immun 2021, 22(1):44-55. [CrossRef]
- Marquet S, Conte I, Poudiougou B, Argiro L, Cabantous S, Dessein H, Burte F, Oumar AA, Brown BJ, Traore A et al.: The IL17F and IL17RA Genetic Variants Increase Risk of Cerebral Malaria in Two African Populations. Infect Immun 2016, 84(2):590-597. [CrossRef]
- Kisia LE, Kempaiah P, Anyona SB, Munde EO, Achieng AO, Ong’echa JM, Lambert CG, Chelimo K, Ouma C, Perkins DJ et al.: Genetic variation in interleukin-7 is associated with a reduced erythropoietic response in Kenyan children infected with Plasmodium falciparum. BMC Med Genet 2019, 20(1):140. [CrossRef]
- Furini AA, Capobianco MP, Storti-Melo LM, Cunha MG, Cassiano GC, Machado RL: Cytokine gene polymorphisms are not associated with anti-PvDBP, anti-PvAMA-1 or anti-PvMSP-119 IgG antibody levels in a malaria-endemic area of the Brazilian Amazon. Malar J 2016, 15(1):374. [CrossRef]
- Cassiano GC, Furini AA, Capobianco MP, Storti-Melo LM, Almeida ME, Barbosa DR, Povoa MM, Nogueira PA, Machado RL: Immunogenetic markers associated with a naturally acquired humoral immune response against an N-terminal antigen of Plasmodium vivax merozoite surface protein 1 (PvMSP-1). Malar J 2016, 15:306. [CrossRef]
- Popa GL, Popa MI: Recent Advances in Understanding the Inflammatory Response in Malaria: A Review of the Dual Role of Cytokines. J Immunol Res 2021, 2021:7785180. [CrossRef]
- Angulo I, Fresno M: Cytokines in the pathogenesis of and protection against malaria. Clin Diagn Lab Immunol 2002, 9(6):1145-1152. [CrossRef]
- Dieye Y, Mbengue B, Dagamajalu S, Fall MM, Loke MF, Nguer CM, Thiam A, Vadivelu J, Dieye A: Cytokine response during non-cerebral and cerebral malaria: evidence of a failure to control inflammation as a cause of death in African adults. PeerJ 2016, 4:e1965. [CrossRef]
- Agak GW, Mouton A, Teles RM, Weston T, Morselli M, Andrade PR, Pellegrini M, Modlin RL: Extracellular traps released by antimicrobial TH17 cells contribute to host defense. J Clin Invest 2021, 131(2). [CrossRef]
- Puerta-Arias JD, Mejia SP, Gonzalez A: The Role of the Interleukin-17 Axis and Neutrophils in the Pathogenesis of Endemic and Systemic Mycoses. Front Cell Infect Microbiol 2020, 10:595301. [CrossRef]
- Dixon B, Lee TJ, Contreras Healey DC, Li J, Goettel JA, Piazuelo MB, Algood HMS: IL-17 Receptor Signaling through IL-17A or IL-17F Is Sufficient to Maintain Innate Response and Control of Helicobacter pylori Immunopathogenesis. Immunohorizons 2022, 6(2):116-129. [CrossRef]
- Evans HG, Gullick NJ, Kelly S, Pitzalis C, Lord GM, Kirkham BW, Taams LS: In vivo activated monocytes from the site of inflammation in humans specifically promote Th17 responses. Proc Natl Acad Sci U S A 2009, 106(15):6232-6237. [CrossRef]
- Banerjee A, Bhattacharya P, Joshi AB, Ismail N, Dey R, Nakhasi HL: Role of pro-inflammatory cytokine IL-17 in Leishmania pathogenesis and in protective immunity by Leishmania vaccines. Cell Immunol 2016, 309:37-41. [CrossRef]
- Hansakon A, Ngamskulrungroj P, Angkasekwinai P: Contribution of Laccase Expression to Immune Response against Cryptococcus gattii Infection. Infect Immun 2020, 88(3). [CrossRef]
- Bunte K, Beikler T: Th17 Cells and the IL-23/IL-17 Axis in the Pathogenesis of Periodontitis and Immune-Mediated Inflammatory Diseases. Int J Mol Sci 2019, 20(14). [CrossRef]
- Nadeem A, Al-Harbi NO, Alfardan AS, Ahmad SF, AlAsmari AF, Al-Harbi MM: IL-17A-induced neutrophilic airway inflammation is mediated by oxidant-antioxidant imbalance and inflammatory cytokines in mice. Biomed Pharmacother 2018, 107:1196-1204. [CrossRef]
- Schon MP, Erpenbeck L: The Interleukin-23/Interleukin-17 Axis Links Adaptive and Innate Immunity in Psoriasis. Front Immunol 2018, 9:1323.
- Li G, Zhang Y, Qian Y, Zhang H, Guo S, Sunagawa M, Hisamitsu T, Liu Y: Interleukin-17A promotes rheumatoid arthritis synoviocytes migration and invasion under hypoxia by increasing MMP2 and MMP9 expression through NF-kappaB/HIF-1alpha pathway. Mol Immunol 2013, 53(3):227-236. [CrossRef]
- Kolls JK, Linden A: Interleukin-17 family members and inflammation. Immunity 2004, 21(4):467-476. [CrossRef]
- Kawaguchi M, Adachi M, Oda N, Kokubu F, Huang SK: IL-17 cytokine family. J Allergy Clin Immunol 2004, 114(6):1265-1273; quiz 1274.
- McGeachy MJ, Cua DJ, Gaffen SL: The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50(4):892-906.
- Helegbe GK, Huy NT, Yanagi T, Shuaibu MN, Kikuchi M, Cherif MS, Hirayama K: Elevated IL-17 levels in semi-immune anaemic mice infected with Plasmodium berghei ANKA. Malar J 2018, 17(1):169.
- Oyegue-Liabagui SL, Bouopda-Tuedom AG, Kouna LC, Maghendji-Nzondo S, Nzoughe H, Tchitoula-Makaya N, Pegha-Moukandja I, Lekana-Douki JB: Pro- and anti-inflammatory cytokines in children with malaria in Franceville, Gabon. Am J Clin Exp Immunol 2017, 6(2):9-20.
- Diop G, Derbois C, Loucoubar C, Mbengue B, Ndao BN, Thiam F, Thiam A, Ndiaye R, Dieye Y, Olaso R et al.: Genetic variants of RNASE3 (ECP) and susceptibility to severe malaria in Senegalese population. Malar J 2018, 17(1):61. [CrossRef]
- Saissy JM, Rouvin B, Koulmann P: [Severe malaria in intensive care units in 2003]. Med Trop (Mars) 2003, 63(3):258-266.
- Takahashi M, Matsuda F, Margetic N, Lathrop M: Automated identification of single nucleotide polymorphisms from sequencing data. J Bioinform Comput Biol 2003, 1(2):253-265. [CrossRef]
- Rodriguez S, Gaunt TR, Day IN: Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol 2009, 169(4):505-514. doi: 10.1093/aje/kwn359.
- Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21(2):263-265. [CrossRef]
- Tregouet DA, Garelle V: A new JAVA interface implementation of THESIAS: testing haplotype effects in association studies. Bioinformatics 2007, 23(8):1038-1039. [CrossRef]
- Penha-Goncalves C: Genetics of Malaria Inflammatory Responses: A Pathogenesis Perspective. Front Immunol 2019, 10:1771. doi: 10.3389/fimmu.2019.01771.
- Pereira VA, Sanchez-Arcila JC, Teva A, Perce-da-Silva DS, Vasconcelos MP, Lima CA, Aprigio CJ, Rodrigues-da-Silva RN, Santos DO, Banic DM et al.: IL10A genotypic association with decreased IL-10 circulating levels in malaria infected individuals from endemic area of the Brazilian Amazon. Malar J 2015, 14:30. [CrossRef]
- Gibbs KD, Schott BH, Ko DC: The Awesome Power of Human Genetics of Infectious Disease. Annu Rev Genet 2022, 56:41-62.
- Malutan AM, Drugan C, Drugan T, Ciortea R, Mihu D: The association between interleukin-4 -590C/T genetic polymorphism, IL-4 serum level, and advanced endometriosis. Cent Eur J Immunol 2016, 41(2):176-181.
- Lang X, Liu W, Hou Y, Zhao W, Yang X, Chen L, Yan Q, Cheng W: IL-17A polymorphism (rs2275913) and levels are associated with preeclampsia pathogenesis in Chinese patients. BMC Med Genomics 2021, 14(1):5.
- Basavaraju U, Shebl FM, Palmer AJ, Berry S, Hold GL, El-Omar EM, Rabkin CS: Cytokine gene polymorphisms, cytokine levels and the risk of colorectal neoplasia in a screened population of Northeast Scotland. Eur J Cancer Prev 2015, 24(4):296-304. [CrossRef]
- Apinjoh TO, Anchang-Kimbi JK, Njua-Yafi C, Mugri RN, Ngwai AN, Rockett KA, Mbunwe E, Besingi RN, Clark TG, Kwiatkowski DP et al.: Association of cytokine and Toll-like receptor gene polymorphisms with severe malaria in three regions of Cameroon. PLoS One 2013, 8(11):e81071. [CrossRef]
- Israelsson E, Maiga B, Kearsley S, Dolo A, Homann MV, Doumbo OK, Troye-Blomberg M, Tornvall P, Berzins K: Cytokine gene haplotypes with a potential effect on susceptibility to malaria in sympatric ethnic groups in Mali. Infect Genet Evol 2011, 11(7):1608-1615.
- Okeyo WA, Munde EO, Okumu W, Raballah E, Anyona SB, Vulule JM, Ong’echa JM, Perkins DJ, Ouma C: Interleukin (IL)-13 promoter polymorphisms (-7402 T/G and -4729G/A) condition susceptibility to pediatric severe malarial anemia but not circulating IL-13 levels. BMC Immunol 2013, 14:15.
- Phelan J, Gomez-Gonzalez PJ, Andreu N, Omae Y, Toyo-Oka L, Yanai H, Miyahara R, Nedsuwan S, de Sessions PF, Campino S et al.: Genome-wide host-pathogen analyses reveal genetic interaction points in tuberculosis disease. Nat Commun 2023, 14(1):549. [CrossRef]
- Mohanty S, Singh US, Mohanty S, Mohanty AK, Pande V, Das A: Evolutionary interplay of single nucleotide polymorphisms at the promoter region of TNF-alpha gene in different clinical outcomes of malaria in India. Infect Genet Evol 2019, 69:107-116. [CrossRef]
- Fedorova L, Khrunin A, Khvorykh G, Lim J, Thornton N, Mulyar OA, Limborska S, Fedorov A: Analysis of Common SNPs across Continents Reveals Major Genomic Differences between Human Populations. Genes (Basel) 2022, 13(8). [CrossRef]
- Miossec P, Korn T, Kuchroo VK: Interleukin-17 and type 17 helper T cells. N Engl J Med 2009, 361(9):888-898. [CrossRef]
- Rahmah Z, Sasmito SD, Siswanto B, Sardjono TW, Fitri LE: Parasitemia Induces High Plasma Levels of Interleukin-17 (IL-17) and Low Levels of Interleukin-10 (IL-10) and Transforming Growth Factor-ss (TGF-ss) in Pregnant Mice Infected with Malaria. Malays J Med Sci 2015, 22(3):25-32.
- Herbert F, Tchitchek N, Bansal D, Jacques J, Pathak S, Becavin C, Fesel C, Dalko E, Cazenave PA, Preda C et al.: Evidence of IL-17, IP-10, and IL-10 involvement in multiple-organ dysfunction and IL-17 pathway in acute renal failure associated to Plasmodium falciparum malaria. J Transl Med 2015, 13:369. [CrossRef]
- Sahu U, Biswas D, Prajapati VK, Singh AK, Samant M, Khare P: Interleukin-17-A multifaceted cytokine in viral infections. J Cell Physiol 2021, 236(12):8000-8019. [CrossRef]
- Scherer EF, Cantarini DG, Siqueira R, Ribeiro EB, Braga EM, Honorio-Franca AC, Franca EL: Cytokine modulation of human blood viscosity from vivax malaria patients. Acta Trop 2016, 158:139-147. [CrossRef]
- Tangteerawatana P, Pichyangkul S, Hayano M, Kalambaheti T, Looareesuwan S, Troye-Blomberg M, Khusmith S: Relative levels of IL4 and IFN-gamma in complicated malaria: association with IL4 polymorphism and peripheral parasitemia. Acta Trop 2007, 101(3):258-265. [CrossRef]
- Domingues W, Kanunfre KA, Rodrigues JC, Teixeira LE, Yamamoto L, Okay TS: Preliminary Report on the Putative Association of Il10 -3575 T/a Genetic Polymorphism with Malaria Symptoms. Rev Inst Med Trop Sao Paulo 2016, 58:30.
- Medina TS, Costa SP, Oliveira MD, Ventura AM, Souza JM, Gomes TF, Vallinoto AC, Povoa MM, Silva JS, Cunha MG: Increased interleukin-10 and interferon-gamma levels in Plasmodium vivax malaria suggest a reciprocal regulation which is not altered by IL-10 gene promoter polymorphism. Malar J 2011, 10:264. [CrossRef]
- Kaur R, Rawat AK, Kumar S, Aadil W, Akhtar T, Narang T, Chopra D: Association of genetic polymorphism of interleukin-17A & interleukin-17F with susceptibility of psoriasis. Indian J Med Res 2018, 148(4):422-426. doi: 10.4103/ijmr.IJMR_1859_16.
- Zacarias JM, Sippert EA, Tsuneto PY, Visentainer JE, de Oliveira e Silva C, Sell AM: The Influence of Interleukin 17A and IL17F Polymorphisms on Chronic Periodontitis Disease in Brazilian Patients. Mediators Inflamm 2015, 2015:147056. doi: 10.1155/2015/147056.
- Long ZW, Yu HM, Wang YN, Liu D, Chen YZ, Zhao YX, Bai L: Association of IL-17 polymorphisms with gastric cancer risk in Asian populations. World J Gastroenterol 2015, 21(18):5707-5718. doi: 10.3748/wjg.v21.i18.5707.
- Wang L, Jiang Y, Zhang Y, Wang Y, Huang S, Wang Z, Tian B, Yang Y, Jiang W, Pang D: Association analysis of IL-17A and IL-17F polymorphisms in Chinese Han women with breast cancer. PLoS One 2012, 7(3):e34400. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).