Submitted:
29 April 2023
Posted:
30 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
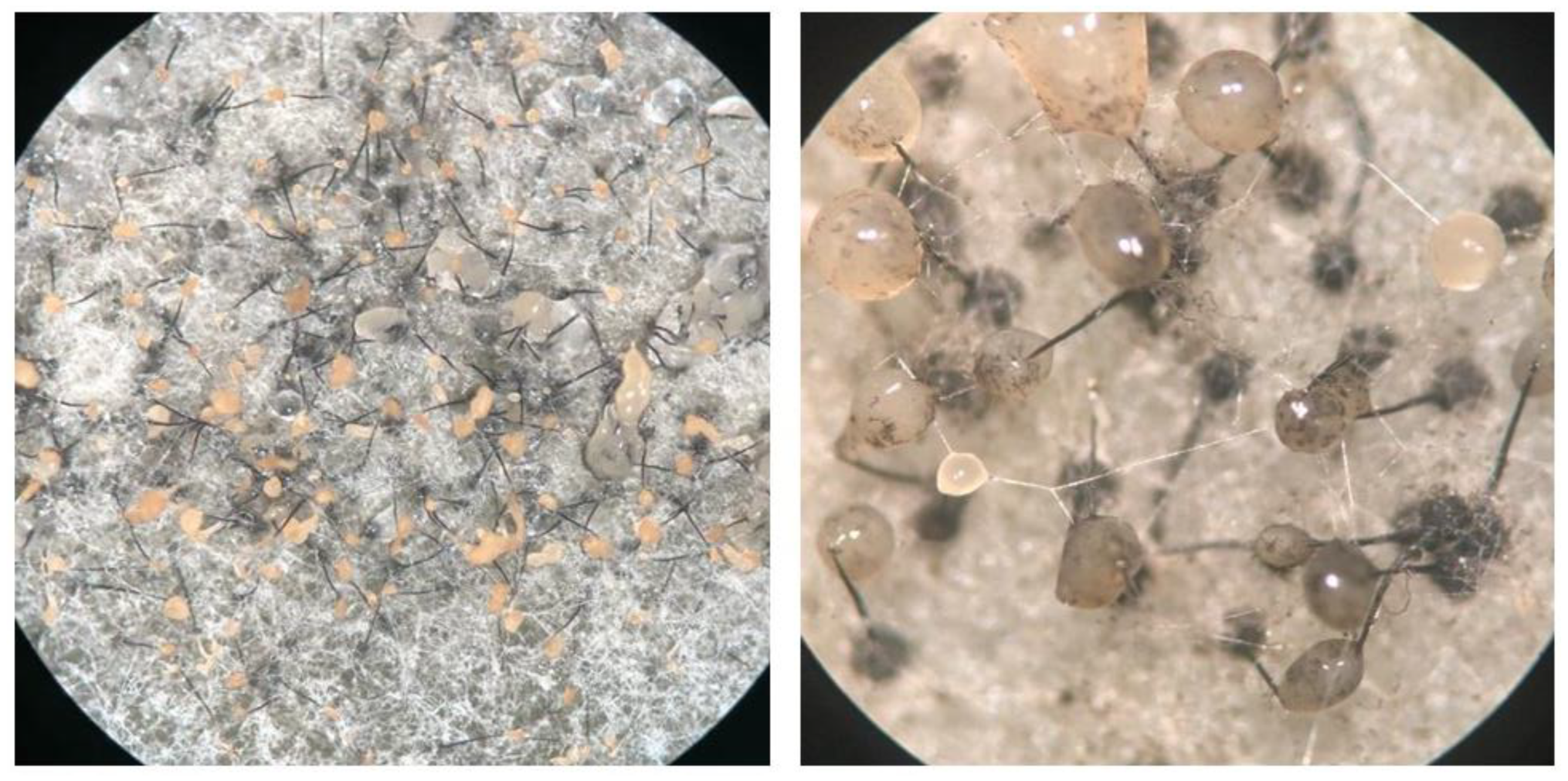
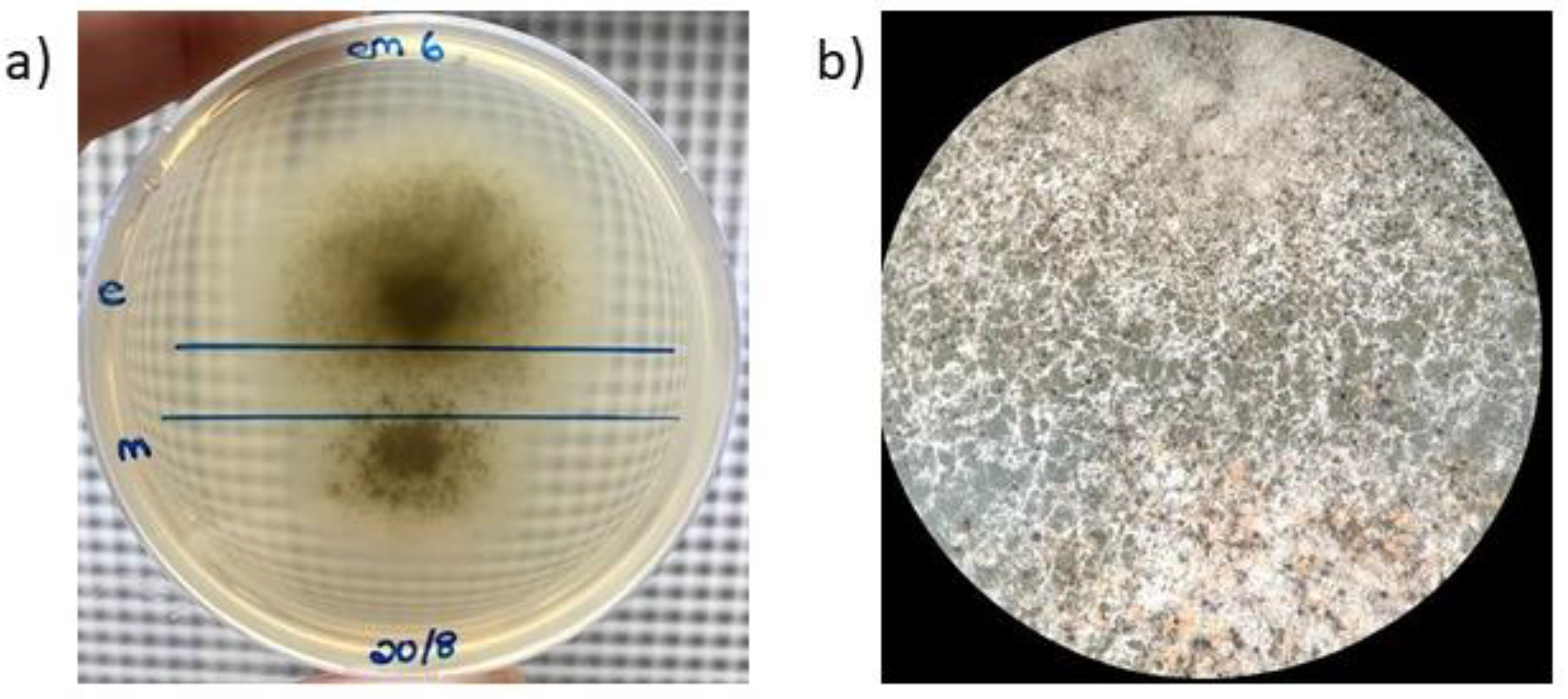
2.1. Interspecific Hybridization of Ceratocystis Isolates
2.2. DNA Extraction and PCR-Based Confirmation of Parental Self-Fertility
2.3. Marker and Primer Design
2.4. Marker Analysis
3. Results
3.1. Interspecific Hybridization of Ceratocystis Isolates
3.2. Development of a PCR-RFLP Method Based on Nuclear Markers
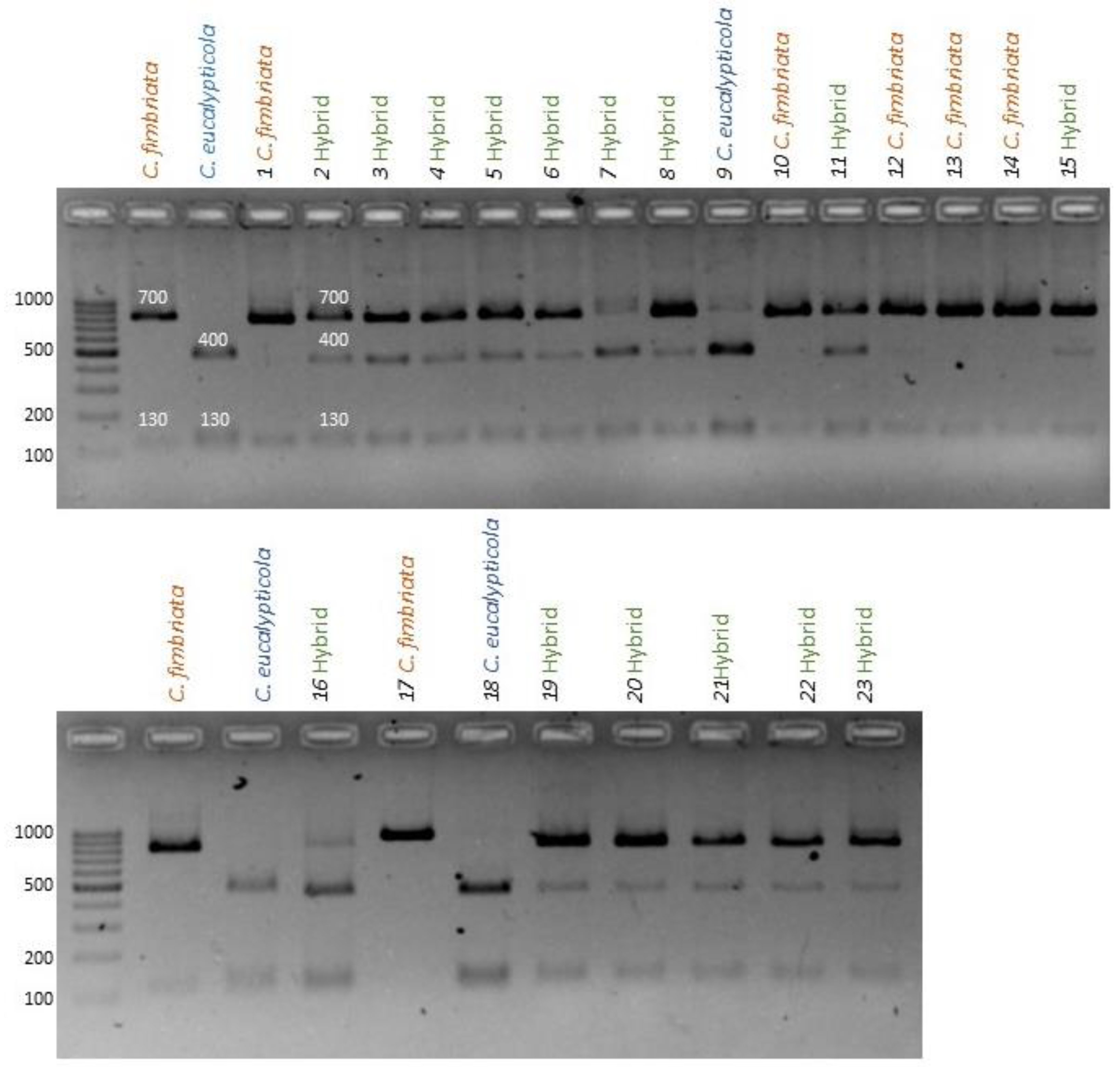
3.3. Identifying Hybrid Cultures Using PCR-RFLP Analysis of Nuclear Markers
3.3.1. C. Fimbriata X C. Eucalypticola
3.3.2. C. Fimbriata X C. Manginecans
3.3.3. C. Manginecans X C. Eucalypticola
3.4. Mitochondrial DNA Inheritance in Hybrid Cultures
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Callaghan, S.; Guest, D. Globalisation, the founder effect, hybrid Phytophthora species and rapid evolution: New headaches for biosecurity. Australas. Plant Pathol. 2015, 44, 255–262. [Google Scholar] [CrossRef]
- Mallet, J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005, 20, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Hessenauer, P.; Fijarczyk, A.; Martin, H.; Prunier, J.; Charron, G.; Chapuis, J.; Bernier, L.; Tanguay, P.; Hamelin, R.C.; Landry, C.R. Hybridization and introgression drive genome evolution of Dutch elm disease pathogens. Nat. Ecol. Evol. 2020, 4, 626–638. [Google Scholar] [CrossRef]
- Samarasinghe, H.; You, M.; Jenkinson, T.S.; Xu, J.; James, T.Y. Hybridization facilitates adaptive evolution in two major fungal pathogens. Genes 2020, 11, 101. [Google Scholar] [CrossRef] [PubMed]
- Feurtey, A.; Stukenbrock, E.H. Interspecific gene exchange as a driver of adaptive evolution in fungi. Annu. Rev. Microbiol. 2018, 72, 377–398. [Google Scholar] [CrossRef]
- Stukenbrock, E.H. The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology 2016, 106, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Harrington, T.C.; McNew, D.L. Partial interfertility among the Ceratocystis species on conifers. Fungal Genet. Biol. 1998, 25, 44–53. [Google Scholar] [CrossRef]
- De Beer, Z.W.; Duong, T.A.; Barnes, I.; Wingfield, B.D.; Wingfield, M.J. Redefining Ceratocystis and allied genera. Stud. Mycol. 2014, 79, 187–219. [Google Scholar] [CrossRef]
- Engelbrecht, C.J.B.; Harrington, T.C. Intersterility, morphology and taxonomy of Ceratocystis fimbriata on sweet potato, cacao and sycamore. Mycologia 2005, 97, 57–69. [Google Scholar] [CrossRef]
- van der Nest, M.A.; Steenkamp, E.T.; Roodt, D.; Soal, N.C.; Palmer, M.; Chan, W.-Y.; Wilken, P.M.; Duong, T.A.; Naidoo, K.; Santana, Q.C.; et al. Genomic analysis of the aggressive tree pathogen Ceratocystis albifundus. Fungal Biol. 2019, 123, 351–363. [Google Scholar] [CrossRef]
- Fourie, A.; van der Nest, M.A.; de Vos, L.; Wingfield, M.J.; Wingfield, B.D.; Barnes, I. QTL mapping of mycelial growth and aggressiveness to distinct hosts in Ceratocystis pathogens. Fungal Genet. Biol. 2019, 131, 103242. [Google Scholar] [CrossRef]
- Naidoo, K.; Steenkamp, E.T.; Coetzee, M.P.; Wingfield, M.J.; Wingfield, B.D. Concerted evolution in the ribosomal RNA cistron. PLoS ONE 2013, 8, e59355. [Google Scholar] [CrossRef] [PubMed]
- Fourie, A.; Wingfield, M.J.; Wingfield, B.D.; Thu, P.Q.; Barnes, I. A possible centre of diversity in South East Asia for the tree pathogen, Ceratocystis manginecans. Infect. Genet. Evol. 2016, 41, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Brawner, J.; Japarudin, Y.; Lapammu, M.; Rauf, R.; Boden, D.; Wingfield, M., J. . Evaluating the inheritance of Ceratocystis acaciivora symptom expression in a diverse Acacia mangium breeding population. South. For. A J. For. Sci. 2015, 77, 83–90. [Google Scholar] [CrossRef]
- Tarigan, M.; Roux, J.; Van Wyk, M.; Tjahjono, B.; Wingfield, M.J. A new wilt and die-back disease of Acacia mangium associated with Ceratocystis manginecans and C. acaciivora sp. nov. in Indonesia. South Afr. J. Bot. 2011, 77, 292–304. [Google Scholar] [CrossRef]
- Al Adawi, A.; Deadman, M.; Al Rawahi, A.; Al Maqbali, Y.; Al Jahwari, A.; Al Saadi, B.; Al Amri, I.; Wingfield, M. Aetiology and causal agents of mango sudden decline disease in the Sultanate of Oman. Eur. J. Plant Pathol. 2006, 116, 247–254. [Google Scholar] [CrossRef]
- Chen, S.; Van Wyk, M.; Roux, J.; Wingfield, M.J.; Xie, Y.; Zhou, X. Taxonomy and pathogenicity of Ceratocystis species on Eucalyptus trees in South China, including C. chinaeucensis sp. nov. Fungal Divers. 2013, 58, 267–279. [Google Scholar] [CrossRef]
- Harrington, T.C.; Huang, Q.; Ferreira, M.A.; Alfenas, A.C. Genetic analyses trace the Yunnan, China population of Ceratocystis fimbriata on pomegranate and taro to populations on Eucalyptus in Brazil. Plant Dis. 2015, 99, 106–111. [Google Scholar] [CrossRef]
- Al Adawi, A.O.; Barnes, I.; Khan, I.A.; Al Subhi, A.M.; Al Jahwari, A.A.; Deadman, M.L.; Wingfield, B.D.; Wingfield, M.J. Ceratocystis manginecans associated with a serious wilt disease of two native legume trees in Oman and Pakistan. Australas. Plant Pathol. 2013, 42, 179–193. [Google Scholar] [CrossRef]
- Van Wyk, M.; Al Adawi, A.O.; Khan, I.A.; Deadman, M.L.; Al Jahwari, A.A.; Wingfield, B.D.; Ploetz, R.; Wingfield, M.J. Ceratocystis manginecans sp. nov., causal agent of a destructive mango wilt disease in Oman and Pakistan. Fungal Divers. 2007, 27, 213–230. [Google Scholar]
- Halsted, B.D.; Fairchild, D.G. Sweet-potato black rot. J. Mycol. 1891, 1–11. [Google Scholar] [CrossRef]
- Halsted, B.D. Some fungus diseases of the sweet potato; New Jersey Agricultural College Experiment Station: 1890; pp. 25–27.
- Li, Q.; Harrington, T.C.; McNew, D.; Li, J.; Huang, Q.; Somasekhara, Y.M.; Alfenas, A.C. Genetic bottlenecks for two populations of Ceratocystis fimbriata on sweet potato and pomegranate in China. Plant Dis. 2016, 100, 2266–2274. [Google Scholar] [CrossRef] [PubMed]
- Paul, N.C.; Nam, S.-S.; Kachroo, A.; Kim, Y.-H.; Yang, J.-W. Characterization and pathogenicity of sweet potato (Ipomoea batatas) black rot caused by Ceratocystis fimbriata in Korea. Eur. J. Plant Pathol. 2018, 152, 833–840. [Google Scholar] [CrossRef]
- Harter, L.L.; Weimer, J. A monographic study of sweet-potato diseases and their control; Washington, D.C., 29; pp. 1–118. 19 April.
- van Wyk, M.; Roux, J.; Nkuekam, G.K.; Wingfield, B.D.; Wingfield, M.J. Ceratocystis eucalypticola sp. nov. from Eucalyptus in South Africa and comparison to global isolates from this tree. IMA Fungus 2012, 3, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Wilken, P.M.; Steenkamp, E.T.; Wingfield, M.J.; De Beer, Z.W.; Wingfield, B.D. DNA loss at the Ceratocystis fimbriata mating locus results in self-sterility. PLoS ONE 2014, 9, e92180. [Google Scholar] [CrossRef] [PubMed]
- Harrington, T.C.; Kazmi, M.R.; Al-Sadi, A.M.; Ismail, S.I. Intraspecific and intragenomic variability of ITS rDNA sequences reveals taxonomic problems in Ceratocystis fimbriata sensu stricto. Mycologia 2014, 106, 224–242. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.S.; Harrington, T.C.; Ferreira, M.A.; Damacena, M.B.; Al-Sadi, A.M.; Al-Mahmooli, I.H.; Alfenas, A.C. Species or genotypes? Reassessment of four recently described species of the Ceratocystis wilt pathogen, Ceratocystis fimbriata, on Mangifera indica. Phytopathology 2015, 105, 1229–1244. [Google Scholar] [CrossRef] [PubMed]
- Fourie, A.; Wingfield, M.J.; Wingfield, B.D.; van der Nest, M.A.; Loots, M.T.; Barnes, I. Inheritance of phenotypic traits in the progeny of a Ceratocystis interspecific cross. Fungal Biol. 2018, 122, 717–729. [Google Scholar] [CrossRef]
- Harrington, T.C.; McNew, D.L. Self-fertility and uni-directional mating-type switching in Ceratocystis coerulescens, a filamentous ascomycete. Curr. Genet. 1997, 32, 52–59. [Google Scholar] [CrossRef]
- Harrington, T.C.; Steimel, J.; Kile, G. Genetic variation in three Ceratocystis species with outcrossing, selfing and asexual reproductive strategies. Eur. J. For. Pathol. 1998, 28, 217–226. [Google Scholar] [CrossRef]
- Lee, D.H.; Roux, J.; Wingfield, B.D.; Wingfield, M.J. Variation in growth rates and aggressiveness of naturally occurring self-fertile and self-sterile isolates of the wilt pathogen Ceratocystis albifundus. Plant Pathol. 2015, 64, 1103–1109. [Google Scholar] [CrossRef]
- Yun, S.H.; Kim, H.K.; Lee, T.; Turgeon, B.G. Self-fertility in Chromocrea spinulosa is a consequence of direct repeat-mediated loss of MAT1-2, subsequent imbalance of nuclei differing in mating type, and recognition between unlike nuclei in a common cytoplasm. PLoS Genet. 2017, 13, e1006981. [Google Scholar] [CrossRef]
- Xu, L.; Jardini, T.M.; Chen, W. Direct repeat-mediated DNA deletion of the mating type MAT1-2 genes results in unidirectional mating type switching in Sclerotinia trifoliorum. Sci. Rep. 2016, 6, 27083. [Google Scholar] [CrossRef]
- Witthuhn, R.C.; Harrington, T.C.; Wingfield, B.D.; Steimel, J.P.; Wingfield, M.J. Deletion of the MAT- 2 mating-type gene during uni-directional mating-type switching in Ceratocystis. Curr. Genet. 2000, 38, 48–52. [Google Scholar] [CrossRef]
- Johnson, J.; Harrington, T.; Engelbrecht, C.J.B. Phylogeny and taxonomy of the North American clade of the Ceratocystis fimbriata complex. Mycologia 2005, 97, 1067–1092. [Google Scholar] [CrossRef]
- Harrington, T.C.; Pashenova, N.V.; McNew, D.L.; Steimel, J.; Konstantinov, M.Y. Species delimitation and host specialization of Ceratocystis laricicola and C. polonica to larch and spruce. Plant Dis. 2002, 86, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Wilken, P.M. Mating in Ceratocystis. University of Pretoria, 2015.
- Damm, U.; Mostert, L.; Crous, P.W.; Fourie, P.H. Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Persoonia 2008, 20, 87–102. [Google Scholar] [CrossRef]
- Wingfield, B.; Fourie, A.; Simpson, M.; Bushula-Njah, V.; Aylward, J.; Barnes, I.; Coetzee, M.; Dreyer, L.; Duong, T.; Geiser, D.; et al. IMA Genome-F 11: Draft genome sequences of Fusarium xylarioides, Teratosphaeria gauchensis and T. zuluensis and genome annotation for Ceratocystis fimbriata. IMA Fungus 2019, 10. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 1999, 132, 365–386. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, W71–W74. [Google Scholar] [CrossRef]
- Kramer, D.; Lane, F.A.; Steenkamp, E.T.; Wingfield, B.D.; Wilken, P.M. Unidirectional mating-type switching confers self-fertility to Thielaviopsis cerberus, the only homothallic species in the genus. Fungal Biol. 2021, 125, 427–434. [Google Scholar] [CrossRef]
- Morard, M.; Ibanez, C.; Adam, A.C.; Querol, A.; Barrio, E.; Toft, C. Genomic instability in an interspecific hybrid of the genus Saccharomyces: A matter of adaptability. Microb. Genom. 2020, 6, mgen000448. [Google Scholar] [CrossRef] [PubMed]
- Feurtey, A.; Stevens, D.M.; Stephan, W.; Stukenbrock, E.H. Interspecific gene exchange introduces high genetic variability in crop pathogen. Genome Biol. Evol. 2019, 11, 3095–3105. [Google Scholar] [CrossRef] [PubMed]
- Steenkamp, E.T.; Wingfield, M.J.; McTaggart, A.R.; Wingfield, B.D. Fungal species and their boundaries matter—Definitions, mechanisms and practical implications. Fungal Biol. Rev. 2018, 32, 104–116. [Google Scholar] [CrossRef]
- Harrison, R.G.; Larson, E.L. Hybridization, introgression, and the nature of species boundaries. J. Hered. 2014, 105, 795–809. [Google Scholar] [CrossRef]
- Ferreira, E.M.; Harrington, T.C.; Thorpe, D.J.; Alfenas, A.C. Genetic diversity and interfertility among highly differentiated populations of Ceratocystis fimbriata in Brazil. Plant Pathol. 2010, 59, 721–735. [Google Scholar] [CrossRef]
- Harrington, T.C.; Thorpe, D.J.; Alfenas, A.C. Genetic variation and variation in aggressiveness to native and exotic hosts among Brazilian populations of Ceratocystis fimbriata. Phytopathology 2011, 101, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Nkuekam, G.K.; Wingfield, M.J.; Roux, J. Ceratocystis species, including two new taxa, from Eucalyptus trees in South Africa. Australas. Plant Pathol. 2013, 42, 283–311. [Google Scholar] [CrossRef]
- Depotter, J.R.L.; Seidl, M.F.; Wood, T.A.; Thomma, B.P.H.J. Interspecific hybridization impacts host range and pathogenicity of filamentous microbes. Curr. Opin. Microbiol. 2016, 32, 7–13. [Google Scholar] [CrossRef]
- Brasier, C. The rise of the hybrid fungi. Nature 2000, 405, 134–135. [Google Scholar] [CrossRef]
- Xu, J. The inheritance of organelle genes and genomes: Patterns and mechanisms. Genome 2005, 48, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Birky, C.W. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annu. Rev. Genet. 2001, 35, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Birky, C.W. Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc. Natl. Acad. Sci. 1995, 92, 11331–11338. [Google Scholar] [CrossRef]
- Xu, J.; Li, H. Current perspectives on mitochondrial inheritance in fungi. Cell Health Cytoskelet. 2015, 7, 143. [Google Scholar] [CrossRef]
- Wilson, A.J.; Xu, J. Mitochondrial inheritance: Diverse patterns and mechanisms with an emphasis on fungi. Mycology 2012, 3, 158–166. [Google Scholar] [CrossRef]
- Reboud, X.; Zeyl, C. Organelle inheritance in plants. Heredity 1994, 72, 132–140. [Google Scholar] [CrossRef]
- Mendoza, H.; Perlin, M.H.; Schirawski, J. Mitochondrial inheritance in phytopathogenic fungi - everything is known, or is it? Int. J. Mol. Sci. 2020, 21, 3883. [Google Scholar] [CrossRef]
- Sun, S.; Fu, C.; Ianiri, G.; Heitman, J. The pheromone and pheromone receptor mating-type locus is involved in controlling uniparental mitochondrial inheritance in Cryptococcus. Genetics 2020, 214, 703–717. [Google Scholar] [CrossRef]
- Barr, C.M.; Neiman, M.; Taylor, D.R. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005, 168, 39–50. [Google Scholar] [CrossRef]
- Xu, J.; Yan, Z.; Guo, H. Divergence, hybridization, and recombination in the mitochondrial genome of the human pathogenic yeast Cryptococcus gattii. Mol. Ecol. 2009, 18, 2628–2642. [Google Scholar] [CrossRef]
- Depotter, J.R.L.; Beveren, v.F.; Moreno, L.; Kramer, H.M.; Chavarro Carrero, E.A.; Fiorin, G.L.; Berg, v.d.G.C.M.; Wood, T.A.; Thomma, B.P.H.J.; Seidl, M.F. The interspecific fungal hybrid Verticillium longisporum displays subgenome-specific gene expression. mBio 2021, 12, e0149621. [Google Scholar] [CrossRef]
- Hénault, M.; Marsit, S.; Charron, G.; Landry, C.R. Hybridization drives mitochondrial DNA degeneration and metabolic shift in a species with biparental mitochondrial inheritance. Genome Res. 2022, 32, 2043–2056. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, J. Mitochondrial genome polymorphisms in the human pathogenic fungus Cryptococcus neoformans. Front. Microbiol. 2020, 11, 706. [Google Scholar] [CrossRef]
- Lee, D.-H.; Roux, J.; Wingfield, B.D.; Wingfield, M.J. Non-Mendelian segregation influences the infection biology and genetic structure of the African tree pathogen Ceratocystis albifundus. Fungal Biol. 2018, 122, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.D.; Bloom, R.J.; Jiang, S.; Durand, H.K.; Dallow, E.; Mukherjee, S.; David, L.A. Measuring and mitigating PCR bias in microbiota datasets. PLoS Comput. Biol. 2021, 17, e1009113. [Google Scholar] [CrossRef] [PubMed]
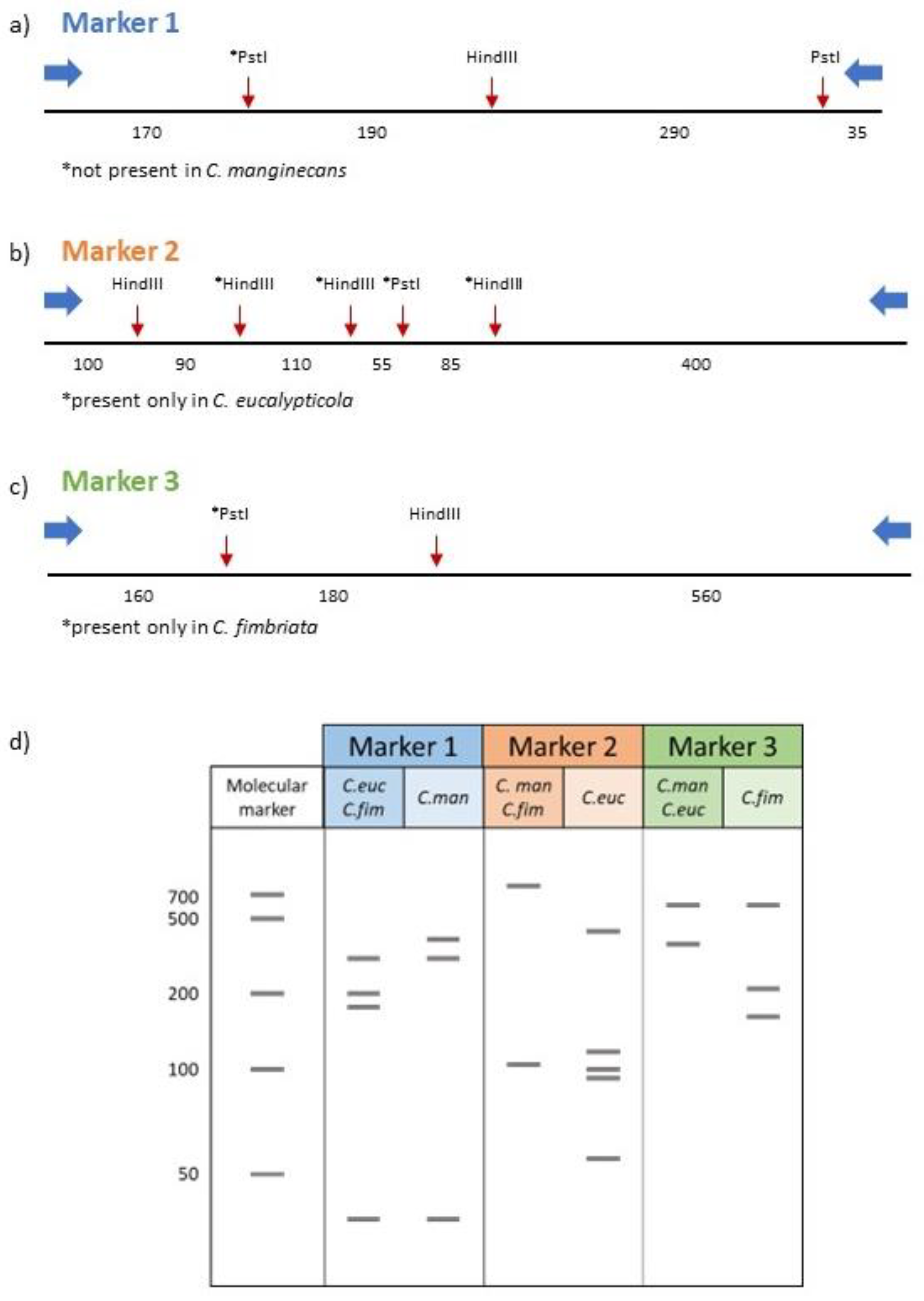
| Primer name* | Sequence (5’ → 3’) | Size (bp) | Notes |
|---|---|---|---|
| marker1_R | TTTCTGCTGTCCACACCTTG | 692 | Nuclear primer set 1. Region amplified from all three species but restriction enzyme (RE) digestion produces unique banding pattern for C. manginecans. |
| marker1_F | TTTCTGCTGTCCACACCTTG | ||
| marker2_R | GACCGCATGGTTGAGGTTAC | 836 | Nuclear primer set 2. Region amplified from all three species but RE digestion produces unique banding pattern for C. eucalypticola. |
| marker2_F | GTTCATGATGCCATCGACAC | ||
| marker3_R | GTGTAGCCGTCGGAAAATGT | 908 | Nuclear primer set 3. Region amplified from all three species but RE digestion produces unique banding pattern for C. fimbriata. |
| marker3_F | TGTTGGATGGGCTGTATTGA | ||
| Cf_Mt_F1 | GAAGTGCCTTCGCTTTAT | 436 | Targets the MAT1-1-1 gene in all three species. |
| Cf_Mt_R1 | GACCGCGATTCTAACCAAAA | ||
| MAT1-2-1F | AAGATGCTCTTTAATACCCACCA | 495 | Targets the MAT1-2-1 gene in all three species. |
| MAT1-2-1R | TGCCGCTAATAAGCTAGGAA | ||
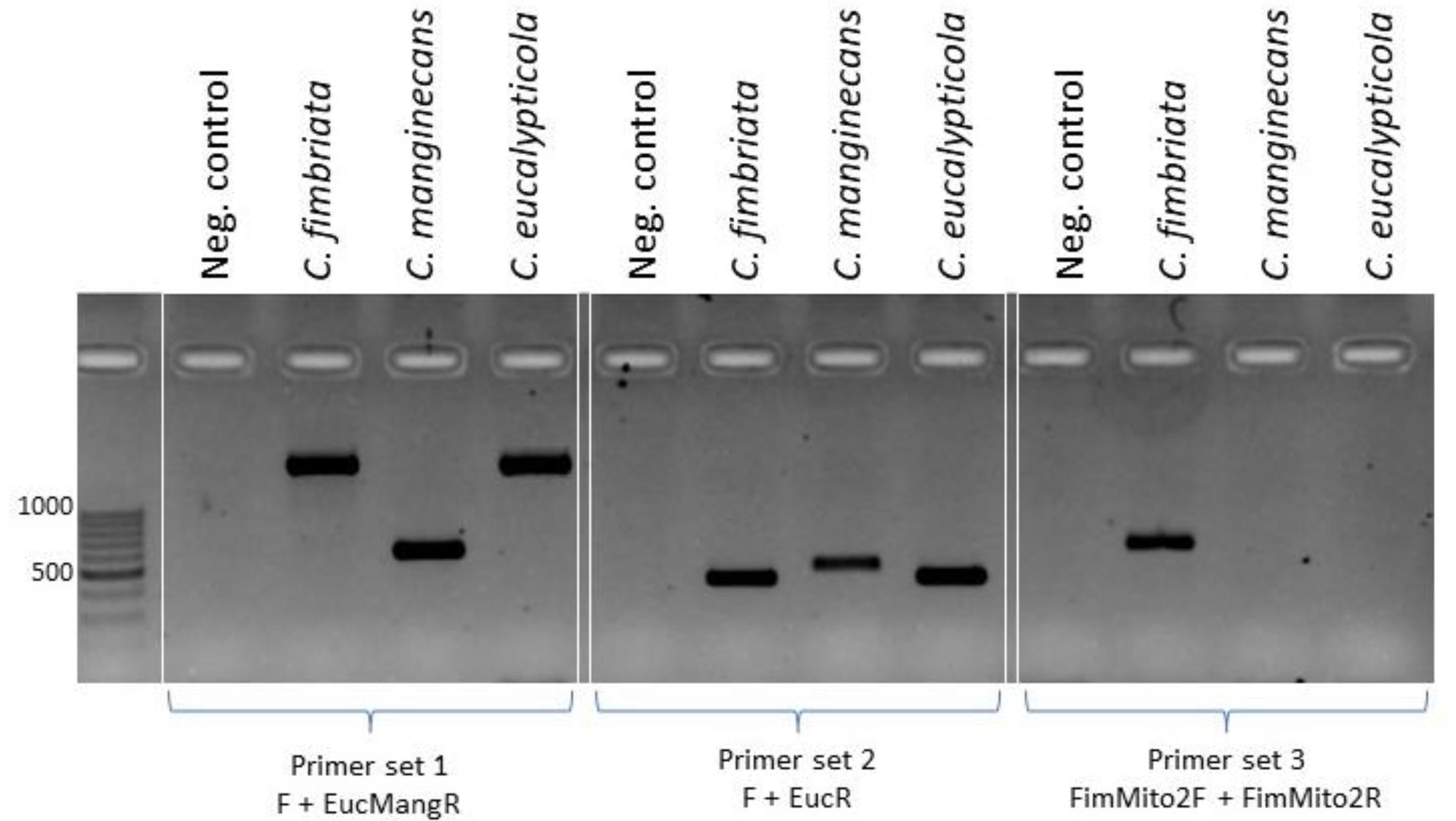
| F | AATTGGATCTTCAACGACTAAAC | 562 1886 1895 |
Mitochondrial primer set 1. Amplicon of 562 in C. manginecans, and amplicons of 1886 and 1895 bp in C. eucalypticola and C. fimbriata. |
| EucMangR | GGGCTCTGTTAGTCTCTGCA | ||
| F | AATTGGATCTTCAACGACTAAAC | 387 387 |
Mitochondrial primer set 2. Amplicons of 387 and 387 bp in C. eucalypticola and C. fimbriata. |
| EucR | TCCGCGATCATCCATTTCTC | ||
| FimMito2F | CCTGCATCTCGTCCTATCGT | 559 | Mitochondrial primer set 3 amplifies only in C. fimbriata. |
| FimMito2R | TCGCCTTTCAAGTTCCATGC | ||
| feMitSeq_F | CATTTGGGGGCTTTTTGTAA | n/a | Primer used to sequence amplicons produced with mitochondrial primer set 1 in order to score the region of variability between C. eucalypticola and C. fimbriata. |
| Interspecific cross | Hybridization | Selfing | Number of cultures containing hybrids | Mitochondrial inheritance in hybrids |
|---|---|---|---|---|
| C. fimbriatax C. eucalypticola | ||||
| Only one nuclear marker | 11 | 24 | 26 of 50 | Both parents |
| Both nuclear markers | 15 | 24 | ||
| Both mitochondrial markers | 6 | n/a | ||
| C. fimbriatax C. manginecans | ||||
| Only one nuclear marker | 8 | 41 | 8 of 49 | Both parents |
| Both nuclear markers | 0 | 41 | ||
| Both mitochondrial markers | 8 | n/a | ||
| C. manginecansx C. eucalypticola | ||||
| Only one nuclear marker | 0 | 42 | 0 of 42 | n/a |
| Both nuclear markers | 0 | 7 | ||
| Both mitochondrial markers | n/a | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





