Submitted:
02 May 2023
Posted:
03 May 2023
You are already at the latest version
Abstract
Keywords:
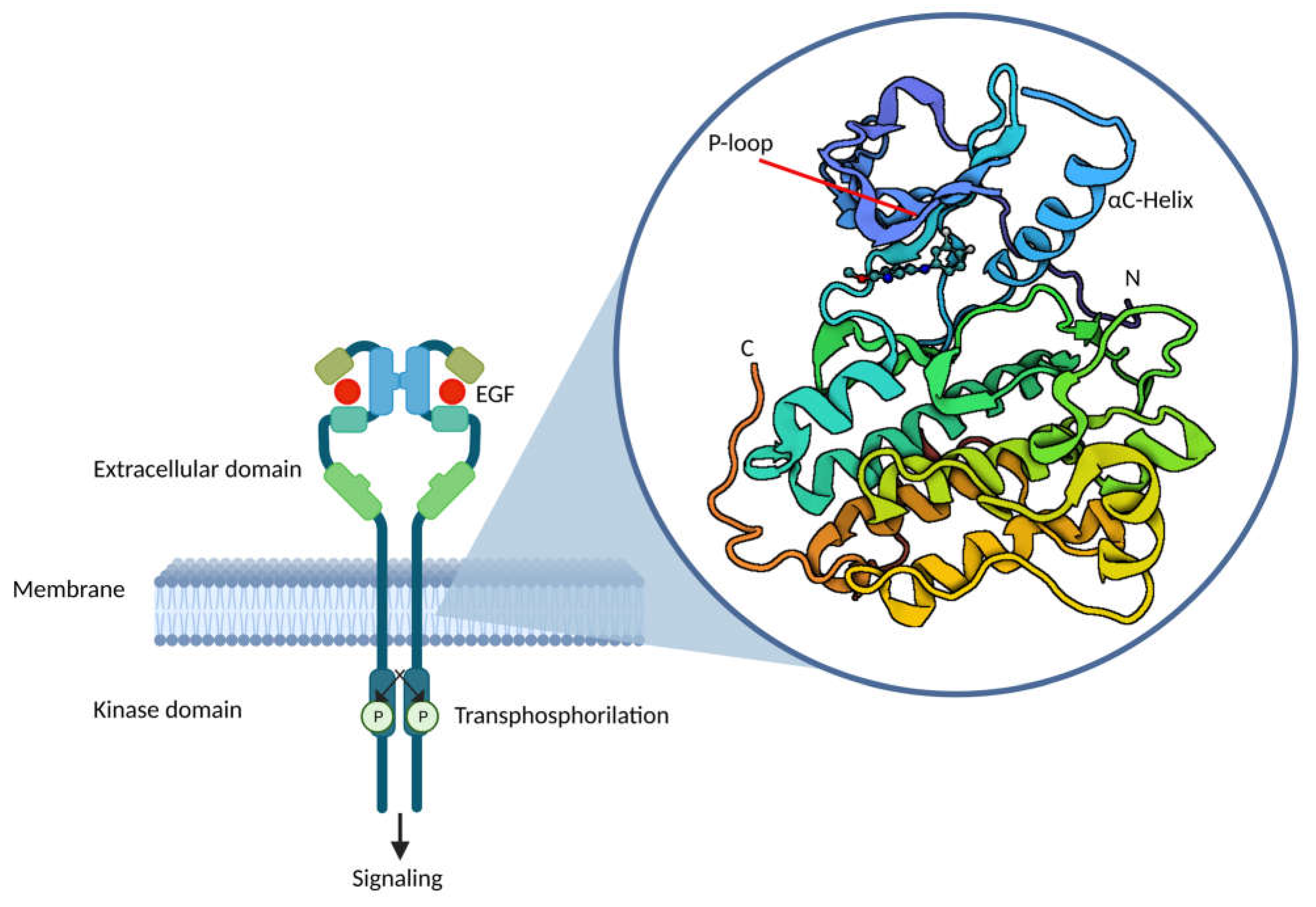
1. Introduction
2. Epidemiology of uncommon EFGR mutation
3. EGFR mutation testing methods
4. From exon-based to structure-based classification of EGFR mutation
5. Compound mutations
5. Treatment activity data of different TKIs
5.1. The more common among uncommon: L861Q, G719X and S768I
5.2. Focus on exon 18
5.3. Focus on exon 19
5.4. Focus on exon 20
5.5. Focus on exon 21
6. Intracranial activity of different EGFR TKIs in uncommon mutations
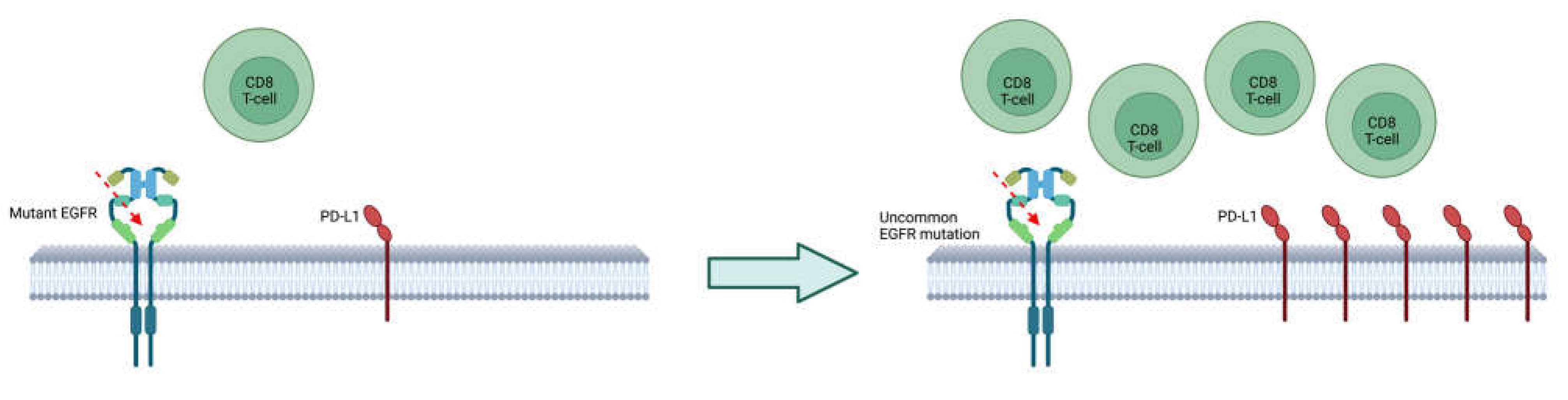
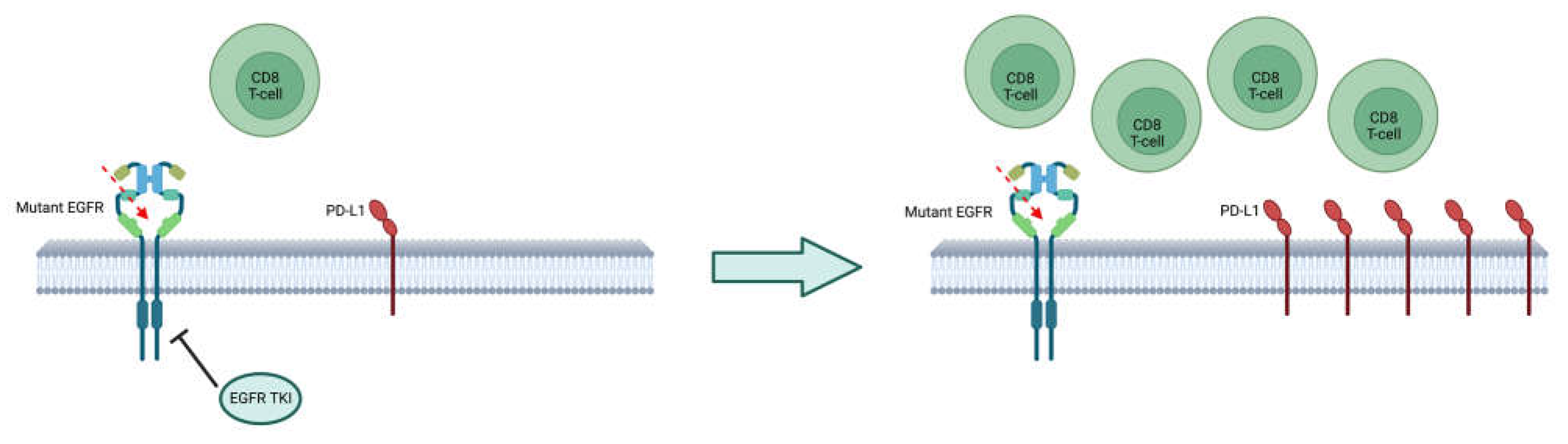
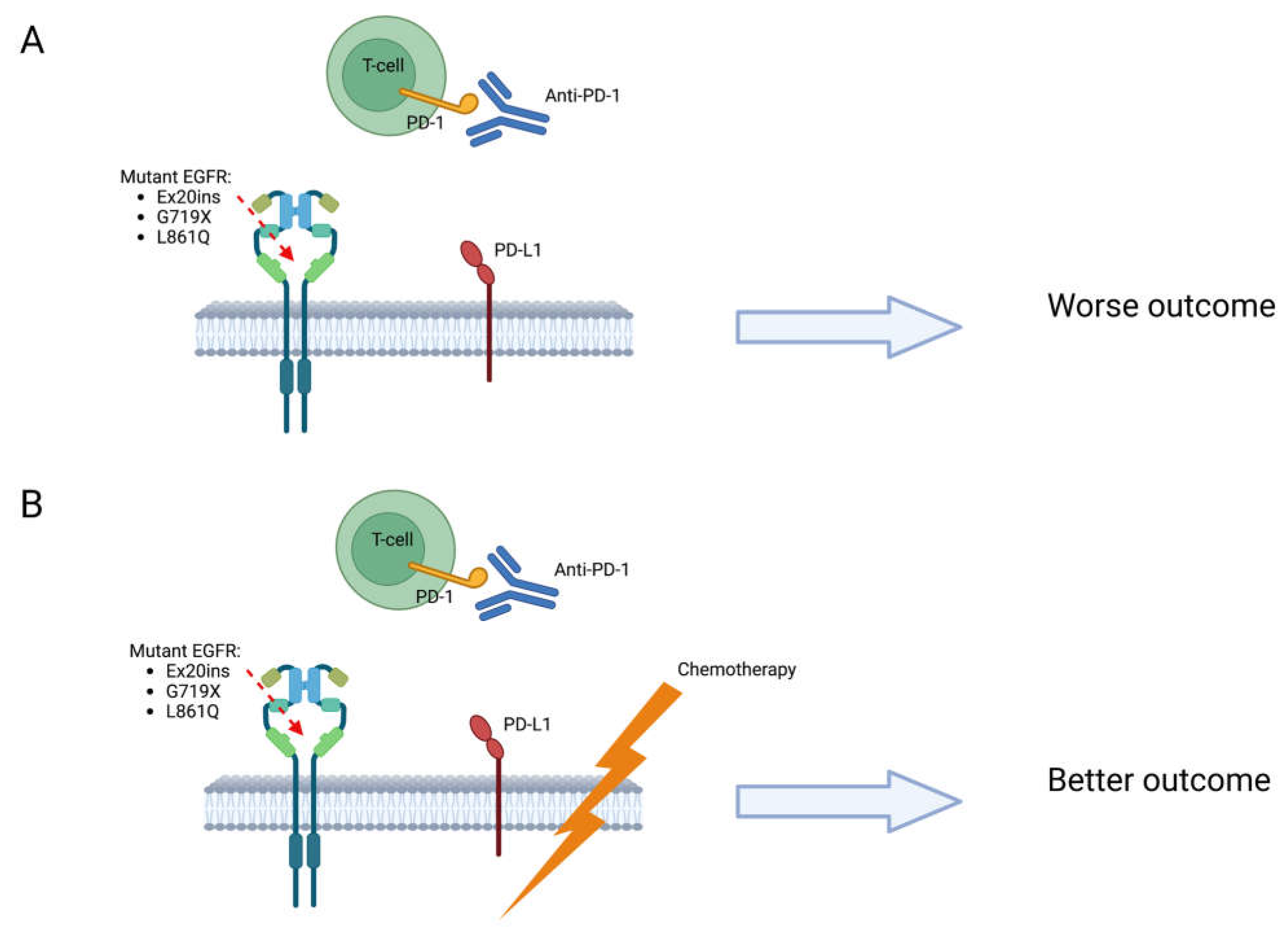
7. Response to immunotherapy and chemoimmunotherapy
8. How to define a treatment sequence
9. Ongoing clinical trials
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F.; Bsc, M.F.B.; Me, J.F.; Soerjomataram, M.I.; et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, F.; Tortora, G. EGFR Antagonists in Cancer Treatment. N. Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef] [PubMed]
- Linardou, H.; Dahabreh, I.J.; Bafaloukos, D.; Kosmidis, P.; Murray, S. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat. Rev. Clin. Oncol. 2009, 6, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-L.; Yuan, J.-Q.; Wang, K.-F.; Fu, X.-H.; Han, X.-R.; Threapleton, D.; Yang, Z.-Y.; Mao, C.; Tang, J.-L. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2016, 7, 78985–78993. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Canepa, H.M.; Bailey, A.S.; Nakayama, S.; Yamaguchi, N.; Goldstein, M.A.; Huberman, M.S.; Costa, D.B. Compound EGFR Mutations and Response to EGFR Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2013, 8, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S. , et al., EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005, 352, 786–792. [Google Scholar] [CrossRef]
- Westover, D.; Zugazagoitia, J.; Cho, B.C.; Lovly, C.M.; Paz-Ares, L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann. Oncol. 2018, 29, i10–i19. [Google Scholar] [CrossRef]
- Evans, M.; O’sullivan, B.; Smith, M.; Hughes, F.; Mullis, T.; Trim, N.; Taniere, P. Large-Scale EGFR Mutation Testing in Clinical Practice: Analysis of a Series of 18,920 Non-Small Cell Lung Cancer Cases. Pathol. Oncol. Res. 2019, 25, 1401–1409. [Google Scholar] [CrossRef]
- John, T.; Taylor, A.; Wang, H.; Eichinger, C.; Freeman, C.; Ahn, M.-J. Uncommon EGFR mutations in non-small-cell lung cancer: A systematic literature review of prevalence and clinical outcomes. Cancer Epidemiology 2022, 76, 102080. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Xu, C.-R.; Hu, C.-P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Shi, J.H.; Märten, A.; et al. Afatinib versus gemcitabine/cisplatin for first-line treatment of Chinese patients with advanced non-small-cell lung cancer harboring EGFR mutations: subgroup analysis of the LUX-Lung 6 trial. OncoTargets Ther. 2018, 11, 8575–8587. [Google Scholar] [CrossRef]
- Ho, G.-F.; Chai, C.-S.; Alip, A.; Wahid, M.I.A.; Abdullah, M.M.; Foo, Y.-C.; How, S.-H.; Zaatar, A.; Lam, K.-S.; Leong, K.-W.; et al. Real-world experience of first-line afatinib in patients with EGFR-mutant advanced NSCLC: a multicenter observational study. BMC Cancer 2019, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.-C.; Lam, D.C.-L.; Tsai, C.-M.; Chen, Y.-M.; Shih, J.-Y.; Aggarwal, S.; Wang, S.; Kim, S.-W.; Kim, Y.-C.; Wahid, I.; et al. Experience from Asian centers in a named-patient-use program for afatinib in patients with advanced non-small-cell lung cancer who had progressed following prior therapies, including patients with uncommon EGFR mutations. Int. J. Clin. Oncol. 2021, 26, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.-H.; Sequist, L.V.; Geater, S.L.; Tsai, C.-M.; Mok, T.S.K.; Schuler, M.; Yamamoto, N.; Yu, C.-J.; I Ou, S.-H.; Zhou, C.; et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Mok, T.; Peters, S.; Popat, S.; Ahn, M.-J.; de Marinis, F. Recent Advances on the Role of EGFR Tyrosine Kinase Inhibitors in the Management of NSCLC With Uncommon, Non Exon 20 Insertions, EGFR Mutations. J. Thorac. Oncol. 2021, 16, 764–773. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; de Marinis, F.; Tu, H.-Y.; Laktionov, K.K.; Feng, J.; Poltoratskiy, A.; Zhao, J.; Tan, E.H.; Gottfried, M.; Lee, V.; et al. Afatinib in EGFR TKI-Naïve Patients with Locally Advanced or Metastatic EGFR Mutation-Positive Non-Small Cell Lung Cancer: A Pooled Analysis of Three Phase IIIb Studies. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Popat, S.; Hsia, T.-C.; Hung, J.-Y.; Jung, H.A.; Shih, J.-Y.; Park, C.K.; Lee, S.H.; Okamoto, T.; Ahn, H.K.; Lee, Y.C.; et al. Tyrosine Kinase Inhibitor Activity in Patients with NSCLC Harboring Uncommon EGFR Mutations: A Retrospective International Cohort Study (UpSwinG). Oncol. 2022, 27, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.-Y.; Ke, E.-E.; Yang, J.-J.; Sun, Y.-L.; Yan, H.-H.; Zheng, M.-Y.; Bai, X.-Y.; Wang, Z.; Su, J.; Chen, Z.-H.; et al. A comprehensive review of uncommon EGFR mutations in patients with non-small cell lung cancer. Lung Cancer 2017, 114, 96–102. [Google Scholar] [CrossRef]
- Russo, A.; Franchina, T.; Ricciardi, G.; Battaglia, A.; Picciotto, M.; Adamo, V. Heterogeneous Responses to Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinase Inhibitors (TKIs) in Patients with Uncommon EGFR Mutations: New Insights and Future Perspectives in this Complex Clinical Scenario. Int. J. Mol. Sci. 2019, 20, 1431. [Google Scholar] [CrossRef]
- Gristina, V.; Malapelle, U.; Galvano, A.; Pisapia, P.; Pepe, F.; Rolfo, C.; Tortorici, S.; Bazan, V.; Troncone, G.; Russo, A. The significance of epidermal growth factor receptor uncommon mutations in non-small cell lung cancer: A systematic review and critical appraisal. Cancer Treat. Rev. 2020, 85, 101994. [Google Scholar] [CrossRef]
- Mehta, A.; Vasudevan, S. Rare epidermal growth factor receptor gene alterations in non-small cell lung cancer patients, tyrosine kinase inhibitor response and outcome analysis. Cancer Treat. Res. Commun. 2021, 28, 100398. [Google Scholar] [CrossRef]
- Graham, R.P.; Treece, A.L.; Lindeman, N.I.; Vasalos, P.; Shan, M.; Jennings, L.J.; Rimm, D.L. Worldwide Frequency of Commonly Detected EGFR Mutations. Arch. Pathol. Lab. Med. 2018, 142, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Pilotto, S.; Passiglia, F.; Pepe, F.; Pisapia, P.; Righi, L.; Listì, A.; Bironzo, P.; Belluomini, L.; Tabbò, F.; et al. Dealing with NSCLC EGFR mutation testing and treatment: A comprehensive review with an Italian real-world perspective. Crit. Rev. Oncol. 2021, 160, 103300. [Google Scholar] [CrossRef] [PubMed]
- Robichaux, J.P.; Le, X.; Vijayan, R.S.K.; Hicks, J.K.; Heeke, S.; Elamin, Y.Y.; Lin, H.Y.; Udagawa, H.; Skoulidis, F.; Tran, H.; et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2021, 597, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Silveira, C.; Janeiro, A.; Malveiro, S.; Oliveira, A.; Felizardo, M.; Nogueira, F.; Teixeira, E.; Martins, J.; Carmo-Fonseca, M. Detection of rare and novel EGFR mutations in NSCLC patients: Implications for treatment-decision. Lung Cancer 2020, 139, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Brodie, S.; Agersborg, S.; Funari, V.A.; Albitar, M. Significant Improvement in Detecting BRAF, KRAS, and EGFR Mutations Using Next-Generation Sequencing as Compared with FDA-Cleared Kits. Mol. Diagn. Ther. 2017, 21, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Hua, P.; Liu, N.; Li, Q.; Zhu, X.; Jiang, L.; Zheng, K.; Su, X. Targeted next-generation sequencing in cytology specimens for molecular profiling of lung adenocarcinoma. 2018, 11, 3647–3655. [Google Scholar]
- Warth, A.; Penzel, R.; Brandt, R.; Sers, C.; Fischer, J.R.; Thomas, M.; Herth, F.J.F.; Dietel, M.; Schirmacher, P.; Bläker, H. Optimized algorithm for Sanger sequencing-based EGFR mutation analyses in NSCLC biopsies. Virchows Arch. 2012, 460, 407–414. [Google Scholar] [CrossRef]
- Liang, C.; Wu, Z.; Gan, X.; Liu, Y.; You, Y.; Liu, C.; Zhou, C.; Liang, Y.; Mo, H.; Chen, A.M.; et al. Detection of Rare Mutations inEGFR-ARMS-PCR-Negative Lung Adenocarcinoma by Sanger Sequencing. Yonsei Med J. 2018, 59, 13–19. [Google Scholar] [CrossRef]
- Mao, L.; Zhao, W.; Li, X.; Zhang, S.; Zhou, C.; Zhou, D.; Ou, X.; Xu, Y.; Tang, Y.; Ou, X.; et al. Mutation Spectrum of EGFR From 21,324 Chinese Patients With Non-Small Cell Lung Cancer (NSCLC) Successfully Tested by Multiple Methods in a CAP-Accredited Laboratory. Pathol. Oncol. Res. 2021, 27, 602726. [Google Scholar] [CrossRef]
- Siqueira, J.F.; Fouad, A.F.; Rôças, I.N. Pyrosequencing as a tool for better understanding of human microbiomes. J. Oral Microbiol. 2012, 4. [Google Scholar] [CrossRef]
- Kanagal-Shamanna, R. Digital PCR: Principles and Applications. Methods Mol Biol 2016, 1392, 43–50. [Google Scholar] [PubMed]
- Gu, J.; Zang, W.; Liu, B.; Li, L.; Huang, L.; Li, S.; Rao, G.; Yu, Y.; Zhou, Y. Evaluation of digital PCR for detecting low-level EGFR mutations in advanced lung adenocarcinoma patients: a cross-platform comparison study. Oncotarget 2017, 8, 67810–67820. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Janku, F.; Jung, B.; Hou, C.; Madwani, K.; Alden, R.; Razavi, P.; Reis-Filho, J.; Shen, R.; Isbell, J.; et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann. Oncol. 2019, 30, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.; Barlesi, F.; Lolkema, M.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Bauml, J.; Viteri, S.; Minchom, A.; Bazhenova, L.; Ou, S.; Schaffer, M.; Le Croy, N.; Riley, R.; Mahadevia, P.; Girard, N. FP07.12 Underdiagnosis of EGFR Exon 20 Insertion Mutation Variants: Estimates from NGS-based Real-World Datasets. J. Thorac. Oncol. 2021, 16, S208–S209. [Google Scholar] [CrossRef]
- Yenerall, P.; Kittler, R.; Minna, J. Structure-based classification of EGFR mutations informs inhibitor selection for lung cancer therapy. Cancer Cell 2021, 39, 1455–1457. [Google Scholar] [CrossRef]
- Kim, E.Y. , et al., Compound EGFR mutation is frequently detected with co-mutations of actionable genes and associated with poor clinical outcome in lung adenocarcinoma. Cancer Biol Ther 2016, 17, 237–245. [Google Scholar] [CrossRef]
- Suda, K.; Sakai, K.; Obata, K.; Ohara, S.; Fujino, T.; Koga, T.; Hamada, A.; Soh, J.; Nishio, K.; Mitsudomi, T. Inter- and Intratumor Heterogeneity of EGFR Compound Mutations in Non–Small Cell Lung Cancers: Analysis of Five Cases. Clin. Lung Cancer 2021, 22, e141–e145. [Google Scholar] [CrossRef]
- Roper, N.; Brown, A.-L.; Wei, J.S.; Pack, S.; Trindade, C.; Kim, C.; Restifo, O.; Gao, S.; Sindiri, S.; Mehrabadi, F.; et al. Clonal Evolution and Heterogeneity of Osimertinib Acquired Resistance Mechanisms in EGFR Mutant Lung Cancer. Cell Rep. Med. 2020, 1, 100007. [Google Scholar] [CrossRef]
- Chen, H.; Cheng, X. Tumor heterogeneity and resistance to EGFR-targeted therapy in advanced nonsmall cell lung cancer: challenges and perspectives. OncoTargets Ther. 2014, 7, 1689–1704. [Google Scholar] [CrossRef]
- Attili, I.; Passaro, A.; Pisapia, P.; Malapelle, U.; de Marinis, F. Uncommon EGFR Compound Mutations in Non-Small Cell Lung Cancer (NSCLC): A Systematic Review of Available Evidence. Curr. Oncol. 2022, 29, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Yu, X. , et al., First-generation EGFR tyrosine kinase inhibitor therapy in 106 patients with compound EGFR-mutated lung cancer: a single institution's clinical practice experience. Cancer Commun (Lond) 2018, 38, 51. [Google Scholar] [PubMed]
- Hata, A.; Yoshioka, H.; Fujita, S.; Kunimasa, K.; Kaji, R.; Imai, Y.; Tomii, K.; Iwasaku, M.; Nishiyama, A.; Ishida, T.; et al. Complex Mutations in the Epidermal Growth Factor Receptor Gene in Non-small Cell Lung Cancer. J. Thorac. Oncol. 2010, 5, 1524–1528. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Jin, B.; Chu, T.; Dong, X.; Yang, H.; Zhang, Y.; Wu, D.; Lou, Y.; Zhang, X.; Wang, H.; et al. EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non-small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: A real-world study in China. Lung Cancer 2016, 96, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Lund-Iversen, M.; Kleinberg, L.; Fjellbirkeland, L.; Helland. ; Brustugun, O.T. Clinicopathological Characteristics of 11 NSCLC Patients with EGFR-Exon 20 Mutations. J. Thorac. Oncol. 2012, 7, 1471–1473. [Google Scholar] [CrossRef] [PubMed]
- Klughammer, B.; Brugger, W.; Cappuzzo, F.; Ciuleanu, T.; Mok, T.; Reck, M.; Tan, E.H.; Delmar, P.; Klingelschmitt, G.; Yin, A.-Y.; et al. Examining Treatment Outcomes with Erlotinib in Patients with Advanced Non–Small Cell Lung Cancer Whose Tumors Harbor Uncommon EGFR Mutations. J. Thorac. Oncol. 2016, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Tian, P.; Chen, B.; Wang, T.; Li, W. The prognostic impact of TP53 comutation in EGFR mutant lung cancer patients: a systematic review and meta-analysis. Postgrad. Med. 2019, 131, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Song, J.; Wang, Y.; Huang, L.; Sun, L.; Zhao, J.; Zhang, S.; Jing, W.; Ma, J.; Han, C. Concurrent Genetic Alterations and Other Biomarkers Predict Treatment Efficacy of EGFR-TKIs in EGFR-Mutant Non-Small Cell Lung Cancer: A Review. Front. Oncol. 2020, 10, 610923. [Google Scholar] [CrossRef]
- Shah, R.; Lester, J.F. Tyrosine Kinase Inhibitors for the Treatment of EGFR Mutation-Positive Non–Small-Cell Lung Cancer: A Clash of the Generations. Clin. Lung Cancer 2020, 21, e216–e228. [Google Scholar] [CrossRef]
- Alanazi, A.; Yunusa, I.; Elenizi, K.; I Alzarea, A. Efficacy and safety of tyrosine kinase inhibitors in advanced non-small-cell lung cancer harboring epidermal growth factor receptor mutation: a network meta-analysis. Lung Cancer Manag. 2021, 10, LMT43. [Google Scholar] [CrossRef]
- Reck, M.; van Zandwijk, N.; Gridelli, C.; Baliko, Z.; Rischin, D.; Allan, S.; Krzakowski, M.; Heigener, D. Erlotinib in Advanced Non-small Cell Lung Cancer: Efficacy and Safety Findings of the Global Phase IV Tarceva Lung Cancer Survival Treatment Study. J. Thorac. Oncol. 2010, 5, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.A.; Rodrigues Pereira, J.; Ciuleanu, T.E.; Tan, E.H.; Hirsh, V.; Thongprasert, S.; Campos, D.; Maoleekoonpiroj, S.; Smylie, M.; Martins, R.; et al. Erlotinib in Previously Treated Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2005, 353, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wu, Y.-L.; Chen, G.; Feng, J.; Liu, X.-Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Douillard, J.-Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; McCormack, R.; Webster, A.; Milenkova, T. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br. J. Cancer 2014, 110, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Kobayashi, K.; Usui, K.; Maemondo, M.; Okinaga, S.; Mikami, I.; Ando, M.; Yamazaki, K.; Saijo, Y.; Gemma, A.; et al. First-Line Gefitinib for Patients With Advanced Non–Small-Cell Lung Cancer Harboring Epidermal Growth Factor Receptor Mutations Without Indication for Chemotherapy. J. Clin. Oncol. 2009, 27, 1394–1400. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.; Wu, Y.-L.; Thongprasert, S.; Yang, C.-H.; Chu, D.-T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Cheng, Y.; Zhou, X.; Lee, K.H.; Nakagawa, K.; Niho, S.; Tsuji, F.; Linke, R.; Rosell, R.; Corral, J.; et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol. 2017, 18, 1454–1466. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, L.; Tan, E.-H.; O’byrne, K.; Zhang, L.; Hirsh, V.; Boyer, M.; Yang, J.-H.; Mok, T.; Lee, K.H.; Lu, S.; et al. Afatinib versus gefitinib in patients with EGFR mutation-positive advanced non-small-cell lung cancer: overall survival data from the phase IIb LUX-Lung 7 trial. Ann. Oncol. 2017, 28, 270–277. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Cho, J.H. , et al., Osimertinib for Patients With Non-Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Multicenter, Open-Label, Phase II Trial (KCSG-LU15-09). J Clin Oncol 2020, 38, 488–495. [Google Scholar] [CrossRef]
- Janning, M.; Süptitz, J.; Albers-Leischner, C.; Delpy, P.; Tufman, A.; Velthaus-Rusik, J.-L.; Reck, M.; Jung, A.; Kauffmann-Guerrero, D.; Bonzheim, I.; et al. Treatment outcome of atypical EGFR mutations in the German National Network Genomic Medicine Lung Cancer (nNGM). Ann. Oncol. 2022, 33, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, C.; Wang, Z.; Hu, Y.; Zhang, G.; Zhang, M.; Zheng, X.; Zhang, X.; Yang, J.; Ma, Z.; et al. Efficacy and long-term survival of advanced lung adenocarcinoma patients with uncommon EGFR mutations treated with 1st generation EGFR-TKIs compared with chemotherapy as first-line therapy. Lung Cancer 2019, 130, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.Y. , et al., A Phase IIIb Open-Label, Single-Arm Study of Afatinib in EGFR TKI-Naive Patients with EGFRm+ NSCLC: Final Analysis, with a Focus on Patients Enrolled at Sites in China. Target Oncol 2022, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.-H.; Schuler, M.; Popat, S.; Miura, S.; Heeke, S.; Park, K.; Märten, A.; Kim, E.S. Afatinib for the Treatment of NSCLC Harboring Uncommon EGFR Mutations: A Database of 693 Cases. J. Thorac. Oncol. 2020, 15, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y. , et al., Treatment of Patients With Non-small-cell Lung Cancer With Uncommon EGFR Mutations in Clinical Practice. Anticancer Res 2020, 40, 5757–5764. [Google Scholar] [CrossRef] [PubMed]
- UPDATES, F. , New Indication Approved for Afatinib in NSCLC. Oncology Times 2018, 40, 18. [Google Scholar]
- Floc'H, N.; Lim, S.; Bickerton, S.; Ahmed, A.; Orme, J.; Urosevic, J.; Martin, M.J.; Cross, D.A.; Cho, B.C.; Smith, P.D. Osimertinib, an Irreversible Next-Generation EGFR Tyrosine Kinase Inhibitor, Exerts Antitumor Activity in Various Preclinical NSCLC Models Harboring the Uncommon EGFR Mutations G719X or L861Q or S768I. Mol. Cancer Ther. 2020, 19, 2298–2307. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Aredo, J.V.; Piper-Vallillo, A.; Huppert, L.; Rotow, J.K.; Husain, H.; Stewart, S.; Cobb, R.; Wakelee, H.A.; Blakely, C.M.; et al. Osimertinib in NSCLC With Atypical EGFR-Activating Mutations: A Retrospective Multicenter Study. JTO Clin. Res. Rep. 2023, 4, 100459. [Google Scholar] [CrossRef]
- Sequist, L.V. , et al., Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non-small-cell lung cancer. J Clin Oncol 2010, 28, 3076–83. [Google Scholar] [CrossRef]
- Heigener, D.F.; Schumann, C.; Sebastian, M.; Sadjadian, P.; Stehle, I.; Märten, A.; Lüers, A.; Griesinger, F.; Scheffler, M. ; for the Afatinib Compassionate Use Consortium (ACUC) Afatinib in Non-Small Cell Lung Cancer Harboring Uncommon EGFR Mutations Pretreated With Reversible EGFR Inhibitors. Oncol. 2015, 20, 1167–1174. [Google Scholar] [CrossRef]
- Costa, D.B.; B. , D. Kinase inhibitor-responsive genotypes in EGFR mutated lung adenocarcinomas: moving past common point mutations or indels into uncommon kinase domain duplications and rearrangements. Transl. Lung Cancer Res. 2016, 5, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y. , et al., EGFR Exon 18 Mutations in Lung Cancer: Molecular Predictors of Augmented Sensitivity to Afatinib or Neratinib as Compared with First- or Third-Generation TKIs. Clin Cancer Res 2015, 21, 5305–5313. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Takeda, M.; Tamiya, A.; Kasai, T.; Atagi, S. Programmed death-ligand 1 expression in uncommon epidermal growth factor receptor mutation-positive non-small-cell lung cancer. Ann. Oncol. 2018, 29, 2262–2263. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.P. , et al., Clinical outcomes in non-small cell lung cancers harboring different exon 19 deletions in EGFR. Clin Cancer Res 2012, 18, 3470–3477. [Google Scholar] [CrossRef]
- Peng, X.; Long, X.; Liu, L.; Zeng, L.; Yang, H.; Jiang, W.; Liao, D.; Li, K.; Wang, J.; Lizaso, A.; et al. Clinical impact of uncommon epidermal growth factor receptor exon 19 insertion-deletion variants on epidermal growth factor receptor-tyrosine kinase inhibitor efficacy in non-small-cell lung cancer. Eur. J. Cancer 2020, 141, 199–208. [Google Scholar] [CrossRef] [PubMed]
- He, M. , et al., EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res 2012, 18, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Leighl, N.; Blackhall, F.; Popat, S.; Kerr, K.; Ahn, M.; Arcila, M.; Arrieta, O.; Planchard, D.; de Marinis, F.; et al. ESMO expert consensus statements on the management of EGFR mutant non-small-cell lung cancer. Ann. Oncol. 2022, 33, 466–487. [Google Scholar] [CrossRef]
- Vyse, S.; Huang, P.H. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 51. [Google Scholar] [CrossRef]
- Vasconcelos, P.E.; Gergis, C.; Viray, H.; Varkaris, A.; Fujii, M.; Rangachari, D.; VanderLaan, P.A.; Kobayashi, I.S.; Kobayashi, S.S.; Costa, D.B. EGFR-A763_Y764insFQEA Is a Unique Exon 20 Insertion Mutation That Displays Sensitivity to Approved and In-Development Lung Cancer EGFR Tyrosine Kinase Inhibitors. JTO Clin. Res. Rep. 2020, 1. [Google Scholar] [CrossRef]
- Hasegawa, H.; Yasuda, H.; Hamamoto, J.; Masuzawa, K.; Tani, T.; Nukaga, S.; Hirano, T.; Kobayashi, K.; Manabe, T.; Terai, H.; et al. Efficacy of afatinib or osimertinib plus cetuximab combination therapy for non-small-cell lung cancer with EGFR exon 20 insertion mutations. Lung Cancer 2018, 127, 146–152. [Google Scholar] [CrossRef]
- Elamin, Y.Y.; Robichaux, J.P.; Carter, B.W.; Altan, M.; Gibbons, D.L.; Fossella, F.V.; Lam, V.K.; Patel, A.B.; Negrao, M.V.; Le, X.; et al. Poziotinib for Patients With HER2 Exon 20 Mutant Non–Small-Cell Lung Cancer: Results From a Phase II Trial. J. Clin. Oncol. 2022, 40, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Elamin, Y.Y.; Robichaux, J.P.; Carter, B.W.; Altan, M.; Tran, H.; Gibbons, D.L.; Heeke, S.; Fossella, F.V.; Lam, V.K.; Le, X.; et al. Poziotinib for EGFR exon 20-mutant NSCLC: Clinical efficacy, resistance mechanisms, and impact of insertion location on drug sensitivity. Cancer Cell 2022, 40, 754–767. [Google Scholar] [CrossRef] [PubMed]
- Prelaj, A.; Bottiglieri, A.; Proto, C.; Russo, G.L.; Signorelli, D.; Ferrara, R.; Galli, G.; De Toma, A.; Viscardi, G.; Brambilla, M.; et al. Poziotinib for EGFR and HER2 exon 20 insertion mutation in advanced NSCLC: Results from the expanded access program. Eur. J. Cancer 2021, 149, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C. , et al., Treatment Outcomes and Safety of Mobocertinib in Platinum-Pretreated Patients With EGFR Exon 20 Insertion-Positive Metastatic Non-Small Cell Lung Cancer: A Phase 1/2 Open-label Nonrandomized Clinical Trial. JAMA Oncol 2021, 7, e214761. [Google Scholar] [CrossRef] [PubMed]
- FDA. EDA grants accelerated approval to mobocertinib for metastatic non-small cell lung cancer with EGFR exon 20 insertion mutations. 2022 , 2022]; Available from: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-mobocertinib-metastatic-non-small-cell-lung-cancer-egfr-exon-20. 10 April.
- FDA. FDA Approves First Targeted Therapy for Subset of Non-Small Cell Lung Cancer. 2022 Accessed , 2022]; Available from: https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-subset-non-small-cell-lung-cancer. 10 April.
- Agency, E.M. Rybrevant. 2022 , 2022]; Available from: https://www.ema.europa.eu/en/medicines/human/EPAR/rybrevant. 10 April.
- Park, K.; Haura, E.B.; Leighl, N.B.; Mitchell, P.; Shu, C.A.; Girard, N.; Viteri, S.; Han, J.-Y.; Kim, S.-W.; Lee, C.K.; et al. Amivantamab in EGFR Exon 20 Insertion–Mutated Non–Small-Cell Lung Cancer Progressing on Platinum Chemotherapy: Initial Results From the CHRYSALIS Phase I Study. J. Clin. Oncol. 2021, 39, 3391–3402. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Sabari, J.K.; Haura, E.B.; Shu, C.A.; Spira, A.; Salgia, R.; Reckamp, K.L.; Sanborn, R.E.; Govindan, R.; Bauml, J.M.; et al. Management of infusion-related reactions (IRRs) in patients receiving amivantamab in the CHRYSALIS study. Lung Cancer 2023, 178, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Janne, P.; Wang, M.; Mitchell, P.; Fang, J.; Nian, W.; Chiu, C.; Zhou, J.; Zhao, Y.; Su, W.; Camidge, D.; et al. OA15.02 Phase 1 Studies of DZD9008, an Oral Selective EGFR/HER2 Inhibitor in Advanced NSCLC with EGFR Exon20 Insertion Mutations. J. Thorac. Oncol. 2021, 18, S874. [Google Scholar] [CrossRef]
- Piotrowska, Z.; Nguyen, D.; Koczywas, M.; Tchekmedyian, N.; Clancy, M.; Witter, D.; Page, A.; Zawel, L.; Yu, H. 1345P Preliminary safety and activity of CLN-081 in NSCLC with EGFR exon 20 insertion mutations (Ins20). Annals of Oncology 2020, 31 (Suppl. S4), S754–S840. [Google Scholar] [CrossRef]
- S. V. Liu, L.C.V., V.H.F. Lee, V.W. Zhu, C.S. Baik, A. Sacher, C.E. McCoach, D. Nguyen, J.Y. Li, J.M. Pacheco, C. Kim, T.F. Burns, E.L. Schenk, N. Leighl, L. Tozzi, D.R. Camidge, LBA61 - First analysis of RAIN-701: Study of tarloxotinib in patients with non-small cell lung cancer (NSCLC) EGFR Exon 20 insertion, HER2-activating mutations & other solid tumours with NRG1/ERBB gene fusions. Annals of Oncology 2020, 31 (suppl_4), S1142–S1215. [Google Scholar]
- De Pas, T.; Toffalorio, F.; Manzotti, M.; Fumagalli, C.; Spitaleri, G.; Catania, C.; Delmonte, A.; Giovannini, M.; Spaggiari, L.; de Braud, F.; et al. Activity of Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors in Patients with Non-small Cell Lung Cancer Harboring Rare Epidermal Growth Factor Receptor Mutations. J. Thorac. Oncol. 2011, 6, 1895–1901. [Google Scholar] [CrossRef]
- Galli, G.; Corrao, G.; Imbimbo, M.; Proto, C.; Signorelli, D.; Ganzinelli, M.; Zilembo, N.; Vitali, M.; de Braud, F.; Garassino, M.C.; et al. Uncommon mutations in epidermal growth factor receptor and response to first and second generation tyrosine kinase inhibitors: A case series and literature review. Lung Cancer 2018, 115, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Massarelli, E.; Johnson, F.M.; Erickson, H.S.; Wistuba, I.I.; Papadimitrakopoulou, V. Uncommon Epidermal Growth Factor Receptor mutations in non-small cell lung cancer and their mechanisms of EGFR tyrosine kinase inhibitors sensitivity and resistance. Lung Cancer 2013, 80, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Guo, T.; Zhou, Y.; Zhao, Y.; Chu, L.; Chu, X.; Yang, X.; Ni, J.; Zhu, Z. Clinical outcomes of advanced non-small cell lung cancer patients harboring distinct subtypes of EGFR mutations and receiving first-line tyrosine kinase inhibitors: brain metastasis and de novo T790M matters. BMC Cancer 2022, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Soria, J.C. , et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Morise, M.; Wakuda, K.; Hataji, O.; Shimokawaji, T.; Takahashi, K.; Furuya, N.; Takeyama, Y.; Goto, Y.; Abe, T.; et al. A multicenter cohort study of osimertinib compared with afatinib as first-line treatment for EGFR-mutated non-small-cell lung cancer from practical dataset: CJLSG1903. ESMO Open 2021, 6, 100115. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhang, J.; Tang, D.; Ye, X.; Li, J.; Mu, N.; Li, Z.; Liu, R.; Xiang, L.; Huang, C.; et al. Tyrosine Kinase Inhibitors Could Be Effective Against Non-small Cell Lung Cancer Brain Metastases Harboring Uncommon EGFR Mutations. Front. Oncol. 2020, 10, 224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Y.; Liu, Z.; Wang, L.; Yao, Y.; Liu, Y.; Hao, X.Z.; Wang, J.; Xing, P.; Li, J. Efficacy of dacomitinib in patients with EGFR-mutated NSCLC and brain metastases. Thorac. Cancer 2021, 12, 3407–3415. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Pu, X.; Jiang, M.; Wang, J.; Li, J.; Li, K.; Xu, Y.; Xu, F.; Chen, B.; Wang, Q.; et al. Dacomitinib induces objective responses in metastatic brain lesions of patients with EGFR-mutant non-small-cell lung cancer: A brief report. Lung Cancer 2021, 152, 66–70. [Google Scholar] [CrossRef]
- CLN-081 Produces Antitumor Activity With Acceptable Safety in Heavily Pretreated EGFR Exon 20 Insertion+ NSCLC. 2022; Available from: https://www.onclive.com/view/cln-081-produces-antitumor-activity-with-acceptable-safety-in-heavily-pretreated-egfr-exon-20-insertion-nsclc.
- Janne, P.A.; Ramalingam, S.S.; Yang, J.C.-H.; Riely, G.J.; Bunn, V.; Jin, S.; Zhou, C.; Camidge, D.R. Mobocertinib (TAK-788) in EGFR exon 20 insertion (ex20ins)+ metastatic non–small cell lung cancer (mNSCLC): Treatment (tx) beyond progressive disease (PD) in platinum-pretreated patients (pts) with and without intracranial PD. J. Clin. Oncol. 2022, 40, 9099–9099. [Google Scholar] [CrossRef]
- Vyse, S. and P.H. Huang, Amivantamab for the treatment of EGFR exon 20 insertion mutant non-small cell lung cancer. Expert Rev Anticancer Ther 2022, 22, 3–16. [Google Scholar] [CrossRef]
- Catherine, A. Shu, K.G., Byoung Chul Cho, Frank Griesinger, James Chih-Hsin Yang, Enriqueta Felip, John Xie, Jun Chen, Janine Mahoney, Meena Thayu, Roland Elmar Knoblauch, Leonardo Trani, Joshua Bauml, CHRYSALIS-2: A phase 1/1b study of lazertinib as monotherapy and in combination with amivantamab in patients with EGFR-mutant NSCLC. J Clin Oncol 2021, 39. [Google Scholar]
- Hastings, K.; Yu, H.; Wei, W.; Sanchez-Vega, F.; DeVeaux, M.; Choi, J.; Rizvi, H.; Lisberg, A.; Truini, A.; Lydon, C.; et al. EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer. Ann. Oncol. 2019, 30, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.-Y.; Zhang, J.-T.; Liu, S.-Y.; Su, J.; Zhang, C.; Xie, Z.; Zhou, Q.; Tu, H.-Y.; Xu, C.-R.; Yan, L.-X.; et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. OncoImmunology 2017, 6, e1356145. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, S.S.; Zaiss, D.M.W. Emerging Role of EGFR Mutations in Creating an Immune Suppressive Tumour Microenvironment. Biomedicines 2021, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Garassino, M.C.; Cho, B.-C.; Kim, J.-H.; Mazières, J.; Vansteenkiste, J.; Lena, H.; Jaime, J.C.; Gray, J.E.; Powderly, J.; Chouaid, C.; et al. Durvalumab as third-line or later treatment for advanced non-small-cell lung cancer (ATLANTIC): an open-label, single-arm, phase 2 study. Lancet Oncol. 2018, 19, 521–536. [Google Scholar] [CrossRef]
- Isomoto, K.; Haratani, K.; Hayashi, H.; Shimizu, S.; Tomida, S.; Niwa, T.; Yokoyama, T.; Fukuda, Y.; Chiba, Y.; Kato, R.; et al. Impact of EGFR-TKI Treatment on the Tumor Immune Microenvironment in EGFR Mutation–Positive Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 2037–2046. [Google Scholar] [CrossRef]
- Kirchner, M.; Kluck, K.; Brandt, R.; Volckmar, A.-L.; Penzel, R.; Kazdal, D.; Endris, V.; Neumann, O.; Seker-Cin, H.; Goldschmid, H.; et al. The immune microenvironment in EGFR- and ERBB2-mutated lung adenocarcinoma. ESMO Open 2021, 6, 100253. [Google Scholar] [CrossRef]
- Lau, S.C.; Fares, A.F.; Le, L.W.; Mackay, K.M.; Soberano, S.; Chan, S.W.; Smith, E.; Ryan, M.; Tsao, M.S.; Bradbury, P.A.; et al. Subtypes of EGFR- and HER2-Mutant Metastatic NSCLC Influence Response to Immune Checkpoint Inhibitors. Clin. Lung Cancer 2021, 22, 253–259. [Google Scholar] [CrossRef]
- Shen, C.-I.; Chao, H.-S.; Shiao, T.-H.; Chiang, C.-L.; Huang, H.-C.; Luo, Y.-H.; Chiu, C.-H.; Chen, Y.-M. Comparison of the outcome between immunotherapy alone or in combination with chemotherapy in EGFR-mutant non-small cell lung cancer. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Mazieres, J.; Drilon, A.; Lusque, A.B.; Mhanna, L.; Cortot, A.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef]
- Yamada, T.; Hirai, S.; Katayama, Y.; Yoshimura, A.; Shiotsu, S.; Watanabe, S.; Kikuchi, T.; Hirose, K.; Kubota, Y.; Chihara, Y.; et al. Retrospective efficacy analysis of immune checkpoint inhibitors in patients with EGFR-mutated non-small cell lung cancer. Cancer Med. 2019, 8, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-H.S.; Wang, C.-C.; Huang, Y.-C.; Pavlidis, S.; Liu, C.-Y.; Ko, H.-W.; Chung, F.-T.; Lin, T.-Y.; Wang, C.-L.; Guo, Y.-K.; et al. Comparison of a combination of chemotherapy and immune checkpoint inhibitors and immune checkpoint inhibitors alone for the treatment of advanced and metastatic non-small cell lung cancer. Thorac. Cancer 2019, 10, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Mok, T.S.K.; Nishio, M.; Jotte, R.M.; Cappuzzo, F.; Orlandi, F.; Stroyakovskiy, D.; Nogami, N.; Rodríguez-Abreu, D.; Moro-Sibilot, D.; et al. Atezolizumab plus bevacizumab and chemotherapy in non-small-cell lung cancer (IMpower150): key subgroup analyses of patients with EGFR mutations or baseline liver metastases in a randomised, open-label phase 3 trial. Lancet Respir. Med. 2019, 7, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Mok, T.; Socinski, M.; Jotte, R.; Lim, D.-T.; Cappuzzo, F.; Orlandi, F.; Stroyakovskiy, D.; Nogami, N.; Rodriguez-Abreu, D.; et al. 1293P IMpower150: Updated efficacy analysis in patients with EGFR mutations. Annals of Oncology 2020, 31 (Suppl. S4), S754–S840. [Google Scholar] [CrossRef]
- Nogami, N.; Barlesi, F.; Socinski, M.A.; Reck, M.; Thomas, C.A.; Cappuzzo, F.; Mok, T.S.; Finley, G.; Aerts, J.G.; Orlandi, F.; et al. IMpower150 Final Exploratory Analyses for Atezolizumab Plus Bevacizumab and Chemotherapy in Key NSCLC Patient Subgroups With EGFR Mutations or Metastases in the Liver or Brain. J. Thorac. Oncol. 2021, 17, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-G.; Shih, J.-Y. Management of acquired resistance to EGFR TKI–targeted therapy in advanced non-small cell lung cancer. Mol. Cancer 2018, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-Y.; Liu, S.-Y.; Wu, Y.-L. Safety of EGFR-TKIs for EGFR mutation-positive non-small cell lung cancer. Expert Opin. Drug Saf. 2020, 19, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. , et al., Comparison of gefitinib, erlotinib and afatinib in non-small cell lung cancer: A meta-analysis. Int J Cancer 2017, 140, 2805–2819. [Google Scholar] [CrossRef]
- Park, K.; Tan, E.-H.; O'Byrne, K.; Zhang, L.; Boyer, M.; Mok, T.; Hirsh, V.; Yang, J.C.-H.; Lee, K.H.; Lu, S.; et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016, 17, 577–589. [Google Scholar] [CrossRef]
- Hochmair, M.J.; Morabito, A.; Hao, D.; Yang, C.-T.; A Soo, R.; Yang, J.C.-H.; Gucalp, R.; Halmos, B.; Wang, L.; Märten, A.; et al. Sequential afatinib and osimertinib in patients with EGFR mutation-positive non-small-cell lung cancer: updated analysis of the observational GioTag study. Futur. Oncol. 2019, 15, 2905–2914. [Google Scholar] [CrossRef]
- Options, C.C. CHRYSALIS-2: Update of Amivantamab + Lazertinib in EGFR-Mutant NSCLC After Progression on Osimertinib and Chemotherapy. 2022; Available from: https://clinicaloptions.com/CE-CME/chrysalis-2-update/100003234.
| Compound mutation | Response to treatment | Proposed treatment |
|---|---|---|
| Combined common EGFR mutations (ex21 p.L858R + ex19del) |
Response rate (RR) ≥ 75% with either 1st or 2nd generation TKIs | 1st or 2nd gen TKIs |
| Combined common (ex21 p.L858R + ex19del) plus uncommon EGFR mutations (any but ex21 p.L858R, ex19del or de novo ex20 p.T790M) |
RR 40–80% and 100% with 1st generation TKIs and afatinib | Afatinib |
| Combined uncommon EGFR mutations | RR 20–70%, ~80% and ~75% with 1st generation TKIs, afatinib and osimertinib, respectively | Afatinib |
| Combined EGFR mutation (any) plus de novo ex20 p.T790M | Primary resistance to first- and second-generation EGFR TKIs; osimertinib (RR 33.3%, DCR 100%) | Osimertinib |
| Ex20 inhibitor | Trial | Toxicity | Response to treatment |
|---|---|---|---|
| Poziotinib | NCT03066206 | Diarrhea 92%, skin rash 90%, oral mucositis 68%, paronychia 68%, dry skin 60% (66% of G3 AEs on EAP) | ORR 32%, mPFS 5.5 m, mOS 19.2 m ORR of 46% and 0% in near (aa A767 to P772) vs far loop ins |
| Mobocertinib | NCT02716116 |
PPP cohort 69% G≥3 AEs 46% SAE EXCLAIM cohort 66% G≥3 AEs 44% SAE |
PPP cohort ORR 28% by IRC and 35% by investigator assessment, mPFS 7.3 m by IRC, mOS 24.0 m EXCLAIM cohort ORR 25% by IRC and 32% by investigator assessment |
| Amivantamab | NCT02609776 | At RP2D 39% G≥3 AEs, 31% SAE | ORR 40%, mPFS 8.3 m |
| Sunvozertinib (DZD9008) | NCT03974022 and CTR20192097 | Most common TEAEs: diarrhea (G3 5.2%) and skin rash (G3 1%) |
ORR 39.3% across all dose levels; dose level of 300 mg once daily, ORR 48.4% and DCR 90.3% |
| CLN-081 (TAS6417) | NCT04036682 | Constipation 8%, diarrhea 8%, dizziness 8%, fatigue 8%, and chest pain 8% | 5 evaluable pts: 2 pts PR, 3 pts SD |
| Tarloxotinib | NCT03805841 | G3 TEAEs: prolonged QTc 34.8%, rash 4.3%, diarrhea 4.3%, increased ALT 4.3% | DCR 60% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





