Submitted:
03 May 2023
Posted:
05 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Parameterization of sulfotyrosine
2.2. Generation of starting structures
2.3. Molecular dynamics simulations
2.4. Peptide synthesis and binding assays
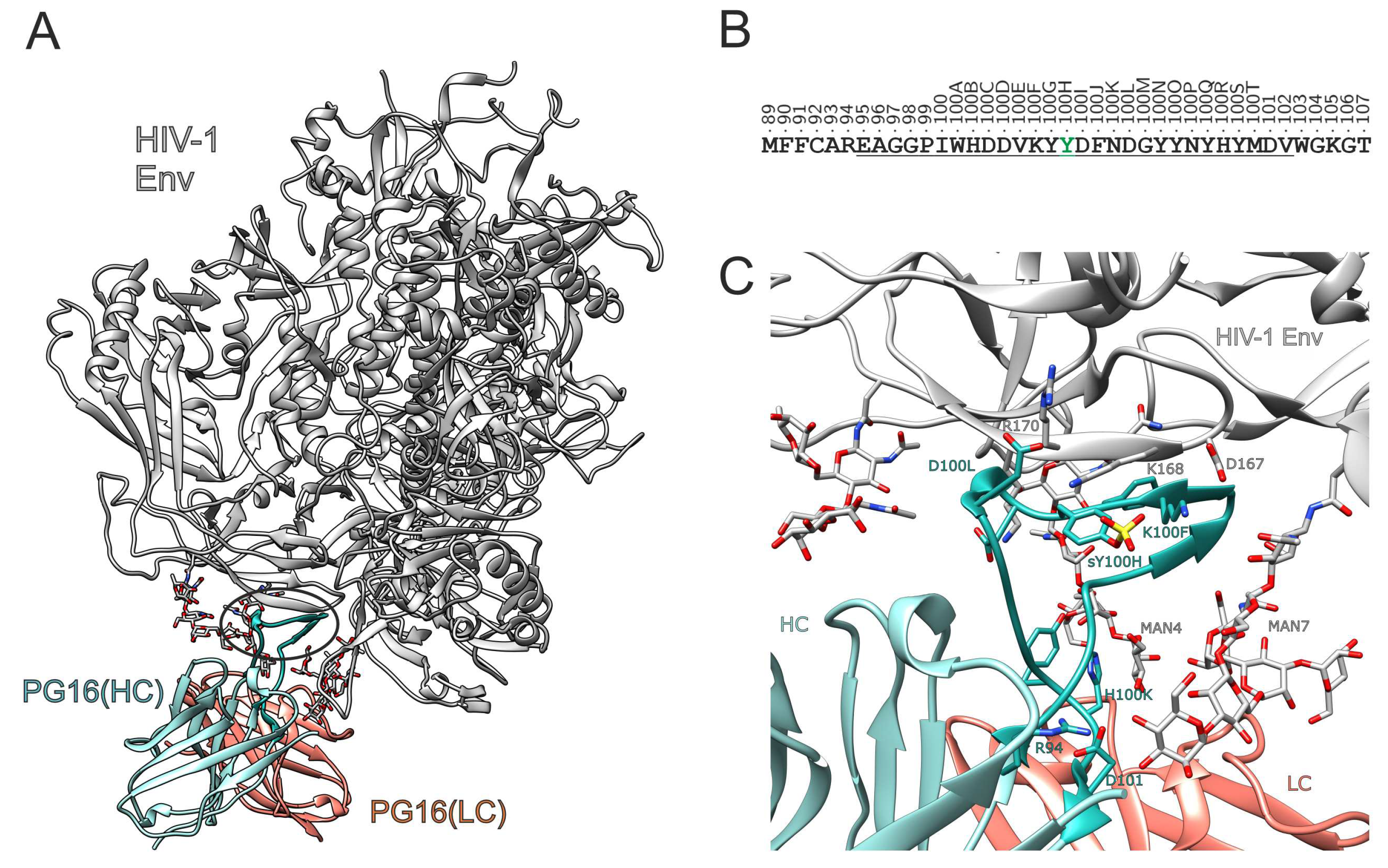
3. Results and Discussion
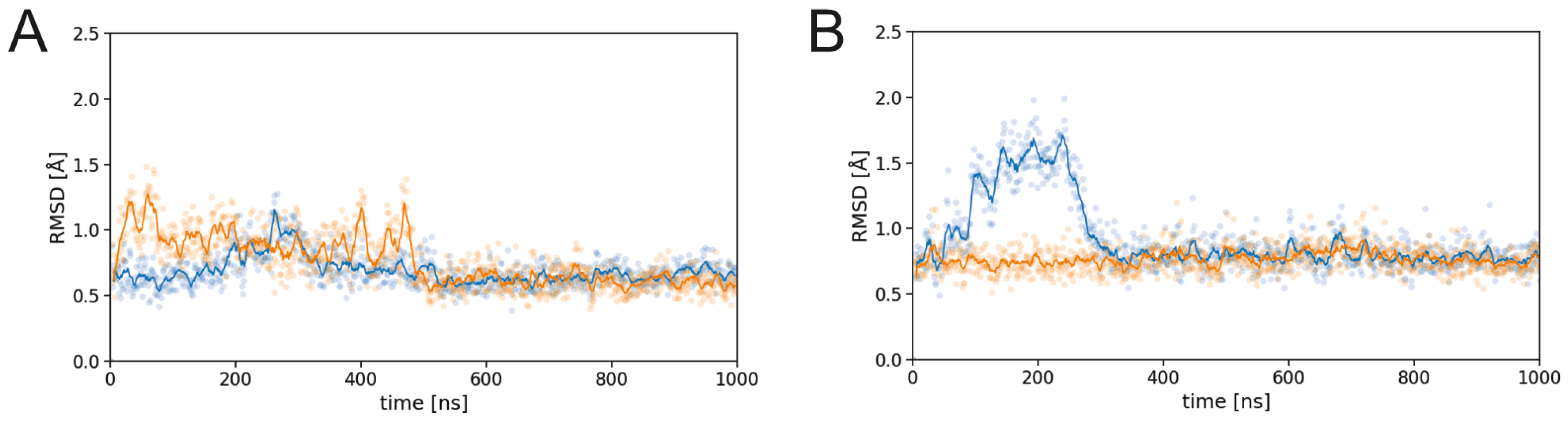
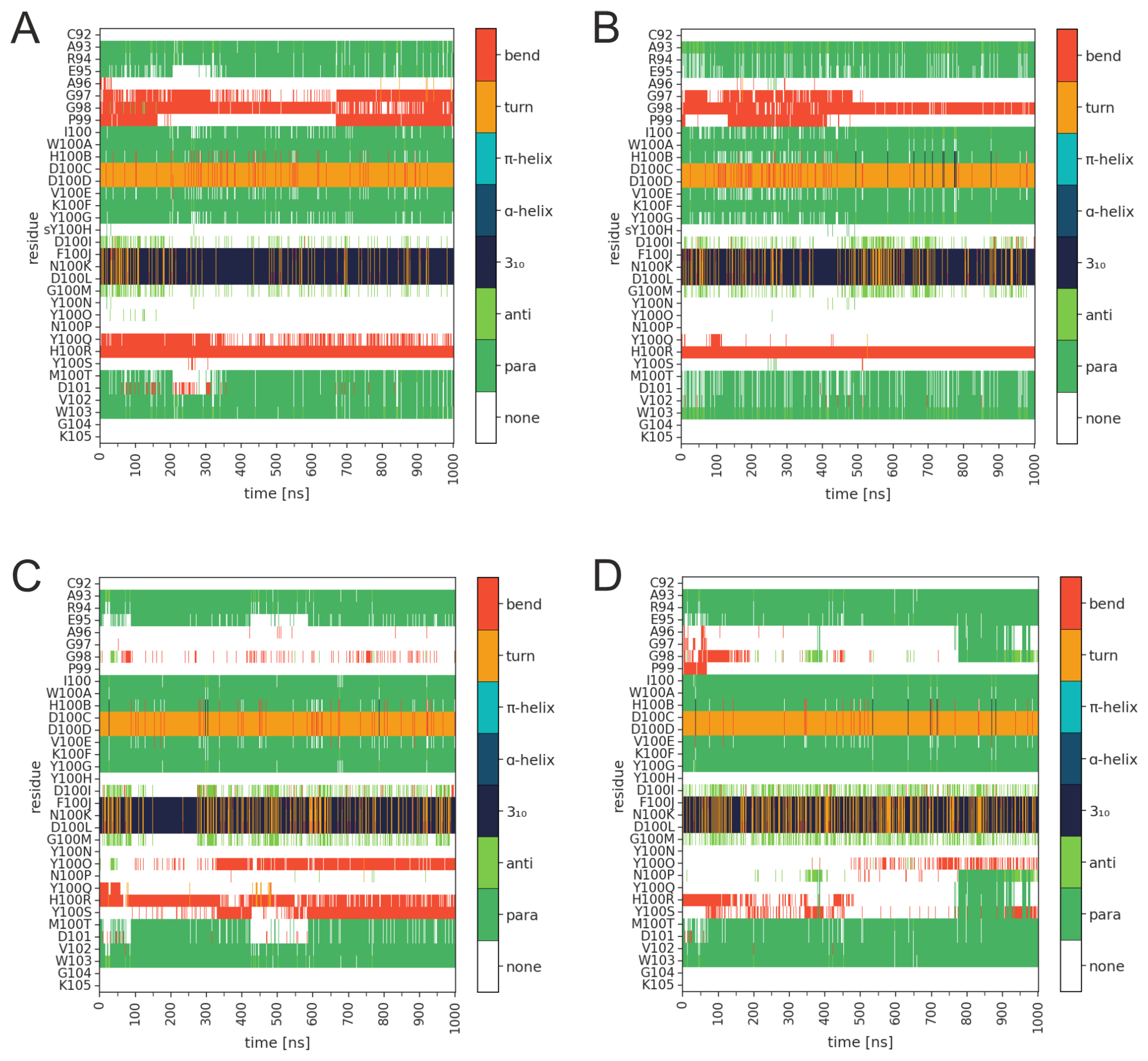
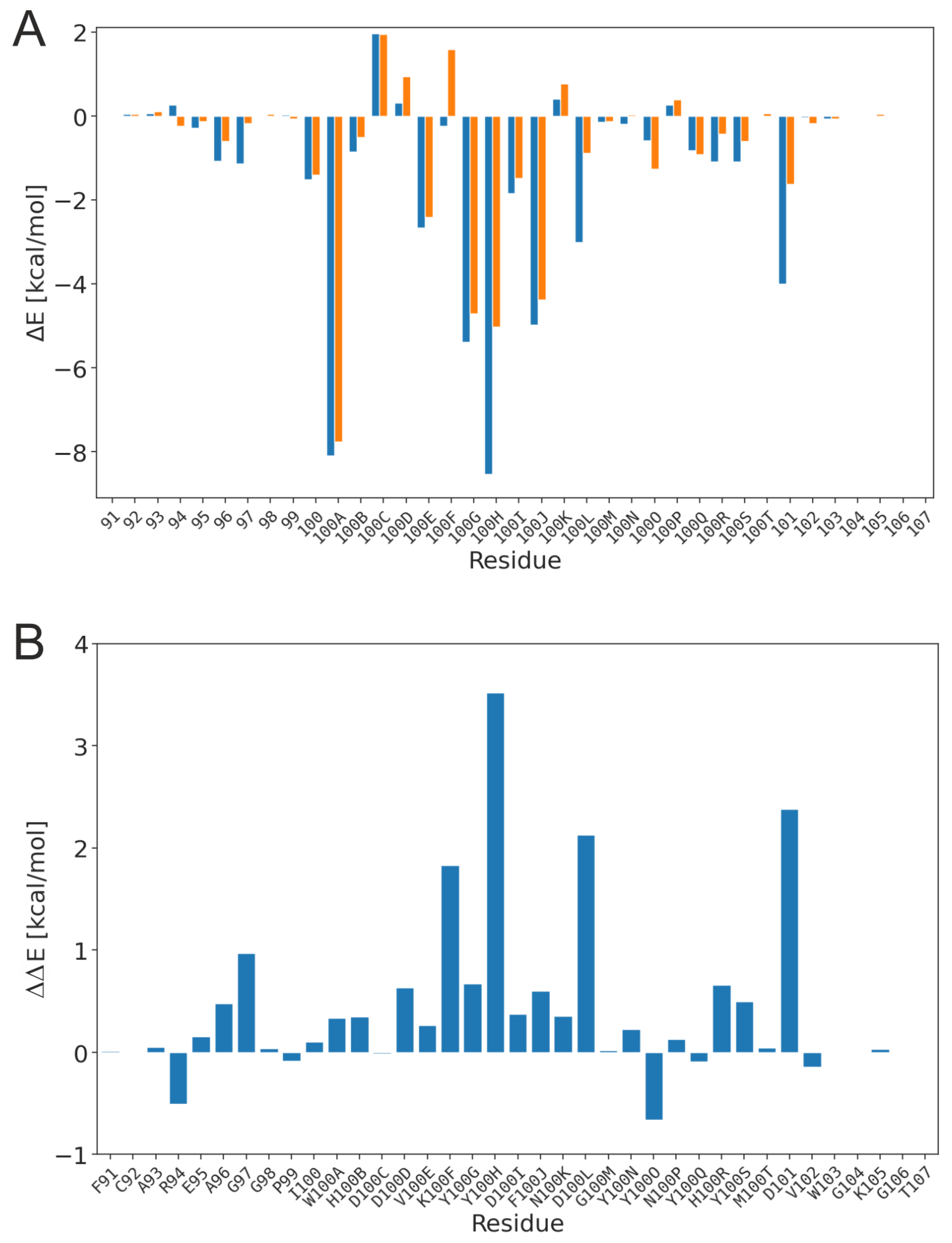
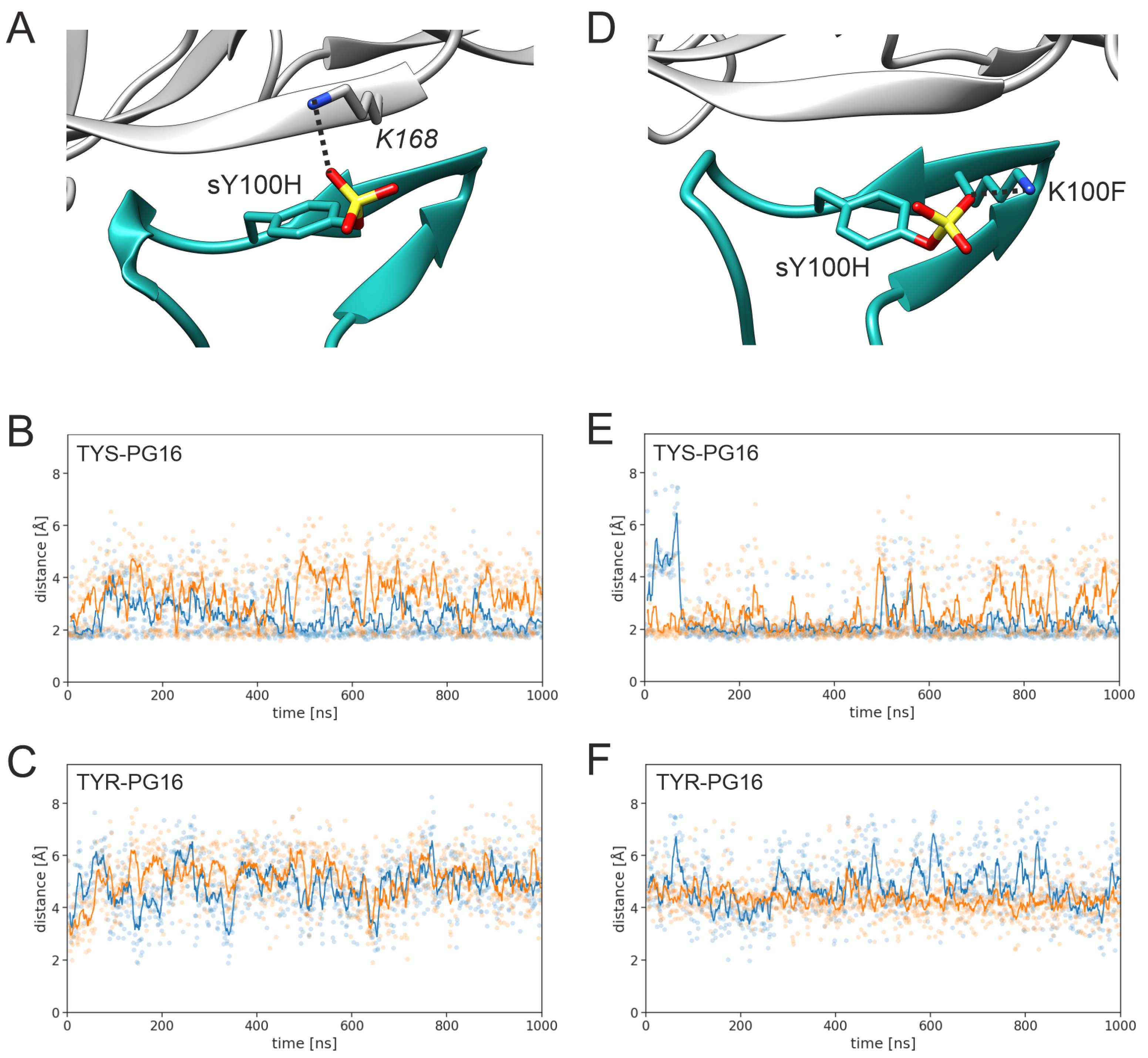
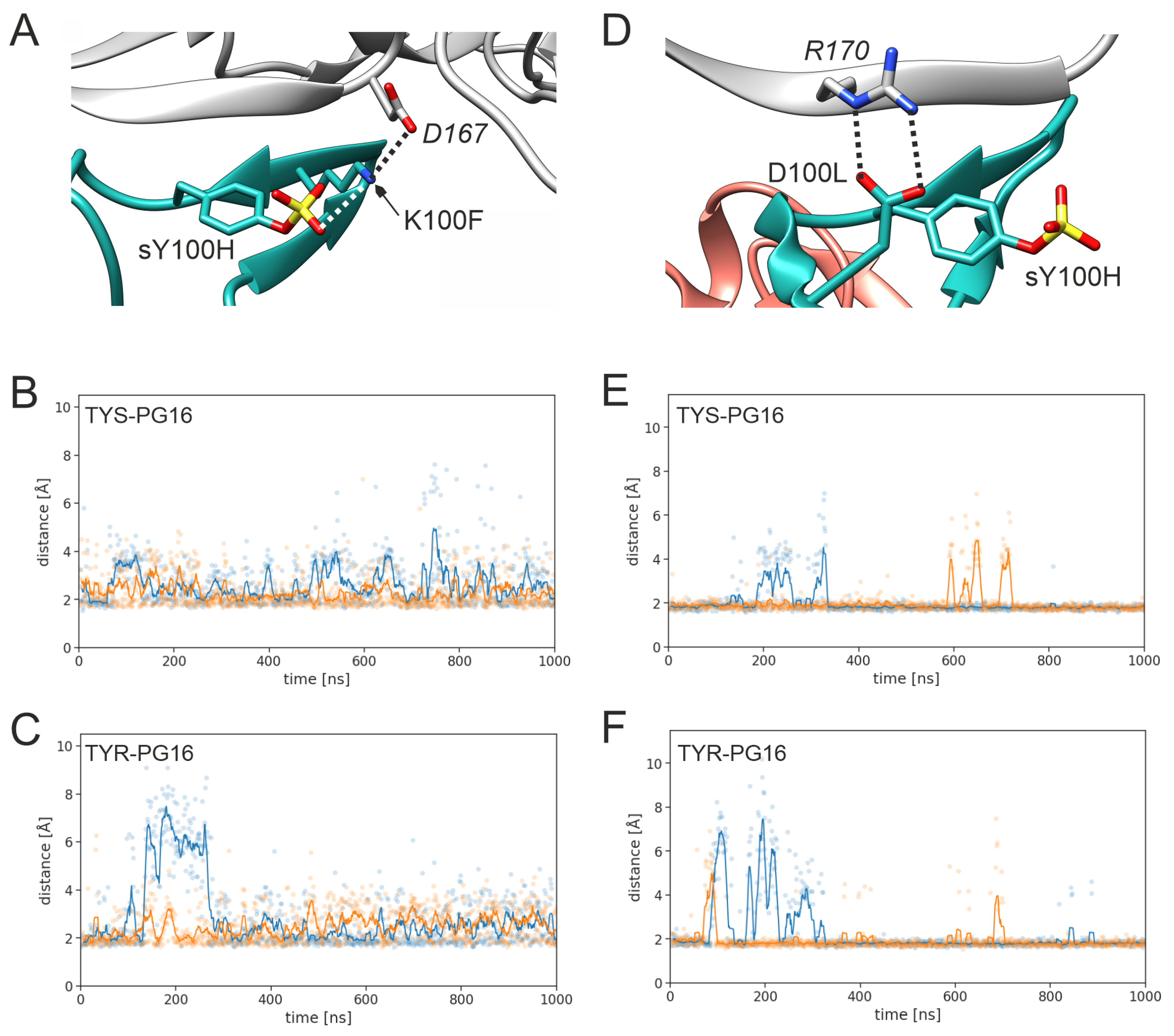
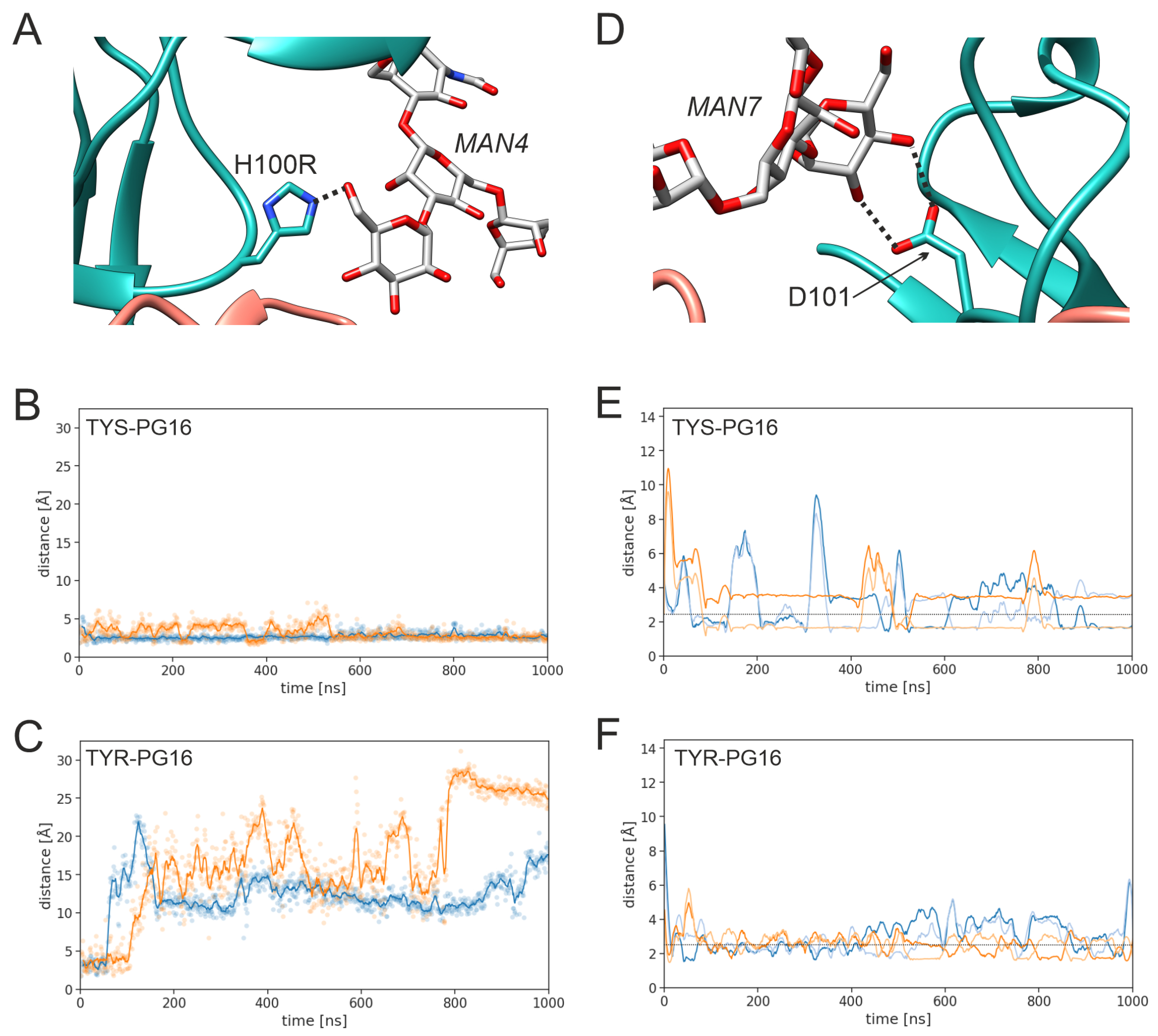
3.1. Effect of sulfation on PG16 dynamics and interactions
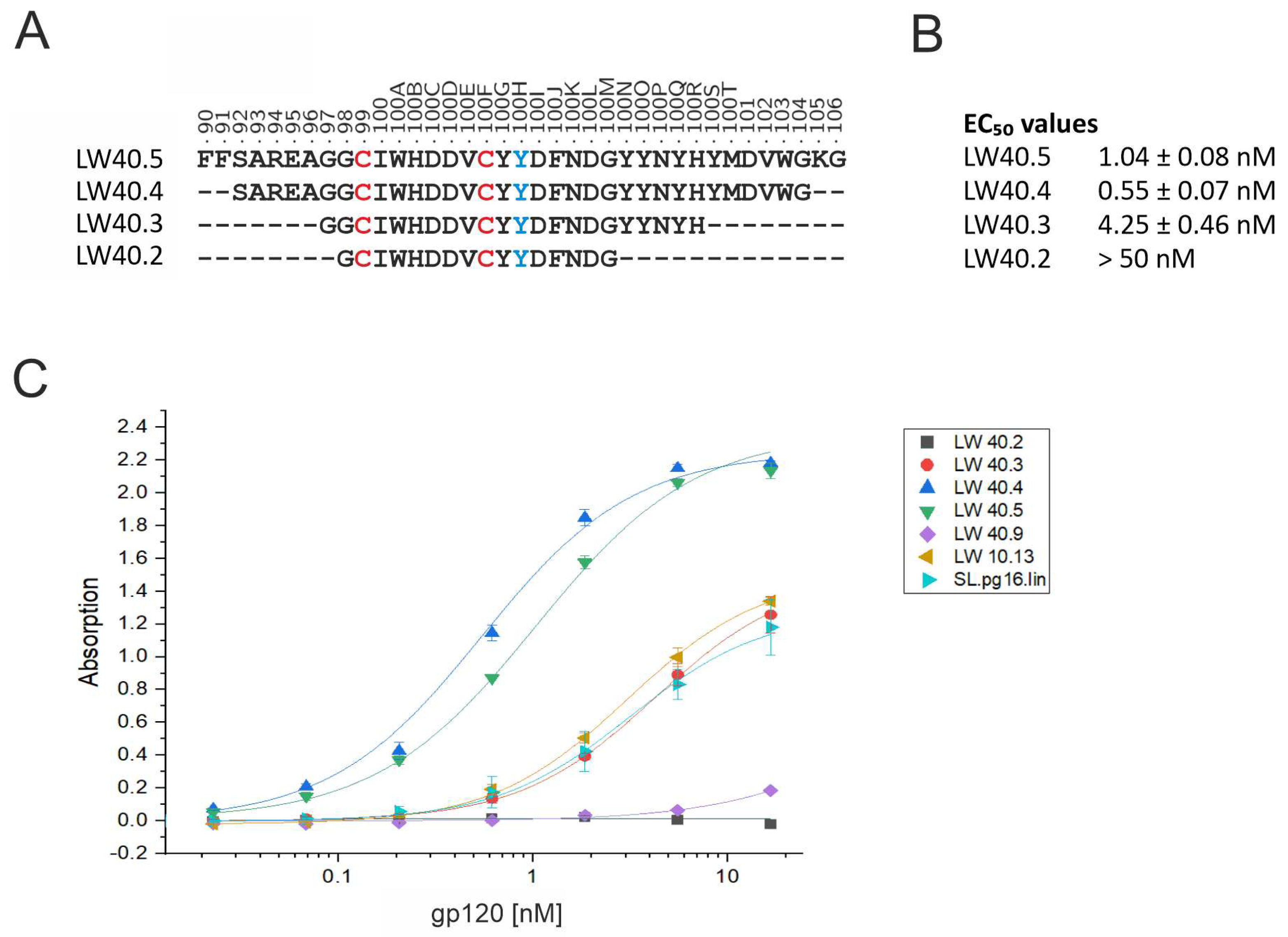
3.2. Design of PG16-derived peptides
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barré-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vézinet-Brun, F.; Rouzioux, C.; others. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.C.; Salahuddin, S.Z.; Popovic, M.; Shearer, G.M.; Kaplan, M.; Haynes, B.F.; Palker, T.J.; Redfield, R.; Oleske, J.; Safai, B.; others. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 1984, 224, 500–503. [Google Scholar] [CrossRef] [PubMed]
- UNAIDS. Homepage — unaids.org. https://www.unaids.org/en, 2023. [Accessed 23-Apr-2023].
- Spencer, D.A.; Shapiro, M.B.; Haigwood, N.L.; Hessell, A.J. Advancing HIV broadly neutralizing antibodies: from discovery to the clinic. Front. Public Health 2021, 9, 690017. [Google Scholar] [CrossRef]
- Walsh, S.R.; Seaman, M.S. Broadly neutralizing antibodies for HIV-1 prevention. Front. Immunol. 2021, 12, 712122. [Google Scholar] [CrossRef] [PubMed]
- Mascola, J.R.; Stiegler, G.; VanCott, T.C.; Katinger, H.; Carpenter, C.B.; Hanson, C.E.; Beary, H.; Hayes, D.; Frankel, S.S.; Birx, D.L.; others. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 2000, 6, 207–210. [Google Scholar] [CrossRef]
- Shibata, R.; Igarashi, T.; Haigwood, N.; Buckler-White, A.; Ogert, R.; Ross, W.; Willey, R.; Cho, M.W.; Martin, M.A. Neutralizing antibody directed against the HIV–1 envelope glycoprotein can completely block HIV–1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 1999, 5, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Hessell, A.J.; Rakasz, E.G.; Poignard, P.; Hangartner, L.; Landucci, G.; Forthal, D.N.; Koff, W.C.; Watkins, D.I.; Burton, D.R. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009, 5, e1000433. [Google Scholar] [CrossRef] [PubMed]
- Scheid, J.F.; Horwitz, J.A.; Bar-On, Y.; Kreider, E.F.; Lu, C.L.; Lorenzi, J.C.; Feldmann, A.; Braunschweig, M.; Nogueira, L.; Oliveira, T.; others. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 2016, 535, 556–560. [Google Scholar] [CrossRef]
- Haußner, C.; Damm, D.; Nirschl, S.; Rohrhofer, A.; Schmidt, B.; Eichler, J. Peptide Paratope Mimics of the Broadly Neutralizing HIV-1 Antibody b12. ChemBioChem 2017, 18, 647–653. [Google Scholar] [CrossRef]
- Haußner, C.; Lach, J.; Eichler, J. Synthetic antibody mimics for the inhibition of protein–ligand interactions. Curr. Opin. Chem. Biol. 2017, 40, 72–77. [Google Scholar] [CrossRef]
- Groß, A.; Hashimoto, C.; Sticht, H.; Eichler, J. Synthetic peptides as protein mimics. Front. Bioeng. Biotechnol. 2016, 3, 211. [Google Scholar] [CrossRef] [PubMed]
- Kozarsky, K.; Penman, M.; Basiripour, L.; Haseltine, W.; Sodroski, J.; Krieger, M. Glycosylation and processing of the human immunodeficiency virus type 1 envelope protein. J. Acquir. Immune Defic. Syndr. 1989, 2, 163–169. [Google Scholar]
- Pejchal, R.; Walker, L.M.; Stanfield, R.L.; Phogat, S.K.; Koff, W.C.; Poignard, P.; Burton, D.R.; Wilson, I.A. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. USA 2010, 107, 11483–11488. [Google Scholar] [CrossRef] [PubMed]
- Pancera, M.; McLellan, J.S.; Wu, X.; Zhu, J.; Changela, A.; Schmidt, S.D.; Yang, Y.; Zhou, T.; Phogat, S.; Mascola, J.R.; others. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 2010, 84, 8098–8110. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Peng, H.; Chen, B.; Harrison, S.C. Cryo-EM structure of full-length HIV-1 Env bound with the Fab of antibody PG16. J. Mol. Biol. 2020, 432, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Kabat, E.A. Sequences of proteins of immunological interest; Number 91 in 1, US Department of Health and Human Services, Public Health Service, National Institute of Health, 1991.
- Stewart, V.; Ronald, P.C. Sulfotyrosine residues: interaction specificity determinants for extracellular protein-protein interactions. J. Bio. Chem. 2022, 102232. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.X.; Doria-Rose, N.A.; Schneck, N.A.; Ivleva, V.B.; Tippett, B.; Shadrick, W.R.; O’Connell, S.; Cooper, J.W.; Schneiderman, Z.; Zhang, B.; others. Tyrosine O-sulfation proteoforms affect HIV-1 monoclonal antibody potency. Sci. Rep. 2022, 12, 8433. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Malins, L.R.; Roche, M.; Sterjovski, J.; Duncan, R.; Garcia, M.L.; Barnes, N.C.; Anderson, D.A.; Stone, M.J.; Gorry, P.R.; others. Site-selective solid-phase synthesis of a CCR5 sulfopeptide library to interrogate HIV binding and entry. ACS Chem. Bio. 2014, 9, 2074–2081. [Google Scholar] [CrossRef]
- Liu, C.C.; Choe, H.; Farzan, M.; Smider, V.V.; Schultz, P.G. Mutagenesis and evolution of sulfated antibodies using an expanded genetic code. Biochemistry 2009, 48, 8891–8898. [Google Scholar] [CrossRef]
- Pancera, M.; Shahzad-ul Hussan, S.; Doria-Rose, N.A.; McLellan, J.S.; Bailer, R.T.; Dai, K.; Loesgen, S.; Louder, M.K.; Staupe, R.P.; Yang, Y.; others. Structural basis for diverse N-glycan recognition by HIV-1–neutralizing V1–V2–directed antibody PG16. Nat. Struct. Mol. Biol. 2013, 20, 804–813. [Google Scholar]
- Rapp, C.; Snow, S.; Laufer, T.; McClendon, C.L. The role of tyrosine sulfation in the dimerization of the CXCR4: SDF-1 complex. Protein Sci. 2013, 22, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Cimbro, R.; Gallant, T.R.; Dolan, M.A.; Guzzo, C.; Zhang, P.; Lin, Y.; Miao, H.; Van Ryk, D.; Arthos, J.; Gorshkova, I.; others. Tyrosine sulfation in the second variable loop (V2) of HIV-1 gp120 stabilizes V2–V3 interaction and modulates neutralization sensitivity. Proc. Natl. Acad. Sci. 2014, 111, 3152–3157. [Google Scholar] [CrossRef] [PubMed]
- Miyanabe, K.; Yamashita, T.; Abe, Y.; Akiba, H.; Takamatsu, Y.; Nakakido, M.; Hamakubo, T.; Ueda, T.; Caaveiro, J.M.; Tsumoto, K. Tyrosine sulfation restricts the conformational ensemble of a flexible peptide, strengthening the binding affinity for an antibody. Biochemistry 2018, 57, 4177–4185. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Bank, R.P.D. RCSB PDB - TYS Ligand Summary Page — rcsb.org. https://www.rcsb.org/ligand/TYS. [Accessed 26-Apr-2023].
- Schaftenaar, G.; Noordik, J.H. Molden: a pre-and post-processing program for molecular and electronic structures. J. Comput.-Aided Mol. Des. 2000, 14, 123–134. [Google Scholar] [CrossRef]
- Khoury, G.A.; Thompson, J.P.; Smadbeck, J.; Kieslich, C.A.; Floudas, C.A. Forcefield_PTM: Ab initio charge and AMBER forcefield parameters for frequently occurring post-translational modifications. J. Chem. Theory Comput. 2013, 9, 5653–5674. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; Li, X.; Caricato, M.; Marenich, A.V.; Bloino, J.; Janesko, B.G.; Gomperts, R.; Mennucci, B.; Hratchian, H.P.; Ortiz, J.V.; Izmaylov, A.F.; Sonnenberg, J.L.; Williams-Young, D.; Ding, F.; Lipparini, F.; Egidi, F.; Goings, J.; Peng, B.; Petrone, A.; Henderson, T.; Ranasinghe, D.; Zakrzewski, V.G.; Gao, J.; Rega, N.; Zheng, G.; Liang, W.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Vreven, T.; Throssell, K.; Montgomery, Jr., J.A.; Peralta, J.E.; Ogliaro, F.; Bearpark, M.J.; Heyd, J.J.; Brothers, E.N.; Kudin, K.N.; Staroverov, V.N.; Keith, T.A.; Kobayashi, R.; Normand, J.; Raghavachari, K.; Rendell, A.P.; Burant, J.C.; Iyengar, S.S.; Tomasi, J.; Cossi, M.; Millam, J.M.; Klene, M.; Adamo, C.; Cammi, R.; Ochterski, J.W.; Martin, R.L.; Morokuma, K.; Farkas, O.; Foresman, J.B.; Fox, D.J. Gaussian˜16 Revision C.01, 2016. Gaussian Inc. Wallingford CT.
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.Y. RED Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed]
- Homeyer, N.; Horn, A.H.; Lanig, H.; Sticht, H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J. Mol. Model. 2006, 12, 281–289. [Google Scholar] [CrossRef]
- D.A. Case.; H.M. Aktulga.; K. Belfon.; I.Y. Ben-Shalom.; J.T. Berryman.; S.R. Brozell.; D.S. Cerutti.; T.E. Cheatham, I.; G.A. Cisneros.; V.W.D. Cruzeiro.; T.A. Darden.; R.E. Duke.; G. Giambasu.; M.K. Gilson.; H. Gohlke.; A.W. Goetz.; R. Harris.; S. Izadi.; S.A. Izmailov.; K. Kasavajhala.; M.C. Kaymak.; E. King.; A. Kovalenko.; T. Kurtzman.; T.S. Lee.; S. LeGrand.; P. Li.; C. Lin.; J. Liu.; T. Luchko.; R. Luo.; M. Machado.; V. Man.; M. Manathunga.; K.M. Merz.; Y. Miao.; O. Mikhailovskii.; G. Monard.; H. Nguyen.; K.A. O’Hearn.; A. Onufriev.; F. Pan.; S. Pantano.; R. Qi.; A. Rahnamoun.; D.R. Roe.; A. Roitberg.; C. Sagui.; S. Schott-Verdugo.; A. Shajan.; J. Shen.; C.L. Simmerling.; N.R. Skrynnikov.; J. Smith.; J. Swails.; R.C. Walker.; J. Wang.; J. Wang.; H. Wei.; R.M. Wolf.; X. Wu.; Y. Xiong.; Y. Xue.; D.M. York.; S. Zhao.; P.A. Kollman. Amber 2022, 2022.
- Gfeller, D.; Michielin, O.; Zoete, V. SwissSidechain: a molecular and structural database of non-natural sidechains. Nucl. Acids Res. 2012, 41, D327–D332. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef]
- Steinbrecher, T.; Latzer, J.; Case, D. Revised AMBER parameters for bioorganic phosphates. J. Chem. Theory Comput. 2012, 8, 4405–4412. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. Chem. Phys. 1998, 79, 926. [Google Scholar] [CrossRef]
- Söldner, C.A.; Horn, A.H.; Sticht, H. Interaction of glycolipids with the macrophage surface receptor Mincle–a systematic molecular dynamics study. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Conrad, M.; Söldner, C.A.; Sticht, H. Effect of Ions and Sequence Variants on the Antagonist Binding Properties of the Histamine H1 Receptor. Int. J. Mol. Sci. 2022, 23, 1420. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Götz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. generalized born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Le Grand, S.; Götz, A.W.; Walker, R.C. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013, 184, 374–380. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham III, T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; others. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Miller III, B.R.; McGee Jr, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA. py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; De Hoon, M.J. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Weißenborn, L.; Richel, E.; Hüseman, H.; Welzer, J.; Beck, S.; Schäfer, S.; Sticht, H.; Überla, K.; Eichler, J. Smaller, Stronger, More Stable: Peptide Variants of a SARS-CoV-2 Neutralizing Miniprotein. Int. J. Mol. Sci. 2022, 23, 6309. [Google Scholar] [CrossRef]
- Torshin, I.Y.; Weber, I.T.; Harrison, R.W. Geometric criteria of hydrogen bonds in proteins and identification ofbifurcated’hydrogen bonds. Protein Eng. Des. Sel. 2002, 15, 359–363. [Google Scholar] [CrossRef]
- Gaschen, B.; Taylor, J.; Yusim, K.; Foley, B.; Gao, F.; Lang, D.; Novitsky, V.; Haynes, B.; Hahn, B.H.; Bhattacharya, T.; others. Diversity considerations in HIV-1 vaccine selection. Science 2002, 296, 2354–2360. [Google Scholar] [CrossRef]
- Zhao, J.; Song, E.; Huang, Y.; Yu, A.; Mechref, Y. Variability in the glycosylation patterns of gp120 proteins from different human immunodeficiency virus type 1 isolates expressed in different host cells. J. Proteome Res. 2021, 20, 4862–4874. [Google Scholar] [CrossRef]
- Jan, M.; Upadhyay, C.; Hioe, C.E. HIV-1 envelope glycan composition as a key determinant of efficient virus transmission via DC-SIGN and resistance to inhibitory lectins. iScience 2019, 21, 413–427. [Google Scholar] [CrossRef]
- Kassler, K.; Meier, J.; Eichler, J.; Sticht, H. Structural basis for species selectivity in the HIV-1 gp120-cd4 interaction: Restoring affinity to gp120 in murine cd4 mimetic peptides. Adv. Bioinformatics 2011, 2011. [Google Scholar] [CrossRef]
- Kondo, H.X.; Kiribayashi, R.; Kuroda, D.; Kohda, J.; Kugimiya, A.; Nakano, Y.; Tsumoto, K.; Takano, Y. Effects of a remote mutation from the contact paratope on the structure of CDR-H3 in the anti-HIV neutralizing antibody PG16. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rajarshi, K.; Khan, R.; Singh, M.K.; Ranjan, T.; Ray, S.; Ray, S. Essential functional molecules associated with SARS-CoV-2 infection: Potential therapeutic targets for COVID-19. Gene 2021, 768, 145313. [Google Scholar] [CrossRef] [PubMed]
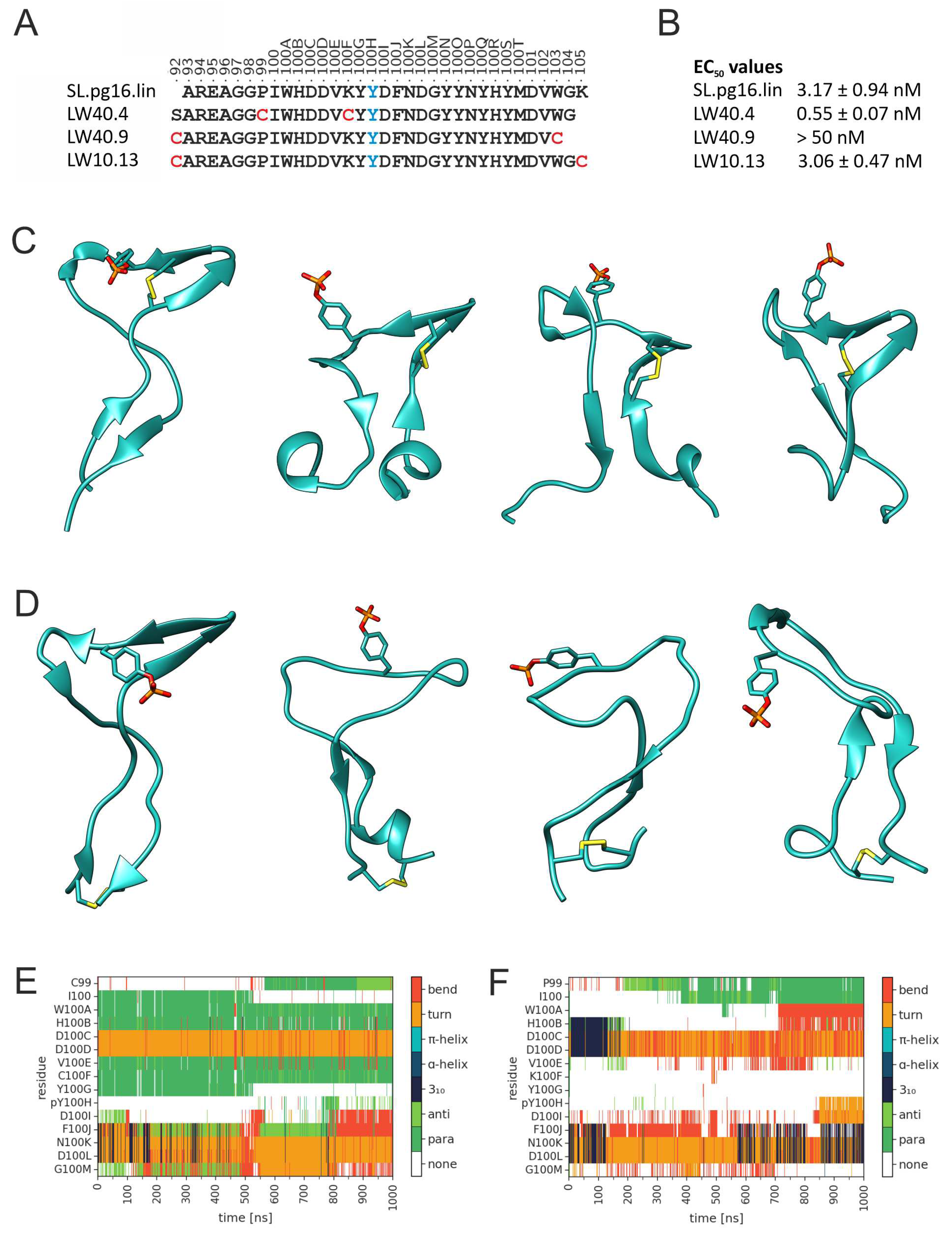
| Peptide | Sequence |
|---|---|
| SL.pg16.lin | Bioa-Aoab-AREAGGPIWHDDVKY(pY)cDFNDGYYNYHYMDVWGK-NH2 |
| LW10.13 | Bio-Aoa-[CAREAGGPIWHDDVKY(pY)DFNDGYYNYHYMDVWGC]-NH2 |
| LW40.02 | Bio-Aoa-G-[CIWHDDVC]-Y(pY)DFNDG-NH2 |
| LW40.03 | Bio-Aoa-GG-[CIWHDDVC]-Y(pY)DFNDGYYNYH-NH2 |
| LW40.04 | Bio-Aoa-SAREAGG-[CIWHDDVC]-Y(pY)DFNDGYYNYHYMDVWG-NH2 |
| LW40.05 | Bio-Aoa-FFSAREAGG-[CIWHDDVC]-Y(pY)DFNDGYYNYHYMDVWGKG-NH2 |
| LW40.09 | Bio-Aoa-[CAREAGGPIWHDDVKY(pY)DFNDGYYNYHYMDVC]-NH2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





