Submitted:
05 May 2023
Posted:
05 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
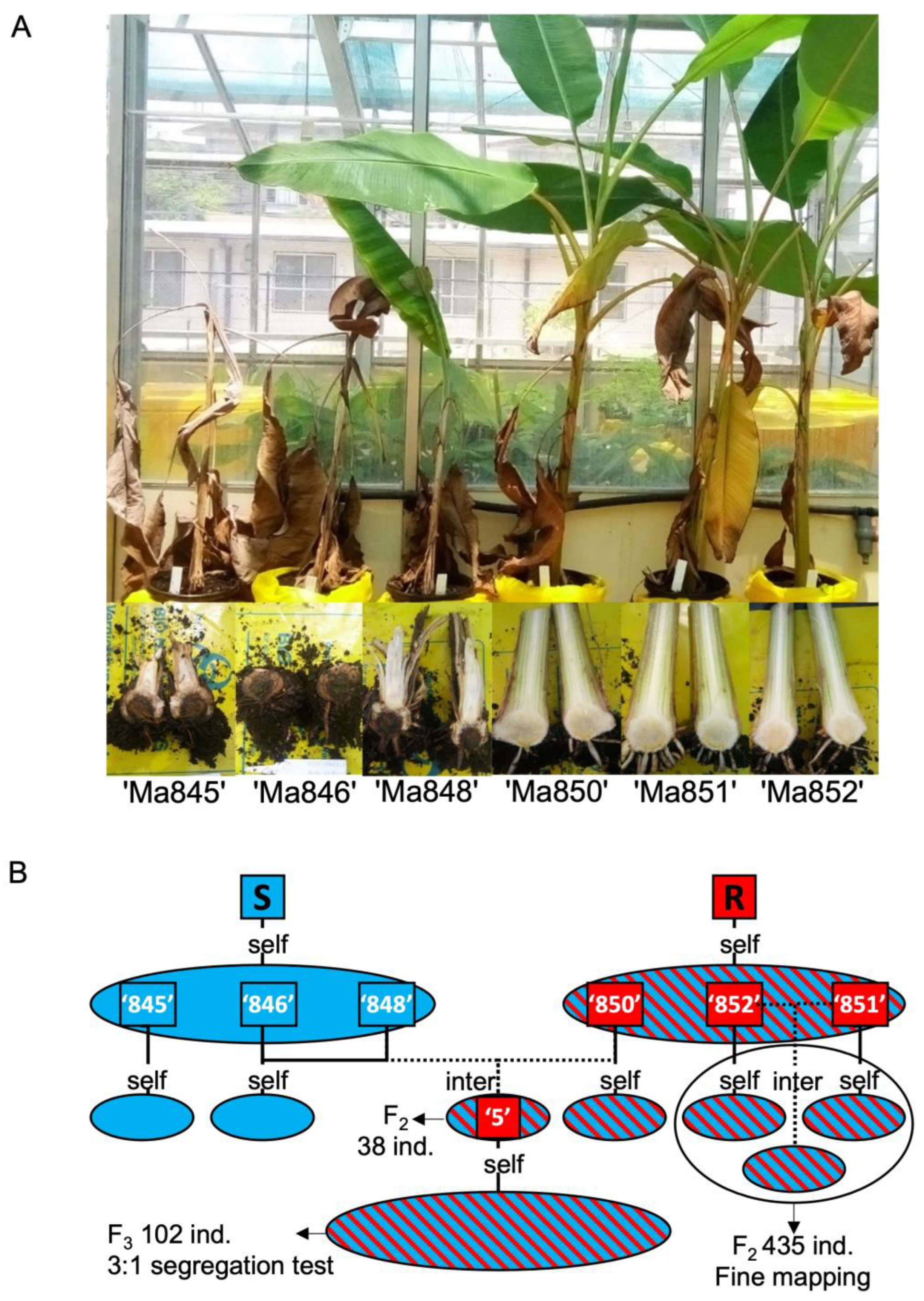
2.1. Foc-STR4 phenotypes and population development
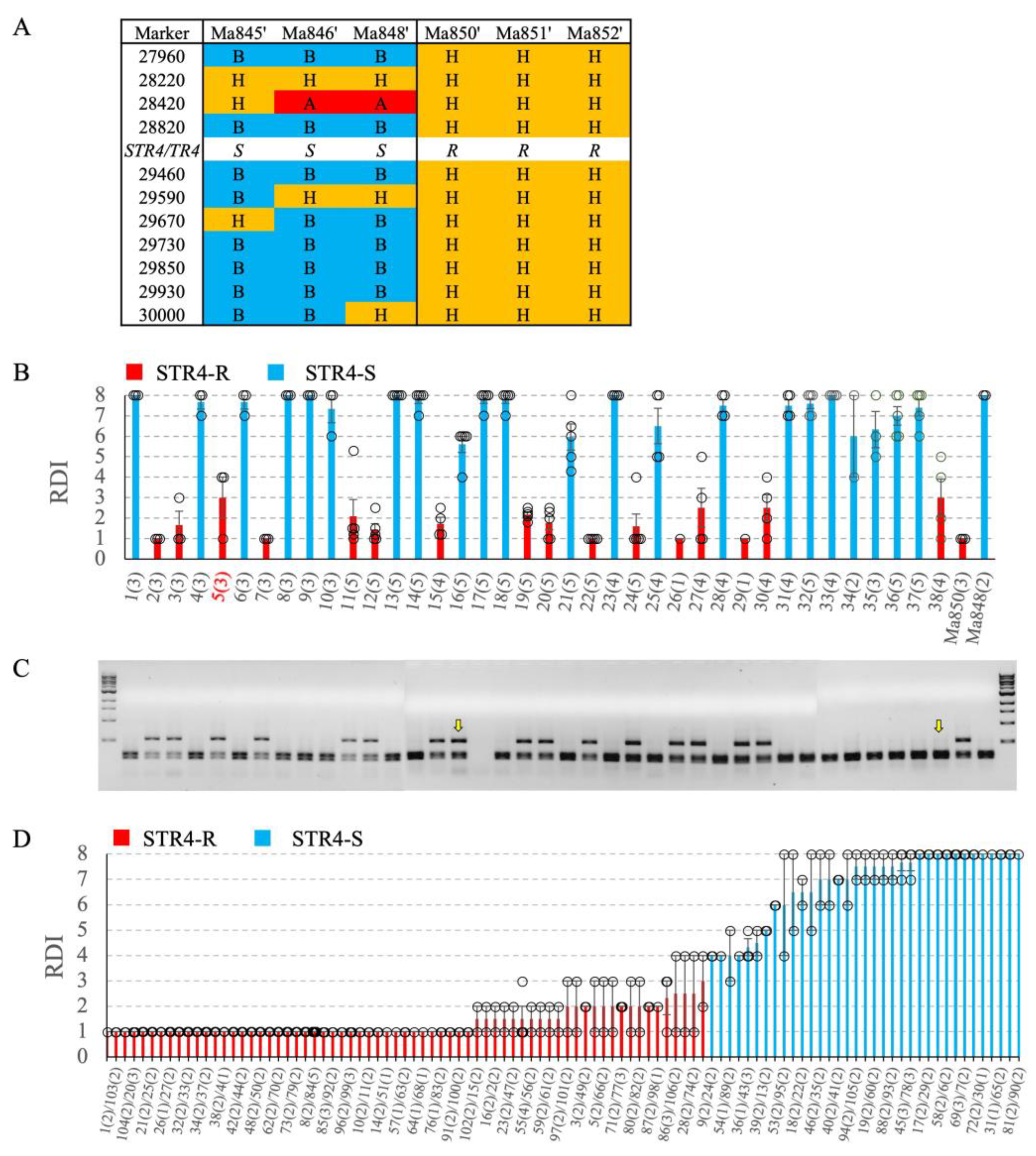
2.2. Marker development
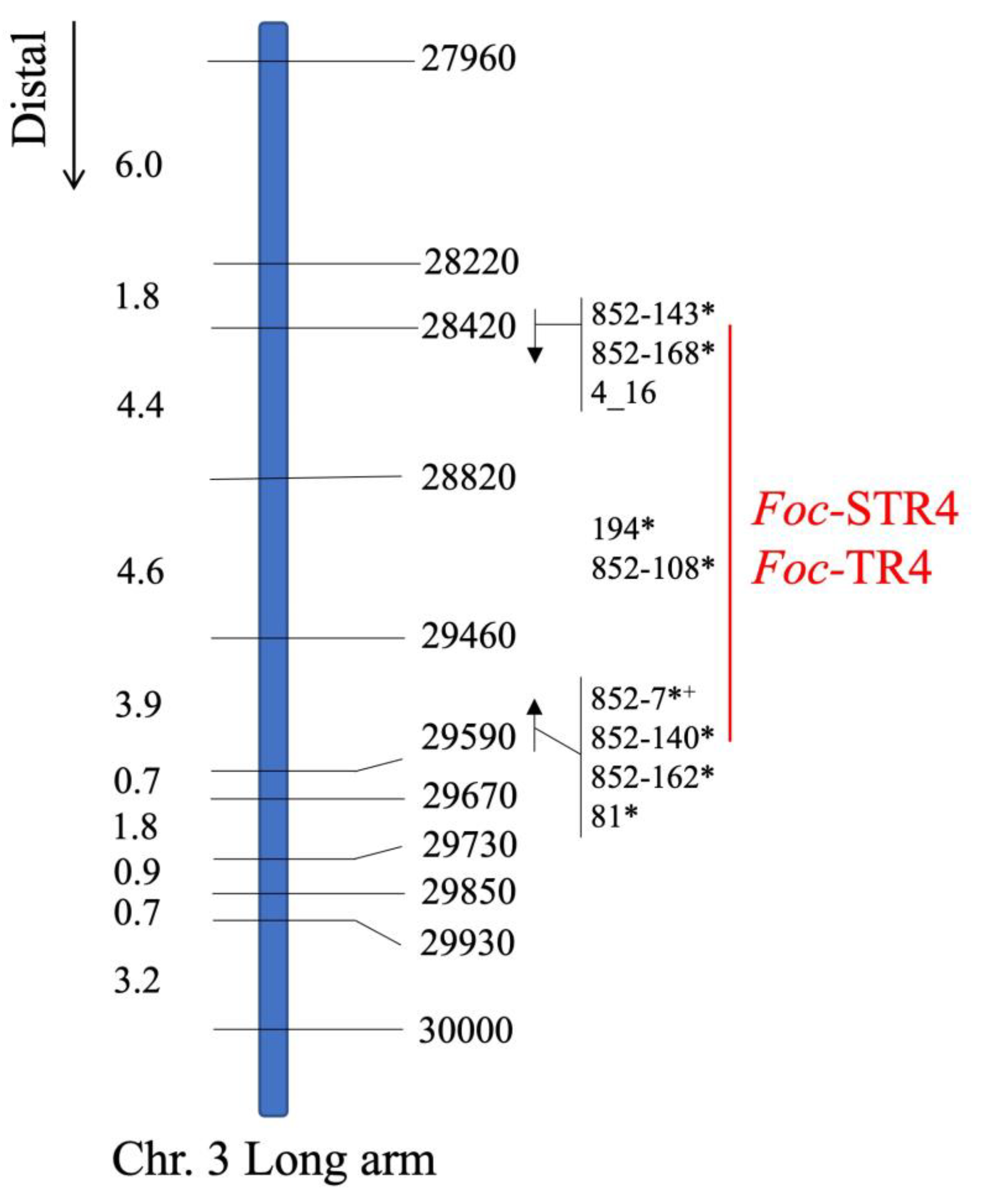
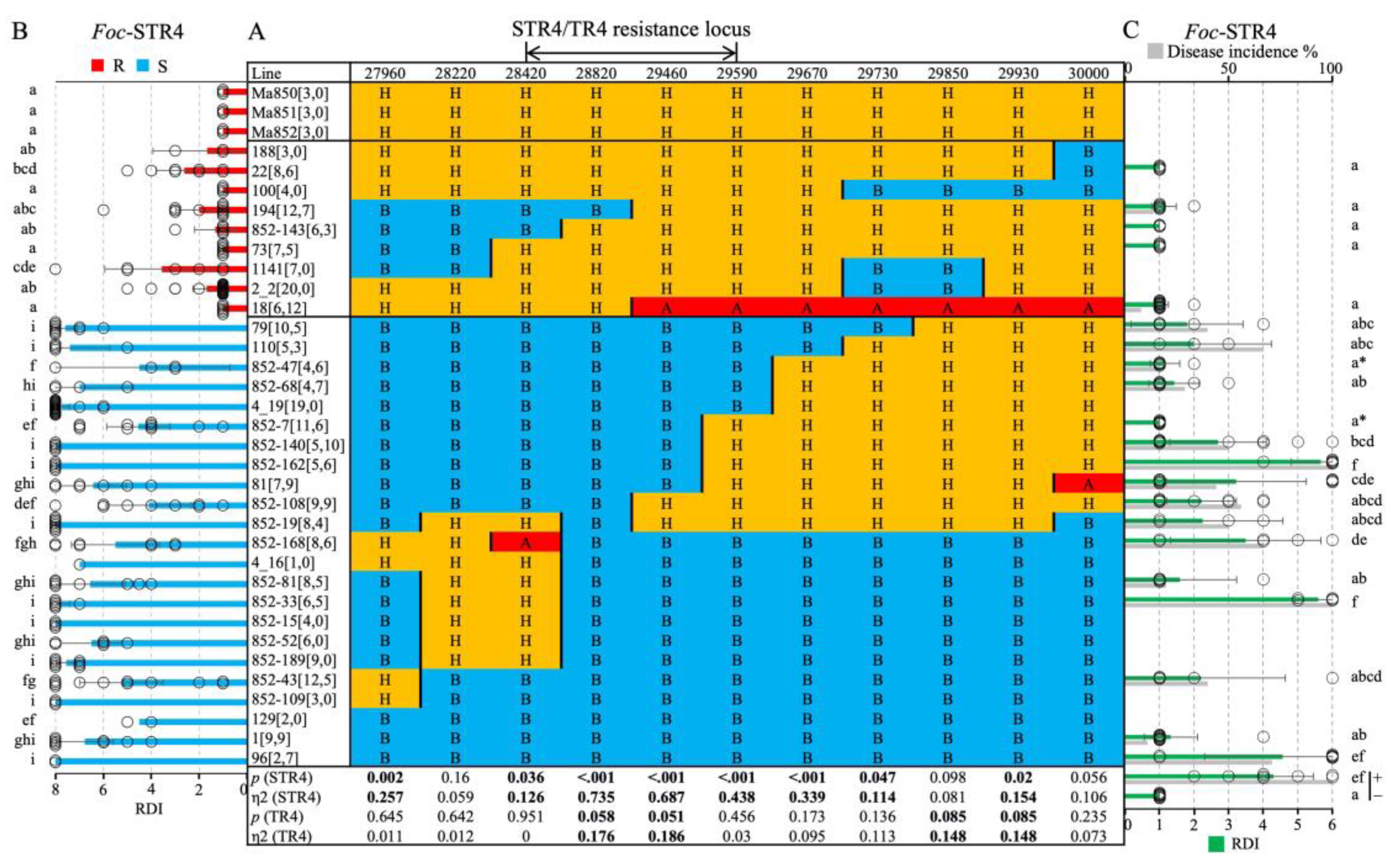
2.3. Genetic mapping
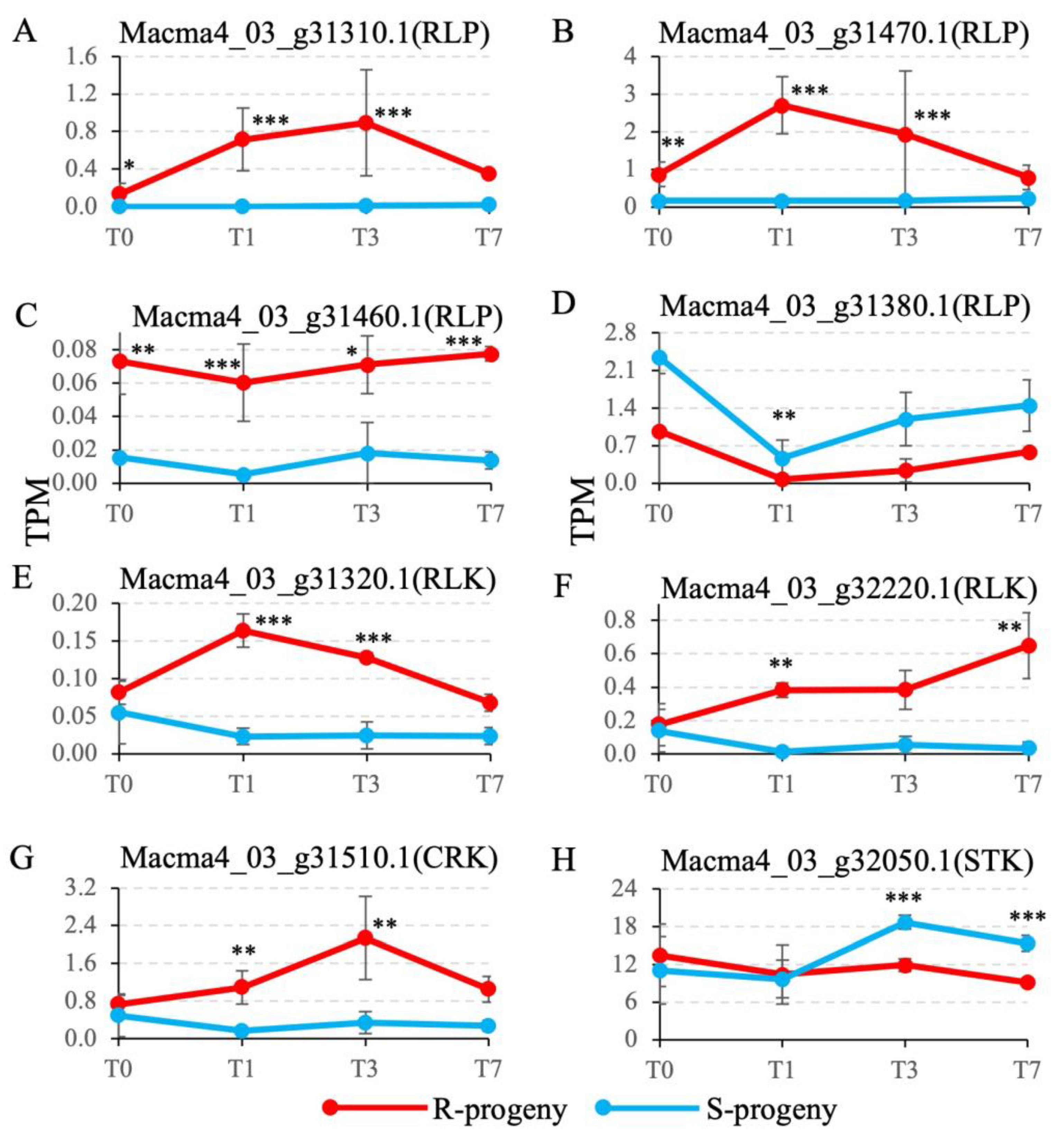
2.4. Candidate R gene expression profiling
2.5. Foc-STR4 resistance and marker validation in ‘population 2’
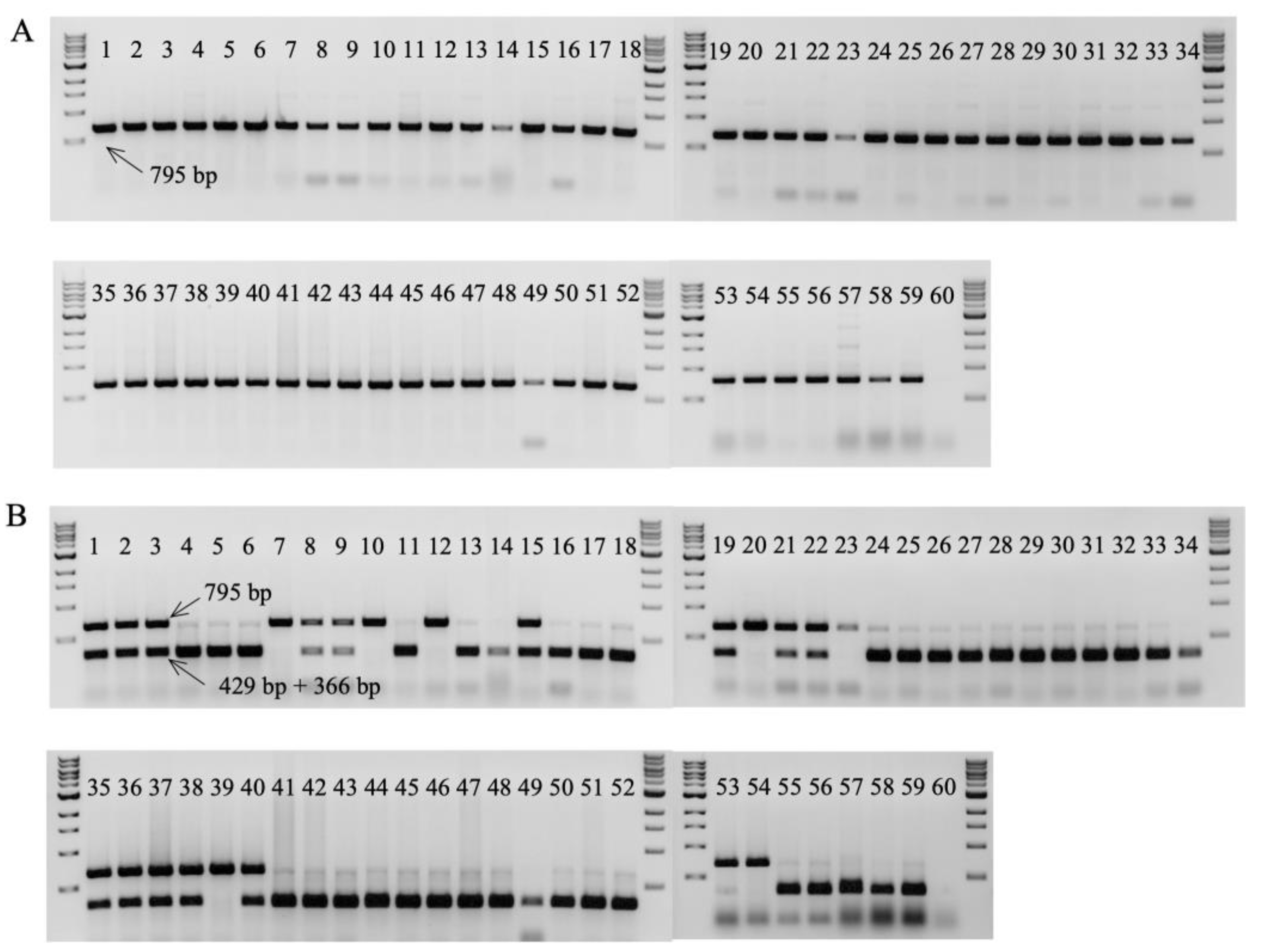
2.6. Validation of Marker 29730 for Marker-assisted selection of TR4 and STR4
3. Discussion
4. Materials and Methods
4.1. Musa acuminata ssp. malaccensis populations
4.2. Fungal isolates
4.3. Foc-STR4 pot trial
4.4. Foc-TR4 pot trial
4.5. Molecular marker development
4.6. DNA extraction and PCR
4.7. Digital gene expression analysis on candidate genes
4.8. Statistical analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perrier, X.; De Langhe, E.; Donohue, M.; Lentfer, C.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.P.; Jenny, C.; et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11311–11318. [Google Scholar] [CrossRef] [PubMed]
- Němečková, A.; Christelová, P.; Čížková, J.; Nyine, M.; Van den Houwe, I.; Svačina, R.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Molecular and cytogenetic study of East African highland banana. Front. Plant Sci. 2018, 9, 1371. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, N.W. The evolution of the bananas. In The Evolution of the Bananas; Longman, 1962. [Google Scholar]
- Simmonds, N.W.; Shepherd, K. The taxonomy and origins of the cultivated bananas. Bot. J. Linn. Soc. 1955, 55, 302–312. [Google Scholar] [CrossRef]
- Sheperd, K. Cytogenetics of the genus Musa. International Network for the Improvement of Banana and Plantain: Montpellier, France, 1999. [Google Scholar]
- Dodds, K. Genetical and cytological studies of Musa: V. Certain edible diploids. J. Genet. 1943, 45, 113–138. [Google Scholar] [CrossRef]
- Rekha, A.; Hiremath, S. Chromosome studies and karyotype analysis of some triploid banana (Musa species) cultivars of AAA genomic group. J. Hortic. Sci. 2008, 3, 30–34. https://jhs.iihr.res.in/index.php/jhs/article/view/590 (accessed on 26 April 2023). [CrossRef]
- Ploetz, R.C. Panama disease: An old nemesis rears its ugly head part 2. The Cavendish era and beyond. Pl. Health Prog. 2005, 23, 1–17. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium wilt of banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef]
- Kistler, H.C.; Alabouvette, C.; Baayen, R.P.; Bentley, S.; Brayford, D.; Coddington, A.; Correll, J.; Daboussi, M.J.; Elias, K.; Fernandez, D.; et al. Systematic numbering of vegetative compatibility groups in the plant pathogenic fungus Fusarium oxysporum. Phytopathology 1998, 88, 30–32. [Google Scholar] [CrossRef]
- Moore, N.; Pegg, K.; Allen, R.; Irwin, J. Vegetative compatibility and distribution of Fusarium oxysporum f. sp. cubense in Australia. Aust. J. Exp. Agric. 1993, 33, 797–802. [Google Scholar] [CrossRef]
- Ploetz, R.C.; Correll, J.C. Vegetative compatibility among races of Fusarium oxysporum f. sp. cubense. Plant Dis. 1988, 72, 325–328. [Google Scholar] [CrossRef]
- Stover, R.H. Fusarium wilt of Banana: Some history and current Status of the Disease. In Fusarium wilt of banana; Ploetz, R.C., Ed.; APS Press: St. Paul, MN, USA, 1990; pp. 1–7. [Google Scholar]
- FAOSTAT. 2023. Available online: https://www.fao.org/faostat/.
- FAO. Banana market review—Preliminary results for 2022. Rome. Available online: https://www.fao.org/3/cc3421en/cc3421en.pdf (accessed on 27 April 2023).
- Pegg, K.G.; Coates, L.M.; O’Neill, W.T.; Turner, D.W. The epidemiology of Fusarium wilt of banana. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, R.; Loganathan, M.; Arthee, R.; Prabakaran, M.; Uma, S. Fusarium wilt: a threat to banana cultivation and its management. CABI Reviews 2020, 1–24. [Google Scholar] [CrossRef]
- Czislowski, E.; Fraser-Smith, S.; Zander, M.; O’Neill, W.T.; Meldrum, R.A.; Tran-Nguyen, L.T.T.; Batley, J.; Aitken, E.A.B. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol. Plant Pathol. 2018, 19, 1155–1171. [Google Scholar] [CrossRef] [PubMed]
- Ordonez, N.; Seidl, M.F.; Waalwijk, C.; Drenth, A.; Kilian, A.; Thomma, B.P.H.J.; Ploetz, R.C.; Kema, G.H.J. Worse comes to worst: bananas and Panama disease—when plant and pathogen clones meet. PLoS Pathog. 2015, 11, e1005197. [Google Scholar] [CrossRef] [PubMed]
- Dita, M.; Barquero, M.; Heck, D.; Mizubuti, E.S.G.; Staver, C.P. Fusarium wilt of banana: current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, W.T.; Henderson, J.; Pattemore, J.A.; O’Dwyer, C.; Perry, S.; Beasley, D.R.; Tan, Y.P.; Smyth, A.L.; Goosem, C.H.; Thomson, K.M.; et al. Detection of Fusarium oxysporum f. sp. cubense tropical race 4 strain in northern Queensland. Australas. Plant Dis. Notes 2016, 11, 33. [Google Scholar] [CrossRef]
- Li, C.; Mostert, G.; Zuo, C.; Beukes, I.; Yang, Q.; Sheng, O.; Kuang, R.; Wei, Y.; Hu, C.; Rose, L. Diversity and distribution of the banana wilt pathogen Fusarium oxysporum f. sp. cubense in China. Fungal Genom. Biol. 2013, 3. [Google Scholar] [CrossRef]
- Hermanto, C.; Sutanto, A.; Jumjunidang; Edison, H.S.; Daniells, J.W.; O’Neill, W.T.; Sinohin, V.G.O.; Molina, A.B.; Taylor, P. Incidence and distribution of Fusarium wilt disease of banana in Indonesia. Acta. Hortic. 2011, 897, 313–322. [Google Scholar] [CrossRef]
- Molina, A.B.; Fabregar, E.; Sinohin, V.G.; Yi, G.; Viljoen, A. Recent occurrence of Fusarium oxysporum f. sp. cubense tropical race 4 in Asia. Acta. Hortic. 2009, 828, 109–116. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense. Phytopathology 2006, 96, 653–656. [Google Scholar] [CrossRef]
- García-Bastidas, F.; Ordóñez, N.; Konkol, J.; Al-Qasim, M.; Naser, Z.; Abdelwali, M.; Salem, N.; Waalwijk, C.; Ploetz, R.C.; Kema, G.H.J. First report of Fusarium oxysporum f. sp. cubense tropical race 4 associated with Panama disease of banana outside Southeast Asia. Plant Dis. 2014, 98, 694. [Google Scholar] [CrossRef] [PubMed]
- Maymon, M.; Sela, N.; Shpatz, U.; Galpaz, N.; Freeman, S. The origin and current situation of Fusarium oxysporum f. sp. cubense tropical race 4 in Israel and the Middle East. Sci. Rep. 2020, 10, 1590. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, R.; Mostert, D.; Gopi, M.; Devi, P.G.; Padmanaban, B.; Molina, A.; Viljoen, A. First detection of Fusarium oxysporum f. sp. cubense tropical race 4 (TR4) on Cavendish banana in India. Eur. J. Plant Pathol. 2019, 154, 777–786. [Google Scholar] [CrossRef]
- Aguayo, J.; Cerf-Wendling, I.; Folscher, A.B.; Fourrier-Jeandel, C.; Ioos, R.; Mathews, M.C.; Mostert, D.; Renault, C.; Wilson, V.; Viljoen, A. First report of Fusarium oxysporum f. sp. cubense tropical race 4 (TR4) causing banana wilt in the Island of Mayotte. Plant Dis. 2021, 105, 219. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, A.; Mostert, D.; Chiconela, T.; Beukes, I.; Fraser, C.; Dwyer, J.; Murray, H.; Amisse, J.; Matabuana, E.L.; Tazan, G.; et al. Occurrence and spread of the banana fungus Fusarium oxysporum f. sp. cubense TR4 in Mozambique. S. Afr. J. Sci. 2020, 116, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Acuña, R.; Rouard, M.; Leiva, A.M.; Marques, C.; Olortegui, J.A.; Ureta, C.; Cabrera-Pintado, R.M.; Rojas, J.C.; Lopez-Alvarez, D.; Cenci, A.; et al. First report of Fusarium oxysporum f. sp. cubense tropical race 4 causing Fusarium wilt in Cavendish bananas in Peru. Plant Dis. 2022, 106, 2268. [Google Scholar] [CrossRef] [PubMed]
- García-Bastidas, F.A.; Quintero-Vargas, J.C.; Ayala-Vasquez, M.; Schermer, T.; Seidl, M.F.; Santos-Paiva, M.; Noguera, A.M.; Aguilera-Galvez, C.; Wittenberg, A.; Hofstede, R.; et al. First report of Fusarium wilt tropical race 4 in Cavendish Bananas caused by Fusarium odoratissimum in Colombia. Plant Dis. 2020, 104, 994–994. [Google Scholar] [CrossRef]
- Chen, A.; Sun, J.; Matthews, A.; Armas-Egas, L.; Chen, N.; Hamill, S.; Mintoff, S.; Tran-Nguyen, L.T.T.; Batley, J.; Aitken, E.A.B. Assessing variations in host resistance to Fusarium oxysporum f sp. cubense Race 4 in Musa species, with a focus on the subtropical race 4. Front. Microbiol. 2019, 10, 1062. [Google Scholar] [CrossRef]
- Warman, N.M.; Aitken, E.A.B. The movement of Fusarium oxysporum f.sp. cubense (sub-tropical race 4) in susceptible cultivars of Banana. Front. Plant Sci. 2018, 9, 1748. [Google Scholar] [CrossRef]
- Ploetz, R.C. Management of Fusarium wilt of banana: A review with special reference to tropical race 4. Crop Prot. 2015, 73, 7–15. [Google Scholar] [CrossRef]
- Salacinas, M.; Meijer, H.J.G.; Mamora, S.H.; Corcolon, B.; Mirzadi Gohari, A.; Ghimire, B.; Kema, G.H.J. Efficacy of disinfectants against tropical race 4 causing Fusarium wilt in Cavendish bananas. Plant Dis. 2022, 106, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Hamill, S.D.; Moisander, J.A.; Smith, M.K. Micropropagation of vegetatively propagated crops: accelerating release of new cultivars and providing an important source of clean planting material. Acta Hortic. 2009, 829, 213–218. [Google Scholar] [CrossRef]
- Bubici, G.; Kaushal, M.; Prigigallo, M.I.; Gómez-Lama Cabanás, C.; Mercado-Blanco, J. Biological control agents against Fusarium wilt of banana. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chaves, N.P.; Staver, C.; Dita, M.A. Potential of Trichoderma asperellum for biocontrol of Fusarium wilt in banana. Acta. Hortic. 2016, 1114, 261–266. [Google Scholar] [CrossRef]
- García-Bastidas, F.A.; Van der Veen, A.J.; Nakasato-Tagami, G.; Meijer, H.J.; Arango-Isaza, R.E.; Kema, G.H. An improved phenotyping protocol for Panama disease in banana. Front. Plant Sci. 2019, 10, 1006. [Google Scholar] [CrossRef] [PubMed]
- Houbin, C.; Chunxiang, X.; Qirui, F.; Guibing, H.; Jianguo, L.; Zehuai, W.; Molina, A.B., Jr. Screening of banana clones for resistance to Fusarium wilt in China. In Proceedings of the 3rd BAPNET meeting: advancing banana and plantain R&D in Asia and the Pacific; 2004; pp. 165–174. [Google Scholar]
- Hwang, S.C.; Ko, W.H. Cavendish banana cultivars resistant to Fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis. 2004, 88, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Mintoff, S.J.; Nguyen, T.V.; Kelly, C.; Cullen, S.; Hearnden, M.; Williams, R.; Daniells, J.W.; Tran-Nguyen, L.T. Banana cultivar field screening for resistance to Fusarium oxysporum f. sp. cubense tropical race 4 in the Northern Territory. J. Fungi 2021, 7, 627. [Google Scholar] [CrossRef]
- Smith, M.; Langdon, P.; Pegg, K.; Daniells, J. Growth, yield and Fusarium wilt resistance of six FHIA tetraploid bananas (Musa spp.) grown in the Australian subtropics. Sci. Hortic. 2014, 170, 176–181. [Google Scholar] [CrossRef]
- Zuo, C.; Deng, G.; Li, B.; Huo, H.; Li, C.; Hu, C.; Kuang, R.; Yang, Q.; Dong, T.; Sheng, O. Germplasm screening of Musa spp. for resistance to Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4). Eur. J. Plant Pathol. 2018, 151, 723–734. [Google Scholar] [CrossRef]
- D’hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef]
- Colasuonno, P.; Marcotuli, I.; Gadaleta, A.; Soriano, J.M. From genetic maps to QTL cloning: An overview for durum wheat. Plants 2021, 10, 315. [Google Scholar] [CrossRef] [PubMed]
- Remington, D.L.; Ungerer, M.C.; Purugganan, M.D. Map-based cloning of quantitative trait loci: progress and prospects. Genet. Res. 2001, 78, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, J.S.; Schwarzacher, T. Domestication, genomics and the future for banana. Ann. Bot. 2007, 100, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Fauré, S.; Noyer, J.L.; Horry, J.P.; Bakry, F.; Lanaud, C.; Gońzalez de León, D. A molecular marker-based linkage map of diploid bananas (Musa acuminata). Theor. Appl. Genet. 1993, 87, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Kayat, F.; Bonar, N.; Waugh, R.; Rajinder, S.; Rahimah, A.R.; Rashid, K.A.; Othman, R.Y. Development of a genetic linkage map for genes associated with resistance and susceptibility to Fusarium oxysporum f. sp. cubense from an F1 hybrid population of Musa acuminata ssp. malaccensis. Acta. Hortic. 2009, 828, 333–340. [Google Scholar] [CrossRef]
- Risterucci, A.M.; Hippolyte, I.; Perrier, X.; Xia, L.; Caig, V.; Evers, M.; Huttner, E.; Kilian, A.; Glaszmann, J.C. Development and assessment of Diversity Arrays Technology for high-throughput DNA analyses in Musa. Theor. Appl. Genet. 2009, 119, 1093–1103. [Google Scholar] [CrossRef]
- Ahmad, F.; Martawi, N.M.; Poerba, Y.S.; de Jong, H.; Schouten, H.; Kema, G.H.J. Genetic mapping of Fusarium wilt resistance in a wild banana Musa acuminata ssp. malaccensis accession. Theor. Appl. Genet. 2020, 133, 3409–3418. [Google Scholar] [CrossRef]
- Hippolyte, I.; Bakry, F.; Seguin, M.; Gardes, L.; Rivallan, R.; Risterucci, A.M.; Jenny, C.; Perrier, X.; Carreel, F.; Argout, X.; et al. A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant Biol. 2010, 10, 65. [Google Scholar] [CrossRef]
- Uwimana, B.; Mwanje, G.; Batte, M.; Akech, V.; Shah, T.; Vuylsteke, M.; Swennen, R. Continuous mapping identifies loci associated with weevil resistance [Cosmopolites sordidus (Germar)] in a triploid banana population. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Chen, A.; Sun, J.; Martin, G.; Gray, L.-A.; Hřibová, E.; Christelová, P.; Yahiaoui, N.; Rounsley, S.; Lyons, R.; Batley, J.; et al. Identification of a major QTL-controlling resistance to the subtropical race 4 of Fusarium oxysporum f. sp. cubense in Musa acuminata ssp. malaccensis. Pathogens 2023, 12, 289. [Google Scholar] [CrossRef] [PubMed]
- Fraser-Smith, S.; Czislowski, E.; Daly, A.; Meldrum, R.; Hamill, S.; Smith, M.; Aitken, E.A.B. Single gene resistance to Fusarium oxysporum f. sp. cubense race 4 in the wild banana Musa acuminata subsp. malaccensis. Acta. Hortic. 2016, 1114, 95–100. [Google Scholar] [CrossRef]
- Christelová, P.; De Langhe, E.; Hřibová, E.; Čížková, J.; Sardos, J.; Hušáková, M.; Van den houwe, I.; Sutanto, A.; Kepler, A.K.; Swennen, R.; et al. Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 2017, 26, 801–824. [Google Scholar] [CrossRef]
- Zhang, L.; Yuan, T.; Wang, Y.; Zhang, D.; Bai, T.; Xu, S.; Wang, Y.; Tang, W.; Zheng, S.-J. Identification and evaluation of resistance to Fusarium oxysporum f. sp. cubense tropical race 4 in Musa acuminata. Pahang. Euphytica 2018, 214, 106. [Google Scholar] [CrossRef]
- Moore, N.Y.; Pegg, K.; Smith, L.; Langdon, P.W.; Bentley, S.; Smith, M.K. Fusarium wilt of banana in Australia. In Banana Fusarium Wilt Management: Towards Sustainable Cultivation; Malaysia, 2001; pp. 64–75. [Google Scholar]
- Li, W.M.; Dita, M.; Wu, W.; Hu, G.B.; Xie, J.H.; Ge, X.J. Resistance sources to Fusarium oxysporum f. sp. cubense tropical race 4 in banana wild relatives. Plant Pathol. 2015, 64, 1061–1067. [Google Scholar] [CrossRef]
- Madalla, N.A.; Swennen, R.; Brown, A.F.; Massawe, C.; Shimwela, M.; Mbongo, D.; Kindimba, G.; Kubiriba, J.; Tumuhimbise, R.; Okurut, A.W.; et al. Yield stability of East African highland cooking banana ‘Matooke’ hybrids. J. Am. Soc. Hortic. Sci. 2022, 147, 334–348. [Google Scholar] [CrossRef]
- Wang, X.; Yu, R.; Li, J. Using genetic engineering techniques to develop banana cultivars with Fusarium wilt resistance and ideal plant architecture. Front. Plant Sci. 2021, 11. [Google Scholar] [CrossRef]
- Martin, G.; Carreel, F.; Coriton, O.; Hervouet, C.; Cardi, C.; Derouault, P.; Roques, D.; Salmon, F.; Rouard, M.; Sardos, J.; et al. Evolution of the banana genome (Musa acuminata) is impacted by large chromosomal translocations. Mol. Biol. Evol. 2017, 34, 2140–2152. [Google Scholar] [CrossRef]
- Rijzaani, H.; Bayer, P.E.; Rouard, M.; Doležel, J.; Batley, J.; Edwards, D. The pangenome of banana highlights differences between genera and genomes. Plant Genome 2022, 15, e20100. [Google Scholar] [CrossRef]
- Rouard, M.; Droc, G.; Martin, G.; Sardos, J.; Hueber, Y.; Guignon, V.; Cenci, A.; Geigle, B.; Hibbins, M.S.; Yahiaoui, N.; et al. Three new genome assemblies support a rapid radiation in Musa acuminata (wild banana). Genome Biol. Evol. 2018, 10, 3129–3140. [Google Scholar] [CrossRef]
- Rouard, M.; Sardos, J.; Sempéré, G.; Breton, C.; Guignon, V.; Van den Houwe, I.; Carpentier, S.C.; Roux, N. A digital catalog of high-density markers for banana germplasm collections. Plants people planet 2022, 4, 61–67. [Google Scholar] [CrossRef]
- Sardos, J.; Rouard, M.; Hueber, Y.; Cenci, A.; Hyma, K.E.; van den Houwe, I.; Hribova, E.; Courtois, B.; Roux, N. A genome-wide association study on the seedless phenotype in banana (Musa spp.) reveals the potential of a selected panel to detect candidate genes in a vegetatively propagated crop. PLoS One 2016, 11, e0154448. [Google Scholar] [CrossRef] [PubMed]
- Martínez, G.; Olivares, B.O.; Rey, J.C.; Rojas, J.; Cardenas, J.; Muentes, C.; Dawson, C. The advance of Fusarium wilt tropical race 4 in Musaceae of Latin America and the Caribbean: Current Situation. Pathogens 2023, 12, 277. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Schachermayr, G.; Keller, B. Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J. 1997, 11, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Dardick, C.; Schwessinger, B.; Ronald, P. Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr. Opin. Plant Biol. 2012, 15, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Bleecker, A.B. Expansion of the Receptor-Like Kinase/Pelle Gene Family and Receptor-Like Proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Yang, Y.; Zheng, H.; Han, X.; Jin, H.; Xiong, Z.; Qian, W.; Xia, L.; Ji, X.; Li, G.; et al. Efficient expression and function of a receptor-like kinase in wheat powdery mildew defence require an intron-located MYB binding site. Plant Biotechnol. J. 2021, 19, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, S.; Deng, Z.; Wang, X.; Chen, T.; Zhang, J.; Chen, S.; Ling, H.; Zhang, A.; Wang, D.; et al. Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. Plant J. 2007, 52, 420–434. [Google Scholar] [CrossRef]
- Lim, C.W.; Yang, S.H.; Shin, K.H.; Lee, S.C.; Kim, S.H. The AtLRK10L1.2, Arabidopsis ortholog of wheat LRK10, is involved in ABA-mediated signaling and drought resistance. Plant Cell Rep. 2015, 34, 447–455. [Google Scholar] [CrossRef]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Hammond-Kosack, K.E.; Tang, S.; Harrison, K.; Jones, J.D. The tomato Cf-9 disease resistance gene functions in tobacco and potato to confer responsiveness to the fungal avirulence gene product avr9. Plant Cell 1998, 10, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Avni, A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 2004, 16, 1604–1615. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Wang, N.; Pi, L.; Li, L.; Duan, W.; Wang, X.; Dou, D. Nicotiana benthamiana LRR-RLP NbEIX2 mediates the perception of an EIX-like protein from Verticillium dahliae. J. Integr. Plant Biol. 2021, 63, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Saintenac, C.; Cambon, F.; Aouini, L.; Verstappen, E.; Ghaffary, S.M.T.; Poucet, T.; Marande, W.; Berges, H.; Xu, S.; Jaouannet, M.; et al. A wheat cysteine-rich receptor-like kinase confers broad-spectrum resistance against Septoria tritici blotch. Nat. Commun. 2021, 12, 433. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Sun, J.; Liu, N.; Sun, X.; Liu, C.; Wu, L.; Liu, G.; Zeng, F.; Hou, C.; Han, S.; et al. A novel cysteine-rich receptor-like kinase gene, TaCRK2, contributes to leaf rust resistance in wheat. Mol. Plant Pathol. 2020, 21, 732–746. [Google Scholar] [CrossRef]
- Acharya, B.R.; Raina, S.; Maqbool, S.B.; Jagadeeswaran, G.; Mosher, S.L.; Appel, H.M.; Schultz, J.C.; Klessig, D.F.; Raina, R. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 2007, 50, 488–499. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Boller, T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Huffaker, A.; Pearce, G.; Ryan, C.A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef]
- Huffaker, A.; Ryan, C.A. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA 2007, 104, 10732–10736. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Schwessinger, B.; Joe, A.; Thomas, N.; Liu, F.; Albert, M.; Robinson, M.R.; Chan, L.J.; Luu, D.D.; Chen, H.; et al. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 2015, 1, e1500245. [Google Scholar] [CrossRef] [PubMed]
- van Dam, S.; Võsa, U.; van der Graaf, A.; Franke, L.; de Magalhães, J.P. Gene co-expression analysis for functional classification and gene–disease predictions. Brief. Bioinform. 2017, 19, 575–592. [Google Scholar] [CrossRef] [PubMed]
- van Westerhoven, A.C.; Meijer, H.J.G.; Seidl, M.F.; Kema, G.H.J. Uncontained spread of Fusarium wilt of banana threatens African food security. PLoS Pathog. 2022, 18, e1010769. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, T.D.M.; Rego, E.C.S.; Alves, G.S.C.; Fonseca, F.C.A.; Cotta, M.G.; Antonino, J.D.; Gomes, T.G.; Amorim, E.P.; Ferreira, C.F.; Costa, M.; et al. Transcriptome profiling of the resistance response of Musa acuminata subsp. burmannicoides, var. Calcutta 4 to Pseudocercospora musae. Int. J. Mol. Sci. 2022, 23, 3589. [Google Scholar] [CrossRef]
- Wilberforce, T.; Batte, M.; Nyine, M.; Tumuhimbise, R.; Alex, B.; Ssali, R.; Talengera, D.; Kubiriba, J.; Lorenzen, J.; Swennen, R.; et al. Performance of NARITA banana hybrds in the preliminary yield trial for three cycles in Uganda. 2017. [Google Scholar] [CrossRef]
- Batte, M.; Nyine, M.; Uwimana, B.; Swennen, R.; Akech, V.; Brown, A.; Hovmalm, H.P.; Geleta, M.; Ortiz, R. Significant progressive heterobeltiosis in banana crossbreeding. BMC Plant Biol. 2020, 20, 489. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, R.; Saraswathi, M.S.; Uma, S.; Loganathan, M.; Backiyarani, S.; Durai, P.; Raj, E.E.; Marimuthu, N.; Kannan, G.; Swennen, R. Identification of sources resistant to a virulent Fusarium wilt strain (VCG 0124) infecting Cavendish bananas. Sci. Rep. 2021, 11, 3183. [Google Scholar] [CrossRef]
- Noordin, N.; Hassan, A.A.; Mahadevan, A.N.M.F.; Ahmad, Z.; Ariffin, S. Lab-based screening using hydroponic system for the rapid detection of Fusarium wilt TR4 tolerance/resistance of banana. In Efficient Screening Techniques to Identify Mutants with TR4 Resistance in Banana: Protocols; Jankowicz-Cieslak, J., Ingelbrecht, I.L., Eds.; Springer: Berlin, Heidelberg, 2022; pp. 79–95. [Google Scholar]
- Wu, Y.; Huang, B.; Peng, X.; Zhang, J. Development of an in vitro hydroponic system for studying the interaction between banana plantlet and Fusarium oxysporum f. sp. cubense. Plant Cell, Tissue Organ Cult. 2021, 146, 107–114. [Google Scholar] [CrossRef]
- Cong, L.L.; Sun, Y.; Wang, Z.; Kang, J.M.; Zhang, T.J.; Biligetu, B.; Yang, Q.C. A rapid screening method for evaluating resistance of alfalfa (Medicago sativa L.) to Fusarium root rot. Can. J. Plant Pathol. 2018, 40, 61–69. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Kidd, B.N.; Singh, K.B. Tools and strategies for genetic and molecular dissection of Medicago truncatula resistance against Fusarium wilt disease. In The Model Legume Medicago truncatula; 2020; pp. 331–339. [Google Scholar]
- Prasath, D.; Matthews, A.; O’Neill, W.T.; Aitken, E.A.B.; Chen, A. Fusarium yellows of ginger (Zingiber officinale Roscoe) caused by Fusarium oxysporum f. sp. zingiberi is associated with cultivar-specific expression of defense-responsive genes. Pathogens 2023, 12, 141. [Google Scholar] [CrossRef]
- Ndayihanzamaso, P.; Mostert, D.; Matthews, M.C.; Mahuku, G.; Jomanga, K.; Mpanda, H.J.; Mduma, H.; Brown, A.; Uwimana, B.; Swennen, R.; et al. Evaluation of Mchare and Matooke bananas for resistance to Fusarium oxysporum f. sp. cubense race 1. Plants 2020, 9, 1082. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, A.; Mahuku, G.; Ssali, R.; Kimunye, J.; Mostert, D.; Ndayihanzamaso, P.; Coyne, D. Banana diseases and pests: field guide for diagnostics and data collection; International Institute of Tropical Agriculture: Ibadan, Nigeria, 2017. [Google Scholar]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Lorenc, M.T.; Hayashi, S.; Stiller, J.; Lee, H.; Manoli, S.; Ruperao, P.; Visendi, P.; Berkman, P.J.; Lai, K.; Batley, J. Discovery of single nucleotide polymorphisms in complex genomes using SGSautoSNP. Biol. 2012, 1, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 2014, 10, 21. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinformatics. 2015, 51, 11.14.11–11.14.19. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.-C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-seq Data from the NCI Patient-Derived Models Repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
| Primer name | Primer sequence (5' to 3') | T (°C) | Frag (bp) | Cut by | Cut sizes (bp) | SNPATG |
|---|---|---|---|---|---|---|
| 27960-SNP1-F1 | GACCAGCAGCAGAAGGACCAGACC | 58 | 764 | BsaI | R:764 | Exon1 |
| 27960-SNP1-R1 | AGAATGAGTGGTATGGGAT | S:394,370 | T1152C | |||
| 28220-SNP8-F1 | CCTGATTGTAAATGGGAAGTTTCTC | 56 | 546 | MnlI | R:292,223,31m | Intron2 |
| 28220-SNP8-R1 | ATCGCCCAGCAGTGATTTGA | S:515,31m | G3100A | |||
| 28420-SNP1-F1 | CAAATATGCTGCTCCATCTG | 54 | 740 | NsiI | R:740 | Intron4 |
| 28420-SNP1-R1 | CTTGGAAGAAACTAACGAGTGT | S:403,337 | A2547G | |||
| 28820-SNP8-F2 | CAGGTAACCATTTAGACTGACAA | 55 | 544 | BstZ17I | R:544 | Exon3 |
| 28820-SNP8-R1 | AATCAAGGAAATAGGGTGGCAC | S:300,244 | C3274T | |||
| 29460-SNP21-F2 | GGATACTTGGACCCTGAGTACCAT | 58 | 344 | XhoI | R:313,31m | Exon4 |
| 29460-SNP21-R1 | CCATCGCTCTCTATTGCTTGC | S:178,135,31m | T6353C | |||
| 29590-SNP1-F1 | GCTCAGATGTCTCAGTCCAGA | 55 | 457 | BstNI | R:457 | Exon1 |
| 29590-SNP1-R1 | CTTCTTCCATCCTCTTCTCC | S:317,140 | A137G | |||
| 29670-SNP8-F1 | AAGAGATGTCATGTTGGTTCATTTG | 56 | 628 | BspCNI | R:628 | Intron5 |
| 29670-SNP8-R1 | CACTCACTCCTGCTATGCGGTTG | S:345,283 | G5078C | |||
| 29730-SNP1-F1 | ATGGCACAGGTGATGTCAGT | 58 | 686 | BcoDI | R:686 | Intron1 |
| 29730-SNP1-R1 | ACTAGATGACTCAGATTAGTAGG | S:359,327 | T544C | |||
| 29730-A-SNP1-F2 | GCAATGAGTACCTCTAAGCA | 56 | 795 | BcoDI | R:795 | Intron1 |
| 29730-A-SNP1-R2 | TAAGTTCTAGTATCAAGTACAA | S:429,366 | T544C | |||
| 29850-SNP13-F2 | CTTGTTCCTGTTACCTATTAG | 56 | 363 | StyI | R:363 | Intron5 |
| 29850-SNP13-R1 | CCTTGTGCCTAGATGCTTGG | S:192,171 | A4287G | |||
| 29930-SNP1-F2 | GTTCACACCCTTGACATCCTA | 54 | 493 | MseI | R:190,64,99m,49m,36m,30m,25m | Intron4 |
| 29930-SNP1-R1 | TAAGCATTCATTAGCAAACGG | S:254,99m,49m,36m,30m,25m | A3401G | |||
| 30000-SNP2-F2 | CTTAAAACTTGGCGGAAGG | 56 | 468 | NsiI | R:251,217 | Exon14 |
| 30000-SNP2-R2 | CTGAAGCACAACTGTCCTTG | S:468 | A6749G |
| ‘DH-Pahang’ v1(GSMUA_Achr3G) | ‘DH-Pahang’ v4(Macma4_03_g) | ‘DH-Pahang’ v4 Position (bp) | Description |
|---|---|---|---|
| 27960 | 30750 | 40,893,205–40,895,172 (-) | MHD domain-containing protein |
| 28220 | 31030 | 41,068,780–41,075,115 (-) | Uncharacterized membrane protein At1g16860-like |
| 28420 | 31200 | 41,183,294–41,197,461 (-) | F-box domain-containing protein |
| 28820 | 31680 | 41,695,490–41,699,989 (+) | Bifunctional nuclease 2 |
| 29460 | 32270 | 42,052,018–42,058,909 (+) | Leaf rust 10 disease-resistance locus receptor-like protein kinase-like 1.3 |
| 29590 | 32440 | 42,138,268–42,142,592 (-) | Pentatricopeptide repeat-containing protein At4g28010 |
| 29670 | 32510 | 42,186,029–42,193,520 (-) | Cycloartenol-C-24-methyltransferase 1 |
| 29730 | 32560 | 42,210,035–42,215,274 (-) | Nuclear transcription factor Y subunit A-1 |
| 29850 | 32690 | 42,283,482–42,289,346 (+) | WRKY transcription factor SUSIBA2 |
| 29930 | 32770 | 42,323,762–42,327,884 (-) | Hypothetical protein |
| 30000 | 32830 | 42,349,497–42,357,604 (-) | Long chain base biosynthesis protein 2d |
| Line | Name (Subspecies/genome) | Accession | 29730 marker locus | Foc-STR4 | Foc-TR4 |
|---|---|---|---|---|---|
| 1a | 'Ma850' (malaccensis) | MRF850 | + (Het) | R [33] | R [33,58] |
| 2a | 'Ma851' (malaccensis) | MRF851 | + (Het) | R [33] | R [58] |
| 3a | 'Ma852' (malaccensis) | MRF852 | + (Het) | R [33] | R [58] |
| 4a | 'Ma845' (malaccensis) | MRF845 | - | n/a | n/a |
| 5a | 'Ma846' (malaccensis) | MRF846 | - | S [33] | n/a |
| 6a | 'Ma848' (malaccensis) | MRF848 | - | S [33] | S [33,58] |
| 7a | 'Pahang' (malaccensis) | MRF1649 | + | R [33] | R [33,45] |
| 8a | 'SH-3362' (AA) | MRF2010 | + (Het) | R [33] | R [33,43] |
| 9a | 'SH-3362' (AA) | MRF2013 | + (Het) | R [33] | R [33,43] |
| 10a | ‘Madang Guadeloupe’(malaccensis) | MRF655 | + | R [33] | R [33] |
| 11a | 'Calcutta 4' (burmannica) | MRF1642 | - | R [33] | R [33,45] |
| 12a | 'SH-3217' (AA) | MRF2005 | + | R [33] | R [33,43] |
| 13a | 'IV9 Calcutta4' (AA) | MRF526 | - | R [33] | R [33] |
| 14a | 'Pisang Jari Buaya' (AA) | MRF1244 | - | R [33] | R [33,45] |
| 15a | 'Ma-ITC0250' (malaccensis) | MRF826 | + (Het) | R [33] | R [33] |
| 16a | 'M61 Guadeloupe' (AA) | MRF654 | - | SS [33] | R [33] |
| 17a | 'CAM-020' (AA) | MRF1657 | - | S [33] | R [33] |
| 18a | 'SH-3142' (AA) | MRF1984 | - | R [33] | R [33,43] |
| 19a | M. a. malaccensis | ITC0399 | + (Het) | n/a | n/a |
| 20a | 'Pahang' (malaccensis) | ITC0609 | + | R [33] | R [33,40,45,60] |
| 21b | 'Pa Musore no2' (M. acuminata spp.) | ITC0668 | + (Het) | n/a | n/a |
| 22b | 'Kluai Pal' (malaccensis) | ITC0979 | + (Het) | n/a | n/a |
| 23b | 'DH Pahang' (malaccensis) | ITC1511 | + | n/a | R [45,46] |
| 24b | M a. malaccensis | ITC0074 | - | n/a | n/a |
| 25b | 'Pa Musore no3' (M. acuminata spp.) | ITC0406 | - | n/a | n/a |
| 26b | 'Pa_Songkhla' (M. acuminata spp.) | ITC0408 | - | n/a | n/a |
| 27b | 'Selangor 2' (malaccensis) | ITC0629 | - | n/a | n/a |
| 28b | 'Pisang Raja Udang' (AA) | ITC0976 | - | n/a | n/a |
| 29b | 'THA018' (malaccensis) | ITC1067 | - | n/a | n/a |
| 30b | 'Pisang Kra' (malaccensis) | ITC1345 | - | n/a | n/a |
| 31b | 'Pisang Serun 403' (malaccensis) | ITC1347 | - | n/a | n/a |
| 32b | 'Pisang Serun 404' (malaccensis) | ITC1348 | - | n/a | n/a |
| 33b | 'Pisang Serun 400' (malaccensis) | ITC1349 | - | n/a | n/a |
| 34b | 'IB-99' | ITC1447 | - | n/a | n/a |
| 35c | 'TMB2×7197-2' (AA) | - | + (Het) | n/a | n/a |
| 36c | '5610S-1' (AA) | - | + (Het) | n/a | n/a |
| 37c | 'SH-3362' (AA) | MUSA214 | + (Het) | R [33] | R [33] |
| 38c | 'Malaccensis 250' (malaccensis) | ITC0250 | + (Het) | R [33] | n/a |
| 39c | 'SH-3217' (AA) | MMC218 | + | R [33] | R [43] |
| 40c | 'SH-3361' (AA) | - | + (Het) | n/a | n/a |
| 41c | 'TMB2×8075-7' (AA) | - | - | n/a | n/a |
| 42c | 'Hutishamba' (AA) | MMC486 | - | n/a | n/a |
| 43c | 'Mshare Laini' (AA) | - | - | n/a | n/a |
| 44c | 'cv. Rose' (AA) | ITC0712 | - | n/a | R [40,41] |
| 45c | 'Mularu' (AA) | MMC465 | - | n/a | n/a |
| 46c | 'Kamunyila' (AA) | MMC479 | - | n/a | n/a |
| 47c | 'Mlelembo' (AA) | ITC1544 | - | n/a | n/a |
| 48c | 'Njuru' (AA) | MMC418 | - | n/a | n/a |
| 49c | 'Kahuti' (AA) | ITC1468 | - | n/a | n/a |
| 50c | 'Mbwazirume' (AAA) | ITC0084 | - | n/a | R [45] |
| 51c | 'Sukari Ndiizi' (AAB) | MMC167 | - | n/a | n/a |
| 52c | 'Nshonowa' (AA) | ITC1466 | - | n/a | n/a |
| 53d | 'FHIA-3' (AABB) | MRF1941 | + (Het) | S [33,61] | SS [33] S [41] R [43] |
| 54d | 'FHIA-25' (AAB) | MRF1960 | + | R [33] | R [33,43,45] |
| 55d | 'FHIA-21' (AAAB) | MRF1205 | - | n/a | S [41],R [45] |
| 56d | 'FHIA-23' (AAAA) | MRF1207 | - | S [33] | SS [33], S [41] |
| 57d | 'GCTCV-119' (AAA) | MRF1860 | - | R [33] | R [33,41] |
| 58d | 'FHIA-2' (AAAB) | MRF1933 | - | S [33,61] | R [33,43] S [41] |
| 59d | 'FHIA-1'/'Goldfinger' (AAAB) | MRF1959 | - | R [33] | R [33,43] S [62] |
| 60d | Musa balbisiana (BB) | MRF1593 | - | S [33] | S [62] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





