Preprint
Communication
Cell-Free Methylated PTGER4 and SHOX2 Plasma DNA as a Biomarker for Therapy Monitoring and Prognosis in Advanced Stage NSCLC Patients
Altmetrics
Downloads
128
Views
37
Comments
0
A peer-reviewed article of this preprint also exists.
This version is not peer-reviewed
Submitted:
04 May 2023
Posted:
05 May 2023
You are already at the latest version
Alerts
Abstract
Notwithstanding the fact that there is some improvement for an earlier detection of patients with lung cancer, the majority of them still present with a late-stage disease at the time of diagnosis. Next to the most frequently used factors affecting the prognosis of lung cancer patients (stage, performance and age) the recent application of biomarkers obtained by liquid profiling gained more acceptance. In our study we aimed to answer these questions: i) is the quantification of free-circulating methylated PTGER4 and SHOX2 plasma DNA an useful method for the therapy monitoring and is this also possible for patients treated with different therapy regimens?, and ii) is this approach possible when blood drawing tubes are used which allow for a delayed processing of blood samples? Baseline values for mPTGER4 and mSHOX2 do not allow for a clear discrimination between different response groups. In contrast the combination of the methylation values for both genes show a clear difference between responders vs non-responders at the time of re-staging. Additionally, blood drawing into tubes stabilizing the sample give researchers more flexibility.
Keywords:
Subject: Biology and Life Sciences - Life Sciences
1. Introduction
The last years had seen some progress for the treatment of advanced stage lung cancer patients, specifically, for patients harboring druggable genetic alterations like activating EGFR gene mutations, mutations in BRAF and MET genes, and variants of ALK and ROS1 [1]. This applies to non-small cell (NSCLC) and small cell (SCLC) lung cancer patients who are treated with targeted or immunotherapeutic agents [2]. Notwithstanding there are different resistance mechanisms in place leading to a therapy failure also for these novel treatments. This underscores the need for the search and establishment of reliable tumor markers for therapy monitoring.
Recently liquid profiling has emerged as a promising tool for a longitudinal analysis of response to a given therapy particularly as it can be performed in real-time. After the first description of cell-free nucleic acids detectable in plasma and serum of humans [3], the group of Anker and Stroun laid the foundation for the current interest in this method [4,5,6,7]. Currently, there are several methods for the analysis of extracellular nucleic acids quantitatively and qualitatively with whole genome sequencing, exon sequencing, genome-wide methylation analysis and others [8,9].
An easier, faster and more straightforward approach is the quantitative measurement of a smaller gene panel of methylated sequences in cell-free DNA with real-time PCR. The usefulness of this approach to demonstrate a relationship between the presence of methylated plasma DNA and therapy response in lung cancer patients has been shown in several papers [10,11,12,13].
Together with Wang et al [14], our group was among the first in demonstrating the potential of a longitudinal analysis of methylated cell-free DNA for therapy monitoring. We demonstrated a good correlation between the longitudinal measurement of extracellular plasma mSHOX2 DNA and the response to cytotoxic treatment in late stage lung cancer patients [15]. In this study we used EDTA tubes which were processed within one to two hours after blood drawing. Previous studies had demonstrated that EDTA blood should not be stored for more that six hours as a longer storage leads to cell lysis and a „contamination“ of cell-free DNA with genomic DNA. In order to allow for an extended storage of blood tubes (including shipping to a remote laboratory) it is necessary to stabilize the blood sample with special additives. Several companies have developed specially designed blood drawing tubes such as the PAXgene Blood ccfDNA tubes which received market approval in 2016.
In this paper we extended our former study as we included more patients from several hospitals (multicenter), and used PAXgene Blood ccfDNA tubes (Qiagen, Germany) for blood sampling. Additionally we used a modified marker panel for the detection of methylated cell-free circulating DNA in the plasma of late-stage lung cancer patients undergoing a systemic therapy.
2. Materials and Methods
2.1. Patients
In total, 96 patients, among them 78 from the DRK Kliniken Berlin-Mitte and 18 from the other participating clinics, with late stage histologically confirmed non-small cell lung cancer (NSCLC) who received a first-line treatment consisting of chemo +/- radiotherapy, anti-EGFR therapy or immunotherapy were enrolled prospectively in the present study. The patients were consecutively referred to the participating clinics for diagnosis and treatment from August 2016 until October 2020. The study was approved by the Ethics Committee of the University Halle/Saale on June 19, 2014 (Reference number 2014-52) and all patients gave informed written consent prior to inclusion in the study. This study was set up as a multicentric trial and the majority of patients were treated at the DRK Kliniken Berlin-Mitte (78 patients). The blood of these 78 patients was drawn in PAXgene Blood ccfDNA tubes (Qiagen, Germany), while EDTA tubes were used for 18 patients treated in the other participating centers. For the evaluation of the therapy response the data from all 96 patients were used while the analysis for the prognostic relevance was limited to the 78 patients enrolled at the DRK Kliniken Berlin-Mitte.
2.2. Plasma preparation
Blood was taken from patients at the time of diagnosis (pre-treatment) and thereafter in intervals of 7 to 10 days until the time of re-staging. The samples which were obtained at the DRK Kliniken Berlin-Mitte were collected in PAXgene Blood ccfDNA tubes and shipped by regular mail or transported via a courier (at room temperature in any case) to the laboratory in Halle/Saale. The range of delay in processing the blood was 0 to 8 days (median 5 days). The blood was spun once at 1500 rpm for 10 min, the supernatant was carefully transferred into a new tube and re-centrifuged at 3500 rpm for 10 min. The cell-free plasma was aliquotted and stored at -80°C before processing. The blood samples drawn at the other three participating study centers were processed as described above and the plasma was stored at -80°C and shipped on dry ice to the laboratory in Halle/Saale.
2.3. DNA isolation, purification and bisulfite conversion
We used the Epi Bis-kits (Epigenomics, Berlin, Germany) according to the protocol of the manufacturer with an initial input of 3.5 mL plasma for the isolation, purification and bisulfite treatment of the DNA. For the detection and quantification of the gene products, a PCR kit developed by Epigenomics for the simultaneous quantification of mSHOX2, mPTGER4 and ß-actin as a reference gene [Weiss et al J Thorac Oncol 2017] was used. All samples were measured in triplicate. The quantification of mSHOX2, mPTGER4 and ß-actin (as the reference gene for the calculation) was performed according to Kneip et al [17]. The quantification of the cell-free methylated sequences was performed after all prospectively collected plasma samples were complete, i.e. making this analysis an observational study.
2.4. Statistical analysis
Distribution of PTGER4 and SHOX2 methylation values is demonstrated as boxplots for all response groups at time of first radiological staging being partial remission (PR), stable disease (SD) and progressive disease (PD). Prediction of therapy response was evaluated in two settings. For detection of progression, PD was compared with SD+PR while for detection of response, PR was compared with SD+PD. Power of discrimination was calculated as area under the curve (AUC) of receiver operating characteristic (ROC) curves and sensitivity at 90% specificity. Significance of statistical differences was calculated by Wilcoxon-test. For combination of markers, decision tree models were established using appropriate cutoffs and marker order and probabilities for therapy response was shown at every node. Survival was evaluated by Kaplan-Meier-curves and log-rank test. All comparisons were performed two-sided and statistical significance was set at p<0.05. Data analysis was performed using R (version 4.2.0; https://www.R-project.org, free software foundation, Inc., USA).
3. Results
All patients received serial sample collection during the treatment with an average of 6 samples per patients. Treatment was performed for a minimum of 6 months; the median follow-up of the patients was 22 months with a range from 6 to 58 months. We evaluated the biomarker values before start of therapy, at re-staging exams and the relative changes were considered and correlated with radiological response to treatment and survival of the patients.
Of those 96 patients with advanced NSCLC, 83 received regular chemo +/- radiotherapy, 9 a combination of chemo- and immunotherapy and 4 patients an immuntherapy only. Thirty four patients responded well to the treatment at staging exams and showed partial remission (PR), 15 had stable disease (SD), and 33 progressive disease (PD). The therapy response of 14 patients is unknown. The median overall survival (OS) was 8.5 months with a range from 1 to 37 months (Table 1).
Table 1.
Clinical data for all enrolled patients.
| Patients | Number | |
| Patients Sex | female | 62 |
| male | 34 | |
| Age | Range | 48 - 81 |
| median | 63 | |
| Smoker | Yes | 41 |
| No | 4 | |
| Ex-smoker | 30 | |
| Unkown | 21 | |
| Histology | NSCLC (unclassified) | 25 |
| Adenocarcinoma | 54 | |
| Squamous cell carcinoma | 17 | |
| Treatment | Chemo +/- Radiotherapy | 83 |
| Chemo + Immunotherapy | 9 | |
| Immunotherapy | 4 | |
| Treatment response | partial remission | 42 |
| stable disease | 23 | |
| progressive disease | 31 |
3.1. Distribution of mPTGER4 and mSHOX2 methylation values in response groups
Before start of therapy (V1), patients who showed a partial remission had slightly lower methylated PTGER4 levels than SD and PD patients, however this difference was not statistically significant. However, at time of re-staging exam (VS), mPTGER4 levels of PR patients were significantly lower than of SD and even more than of PD patients – and also the ratio of mPTGER4 VS/V1 was significantly different between the response groups (Figure 1A–C). Similar tendencies were observed for SHOX2 methylation with no significant differences between response groups for the pre-therapeutic assessment and lower values for PR and SD than PD patients. In contrast and comparable to the mPTGER4 values, the differences seen for the ratio of mSHOX2 at VS and VS/V1 values reached a low significance level (Figure 1D–F).
3.2. Differentiation of patients according to their therapy response
The power of discrimination between patients with good and poor response to therapy was objectified by areas under the curves (AUC) of receiver operating characteristic (ROC) curves.
For the comparison of patients with progression (PD) versus no progression (PR+SD), AUCs for mPTGER4 were AUC=0.60 (V1), AUC=0.72 (VS), and AUC=0.66 (VS/V1) and for mSHOX2 AUC=0.57 (V1), AUC=0.67 (VS), and AUC=0.71 (VS/V1). Sensitivities for the detection of progression at a 90% specificity vs. non-progression were for mPTGER4 0.60 at V1, 0.74 at VS and 0.65 for VS/V1 (Figure 2 A-C) as well as for mSHOX2 0,59 at V1, 0.64 at VS and 0.69 for VS/V1, respectively (Figure 2 D-F). For the comparison of patients with remission (PR) at a 90% specificity versus no remission (SD+PD), AUCs for mPTGER4 were AUC=0.60 (V1), AUC=0.74 (VS), and AUC=0.65 (VS/V1) (Figure 3 A-C) and for mSHOX2 AUC=0.59 (V1), AUC=0.64 (VS), and AUC=0.69 (VS/V1) (Figure 3 D-F).
3.3. Combination of markers in decision trees
In order to maximize the information for a therapy prediction, markers were combined in a decision tree model by use of appropriate cutoffs and marker order. In using this model we demonstrated that patients with low mSHOX2 ratio (VS/V1) and low mPTGER4 levels at the time of staging (VS) had a high chance of achieving PR at staging exams (Figure 4, Node 3). In contrast, patients with high mSHOX2 ratio (VS/V1) and high mPTGER4 ratio (VS/V1) had a high probability of demonstrating a progressive disease (Figure 4, Node 7). Patients with either low mSHOX2 ratio (VS/V1) and high mPTGER4 (VS) levels or alternatively high mSHOX2 ratio (VS/V1) and low mPTGER4 ratio (VS/V1) ranged intermediate (Figure 4, Nodes 4 and 6).
3.4. Relevance of PTGER4 and SHOX2 for prognosis
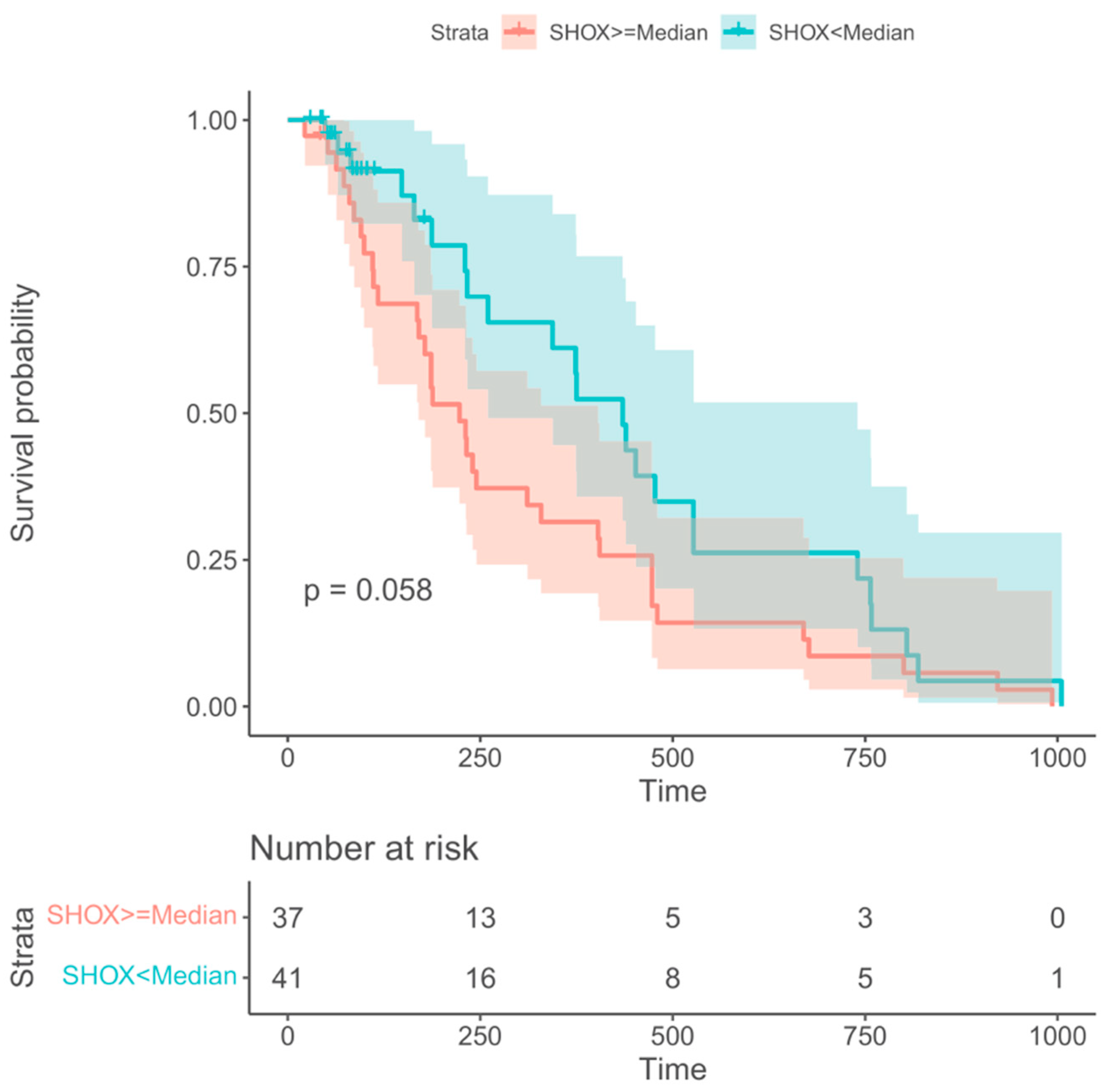
Information regarding overall survival (OS) was available from 78 out of the 96 NSCLC patients. For prediction of OS only SHOX2 methylation at the time of staging (VS) showed borderline significant prognostic information. All other markers and time points did not. Patients with SHOX2 methylation levels below the median had a tendency to longer survival with a median OS of 11 months as compared with patients with SHOX2 methylation levels above the median who had a median OS of 8 months. This difference showed a trend but was not statistically significant with a value of p=0.058 (Figure 5).
4. Discussion
According to the latest US cancer statistics 46% of the newly diagnosed lung cancer patients demonstrate a late stage disease [18]. Furthermore, a recent survey showed that almost 49% of 210 000 newly diagnosed lung cancer patients died within 2 months after receiving their diagnosis [19]. Thus it is of utmost importance to differentiate between patients with a high risk for early death and patients with a better prognosis.
Since our group and other researchers demonstrated that the detection and quantification of methylated ctDNA is a useful approach for the therapy monitoring in lung cancer patients [14,15,16], more papers on this subject were published [20,21]. Additionally, the incorporation of liquid profiling as an additional clinical tool for the diagnosis, treatment stratification, detection of resistance mechanisms and as a prognostic indicator in lung cancer patients has been shown [22,23,24,25].
In order to confirm and to extend the results of our original analysis [15] and to demonstrate the robustness of the method we performed the present study. Apart from increasing the number of patients, we included patients who received different treatment regimens, i.e. chemotherapy with/without radiotherapy, chemotherapy plus immunotherapy and immunotherapy exclusively. In addition we changed the pre-analytical process and applied a modified marker panel.
It is known that blood drawn into EDTA tubes should not be stored for more than 4-6 hours before being processed [26]. A delay of more than 6 hours can lead to irregular results due to lysis of blood cells and "contamination" of cell-free DNA with genomic DNA. Several studies had demonstrated that a storage of blood drawn into PAXgene Blood ccfDNA Tubes for up to 7 days does not change the quantity and quality of plasma DNA [27,28]. Additionally, we applied a modified marker panel which included the detection of cell-free methylated SHOX2 and PTGER4 plasma DNA [27]. All of these factors plus a different patient population might explain the different results obtained in this study as compared to our previous paper [15].
It is interesting to note that the baseline values for mPTGER4 and mSHOX2 do not allow for a clear discrimination between different response groups (Figure 1). In contrast, the methylation values for both genes show a clear difference between responders vs non-responders at the time of re-staging (Figure 1). This observation still holds true when the ratios of the methylation values (VS/V1) for both genes are plotted (Figure 1 box E and F). This data corroborates our initial observation [15] that the methylation values at the time of diagnosis do not allow a differentiation between the two groups while this is possible at the time of re-staging (i.e. 8 to 12 weeks after therapy start). At the time this study was commenced, Epigenomics AG (Berlin, Germany) had introduced a modified kit for the DNA methylation analysis. This new kit included mSHOX2 as well as a second marker, mPTGER4 [27]. Currently, no comparative data exists for the two kits in a head-to-head approach. However, promising results have been published that advocate for the inclusion of a second marker. Indeed, the grouping of patients with low mSHOX2 ratio (VS/V1) plus low mPTGER4 levels at the time of re-staging (VS) in decision trees allowed the discrimination of patients responding to the therapy from non-responding patients (Figure 3). When patients with a high mSHOX2 ratio (VS/V1) and high mPTGER4 levels at the time of staging (VS) were combined we were able to differentiate patients not responding to the therapy but with less statistical power. When the methylation levels of both genes at the time of re-staging were used for Kaplan-Meier curves only mSHOX2 demonstrated a trend for statistical significance but not mPTGER4 (Figure 4). SHOX2 belongs to the homeobox family and is coding for a transcriptional regulator which is involved in pattern formation in both invertebrate and vertebrate species. In contrast PTGER4 is a protein coding gene and belongs to the G-protein coupled receptor family. As for both genes no role in the development of lung cancer has been described so far, we assume that only the former has a functional relationship in lung cancer patients.
In conclusion, our findings demonstrate that quantifying of extracellular free-circulating methylated DNA in plasma could be a valuable tool to monitor the response of lung cancer patients undergoing various treatment regimens. The use of specialized blood drawing tubes to stabilize samples at room temperature allows the collection and transport to a remote laboratory without cooling. This approach gives researchers the possibility for enrolling a large number of patients in future studies to confirm the validity of our current findings.
Author Contributions
E.A, D.R., S.E., G.T., J.K., C.S. and C.G., contributed to data acquisition. M.F., J.K., F.K., S.H. and B.S. contributed to the data analysis and writing of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
No funding was received specifically for this work. The supporters of this project had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and has been approved by the Institutional Review Board (IRB) at the University Hospital of Halle/Saale. Informed consent (written) was obtained from all donors (Reference number 2014-52).
Informed Consent Statement
Informed written consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Epigenomics AG who supported this project by supplying us with isolation and detection kits and Qiagen who supplied us with PAXgene ccfDNA tubes.
Conflicts of Interest
Michael Fleischhacker and Bernd Schmidt are coinventors of the patent: “Methods for assessing the treatment response of cancer patients and for treating cancer patients by analysing CpG methylation” (Application Number: 62076674, patent pending). There are no further patents, products in development or marketed products to declare. This does not alter the authors’ adherence to all the publication policies on sharing data and materials.
References
- Fois, S.S.; Paliogiannis, P.; et al. Molecular Epidemiology of the Main Druggable Genetic Alterations in Non-Small Cell Lung Cancer. Int J Mol Sci 2021, 22, 612. [Google Scholar] [CrossRef] [PubMed]
- S3-Leitlinie Prävention, Diagnostik, Therapie und Nachsorge des Lungenkarzinoms.
- Available online: https://www.leitlinienprogramm-onkologie.de/leitlinien/lungenkarzinom/ (accessed February 2023).
- Mandel, P.; Metais, P. Les Acides nucleiques du plasma sangiun chez l'Homme. C R Seances Soc Biol Fil 1948, 142, 241–3. [Google Scholar]
- Stroun, M.; Anker, P.; et al. Circulating nucleic acids in higher organisms. Int Rev Cytol 1977, 51, 1–48. [Google Scholar] [PubMed]
- Stroun, M.; Anker, P.; et al. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol 1987, 23, 707–12. [Google Scholar] [CrossRef] [PubMed]
- Stroun, M.; Anker, P.; et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989, 46, 318–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Q.; Stroun, M.; et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med 1996, 2, 1033–5. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, L. Cell-Free DNA Methylation Profiling Analysis— Technologies and Bioinformatics. Cancers 2019, 11, 1741. [Google Scholar] [CrossRef] [PubMed]
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Molecular Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- Barault,, L.; Amatu, A.; et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Annals Oncology, 2015, 26, 1994–9. [Google Scholar]
- Huang, G.; Krocker, J.D.; et al. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin Chem Lab Med 2014, 52, 899–909. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; et al. Current status of ctDNA in precision oncology for hepatocellular carcinoma. J Exp Clin Canc Res 2021, 40, 140. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulou, M.; Karaglani, M.; et al. Circulating cell-free DNA in breast cancer: size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene 2018, 38, 3387–3401. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, B.; et al. Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clin Epigenetics 2015, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.; Beyer, J.; et al. Quantification of Cell-Free mSHOX2 Plasma DNA for Therapy Monitoring in Advanced Stage Non-Small Cell (NSCLC) and Small-Cell Lung Cancer (SCLC) Patients. PLOS One 2015, 10, e0118195. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.; Schlegel, A.; et al. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J Thorac Oncol 2017, 12, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Kneip, C.; Schmidt, B.; et al. SHOX2 DNA Methylation Is a Biomarker for the Diagnosis of Lung Cancer in Plasma. J Thorac Oncol 2011, 6, 1632. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; et al. Cancer statistics, 2022. CA Cancer J Clin 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Globus, O.; Sagie, S.; et al. et al. Early death after a diagnosis of metastatic solid cancer – raising awareness and identifying risk factors from the SEER database. medRxiv 2023.
- Ponomaryova, A.A.; Rykova, E.Y.; et al. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 2013, 81, 397–403. [Google Scholar] [CrossRef]
- Janke, F.; Angeles, A.K.; et al. Longitudinal monitoring of cell-free DNA methylation in ALK-positive non-small cell lung cancer patients. Clin Epigenetics 2022, 14, 163. [Google Scholar] [CrossRef]
- Metzenmacher, M.; Hegedues, B.; et al. Combined multimodal ctDNA analysis and radiological imaging for tumor surveillance in Non-small cell lung cancer. Transl Oncology 2022, 15, 101279. [Google Scholar] [CrossRef]
- Pisapia, P.; Malapelle, U.; et al. Liquid Biopsy and Lung Cancer. Acta Cytol 2019, 63, 489–96. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.; Thompson, J.C.; et al. Plasma Cell-Free DNA Genotyping: From an Emerging Concept to a Standard-of-Care Tool in Metastatic Non-Small Cell Lung Cancer. 2021, 26, e1812-e1821.
- Shields, M.D.; Chen, K.; et al. Making the Rounds: Exploring the Role of Circulating Tumor DNA (ctDNA) in Non-Small Cell Lung Cancer. Int J Mol Sci 2022, 23, 9006. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Klotzek, S.; et al. Changes in Concentration of DNA in Serum and Plasma during Storage of Blood Samples. Clin Chem 2003, 49, 1028–9. [Google Scholar] [CrossRef] [PubMed]
- Warton, K.; Yuwono, N.L.; et al. Evaluation of Streck BCT and PAXgene Stabilised Blood Collection Tubes for Cell- Free Circulating DNA Studies in Plasma. Mol Diagn Ther 2017, 21, 563–70. [Google Scholar] [CrossRef]
- Schmidt, B.; Reinicke, D.; et al. Liquid biopsy - Performance of the PAXgene® Blood ccfDNA Tubes for the isolation and characterization of cell-free plasma DNA from tumor patients. Clin Chim Acta 2017, 469, 94–8. [Google Scholar] [CrossRef]
Figure 1.
Boxplots for the distribution of methylated PTGER4 (A-C) and SHOX2 (D-F) before start of therapy (V1), at time of first radiological staging (VS) and the ratio between both time points (VS/V1) for patients with partial remission (PR), stable disease (SD) and progressive disease (PD) in staging exams.
Figure 1.
Boxplots for the distribution of methylated PTGER4 (A-C) and SHOX2 (D-F) before start of therapy (V1), at time of first radiological staging (VS) and the ratio between both time points (VS/V1) for patients with partial remission (PR), stable disease (SD) and progressive disease (PD) in staging exams.
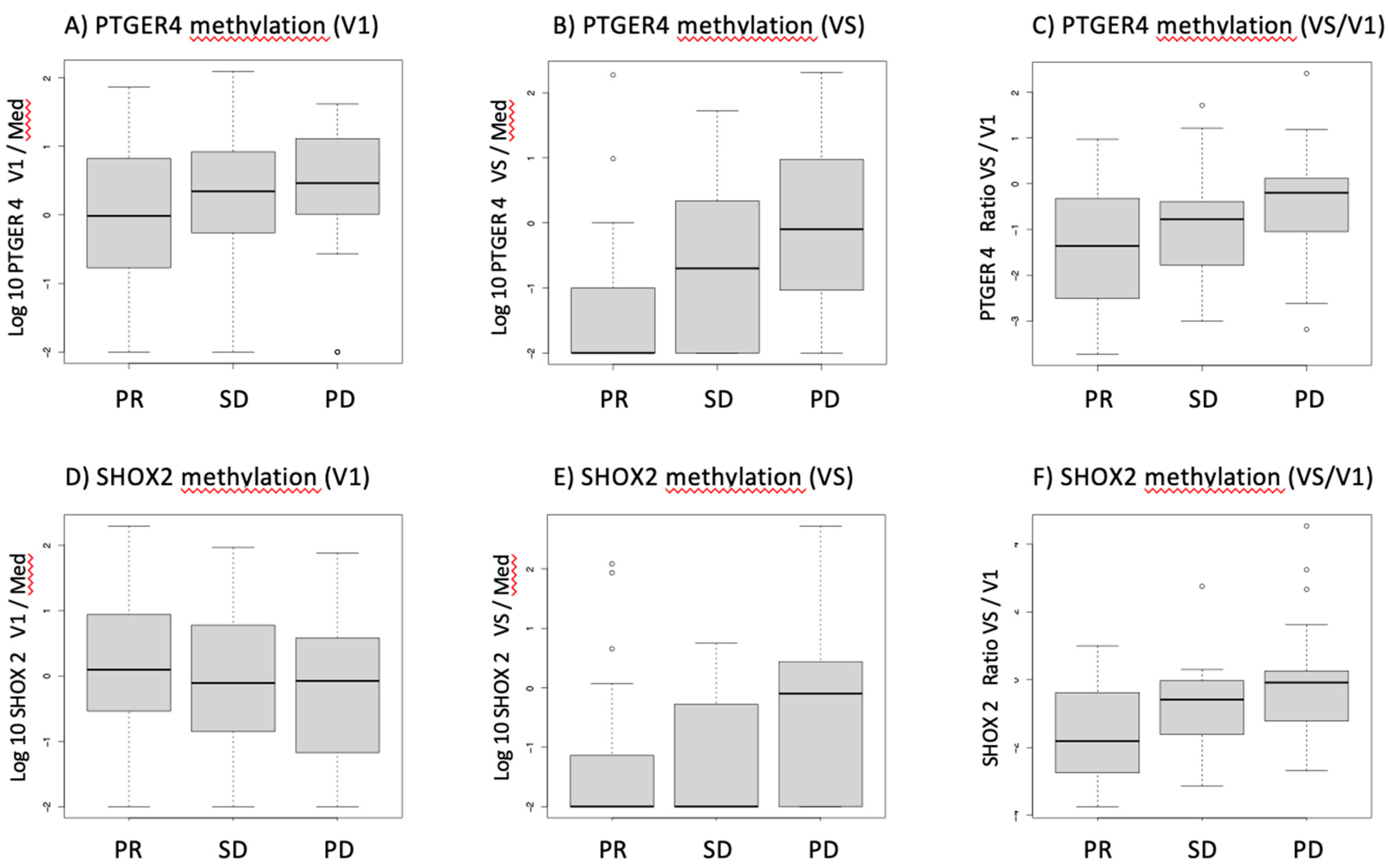
Figure 2.
Receiver-operating characteristic (ROC) curves for the discrimination of patients with progressive disease (PD) from patients with no progression (stable disease + partial remission) for methylated PTGER4 (A-C) and SHOX2 (D-F) before start of therapy (V1), at time of first radiological staging (VS) and the ratio between both time points (VS/V1).
Figure 2.
Receiver-operating characteristic (ROC) curves for the discrimination of patients with progressive disease (PD) from patients with no progression (stable disease + partial remission) for methylated PTGER4 (A-C) and SHOX2 (D-F) before start of therapy (V1), at time of first radiological staging (VS) and the ratio between both time points (VS/V1).
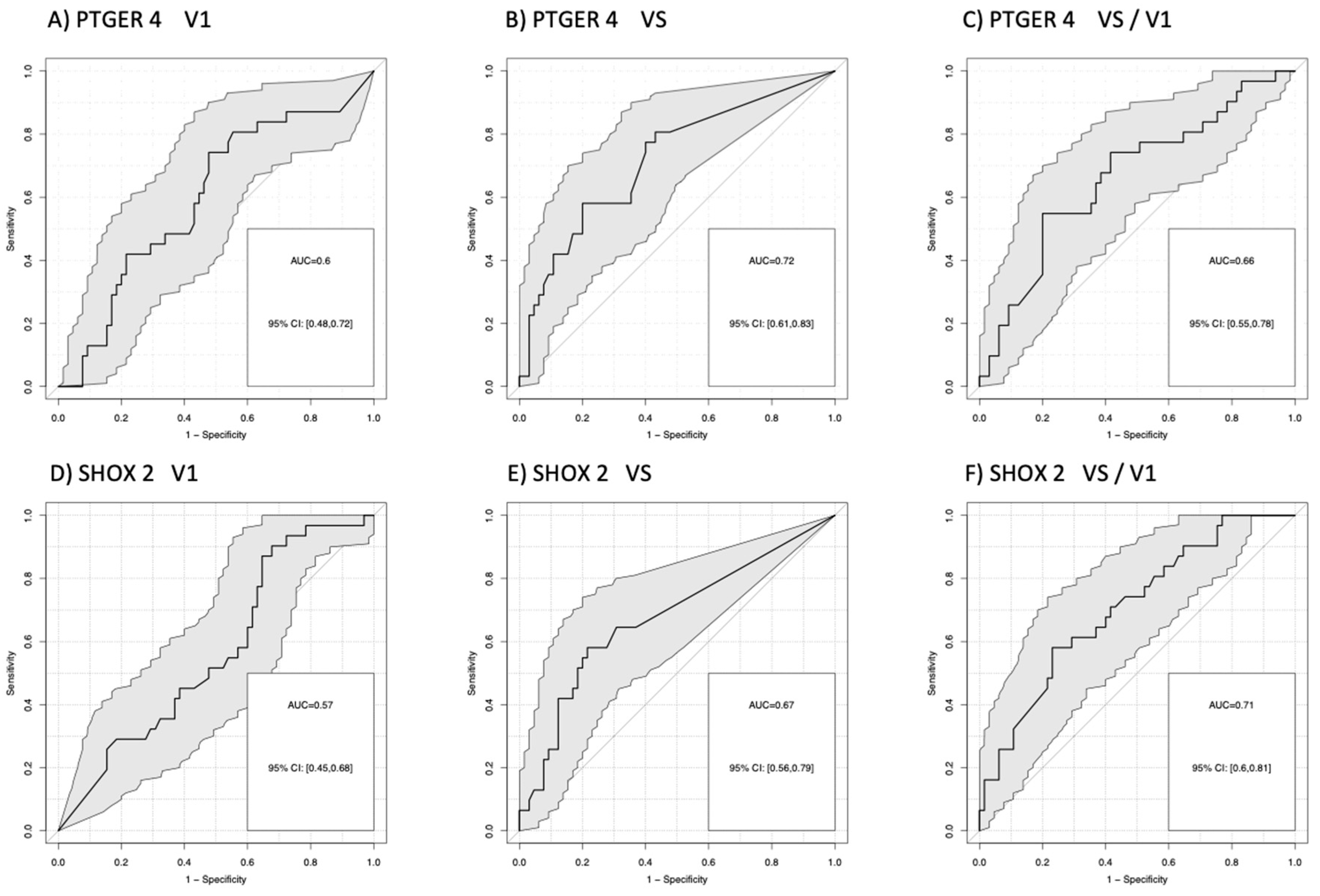
Figure 3.
Receiver-operating characteristic (ROC) curves for the discrimination of patients with remission (PR) from patients with no remission (stable + progressive disease) for methylated PTGER4 (A-C) and SHOX2 (D-F) before start of therapy (V1), at time of first radiological staging (VS) and the ratio between both time points (VS/V1).
Figure 3.
Receiver-operating characteristic (ROC) curves for the discrimination of patients with remission (PR) from patients with no remission (stable + progressive disease) for methylated PTGER4 (A-C) and SHOX2 (D-F) before start of therapy (V1), at time of first radiological staging (VS) and the ratio between both time points (VS/V1).
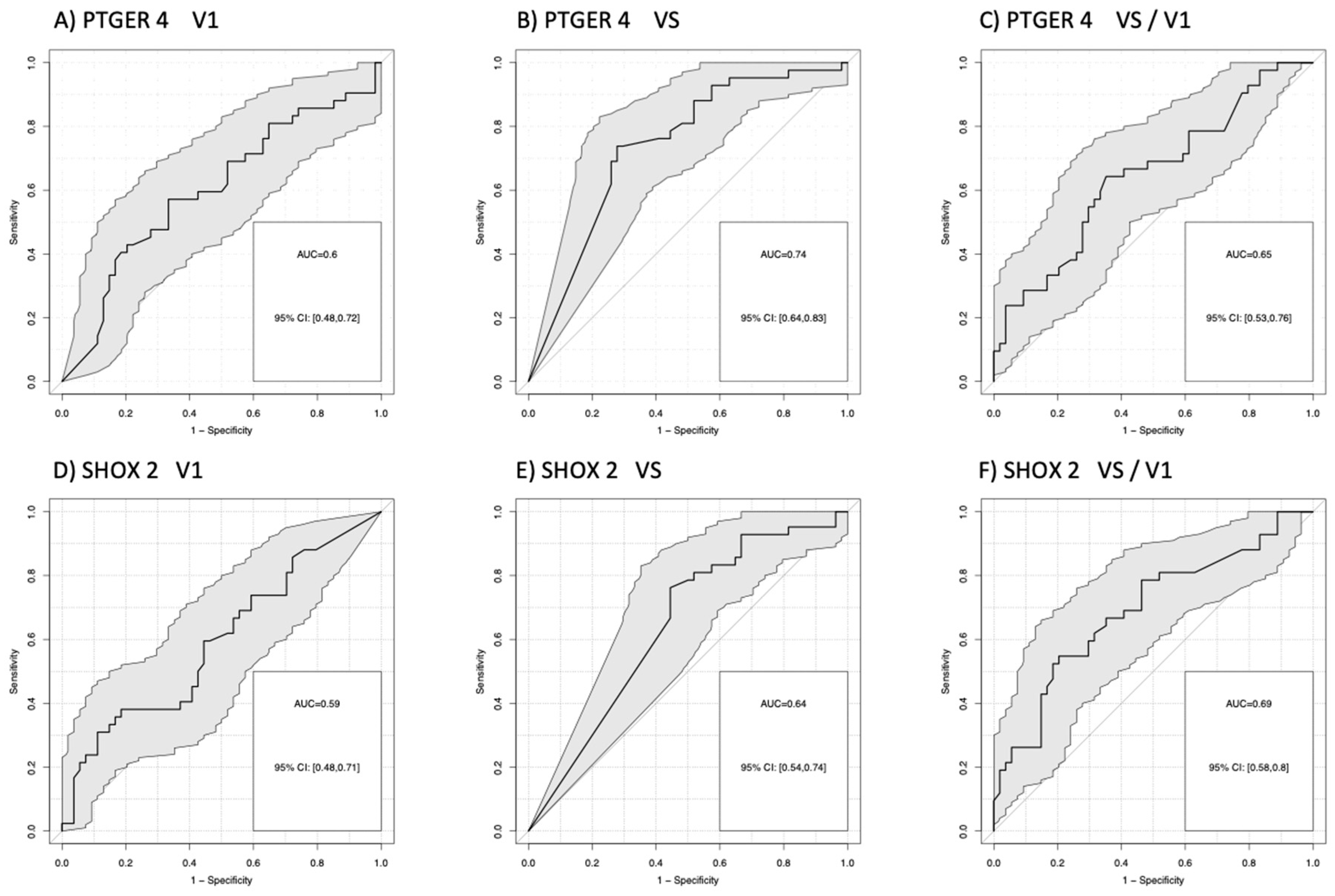
Figure 4.
Decision tree model using combination of markers mSHOX2 ratio (VS/V1), mPTGER4 at staging (VS) and mPTGER4 ratio (VS/V1) for best prediction of therapy response. Patients with low mSHOX2 VS/V1 and low mPTGER4 VS had a high chance of achieving partial remission (R) whereas patients with high mSHOX2 VS/V1 and high mPTGER4 VS/V1 had a high probability for progressive disease (P). A) shows the decision rules and B) the distribution of responses at every decision node.
Figure 4.
Decision tree model using combination of markers mSHOX2 ratio (VS/V1), mPTGER4 at staging (VS) and mPTGER4 ratio (VS/V1) for best prediction of therapy response. Patients with low mSHOX2 VS/V1 and low mPTGER4 VS had a high chance of achieving partial remission (R) whereas patients with high mSHOX2 VS/V1 and high mPTGER4 VS/V1 had a high probability for progressive disease (P). A) shows the decision rules and B) the distribution of responses at every decision node.
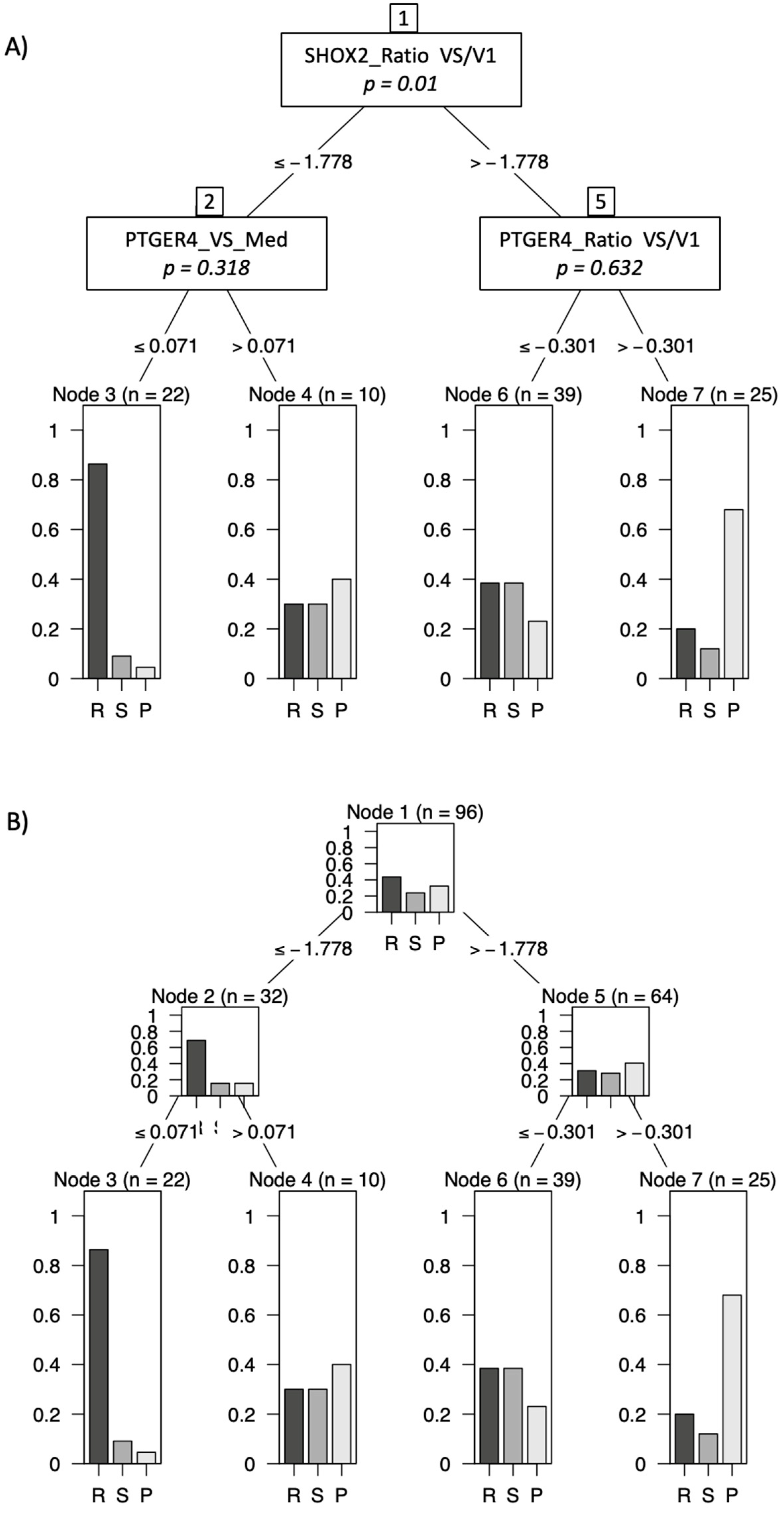
Figure 5.
Kaplan-Meier survival curves for the mSHOX2 at time of staging (VS) showing borderline longer survival for patients with low levels (green) than those with high levels (red).
Figure 5.
Kaplan-Meier survival curves for the mSHOX2 at time of staging (VS) showing borderline longer survival for patients with low levels (green) than those with high levels (red).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Copyright: This open access article is published under a Creative Commons CC BY 4.0 license, which permit the free download, distribution, and reuse, provided that the author and preprint are cited in any reuse.
MDPI Initiatives
Important Links
© 2024 MDPI (Basel, Switzerland) unless otherwise stated







