1. Introduction
The production of
Macrobrachium rosenbergii has reached an impressive 290,708 tons globally in 2021 [
1]. China is a prominent producer, having produced a total of 171,263 tons in 2021 as reported by the "China Fisheries Yearbook," with Guangdong, Jiangsu, and Zhejiang provinces contributing over 92% of the national total. However, the diversity and complexity of diseases affecting
M. rosenbergii have resulted in substantial losses for farmers in recent years. These pathogenic microorganisms that invade
M. rosenbergii include
M. rosenbergii nodavirus (MrNV) and extra-small virus (XSV) [
2], infectious precocity virus (IPV) [
3], decapod iridescent virus 1 (DIV1) [
4], and
Citrobacter freundii [
5], etc.
Aeromonas veronii is a gram-negative bacterium that has caused the mortality of various aquatic animals, such as Nile tilapia (
Oreochromis niloticus) [
6], yellow catfish (
Pelteobagrus fulvidraco) [
7], and
Odontobutis potamophila [
8], etc. Our previous research discovered a dominant strain named WSQ-1, which has caused the mass mortality of adult
M. rosenbergii in certain farms in Gaoyou city of Jiangsu Province. This strain was isolated from the hepatopancreas of dying
M. rosenbergii and identified as
A. veronii. The challenge test, amplification of virulence genes, and histopathological examination of the hepatopancreas all demonstrated that the strain was extremely pathogenic [
9].
As an invertebrate,
M. rosenbergii mainly relies on innate immunity to recognize and resist invading pathogens [
10]. Among these tissues, the hepatopancreas is closely related to nutrient metabolism regulation [
11], the hemolymph plays a crucial role in the host immune response, including the recognition and phagocytosis of pathogenic bacteria [
12,
13]. The gills are more prone to damage and physiological responses compared to other organs, as they participate in the absorption of pathogens into the hemolymph to eliminate them [
14,
15].
The current research conducted a comparative transcriptomic analysis on the hepatopancreas of M. rosenbergii infected with A. veronii at different time points (6, 12, and 24 post-injection). Additionally, four immune-related genes were identified from the transcriptomic data, and their expression levels were analyzed in the hepatopancreas, hemolymph, and gills at different post-infection time points. These findings may provide insights into the host response to A. veronii infection and can be used as a guide for A. veronii prevention and treatment.
2. Materials and Methods
2.1. Experimental prawns and A. veronii preparation
The healthy prawns, which had an average body weight of 3.11±0.49 g and were collected from the M. rosenbergii breeding base in Jiaxin, Zhejiang Province, China, were used in the challenge experiment. Before being subjected to experimental manipulation, all prawns were acclimatized for one week in the recirculating water barrel system of Yuya Technology (Huzhou) Co. LTD (Zhejiang, China). While the water temperature was maintained at 28±2 ℃ and the dissolved oxygen concentration was above 5 mg/L, the prawns were fed twice daily but were not fed during the injection trial. Then 180 prawns were split into two groups (n=90) (the challenge groups and the control groups) at random, and each group was further separated into three parallel groups so that there were 30 prawns in each 60-L plastic container. Random inspections of the prawns were conducted before the injection trial to ensure that they were free of any bacterial infections.
In accordance with previous test’ median lethal dosage (LD50) of 72 hours, the strain WSQ-1 (A. veronii) was first inoculated the tryptic soybean peptone liquid medium (TSB), then placed in a 28 °C shaker with shaking at 180 rpm for 18 h. Lastly, the turbidity was adjusted to 2.50×106 CFU/mL.
2.2. Sample collection
In the experiment, the challenge groups received an intramuscular injection of 50 μL of A. veronii, whereas the control groups were injected with sterilized PBS in the same amount. After 6, 12, and 24 hpi, prawns from the two groups were randomly selected, and the hepatopancreas and gills were temporarily stored in liquid nitrogen and kept at−80 °C in the cryogenic refrigerator. Every three of the same tissues were mixed into one sample at different time points, with three biological replicates. Significantly, the treatment of the hemocytes was particularly important. Briefly, the fresh hemocytes and TRIzol were first added to the cryotube in a 1:3 ratio to lyse the blood cells, and the mixture was then shocked for 1-2 minutes until the floccule was completely cracked. Finally, the mixture was incubated at room temperature for 5 minutes to completely decompose the ribosome and then stored at −80 °C.
2.3. Total RNA extraction and Illumina sequencing
The TRIzol reagent (Invitrogen, USA) was used to extract the total RNA from 18 hepatopancreas samples at 6, 12, and 24 hpi in accordance with the manufacturer’s instructions. The challenge groups were named as AV6_1, AV6_2, ADV_3, AV12_1, AV12_2, AV12_3, AV24_1, AV24_2, AV24_3, whereas the control groups were marked as C6_1, C6_2, C6_3, C12_1, C12_2, C12_3, C24_1, C24_2, C24_3, separately. These libraries were obtained after the whole RNA were first screened and purified using the AMPure XP technology (Beckman Coulter, Beverly, USA). Following these libraries’ initial Qubit 2.0 quantification, they were diluted to 1.5 ng/uL and identified using an Agilent 2100 bioanalyzer. These libraries’ quality was further ensured by using qRT-PCR to precisely measure their effective concentration. The libraries construction and sequencing were completed using the Illumina NovaSeq 6000.
2.4. De Novo assembly, annotation, and classification
To ensure the accuracy and dependability of data analysis, some raw data, such as reads with adapters, low-quality, and N bases, were filtered away to get clean reads. Next, GC content and Q30 were computed with clean data by fastp (version 0.19.7). Trinity was used to assemble the transcripts [
16], and BUSCO software was used to evaluate the correctness and completeness of the transcripts [
17]. Finally, all of the transcripts were classified and annotated using the following 7 databases: Nr, Nt, Pfam, COG, Swiss-Prot, KO, and GO.
2.5. DEGs, KEGG, and GO enrichment analysis
The DESeq2 package (version 1.20.1) [
18] was used in this research to screen DEGs for significantly differential gene expression. In addition, we set three comparisons between challenge and control groups after 6, 12, and 24 hpi (i.e., AV6 vs. C6, AV12 vs. C12, and AV24 vs. C24); next, the immune-related DEGs were collected and analyzed for GO and KEGG enrichment using GOseq and KOBAS software packages, separately [
19].
2.6. Validation of immune-related DEGs by RT-qPCR
10 immune-related DEGs were chosen at random for quantitative real-time PCR (qRT-PCR) analysis in order to verify the credibility of the transcriptome sequencing data. Primer 5.0 software was used to design the primers (
Table 1), which were then synthesized by the company. The cDNA used for qRT-PCR was synthesized by reverse transcription using cDNA Synthesis Supermix (TransGen Biotech, Beijing, China), while the 18S rRNA was used as the reference gene for the qRT-PCR. In addition, amplifications were performed in a 10 µL reaction system with 0.8 µL of cDNA, 3.4 µL of ddH2O, 5 µL of TB Green Premix Ex Taq II (2×), and 0.4 µL each PCR forward primer and reverse primer. The PCR procedure was pre-denaturation at 95 °C for 3 min, followed by 39 cycles of denaturation at 95 °C for 5 s and annealing at 59°C for 30 s. When the temperature rose from 65℃ to 95℃, the melting curve increased by 0.5℃ every 5 s. Each qRT-PCR assay was carried out three biological replicates and three technique replicates. Ultimately, the 2
−ΔΔCT method was used to evaluate relative mRNA expression.
2.7. Expression patterns of four key immune-related DEGs in different tissues
To further analyze the relative mRNA expression in different tissues and time points, four immune-related DEGs, including p21-activated kinase 1(PAK1), inhibitor of nuclear factor kappa-B kinase subunit alpha (IKKα), glycogen synthase kinase 3 beta (GSK3β), and protein Toll (TOLL), were selected and anticipated to be engaged in the immunological response of the tissues. The collected samples at 6, 12, and 24 hpi were used to extract RNA and synthesize cDNA based on the previous methods, and all qRT-PCR reactions were done in triplicate (three biological replicates and three technique replicates). Lastly, the 2-ΔΔCT approach was used to calculate the relative mRNA expression.
2.8. Statistical analysis
SPSS 25.0 software (IBM Corp., Armonk, NY, USA) was used to detect statistical significance for differences in gene expression levels between the challenge and control groups at three time points after infection in three tissues. Then create a chart with GraphPad Prism 9.0 software (GraphPad Software Inc., San Diego, CA, USA).
3. Results
3.1. Transcript assembly, gene functional annotation, GO and KEGG classification
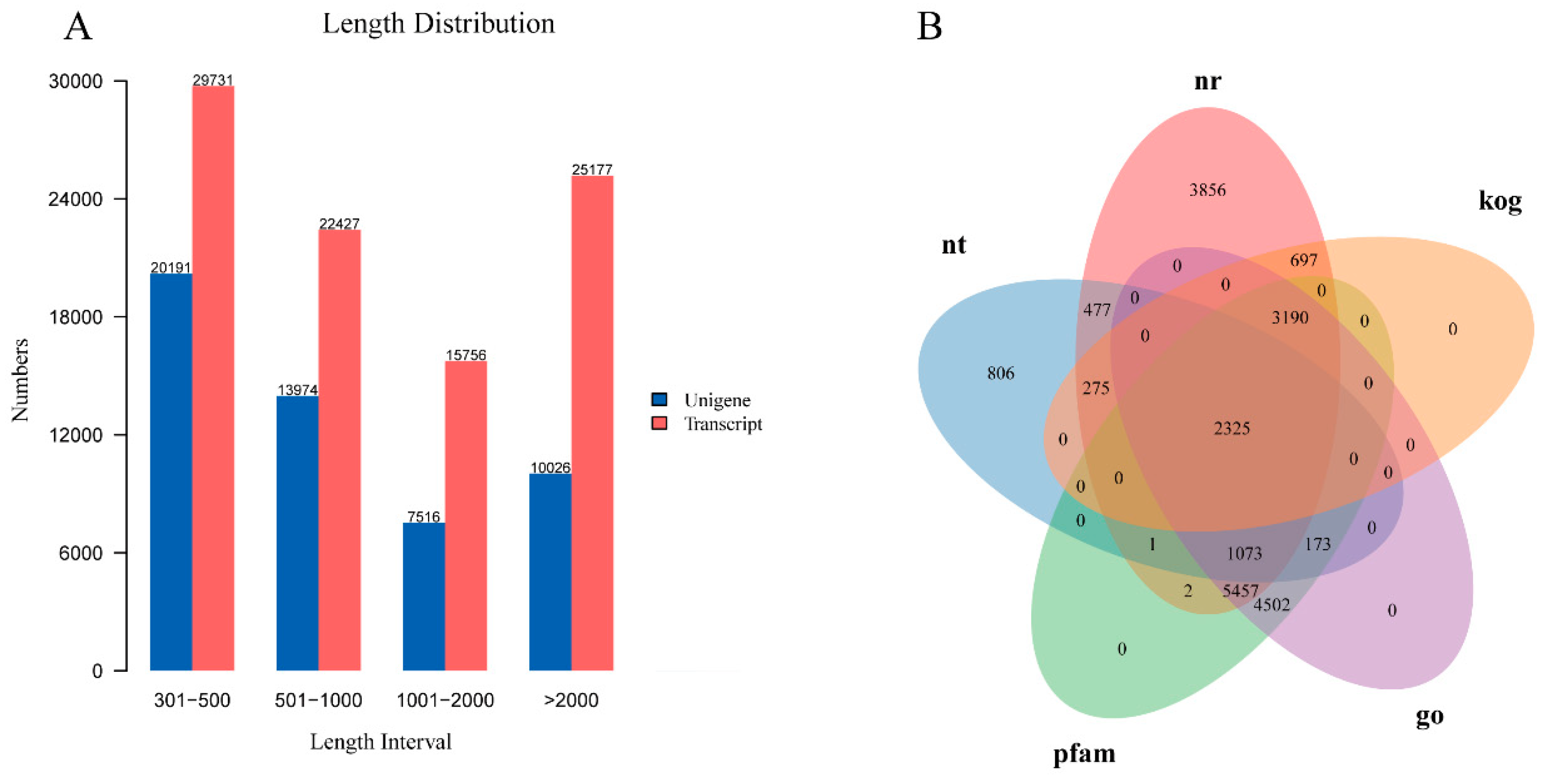
There were totally 22248733-24166064 raw reads in 18 libraries, and 21,489,762-23,722,890 clean reads were obtained after quality control (
Table S1). The Q30 percentages’ average values were 93.35%. Using the Trinity software, 93,091 transcripts were produced, with an average length of 1,716 bp. Finally, 51,707 unigenes were obtained, with sizes ranging from 301 bp to 28,851 bp and the N50 length of 2,808 bp (
Figure 1A).
Seven public databases were used to annotate the functions of all unigenes: 17,353 (33.56%) in NR, 5,130 (9.92%) in NT, 7,354 (14.22%) in KEGG, 12,481 (24.13%) in SwissProt, 16,723 (32.34%) in Pfam, 16,720 (32.33%) in GO, and 6,487 (12.54%) in KOG, respectively (
Table S2). In addition, 2,325 genes were coexisting functional genes annotated in the 5 databases, as demonstrated in the Venn diagram of Nr, Nt, Pfam, GO, and KOG annotations in
Figure 1B.
KEGG is used to comprehend the sophisticated functions of biological systems. In this study, 861 unigenes were associated with “signal transduction”, 618 unigenes with “transport and catabolism”, and 366 unigenes with the “immune system” (
Figure S1.A). Meanwhile, GO analysis is also important. 16,720 unigenes were classified as molecular function, cellular component, and biological process, with 12, 5, and 25 subcategories, respectively. Among the biological processes, 45 unigenes were associated with “antioxidant activity” (GO:0016209), 173 unigenes with “immune system process” (GO:0002376), and 2,526 unigenes were associated with “response to stimulus” (GO:0050896), respectively (
Figure S1.B).
3.2. Identification of DEGs related to A. veronii infection
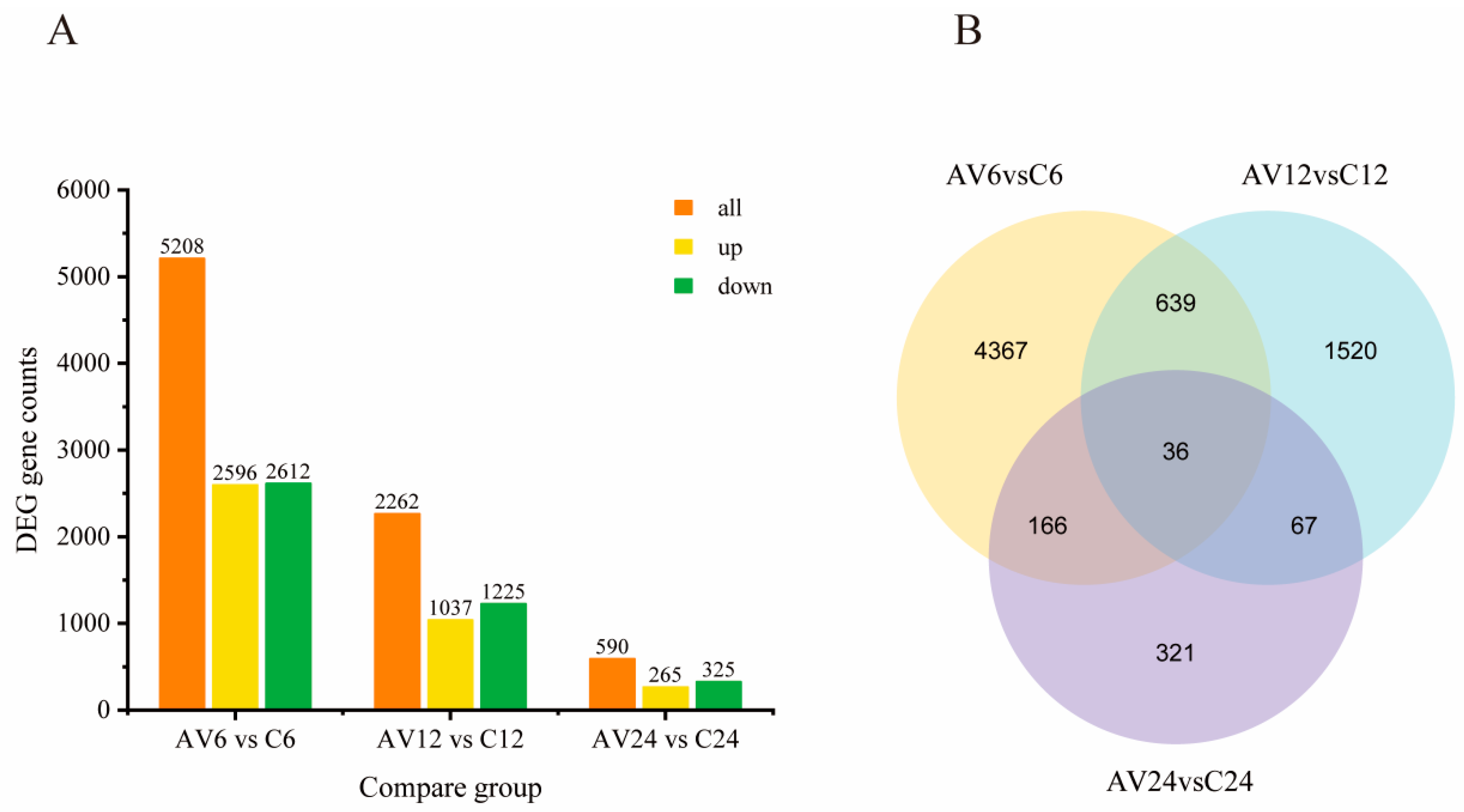
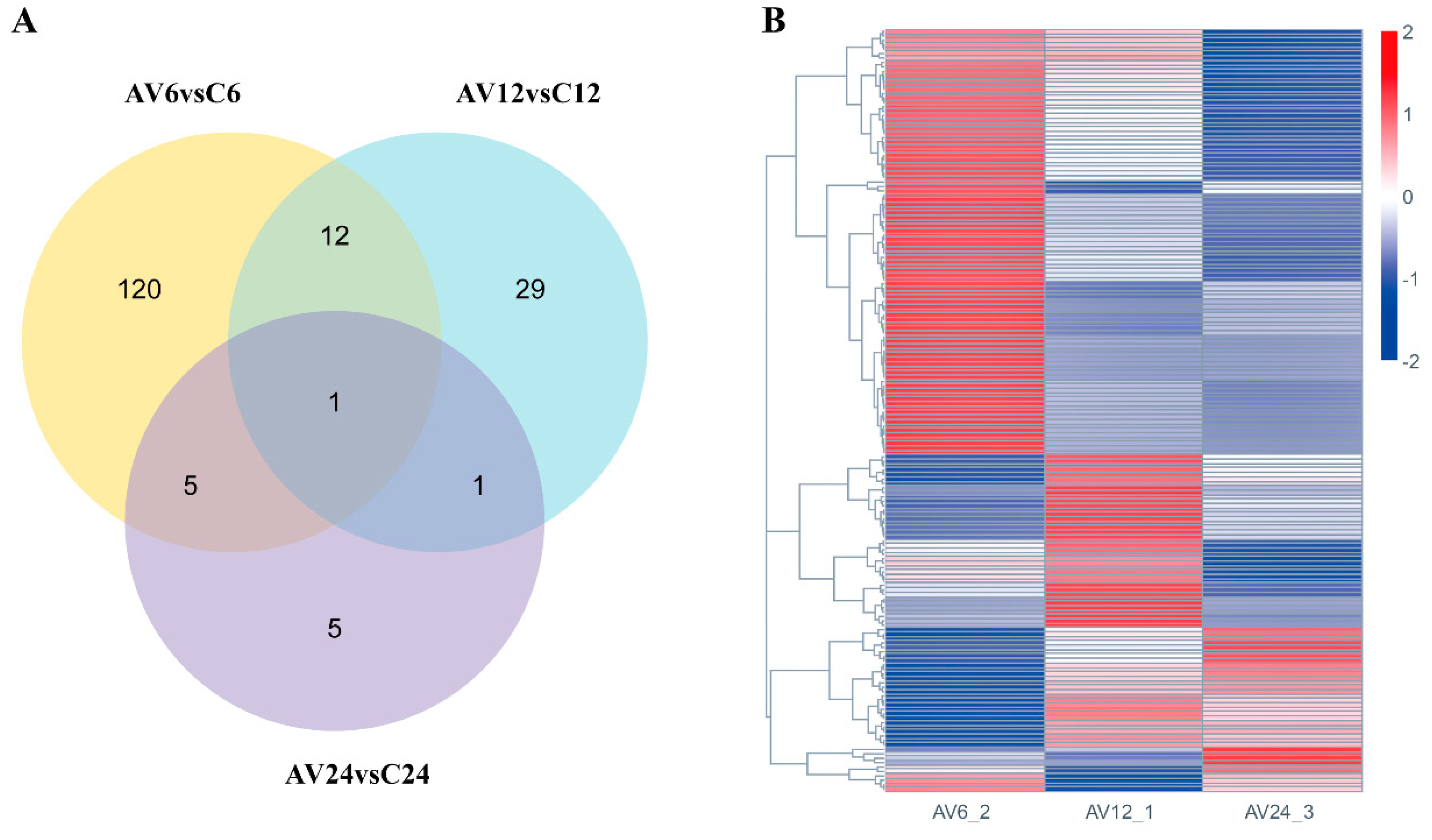
In the current study, 8,060 DEGs (
Table S3) were detected in three transcriptome comparisons between challenge and control groups (AV6 vs. C6, AV12 vs. C12, and AV24 vs. C24), among which 5,208 DEGs (containing 2,596 up-regulated and 2,612 down-regulated genes) were involved at the early stage of the challenge (AV6 vs. C6). The DEGs reduced dramatically with the extension of the challenge time, with 2,262 genes (1,225 down-regulated and 1,037 up-regulated) and 590 genes (325 down-regulated and 265 up-regulated) differentially expressed, respectively, at 12 and 24 hpi (
Figure 2A). The Venn diagram showed that 36 DEGs coexisted in three comparable groups (
Figure 2B).
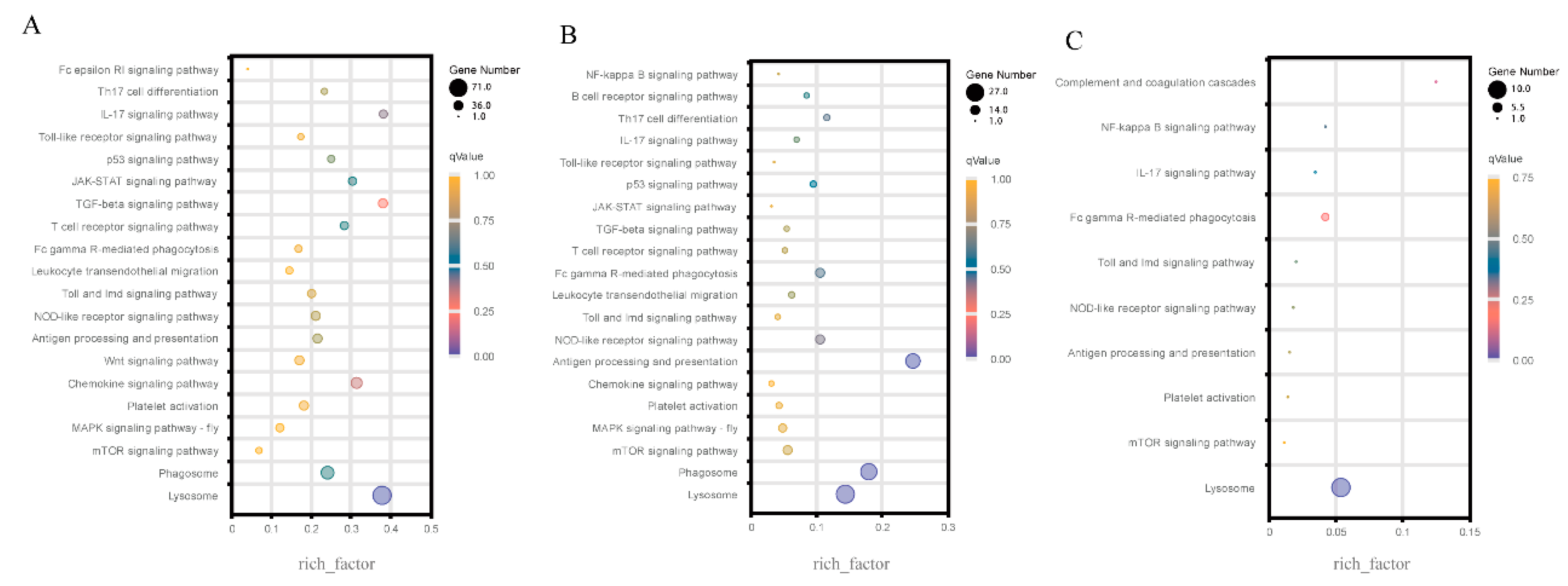
3.3. KEGG enrichment of the immune-related DEGs
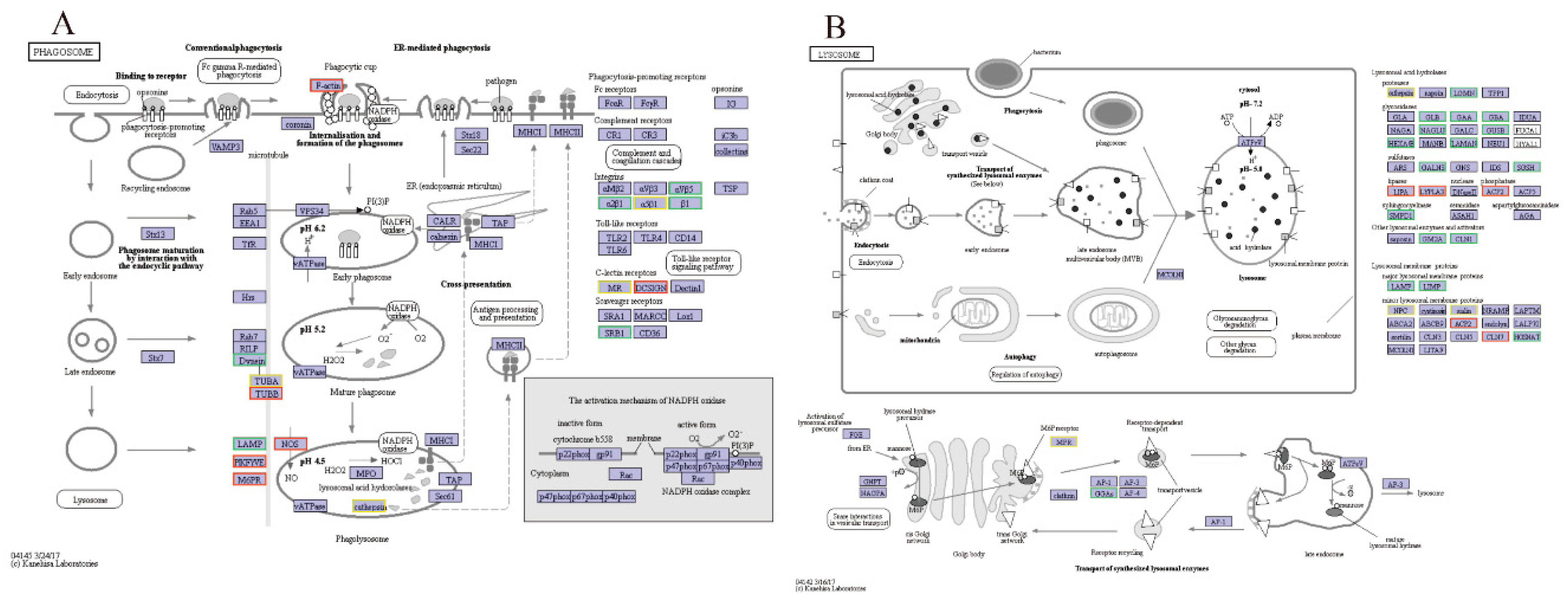
At 6, 12, and 24 hpi, KEGG enrichment of immune-related DEGs in different immune-related pathways was analyzed between the challenge group and the control group. For the three transcriptome comparisons, 28 important immune-related pathways were chosen (
Table S4). As shown in
Figure 3, the lysosome pathway was substantially enriched by all three comparisons, with the highest number of DEG in the top-20 immune-related pathways. Except for
Figure 3C, both comparison groups of AV6 vs. C6 (
Figure 3A) and AV12 vs. C12 (
Figure 3B) greatly enriched the phagosome pathway. A total of 6 up-regulated genes were detected at 6 hpi, including actin gamma 1
(F-actin), C-type lectin domain family 4 member L/M (
DCSIGN), tubulin beta (
TUBB), 1-phosphatidylinositol-3-phosphate 5-kinase (PIKFYVE), cation-dependent mannose-6-phosphate receptor (
M6PR) and nitric-oxide synthase (
NOS1) in phagosome pathway (
Figure 4A), and 4 up-regulated genes at 6 hpi were detected in lysosome pathway, including lysosomal acid lipase (
LIPA), lysophospholipase III (LYPLA3), lysosomal acid phosphatase (
ACP2) and ceroid-lipofuscinosis neuronal protein 7 (
CLN7) (
Figure 4B). Other immune-related pathways, such as the MAPK signaling pathway-fly, the chemokine signaling pathway, antigen processing and presentation, etc., were also abundant in the DEGs.
What is more, Venn diagram analysis showed that 173 immune-related DEGs were detected based on the aforementioned 28 immune-related pathways, and there were 138, 43, and 12 immune-related DEGs, respectively, at 6, 12, and 24 hpi compared to the control groups (
Figure 5A). Furthermore, in the hierarchical cluster analysis, 173 immune-related DEGs in AV6 showed significant differences compared to AV12 and AV24, indicating that the majority of the 173 immune-related DEGs were relatively highly expressed at 6 hpi, then the number of DEGs was drastically reduced at 12 and 24 hpi; some genes’ expression increased at 12 hpi while being relatively low expressed at 6 and 24 hpi; a few genes were highly expressed at 24 hpi compared with 6 and 12 hpi (
Figure 5B).
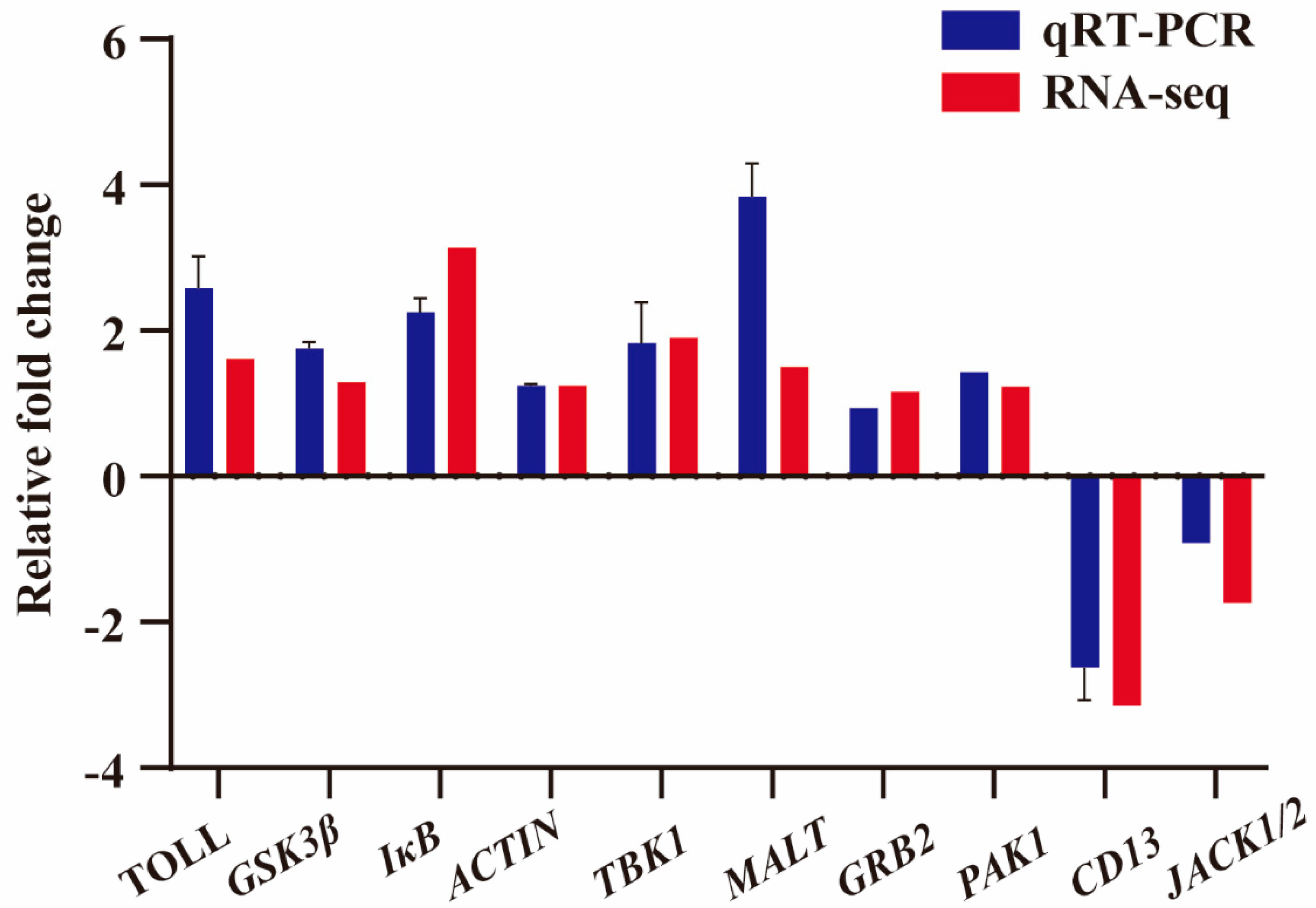
3.4. Validation of DEGs by qRT-PCR
To further confirm the gene expression from RNA-seq data, 10 immune-related DEGs, including
TOLL,
GSK3β,
IκB,
ACTIN,
TBK1,
MALT,
GRB2,
PAK1,
CD13, and
JACK1/2, were selected for qRT-PCR verification at random. The results showed that qRT-PCR had an identical expression tendency to the RNA-seq data, which confirmed the expression of immune-related DEGs in the RNA-seq data (
Figure 6).
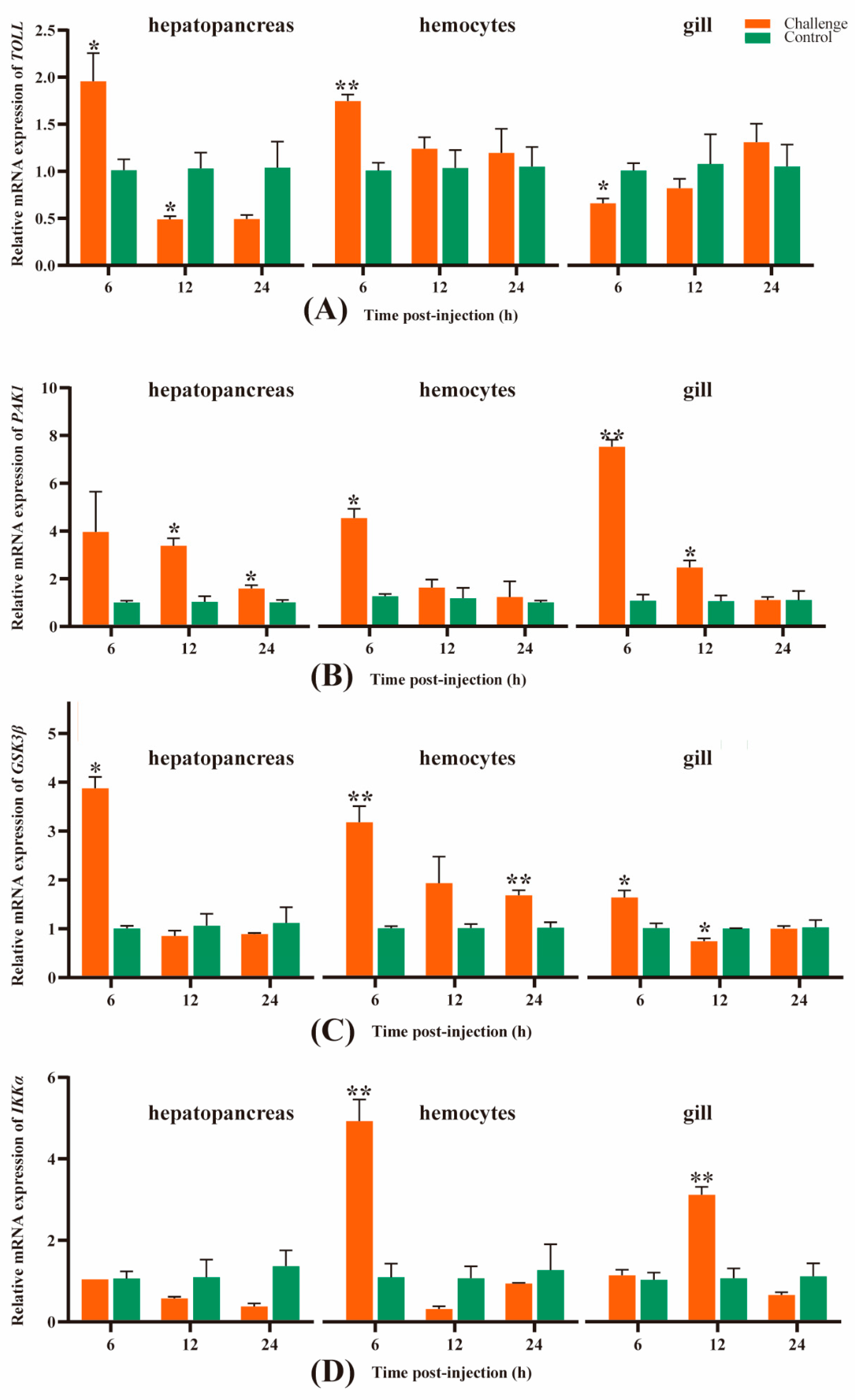
3.5. Temporal and spatial expression levels of four key immune-related genes in M. rosenbergii
To further explore the gene expression in different tissues (hepatopancreas, hemocytes, and gill), four immune-related DEGs, including TOLL, PAK1, GSK3β, and IKKα, were screened from the KEGG pathways, in which these genes overlapped each other.
As seen in
Figure 7A, the
TOLL gene’s expression level in the hepatopancreas of the challenge groups at 6 hpi was significantly higher than that in the control group (P < 0.05), but then sharply decreased at 12 hpi, significantly lower than that in the control group (P < 0.05), and almost kept successively low levels at 24 hpi, indicating that this gene was remarkable up-regulated at 6 hpi, whereas it became down-regulated at 12 and 24 hpi. Similar to the hepatopancreas, the expression trend of
TOLL in the hemocytes of the challenge group was significantly higher than that in the control group (P < 0.01) at 6 hpi, then slightly dropped at 12 hpi and remained similar at 24 hpi, still with a higher expression level than the control group, though the difference was not significant. Therefore, the expression of
TOLL was up-regulated in the hemocytes after
A. veronii infection. Unlike the hepatopancreas and hemocytes, in the gill, the expression of
TOLL was down-regulated at 6 and 12 hpi, significantly lower than the control group at 6 hpi (P < 0.05), then slightly increased at 12 hpi, and became up-regulated at 24 hpi.
In the hepatopancreas, the
PAK1 gene’ expression in the challenge group at 6 hpi was higher than that in the control group, then slightly decreased at 12 hpi and dropped dramatically at 24 hpi, but both of them were significantly higher than the control group (P < 0.05), indicating that the
PAK1 gene was remarkably up-regulated expressed at the three time points, despite the continuing decline at 12 and 24 hpi. A similar trend of
PAK1 gene expression also appeared in the hemocytes and gill, except that the expression level of the
PAK1 gene was slightly lower than that in the control group at 24 hpi in the gill and became down-regulated (
Figure 7B).
In the hepatopancreas and gill, the expression trend of the
GSK3β gene was similar to the
TOLL gene expression in the hepatopancreas, which was significantly higher than that in the control group (P < 0.05), but precipitous decline at 12 and 24 hpi; both of the expression levels at the two points were slightly lower than those in the control group, indicating that the
GSK3β gene was up-regulated at 6 hpi, whereas it became down-regulated at 12 and 24 hpi. What is more, the expression trend of the
GSK3β gene in the hemocytes was similar to that of the
PAK1 gene expression in the hepatopancreas and hemocytes, which was higher than that in the control group at 6 hpi (P < 0.01) but sharply decreased at 12 hpi, in spite of the fact that the difference was not significant compared to the control group. Following that, it maintained a slight decline at 24 hpi, but was significantly higher than that in the control group (P < 0.01), revealed that this gene was up-regulated expressed at all three points (
Figure 7C).
The expression level of the
IKKα gene in the hepatopancreas of the challenge groups at 6 hpi decreased slightly and maintained a sustained decline at 12 and 24 hpi, and all of them were lower than the control group, which revealed this gene remained down-regulated throughout the challenge. Unlike the hepatopancreas, the expression level in the hemocytes was remarkably higher than the control group at 6 hpi (P < 0.01), but decreased sharply at 12 hpi and was lower than the control group. After that, it increased slightly at 24 hpi but was still lower compared to the control. These results revealed the gene
IKKα was up-regulated at 6 hpi and became down-regulated at 12 and 24 hpi. In the gill, the
IKKα gene in the challenge groups was slightly higher at 6 hpi than the control group and was remarkably higher compared to the control group at 12 hpi (P < 0.01), but then sharply declined at 24 hpi and was lower than the control group, indicating the
IKKα gene was up-regulated at 6 and 12 hpi but became down-regulated at 24 hpi (
Figure 7D).
Figure 7.
Gene expression analysis of the TOLL (A), PAK1 (B), GSK3β (C) and IKKα (D) in M. rosenbergii hepatopancreas, hemocytes and gill at 6, 12, 24 hpi, respectively. Data shown are the mean of triplicates ± SD. The asterisk represents significant differences between the challenge and control groups at different time (P < 0.05). The orange column and the green column represent challenge group and control group, respectively.
Figure 7.
Gene expression analysis of the TOLL (A), PAK1 (B), GSK3β (C) and IKKα (D) in M. rosenbergii hepatopancreas, hemocytes and gill at 6, 12, 24 hpi, respectively. Data shown are the mean of triplicates ± SD. The asterisk represents significant differences between the challenge and control groups at different time (P < 0.05). The orange column and the green column represent challenge group and control group, respectively.
4. Discussion
4.1. Phagosome and lysosome pathway analysis
Phagocytosis plays an important role in the innate immunity of
M. rosenbergii [
13,
20]. The phagocytosis process was the particle binding to the cell surface, forming an endocytic, becoming a phagolysosome, and finally digesting within the phagolysosome [
21].
As shown in
Figure 4A, phagocytosis is driven by the recombination of
F-actin, which stimulates the diffusion of pseudopod around the particle and produces phagosomes that are closely apposed to the particle [
22].
F-actin was up-regulated in
M. rosenbergii-infected group at 6 hpi, which might promote the production of phagosomes. C-type lectin receptors (CLRs) plays an important role in autoimmunity, allergy, homeostasis, and anti-microbial host defense [
23], and one of the functions is to eliminate pathogens by regulating phagosome maturation in macrophages [
24]. Up-regulation of the
DCSIGN gene in the challenge group at 6 hpi might indicate promoting the phagosome maturation. In addition, V-type proton ATPase can clear the pathogens in phagosome by increasing hydrogen ion (H+) concentration [
25]. In the present study, the
ATPase gene was down-regulation at 12 hpi compared to the control group, which might suggest a decreased phagocytic capacity of the phagosome in infected individuals compared to healthy
M. rosenbergii, and this result is similar to Zhang et al.’s (2019) study which found a significant down-regulation in diseased sea urchins (Strongylocentrotus intermedius) [
20].
Crustaceans’ innate immunity depends heavily on lysosomes, which mostly degrade hazardous compounds by phagocytosis and endocytosis. Additionally, several lysosomal hydrolytic enzymes, including lipase, proteases, etc., show their greatest enzyme activity at low pH levels [
26].
As shown in
Figure 4B, LIPA is a very important hydrolase, and reducing its activity can lead to symptoms such as hypercholesterolemia and hepatomegaly [
27].
LYPLA3 is a transacylase that may play a specific role in lysosomes [
28]. It is reported that
LYPLA3’s loss increased atherosclerosis in apolipoprotein E-deficient mice [
29].
ACP2 is the most important lysosomal enzyme in crustacean defense, transferring phosphate groups and catalyzing the hydrolysis of phosphorylated proteins [
30]. In addition, loss of
CLN7 will impaire mTOR reactivation and loss of soluble lysosomal proteins [
31]. These genes (
LIPA,
LYPLA3,
ACP2,
CLN7) were up-regulated in the
M. rosenbergii infection group at 6 hpi, suggesting they may improve lysosome activity to defend against
A. veronii invasion.
4.2. Important immune-related genes involved in the immune response
Toll receptor is typically composed of transmembrane regions, and intracellular interleukin hormone receptor domains, and extracellular leucine repeat [
32]. It is the key vector in the immune signal transduction [
33]. In the present study, the expression of
Toll continuously increased in gills for 24 hours and remained highly expressed in hemocytes and hepatopancreas at 6 hpi, indicating that the
Toll gene is involved in the immune response against
A. veronii infection. In the process of attacking
M. rosenbergii with viruses (WSSV), Feng et al. found (2016) that
Toll gene was uniformly up-regulated within 24-48 hours, thus it was speculated that toll gene played a positive role in the fight against viruses. [
34]. The result is similar to the present study that the
Toll expression remained consistently up-regulated in three tissues after injection of
A. veronii, although it varied across tissues and at different times.
PAK1 is the first protein kinase to be discovered and is extensively expressed in eukaryotic tissues while being crucial for biological processes [
35]. In this study,
PAK1 expression significantly enhanced at 6 hpi in three tissues, then decreased at 12 and 24 hpi successively. The expression level at all time points was higher than the control group. It indicated
PAK1 was closely related to immune response in
M. rosenbergii against
A. veronii. This result was similar to the study of Ren et al. (2019), who detected
PAK1 in all tissues of infected sea cucumbers (
Apostichopus japonicus) and the highest expression was found in coelom. After the silencing of
PAK1 gene, lysozyme in coelom was significantly reduced, thus it was speculated that
PAK1 was involved in immune response against bacteria [
36].
GSK-3β plays an important role in innate immunity, phosphorylating and inhibiting glycogen synthase [
37]. Ruan et al. (2018) found that down-regulation of
GSK3β can inhibit WSSV infection, suggesting that it may promote WSSV clearance in
Litopenaeus vannamei by mediating cell apoptosis [
38]. Zhang et al. (2020) also found that
GSK3β negatively affects
L. vannamei when it is infected with WSSV by mediating feedback regulation of the NF-kB pathway. Contrary to the above study,
GSK3β expression in three tissues of
M. rosenbergii remarkably enhanced at 6 hpi in the present study, possibly because these infected individuals were greatly damaged and died in large numbers. The expression of
GSK3β returned to normal at 12 and 24 hpi might be generated by an effective immune response in the surviving shrimp.
IKK (IκB kinase) consists of IKKα, IKKβ, and IKKγ, IKKα and IKKβ are involved in the phosphorylation of IκB and the activation of NF-κB, while IKKγ is in charge of the IKKβ and IKKα’s recruitment [
39]. NF-κB is a transcription factor that is essential for cell cycle, immunity and inflammation, etc. [
40,
41]. NF-κB proteins are usually inactivated by attaching to the IκB (the κB inhibitor) [
42]. IKK will be activated when cells are stimulated from outside such as virus [
43], and the NF-κB protein will be released, leading to inflammatory and immune responses. In this study, the expression of
IKKα was significantly upregulated in the hemocytes at 6 hpi and in gills at 12 hpi in the challenged group, suggesting a severe immune response in the prawns at these time points. However, the expression of
IKKα was downregulated at 24 hpi, indicating a relief in the symptoms of the surviving prawns. The expression of
IKKα in the hepatopancreas kept decreasing, suggesting that the hepatopancreas might be an essential immune organ that responds positively to the stimulus from
A. veronii.
5. Conclusions
In the present study, a total of 51,707 high-quality unigenes and 8060 significant DEGs were obtained from the hepatopancreas at 6, 12, and 24 hpi. 173 key immune-related DEGs were enriched into 28 immune-related pathways by KEGG pathway analysis. The expression of immune-related DEGs in the Lysosome and Phagosome pathways provided insights into the transcriptional regulation caused by bacterial infection in M. rosenbergii. Moreover, the diverse expression patterns of the screened immune-related genes (TOLL, PAK1, GSK3β, and IKKα) in different tissues (hepatopancreas, hemocytes and gill) also revealed the immunity response mechanism between M. rosenbergii and A. veronii. Taken together, our findings provide new insights for further research on disease-resistance breeding of M. rosenbergii.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org., Figure S1: (A) KEGG annotation of unigenes. (B) GO annotation of the unigenes; Table S1: Purified transcriptome data from 18 samples; Table S2: Unigenes annotation information in database; Table S3: The details of DEGs in three transcriptome comparisons (AV6 vs. C6, AV12 vs. C12, and AV24 vs. C24); Table S4: The information on 28 immune-related KEGG pathways enriched with the DEGs.
Author Contributions
Conceptualization, Q.T.and X.P.; methodology, S.Y.; software, X.Y.; validation, Z.Z. and H.T; formal analysis, X.L.; investigation, Z.X.; resources, G.Y.; project administration, Q.T.; funding acquisition, G.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Earmarked Fund for the China Agriculture Research System (CARS-48), key scientific and technological grant of Zhejiang for breeding new agricultural (Aquaculture) varieties (2021C02069-4-3), and the major research & development program (modern agriculture) of Jiangsu province (BE2019352).
Institutional Review Board Statement
This study was conducted according to the Guide for Laboratory Animals developed by the Ministry of Science and Technology (Beijing, China). The experimental protocol for animal care and tissue collection was approved by Huzhou University (the ethical approval code: 20190625).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All supporting data are included within the main article and all supporting data are included within the main article and its supplementary files.
Acknowledgments
Thanks for Qi Shen, Haoyu Lou and Tianci Kong’s help in the experiment.
Conflicts of Interest
All authors declare no conflict of interest.
References
- Fao, F.; Bondad-Reantaso, M.G.; Arthur, J.R. FAO Fisheries and Aquaculture Report NFIA/R1333 (En); FAO: Rome, Italy, 2021.
- Chen, K.F.; Maran, S.; Tan, W.S.; Ong, L.K.; Abidin, S.A.Z.; Othman, I.; Tey, B.T.; Lee, R.F.S. Meta-analysis of studies on pro tection provided by different prophylactic agents, their routes of administration and incubation times against nodavirus infection in Macrobrachium rosenbergii. Aquaculture. 2023, 565. [CrossRef]
- Zhao, C.; Miu, Q.; Liu, S.; Zhou, D.; He, X.; Pang, J.; Weng, S.; He, J. Detection methods, epidemiological investigation, and host ranges of infectious precocity virus (IPV). Aquaculture. 2023, 562. [CrossRef]
- Qian, Q.; Zhou, Y.; Chen, Z.; Zhu, Y.; Xu, J.; Gao, X.; Jiang, Q.; Wang, J.; Zhang, X. Pathogenesis and complete genome sequence of Decapod iridescent virus 1 (DIV1) associated with mass mortality in Macrobrachium rosenbergii. Aquaculture. 2023, 566. [CrossRef]
- Zhao, C.; Wen, H.; Huang, S.; Weng, S.; He, J. A Novel Disease (Water Bubble Disease) of the Giant Freshwater Prawn Macrobrachium rosenbergii Caused by Citrobacter freundii: Antibiotic Treatment and Effects on the Antioxidant Enzyme Activity and Immune Responses. Antioxidants. 2022, 11. [CrossRef]
- Dong, H.T.; Techatanakitarnan, C.; Jindakittikul, P.; Thaiprayoon, A.; Taengphu, S.; Charoensapsri, W.; Khunrae, P.; Rattanarojpong, T.; Senapin, S. Aeromonas jandaei and Aeromonas veronii caused disease and mortality in Nile tilapia, Oreochromis niloticus (L.). J. Fish. Dis. 2017, 40, 1395-1403. [CrossRef]
- Zhai, W.; Wang, Q.; Zhu, X.; Jia, X.; Chen, L. Pathogenic infection and microbial composition of yellow catfish (Pelteobagrus fulvidraco) challenged by Aeromonas veronii and Proteus mirabilis. Aquaculture and Fisheries 2023, 8, 166-173. [CrossRef]
- Liu, G.; Li, J.; Jiang, Z.; Zhu, X.; Gao, X.; Jiang, Q.; Wang, J.; Wei, W.; Zhang, X. Pathogenicity of Aeromonas veronii causing mass mortalities of Odontobutis potamophila and its induced host immune response. Fish. Shellfish. Immunol. 2022, 125, 180-189. [CrossRef]
- Peng, X.; Tu, H.H.; Luo, J.P.; Zhong, Z.X.; Lan, X.; Tang, Q.Y.; Yi, S.K.; Xia, Z.L.; Cai, M.Y.; Yang, G.L. Isolation, identification and virulence gene analysis of pathogenic Aeromonas veronii in Microbrachium rosenberdii and its histopathological observation. Acta Hydrobiologica Sinica. 2023, 47 (6), 1-12. [CrossRef]
- Kumaresan, V.; Palanisamy, R.; Pasupuleti, M.; Arockiaraj, J. Impacts of environmental and biological stressors on immune system of Macrobrachium rosenbergii. Rev. Aquacult. 2017, 9, 283-307. [CrossRef]
- Wang, Y.; Wang, B.; Liu, M.; Jiang, K.; Wang, M.; Wang, L. Comparative transcriptome analysis reveals the different roles between hepatopancreas and intestine of Litopenaeus vannamei in immune response to aflatoxin B1 (AFB1) challenge. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2019, 222, 1-10. [CrossRef]
- Zhang, X.; Zhang, M.; Zheng, H.; Ye, H.; Zhang, X.; Li, S. Source of hemolymph microbiota and their roles in the immune system of mud crab. Dev. Comp. Immunol. 2020, 102, 103470. [CrossRef]
- Gao, X.; Jiang, Z.; Zhang, S.; Chen, Q.; Tong, S.; Liu, X.; Jiang, Q.; Yang, H.; Wei, W.; Zhang, X. Transcriptome analysis and immune-related genes expression reveals the immune responses of Macrobrachium rosenbergii infected by Enterobacter cloacae. Fish. Shellfish. Immunol. 2020, 101, 66-77. [CrossRef]
- Wang, D.-L.; Zuo, D.; Wang, L.-M.; Sun, T.; Wang, Q.; Zhao, Y.-L. Effects of white spot syndrome virus infection on immuno-enzyme activities and ultrastructure in gills of Cherax quadricarinatus. Fish. Shellfish. Immunol. 2012, 32, 645-650. [CrossRef]
- Duan, Y.; Zhang, J.; Dong, H.; Wang, Y.; Liu, Q.; Li, H. Oxidative stress response of the black tiger shrimp Penaeus monodon to Vibrio parahaemolyticus challenge. Fish. Shellfish. Immunol. 2015, 46, 354-365. [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644-652. [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015, 31(19), 3210-3212. [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 2014, 15, 550. [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome. Biol. 2010, 11(2), R14. [CrossRef]
- Zhang, W.; Lv, Z.; Li, C.; Sun, Y.; Jiang, H.; Zhao, M.; Zhao, X.; Shao, Y.; Chang, Y. Transcriptome profiling reveals key roles of phagosome and NOD-like receptor pathway in spotting diseased Strongylocentrotus intermedius. Fish. Shellfish. Immunol. 2019, 84, 521-531. [CrossRef]
- Torunn, T.E.; Løvdal, T.; Berg, T. Phagosome dynamics and function. Bioessays. 2000, 22 (3), 255e263.
- Swanson, J.A.; Baer, S.C. Phagocytosis by zippers and triggers. Trends. Cell. Biol. 1995, 5, 89-92.
- Dambuza, I.M.; Brown, G.D. C-type lectins in immunity: recent developments. Curr. Opin. Immunol. 2015, 32, 21-27. [CrossRef]
- Lee, W.B.; Yan, J.J.; Kang, J.S.; Kim, L.K.; Kim, Y.J. Macrophage C-type lectin is essential for phagosome maturation and acidification during Escherichia coli-induced peritonitis. Biochem. Biophys. Res. Commun. 2017, 493, 1491-1497. [CrossRef]
- Nishi, T.; Forgac, M. The vacuolar (H+)-ATPases--nature's most versatile proton pumps. Nat. Rev. Mol. Cell. Biol. 2002, 3, 94-103. [CrossRef]
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell. Sci. 2016, 129, 4329-4339. [CrossRef]
- Zhang, B.; Porto, A.F. Cholesteryl ester storage disease: protean presentations of lysosomal acid lipase deficiency. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 682-685. [CrossRef]
- Hiraoka, M.; Abe, A.; Shayman, J.A. Cloning and characterization of a lysosomal phospholipase A2, 1-O-acylceramide synthase. J. Biol. Chem. 2002, 277, 10090-10099. [CrossRef]
- Taniyama, Y.; Fuse, H.; Satomi, T.; Tozawa, R.; Yasuhara, Y.; Shimakawa, K.; Shibata, S.; Hattori, M.; Nakata, M.; Taketomi, S. Loss of lysophospholipase 3 increases atherosclerosis in apolipoprotein E-deficient mice. Biochem. Biophys. Res. Commun. 2005, 330, 104-110. [CrossRef]
- Du, J.; Zhu, H.; Liu, P.; Chen, J.; Xiu, Y.; Yao, W.; Wu, T.; Ren, Q.; Meng, Q.; Gu, W.; et al. Immune responses and gene expression in hepatopancreas from Macrobrachium rosenbergii challenged by a novel pathogen spiroplasma MR-1008. Fish. Shellfish. Immunol. 2013, 34, 315-323. [CrossRef]
- Danyukova, T.; Ariunbat, K.; Thelen, M.; Brocke-Ahmadinejad, N.; Mole, S.E.; Storch, S. Loss of CLN7 results in depletion of soluble lysosomal proteins and impaired mTOR reactivation. Hum. Mol. Genet. 2018, 27, 1711-1722. [CrossRef]
- Kowalski, E.J.A.; Li, L. Toll-Interacting Protein in Resolving and Non-Resolving Inflammation. Front. Immunol. 2017, 8, 511. [CrossRef]
- Ou, J.; Chen, H.; Liu, Q.; Bian, Y.; Luan, X.; Jiang, Q.; Ji, H.; Wang, Z.; Lv, L.; Dong, X.; et al. Integrated transcriptome analysis of immune-related mRNAs and microRNAs in Macrobrachium rosenbergii infected with Spiroplasma eriocheiris. Fish. Shellfish. Immunol. 2021, 119, 651-669. [CrossRef]
- Feng, J.; Zhao, L.; Jin, M.; Li, T.; Wu, L.; Chen, Y.; Ren, Q. Toll receptor response to white spot syndrome virus challenge in giant freshwater prawns (Macrobrachium rosenbergii). Fish. Shellfish. Immunol. 2016, 57, 148-159. [CrossRef]
- Ke, Y.; Wang, X.; Jin, X.Y.; Solaro, R.J.; Lei, M. PAK1 is a novel cardiac protective signaling molecule. Front. Med. 2014, 8, 399-403. [CrossRef]
- Ren, L.J.; Li, K.Q.; Zhang, Y.Y.; Wang, y.; Yu, Y.; Cheng, Y.; Lin, K.; Song, J.; Chang, Y.Q. Isolation of a new PAK1 Gene from Sea Cucumber (Apostichopus japonicus) and its Expression analysis and function characterization J. Ocea. Univ. China. 2019, 18 (5), 1147-1157. [CrossRef]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114-131. [CrossRef]
- Ruan, L.; Liu, H.; Shi, H. Characterization and function of GSK3β from Litopenaeus vannamei in WSSV infection. Fish. Shellfish. Immunol. 2018, 82, 220-228. [CrossRef]
- Solt, L.A.; May, M.J. The IκB kinase complex: master regulator of NF-κB signaling. Immunol. Res. 2008, 42, 3-18. [CrossRef]
- Viatour, P.; Merville, M.P.; Bours, V.; Chariot, A. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends. Biochem. Sci. 2005, 30, 43-5. [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal. Transduct. Target. Ther. 2017, 2, 17023. [CrossRef]
- Ferreiro, D.U.; Komives, E.A. Molecular mechanisms of system control of NF-κB signaling by IκBα. Biochemistry. 2010, 49, 1560-1567. [CrossRef]
- Liu, F.; Xia, Y.; Parker, A.S.; Verma, I.M. IKK biology. Immunol. Rev. 2012, 246, 239-253. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).