Introduction
The cell life-based evolution has created immensely complex molecular information structures based on the RNA activity(Delihas, 2006; Higgs and Lehman, 2015). RNA is capable of programming accurate and diverse cell function across a multitude of body plans and “re-wireable” organs of intricate cellular arrangement such as the brain. The role of the RNA has been indispensable in the refined differentiation processes(Mattick and Amaral, 2022). Yet, the pre-cell “chemical” or “abiotic” selection and enhancement of “life”-related molecular functions remains as most enigmatic(Joyce, 1989; Lazcano and Miller, 1996; Cech, 2012). Likely, it will never be possible to know all of the circumstances of that time with certainty, which justifies the use of our current knowledge in extrapolations of the possible evolutionary paths. We indeed have very little idea in what chemophysical environment this initial evolution has occurred in and where it took place. Judging from the number of energy-consuming proteins that facilitate “semi-molten” and apparently “working” state of RNA preventing its excessive structural collapse in the current life(Lüking et al., 1998; Rajkowitsch et al., 2007; Van Treeck and Parker, 2018), perhaps it was a hotter place(Oparin, 1953), or a one with a somewhat lesser emphasis on hydrophobic interactions, compared to the existing major molecular environments. Intriguingly, this early evolution has now been also confidently linked to the function of RNA.
It is fascinating how RNA has carried through some of the most demanding, complex and energy-dependent functions, without being overly replaced by the proteins. However, the current complexity of live systems unlikely can be imagined without the protein function. Thus, outlining the possibilities of the ribosome emergence can clarify some aspects of the origin of life, and is necessary for the understanding of the path of chemical and biological evolution.
In a stark contrast to most other known enzymatic systems, the ribosome as we know it operates in a highly specific way on a broad diversity of the substrates, including highly unspecific substrates such as messenger (m)RNA. Large structural features of these substrates, such as overall shape and charge distribution in transfer (t)RNAs, are used to outclass smaller but still discriminating and important features, such as the selected and attached amino acid residue, to achieve these functions. Furthermore, ribosome is completely programmable with the mRNA code, which represents its nearly completely unspecific, in regard to the sequence, substrate. Over 100 transfer RNAs are interacting with multiple aminoacyl-tRNA synthetases (ARSes) outside of the ribosome to complete the action(Schimmel, 1987; Beuning and Musier-Forsyth, 1999; Ibba and Söll, 2000; Gomez and Ibba, 2020). One of the main questions of biology is how such an interconnected and deeply self-dependent system could have emerged. For such a complex system, can we hypothesise a chemical or biological path of the ribosomal evolution that would not start with a complicated and coincidental aboriginal appearance.
We can rely on our current knowledge of the ribosome structure and functionality, and try outlining its possible functions in a (presumably) much simpler primordial environment. Using this approach, we can investigate a parsimonious route to resolve the notable challenges to the RNA. One critical challenge is the (possible) initial detachment of “live” systems from Darwinian evolution, and subsequent transition into it. Here we provide a perspective on a smooth, uninterrupted evolutionary thread of the ribosome (and consequently, life), in best adherence to the principles of Jacob’s interpretation of Darwinism(Jacob, 1977; Bowman et al., 2015).
Early link of replication with the mechanisms of replication enhancement
An apparent requirement of any sequential improvement, and a de facto definition of the evolution process is a step-wise generation of close copies of the original system. As such, it is impossible to detach the emergence of the archetypal ribosome (protoribosome in the context of this discussion) from the function of RNA replication. The protoribosome has thus been either directly part of the replicase structure, or co-evolved and accelerated improvement of the replication. The protoribosome thus was, one way or another, a part or function of a “riboreplisome”. As unfolded further, we can approach an understanding why riboreplisome could be a fundamental driving principle of the emergence and evolution of the cellular life.
A simple RNA ribozyme replicase-based, RNA plus and minis strand alteration is almost invariably suggested as one of the most straightforward possibilities to consider in self-sustained RNA system(Ichihashi and Yomo, 2016). Here we can strongly rely on a possibility of an emergence of RNA ligase-based ribozyme, as has been shown in synthetic evolution experiments to achieve partial and also more complete polymerisation(Ekland and Bartel, 1996; Johnston et al., 2001; Wochner et al., 2011; Attwater et al., 2018; Tjhung et al., 2020). Importunity, the original RNA ligase in this case was also artificially selected by using random short RNA sequences rather than derived from a natural pre-existing ribozyme, which proves a defined possibility of its random occurrence(Bartel and Szostak, 1993; Ekland et al., 1995). While we certainly know that RNA can catalyse RNA-depended replication and the possible aboriginal RNA protoreplisome may have been fully self-contained(Root-Bernstein and Root-Bernstein, 2015; Tjhung et al., 2020), what could have been the major constraints for such a system? It is possible to name a few.
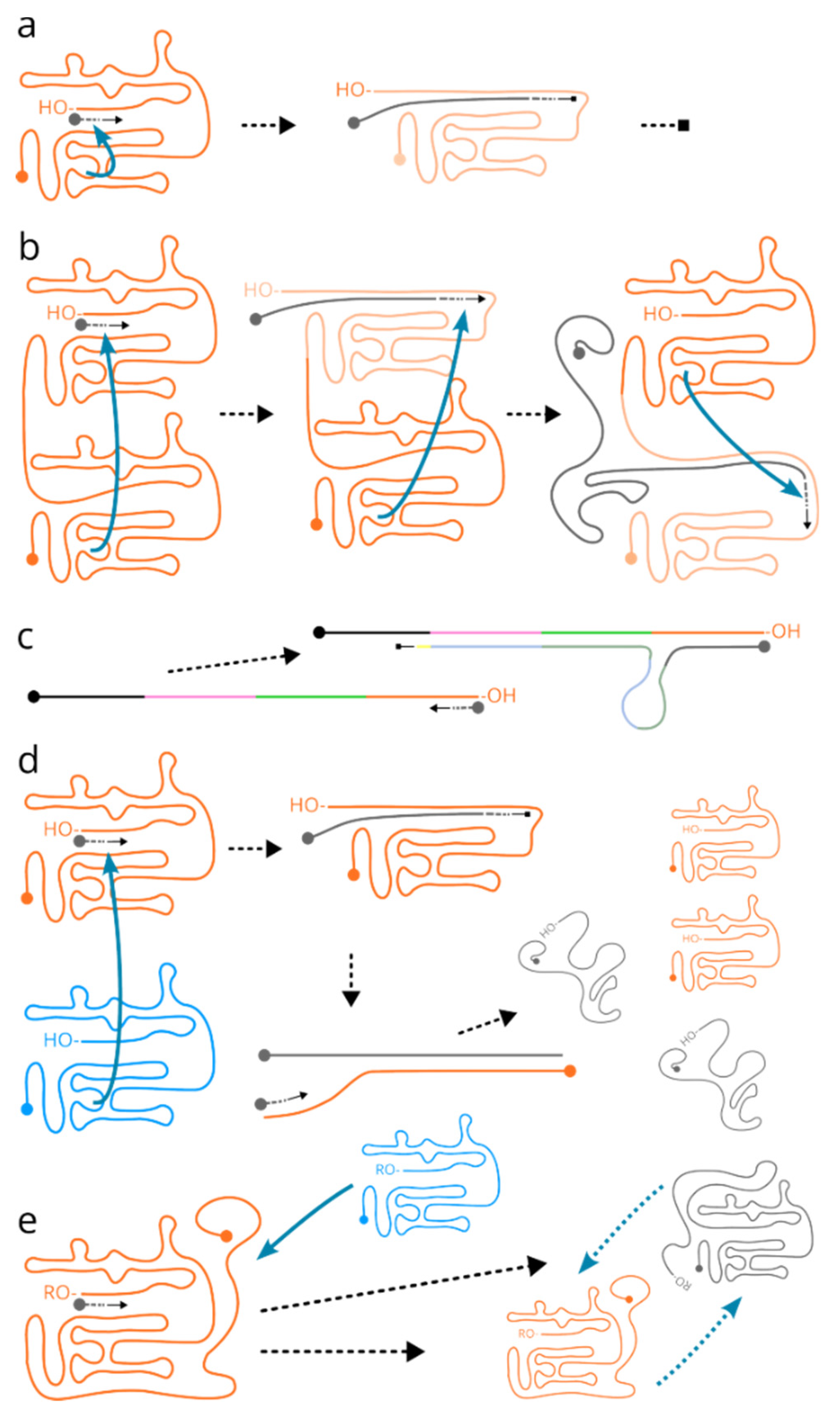
First, complete self-replication in a fully individualised, single-molecule sense is problematic, while not entirely impossible. Inevitably at some point during the replication the polymerase domain would need to be unfolded and replicated too (
Figure 1a), in which case it cannot be a simple
in cis self-replication. Thus, the possible solutions are that the riboreplisome would have two replication domains, handing in the substrate from one to another to maintain the full-length information (
Figure 1b). This mechanism explains an efficient generation of the reverse complement (“minus”) strands, but still requires
in trans interactions later on to convert these into the “positive” strands once again (with some exceptions as outlined further). Another variant of self-multiplication could be tolerant towards incomplete replication by containing multiple copies of the replicase, which could be used in a slippage-like mechanism with relaxed control to produce longer versions that would not functionally suffer from incomplete replication(Canceill et al., 1999; Zhang et al., 2020) (
Figure 1c). The alternative is that replication is performed always
in trans (
Figure 1d,e). Single-molecule
in cis replication has the advantages of “built-in” containment and thus is resistant to dilution, but there can be substantial difficulties such as self-inactivation by forming a duplex with the negative strand. Clearly, some cycling of external conditions, particularly temperature, could have partially resolved duplex-induced shutdown, but it would put replication in dependence of the required environmental factor cycling, which presents a clear disadvantage against any other system that does not have such a requirement.
In trans replication, on the other hand, is a potent way of decomplicating replication chemistry and structure, but requires a close interaction with a molecular “partner”. Thus,
in trans-catalysed replication of the riboreplisome, while more likely due to its simplicity, would require a special means of “substrate” interaction and capture. It is quite conceivable that both
in cis and
in trans variants existed in parallel for some time.
Second, as there is no strict evolutionary pressure to keep the replication-enhancing protoribosome component of the riboreplisome in both strands, it is safe to assume it could have emerged and was preserved in just one type of these (
Figure 1d). Finally, an interesting hack to the situation with the “positive” and “negative” riboreplisome strands is the possibility of the protoreplisome to be an inverted copy (
Figure 1e). Such inverted copy would be also active in the “negative” strand, which in this case provides a solution to the maintenance of an interacting partner even under the conditions of low concentration. We find many evidences of a similarly-arranged systems across the current life, such as in inverted terminal repeats of the DNA viruses. Such riboreplisome appears as fully self-contained and resistant to any dilutions, and thus offers a clear and powerful evolutionary advantage. Because the enzymatic products of the protoribosome component would at some point of the evolution be in any case released into the solution (see later in “
Emergence of the current ribosomal organisation as the possible converging point”), even in this scenario it is likely that only one of the strands encoded the active protoribosome, offering a generational cycle to the system, as well as a first “gene” that needs to be “expressed” and which expression can be controlled in adjustment to environmental demands by selecting which of the partner strands has a more active replicase. It is fascinating that these purely molecular interactions bear a striking resemblance to the different aspects of reproduction, including “sexes”, cases of “hermaphroditism” and “alteration of generations”, in different combinations, as well as feedback-based cellular gene control.
As we favour the in trans (in single-molecule sense) replication, we assume that there was some selection towards developing of means of binding the replication partner in the replication catalytic centre, along with replication priming, which could have occurred most easily by nucleotide complementarity interactions, and is a feature which importance will further become more apparent. Interestingly, indeed it has been shown that such interactions appear very quickly during in vitro evolution of self-replicating RNA ribozymes, and bear notable and fascinating resemblence of the Shine-Dalgarno:anti-Shine-Dalgarno sequence interactions(Wochner et al., 2011).
Theories of ribosomal organisation and emergence
An array of works is now available in the literature theorising on the origin and evolution of the ribosome, and providing experimental proofs to certain concepts and paths of its possible emergence. In the early endeavours, it has been noted that the precursors to the translational machinery would have been far less complex and far more error-prone(Woese, 1965), and that it is likely that no “activating enzymes” (aminoacyl-tRNA synthetases) existed(Woese et al., 1966). It further has been noted that the current state of the translational machinery remained virtually unaltered for at least about a billion of years, and the early “translation” would involve a limited set of active marcomolecules (that is, RNAs), perhaps just a single one(Woese et al., 1966). The most interesting thought, in light of the currently-remaining problems, is that “translation” may not have started as a strictly templated process in the classical understanding, and thus there has been no distinction between the functions of the rRNA, mRNA and tRNA(Woese et al., 1966).
It has been soon noted that prokaryotic and eukaryotic rRNAs are distinct with eukaryotic rRNA being larger, and this size difference has been linked to the larger systemic complexity of eukaryotes(Loening, 1968). Ribosomal protein gene dispersal and sequential inactivation in symbiotic prokaryotes (proto-mitochondria and proto-palstids) were suggested as part of the eukaryotic genome and ribosome evolution(Bogorad, 1975). Additional rRNA chain breaks, such as the “hidden break” in 28S rRNA of certain cladi (e.g. insects) were described – however the functional significance of this phenomena is still uncertain(Ishikawa, 1977; Natsidis et al., 2019; Lake, 1983). Quite notably it was recognised early on that ribosomal proteins diverge substantially between the domains of life, and while there might be about 3,000 amino acids from the structurally-invariable ribosomal protein “core” (which nonetheless not always are conserved in sequence), there are >6,000 amino acids that comprise domain-variable or unique sequences(Wittmann, 1980; Nierhaus et al., 1987; Taylor et al., 2009; Melnikov et al., 2018).
Linking large-scale features of the ribosome to the divergence of the domains of life, it was suggested that the original “paleocyte” could contain eukaryotic and archaeal SSU “beak” but not eukaryotic “lobes”, with the beak subsequently lost in eubacteria(Lake et al., 1982). In the early works a high overall evolutionary flexibility of certain rRNA:protein interacting regions of the ribosome was identified, for example it was proposed that protein partners of the 5S rRNA underwent fusions and splitting over the evolution while, however, keeping the overall molecular weight of the complex relatively consistent. These 5S rRNA:protein transformations also pointed to the likely relationship of early eukaryotes with extreme halophiles(Nazar et al., 1982). Furthermore, it was noticed that symbiotic (mitochondrial) ribosomal proteins, and thus the ribosomes, evolve at a much higher rate than the cytoplasmic counterparts – and can be one of the more rapid genetic respondents of environmental adaptation(Pietromonaco et al., 1986; Wittmann-Liebold, 1986).
The initial structural and phylogenetic advances in the understanding of ribosome, and the deciphering and initial analyses of the genetic code, were firmly summarised in a view that postulated “spontaneous” interaction of short RNA fragments with functions resembling mRNA and tRNA(Woese, 1965, 1970; Woese et al., 1966; Crick et al., 1976; Clark, 1987). So, the “proto-mRNA” would interact with the “proto-tRNA”, whereas “proto-rRNA” fragments would emerge, over time, as the catalysts of this process(Clark, 1987). Furthermore, the in vitro reconstitution studies have identified ribosomal proteins that can initiate rRNA binding, such as L3 and L24 of the bacterial LSU, also providing a pathway for the understanding of sequential “proteinisation” of the ribosome(Nierhaus et al., 1987). Perhaps, one of the most striking findings has been the proposal that several “core” proteins involved in the modulation of the accuracy of the ribosome (S4, S5 and S12 in bacteria vs. S4, S13 and S28 in eukaryotes) were established very early, over 2 billion years ago, and then co-evolved their sequences while preserving key structural interactions(Alksne et al., 1993). This can be a rather conservative estimate, as early cyanobacteria imprints are dated at 3-3.5 billion years ago, suggesting a complete ribosomal function by then(Schopf and Packer, 1987; Pflug, 1986; Garrett, 1999).
With more advances and insight into the accurate structure of the ribosome and its dynamics, role of the ribosomal proteins has been clearly separated into the folding nucleation and guidance function, and translational enhancement function(Ramakrishnan and White, 1998; Shajani et al., 2011; Henras et al., 2015; Davis et al., 2016; Parker and Karbstein, 2023). An intriguing observation here has been that the rRNA folding paths can be concurrent “streams” of differentially-staged binding events, with potential for re-tuning depending on the ongoing homeostatic demands(Davis et al., 2016; Dong et al., 2023). Thus, there is no, and perhaps has not been, a strict requirement for rRNA folding sequence – a strong indication that folding of an early but relatively complete proto-rRNA could take place with little or no assistance and with acceptable success rate. Nonetheless, a fascinating observation was made that the more ancient parts of the ribosome, such as the peptidyl transferase centre (PTC), are folded last(Dong et al., 2023).
Upon the accurate description of the ribosomal structure, a new type of deep sutructure-phylogenetic sequence analysis has become possible, tracing the ribosomal functional centres back in the evolutionary timeline. A “coded character” approach was suggested, whereby rRNA nucleotides are represented reflective of their structural state (helices, stems, hairpins, loops etc.) and then analysed using phylogenetic methods(Caetano-Anollés, 2002). Notably, it was confirmed that the peptidyl-tRNA (P) site is the most ancient structure of the ribosome, along with the structures responsible for the subunit interaction and intersubunit dynamics(Caetano-Anollés, 2002). The rRNA regions responsible for tRNA interaction and operation have shown signs of later co-evolution and traceable sequential adjustment that occurred in the rRNA of both subunits of the ribosome concurrently, providing additional evidence towards the structural link between tRNA translocation and intersubunit movement(Caetano-Anollés, 2002). Importantly, this tracing of conserved structures and interactions suggested gradual simplification and “channelling” of the evolutionary changes, whereby the initial diversity of structures and interactions would be gradually simplified, including size optimisations and non-orthologous function replacement, and structures streamlined in relatively independent “units” to enable modularity and subsequent independent evolution(Caetano-Anollés, 2002; Roberts et al., 2008).
Further observations from the structure-cladistic approach, including other complex molecular systems such as ribonulcease P, have led to a broader theory of high initial proteomic complexity, to the point of considering a protein-first or high-protein-diversity initial ancestor in a “protein interaction world” hypothesis(Andras and Andras, 2005; Sun and Caetano-Anollés, 2010; Kim and Caetano-Anollés, 2011, 2012; Harish and Caetano-Anollés, 2012; Caetano-Anollés and Caetano-Anollés, 2017), somewhat reminiscent of the very early, as well as much more recent ideas of protein-based life and in a challenge to the RNA world hypothesis(Andras and Andras, 2005; Ikehara, 2014; Walker et al., 2017). While intriguing, the ideas of diverse-protein prebiotic precursors do not offer strong evolutionary principles of development (see further in “Emergence of the current ribosomal organisation as the possible converging point”). Nonetheless, the structure-cladistic works have consistently identified several important facts, including that (using prokaryotic nomenclature): (1) ancient parts of the ribosome include structures surrounding tRNA and PTC, such as h11, h44, H76, H38, H41, H42, that are spatially situated nearby each other in the current ribosome, (2) most ancient proteins are S12 and S17 located in the vicinity of h44, (3) ribosomal proteins are clearly separated into groups along the stages of ribosomal evolution with proteins related to factor-enhanced elongation well-isolated from the others, (4) ribosome evolution progressed from mRNA decoding/operations to factor-assisted polymerisation and then formation of modern PTC, and (5) large part of ribosomal evolution revolved around interactions with tRNAs, and tRNA translocation ability has formed before the PTC(Smith et al., 2008; Harish and Caetano-Anollés, 2012). It is intriguing that in many of these analyses there are multiple evidence towards an archaea-like common ancestor of the cellular life(Sun and Caetano-Anollés, 2010; Kim and Caetano-Anollés, 2011, 2012), although approaches based on amino acid usage biases among ribosomal proteins themselves (as arguably traceable relics of the earliest proteins) sometimes point towards a more bacterial-like ancestor(Fournier and Gogarten, 2010).
Potentially triggered by the structure-phylogenetic works and ability to model and predict RNA structures from shorter fragments, a set of ideas has developed over possible “mosaic” nature of the ribosome precursor, which could be a function of several short RNA fragments(Agmon et al., 2005, 2006; Agmon, 2009; Agmon et al., 2009). Proto-ribosome resulting from dimerisation of compact RNA derived from the current ribosomal PTC was first suggested, using a combination of resolved three-dimensional native and predicted secondary structures of the respective RNA fragments(Agmon, 2009). This was followed by a suggestion that proto-ribosome would not initially operate on tRNAs but less complex substrates, and thus removal of the tRNA-binding sites can be justified – leaving a symmetric structure around PTC composed of stem-elbow-stem structures akin to tRNA fold(Agmon et al., 2009; Belousoff et al., 2010; Davidovich et al., 2010; Krupkin et al., 2011; Agmon, 2012). A further dissection of the long LSU RNA using principles of modularity and structural invariance upon elimination of parts has also suggested build-up of rRNA around the H48, H49 and H50 of the PTC, in at least 5 distinct steps of expansion, although this analysis excluded other rRNAs from consideration(Bokov and Steinberg, 2009). These more ancient – more PTC-central principles have led to a presentation of ribosome as a “fossil onion”, where layers to the periphery of the PTC can be “peeled off” to expose its more ancient parts(Hsiao et al., 2009). Overall, the structure conservation-based methods emphasised compactness of the RNA fragments constituting the PTC and the tRNAs, all about 100 nt or less, leading to a notion that protein synthesis may have co-occurred with or even predated efficient replication(Fox, 2010).
Recent works have focused on the further development of the hypothesis of a proto-ribosome emergence as a composite of small RNAs, and have been linking the minimal PTC and ribosomal proteins with the deeper insights into the workings of the genetic code. It was suggested that the initial code could have been 2-nucleotide-based (as compared to triplet) and relied on strong interactions (G, C), encoding just glycine, alanine, proline and diamino proprionic “abiotic” amino acids(Jukes, 1967; Crick, 1968; Eigen and Schuster, 1979; Trifonov, 2000; Hartman and Smith, 2014). Subsequent initial expansion included A (G, C, A) in the still 2-nucleotide-based code to allow for the glutamate and glutamine, histidine, ornitine and aspartate and asparagine encoding. Later, all-base triplet code would introduce termination codons and hydrophobic and aromatic amino acids with more complicated metabolism and stronger hydrophobic core-forming capacity(Hartman and Smith, 2014). A “descending thermostability” hypothesis, also integrated into a circular representation of the evolved genetic code principles, also supports progression from the shorter but strongly typed codons to longer but weak codons, whereby lack of the selective interaction force has been supplemented with more complex multi-point interactions of tRNAs with the other components(Trifonov, 2000; Grosjean and Westhof, 2016). The descending thermostability theory has been further substantiated by increasing structural capacitance hypothesis, whereby initial code attributions using strong codon:anticodon interactions were coding peptides with relatively weak structure-forming capacity, facilitating disordered-to-ordered structure selection and co-evolution of the structural capacitance of the proteome with the ribosome(Buckle and Buckle, 2019).
A view has now been formed about gradual expansion of the PTC-like core of the ribosome by inclusion of newer segments (and functionalities) of rRNA, summarised in very detailed structure-evolutionary studies(Petrov et al., 2014, 2015). A term “accretion” to the “frozen” or “common” core was proposed to this rRNA acquisition process(Petrov et al., 2014). Although there is still much of a debate about the methodology and accuracy of the exact implementation in phylogenetic tracing, and the possibility and mechanisms of the outward-directed expansion of the rRNA structures as a universal principle(Caetano-Anollés and Caetano-Anollés, 2017; Farías-Rico and Mourra-Díaz, 2022), the centres of rRNA that are marked as earliest generally correspond across the techniques. Concurrently to one or another variant of rRNA evolution, it was shown that ribosomal proteins exhibit ancient globular cores while their non-globular extensions are used to adapt the rRNA to the altering surroundings(Melnikov et al., 2018).
Overall, we have by now an excellently-evidenced and well-developed set of theories for plausible and likely path of ribosomal evolution starting with few tRNAs, rudimentary code and ribosome catalytic capacity. Regarding the early ribosomal evolution and emergence, we can summarise that while there are at least three different views on the emergence of the ribosome and the genetic code, namely the RNA world hypothesis(Gilbert, 1986), the protein world hypothesis(Kurland, 2010) and the mixed RNA:protein co-emergence and co-evolution(Brandman et al., 2012; Harish and Caetano-Anollés, 2012; Bowman et al., 2015; Tagami and Li, 2023), it is agreed that RNA is better suited for programmable protein synthesis(Bernhardt and Tate, 2015). All of these theories, nonetheless, do not provide a reasonable link between the feature occurrence and feature selection(Bowman et al., 2015), which is outlined in the subsequent section.
Emergence of the current ribosomal organisation as the possible converging point
As far as the programmed amino acid polymerisation into a protein is concerned, by now it is well-established that the ribosome is an RNA machine(Moore and Steitz, 2002; Steitz and Moore, 2003; Agmon et al., 2006; Ramakrishnan, 2009). Ribosomal RNA has a capacity to self-fold, and drive the self-assembly of the ribosome, while PTC is a ribozyme with conserved nucleotide structures(Polacek and Mankin, 2005; Agmon et al., 2006). Ribosomal proteins are virtually absent from the PTC(Ban et al., 2000; Wimberly et al., 2000; Hsiao and Williams, 2009; Fox, 2010; Krupkin et al., 2011). Early experiments have proven the possibility of a “factor-free” translation, whereby the most immediate protein-based actors of the translation cycle (the translation elongation factors EF-Tu and EF-G) were found not strictly required for the peptide production as such(Gavrilova et al., 1976). Thus, all of the functionally important critical components of translation are made out of RNA: the rRNA, tRNA and mRNA. For some reason this RNA-based core system has not been replaced with a likely more efficient protein-based system even after all the elapsed time, although multiple translation factor proteins now control and efficiate the process. This RNA-first, protein-second feature is important as it makes it clear that protein is the evolutionary product of the ribosome. There may have been no purpose-made (programmed) proteins yet when the riboreplisome was emerging.
Several lines of new experimental evidence were obtained recently that approve important possibilities within the early ribosome. Firstly, it has been demonstrated that short fragments of rRNA PTC capable of interaction with each other (and in certain cases of dimerisation) can facilitate formation of a “peptide bond” between CCA-phenylalanine-biotin conjugate and C-puromycin acting as peptide-bound “P-” and pre-charged “aminoacylated” “A-site” carriers, respectively(Bose et al., 2022). This is a very important observation that solidifies the plausibility of RNA-only ribosomal precursor and also highlights the relative simplicity of components required to achieve a measurable degree of facilitation. Reaction yield with the short PTC RNA constructs was just about 200 times less than with native E. coli LSU used as a control, but the fragments themselves were also hundreds of times shorter than ribosomal RNA, with the most efficient fragment used being just 67 nt long(Bose et al., 2022).
Secondly, it has been shown that ribosome can, in principle, function as a covalently-single RNA molecule(Schmied et al., 2018; Aleksashin et al., 2019). This is a very important observation not just because it resolves the fundamental question if splitting in non-covalently associated subunits is necessary for the ribosome function (which it is not), but also because it allows to hypothesise a single RNA precursor of the ribosome, and much simplifies the imaginable beginning of the genetic code story. The currently most-optimal single-RNA “tethered” ribosome was even able to fully support cell function, albeit with lower efficiency. Although, the lower tethered ribosome efficiency has rather been linked to the inoptimality of the folding and maturation (which did not co-evolve to support the tethered structure) and not translation efficiency per se. The tethered ribosome argument fully addresses a notion if subdivion into the subunits is principally required for the programmed protein synthesis function. If even current, long-evolved, complex and quite specialised in function ribosome can still operate in a cognate way as a single RNA, ribosome indeed could have been a single RNA molecule in the beginning. Tethered ribosomes also showed a considerable capacity of genetic alteration and have been used as a vehicle for targeted multiplex editing and function evolution, such as antibiotic resistance with suggestion for abiotic amino acid function engineering(Radford et al., 2022). The remaining question is then why the ribosome is kept divided into the subunits. This may be a “side effect” of early evolution that was “channelled” into the later stages so quickly that returning to a single-molecule rRNA became virtually impossible. That is, the ribosome divided in the subunits was optimised too fast that any step back towards reduction of the subunits into a singular entity would be competitively impossible. Thus, here it is proposed that current rRNA subdivision in individual molecules bears historic remnants of the rRNA evogenesis, perhaps since the separation of the riboreplisome into rRNA, tRNA and mRNA triade, rather than has a functional meaning.
Thirdly, there is an emerging theoretical and experimental evidence towards a relative ease of ribozyme-facilitated amino acid and peptide reactions, and the formation of RNA-protein hybrid molecules with different chemistry. Classical works directly addressed and definitively proved the possibility of creating a 2-component ribozyme system for charging of tRNAs through amino acid transfer. For example, it was first demonstrated that a selection of a ribozyme that would perform methionine analogue transfer from a 5’P-CAACCA-Met tRNA-like fragment to its 5’-phosphate group is possible, and it was suggested that evolutionary rRNA precursor could have a 3’-OH to 3’-OH acyl-transferase activity such as to transfer that group from a self-aminoacylated RNA to another RNA(Lohse and Szostak, 1996). This transfer activity was then experimentally confirmed by further incorporating a short tRNAfMet complementary sequence allowing interaction of the tRNA 3’ end with the ribozyme 5’-proximal region, which resulted in the tRNA charging by the amino acid. It was also shown that evolution towards specific amino acid recognition is possible in such a system(Lee et al., 2000). It was also demonstrated that a fully self-aminoacylating catalytic tRNA is possible based on these principles(Saito et al., 2001; Murakami et al., 2003). Recently, a “prebiotic” coupling of RNA and proteins using diverse chemistry, including coupling to the bases, was demonstrated as well as evidence provided towards RNA function “enhancement” by short cationic peptides(Castelvecchi, 2022; Müller et al., 2022; Tagami and Li, 2023). Critically, it has been shown that the hybrid RNA-amino acid molecules, such as arising from self-aminoacylation, are capable of forming much more active ribozymes and can themselves become catalysis of subsequent evolution(Radakovic et al., 2022).
Interestingly, most or all of the current theories of ribosomal evogenesis propose a multi-component co-evolution, whether based on an initial mix of proto-tRNAs, or proto-tRNAs and proto-rRNAs, or all three proto-t, -r and -mRNAs. In many cases, presence of co-evolving coded or non-coded peptides is proposed. One of the main arguments of these propositions has been that early replicative processes were inefficient and that they could produce a lot of “noise”, allowing for a spurious generation of short RNA fragments that eventually had a contributing function. It is difficult to fully accept this point of view. Firstly, ribozyme-based replication may not be inaccurate even after limited rounds of evolution(Johnston et al., 2001; Tjhung et al., 2020). Second, transcription error-based functional gain is, by definition, not individually heritable and thus not a subject of natural selection (in contrast to replication and not dismissing the fact that replicative error rate can itself be advantageous). Third, it is difficult to imply cell-like coacervation and restriction of activity of the molecules to their genetic origin at the stage of pre-cell “life”, which again detaches the possible gain in function from the genetic information carrier. Fourth, mechanisms by which individual proto-rRNA fragments would then genetically assemble into much longer rRNA chains are not fully explained. Overall, these views, while providing important solutions to multiple different areas of the ribosomal emergence and the emergence of life as a genetic system, contain a considerable degree of Lamarkism which – while not totally impossible in the more complicated life systems as we now can envisage from the epigenome and eptarnscriptome function – can be too far a stretch for the simple, clean early steps of genetic evolution.
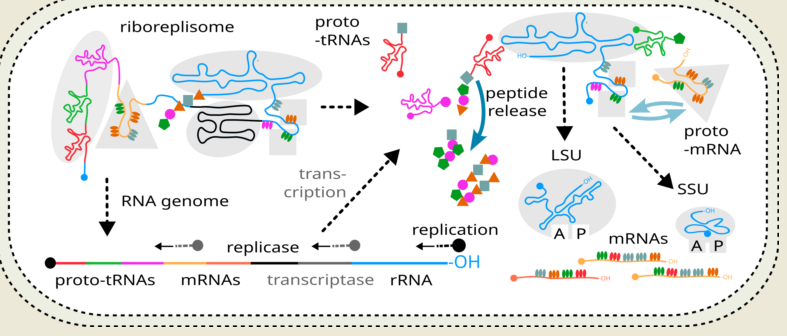
We combine the above-mentioned facts and limitations to propose an early existence of a riboreplisome, an RNA molecule that had features of a complete organism at least from the genetic standpoint. We suggest the following key steps in the emergence and early evolution of the riboreplisome, some of which are linked to creating pre-requisites towards a competitive advantage of genetic material compartmentalisation and cellular life (
Figure 2 and
Figure 3).
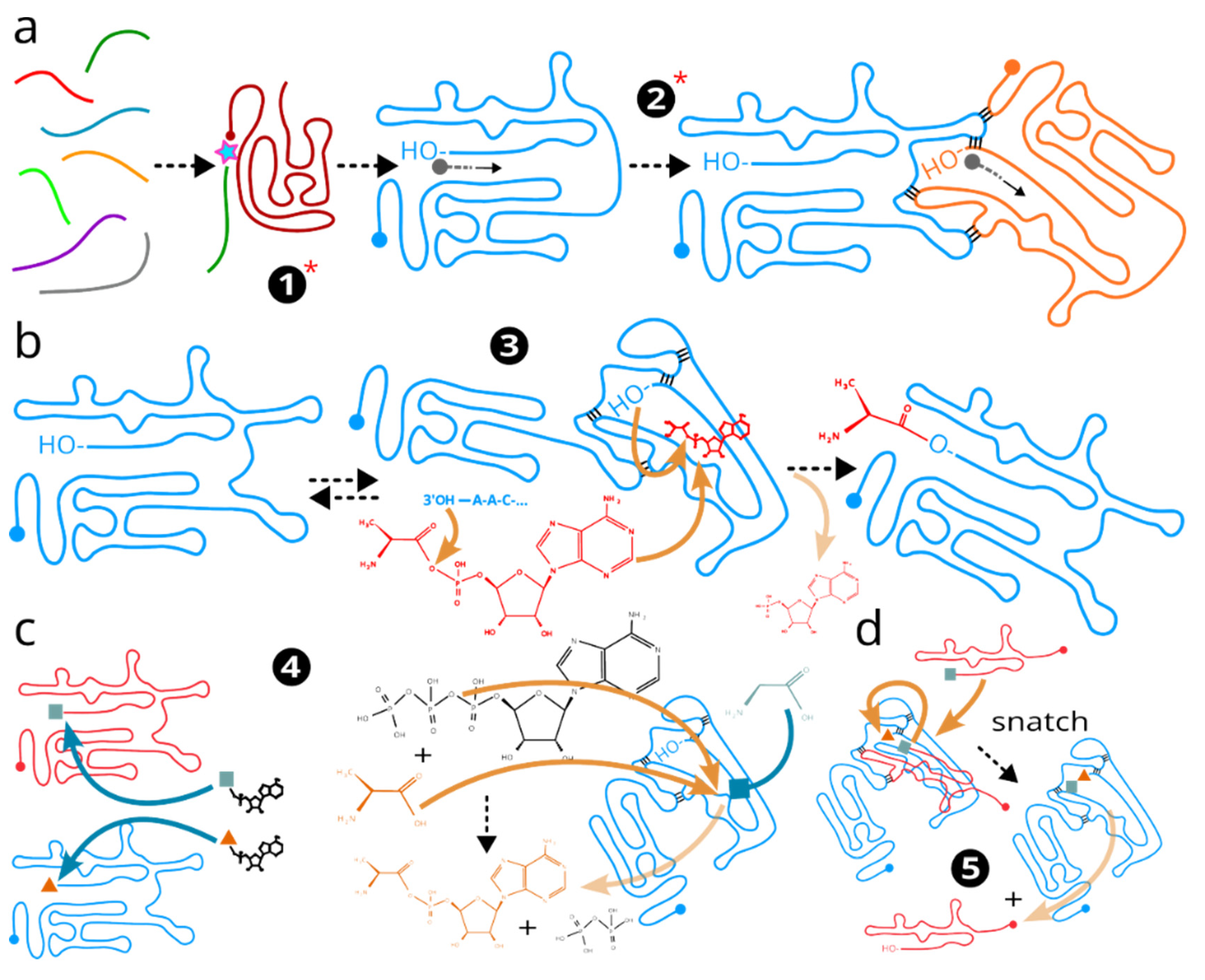
(1) Small oligomeric RNA nucleotides spontaneously develop ligation and then limited templated polymerisation activity, a plausibility of which has been experimentally demonstrated(Bartel and Szostak, 1993; Ekland et al., 1995; Attwater et al., 2018) (
Figure 2a; left). This process have likely occurred multiple times without subsequent development, before accidental acquisition of other ordered features as follows.
(2) Templated polymerisation activity becomes more ordered by limited baseparining-induced interactions with the replication “partner”, using replication topology from or similar to discussed in the “
Early link of replication with the mechanisms of replication enhancement” and
Figure 1(Tjhung et al., 2020). This forms RNA ribozyme “replisome” and, importantly, restricts the generic copying by allowing mostly copying of “self” and not “others” (
Figure 2a; right). From now on, Darwinian natural selection and evolution in the classical sense are possible and continue uninterruptedly.
(3) As the RNA ribozyme replisome has an ability to interact with its replication substrate 3’ end and has the RNA single-strand operating capacity, it also can interact with its own 3’ end, and develops ability to modify (“charge”) it with different chemistry, including amino acids(Lee et al., 2000). The charging occurs in bind-and-release cycles, while using environmentally-available amino-acid-activated nucleotides, including the aminoacyl-AMP (
Figure 2b). In the
Discussion, we speculate about the possible evolutionary driving forces behind this acquisition. This forms the earliest “riboreplisome”, which we can define as RNA ribozyme capable of replication plus at least one more type of self-modification chemistry, such as involving its 3’ end, and which eventually converges on aminoacylation. Riboreplisome overtakes RNA ribozyme replisome at this point and notably has all features of a complete genetic system as it has the copied genetic material, copying mechanism and additional independent “features” that can define copying efficiency and be a subject of genetic selection.
(4) Riboreplisomes develop specialisation and perhaps, even allosteric regulation, towards more specific aminoacylation (
Figure 2c). A plausibility of such process has been demonstrated with self-aminoacylating tRNA ribozymes(Murakami et al., 2003). By using amino acids more selectively, the riboreplisomes could adapt to the environment better. As riboreplisomes may started to deplete the energised precursors (aminoacyl-AMPs or other nucleotides), they further evolved their catalytic centre to pre-charge ATP or its functional analogue with amino acids that they need most (rather than those that their “competitors” need), a reaction that is, in fact, somewhat similar to the 3’ end aminoacylation from aminoacyl-AMP. This reaction may have involved the riboreplisome’s 5’ end(Lee et al., 2000).
(5) Now that riboreplisomes can purposefully produce aminoacylation of different types, a possibility of a combinatorial aminoacylation arises. For example, due to the chemical degradation processes, terminated or unprocessive replication, riboreplisome world begins to accumulate 3’ end fragments that eventually get charged with amino acids. Using these fragments, or perhaps sometimes “snatching” from the 3’ ends of the other riboreplisomes, riboreplisome transfers these additional aminoacids to its 3’ end (
Figure 2d). As the pre-charging of the available RNA 3’ ends is already somewhat specific, riboreplisome develops interaction specificity towards discriminated binding of the new substrates nearby its main catalytic centre, based on its peptide preferences. This, essentially, completes a primordial PTC.
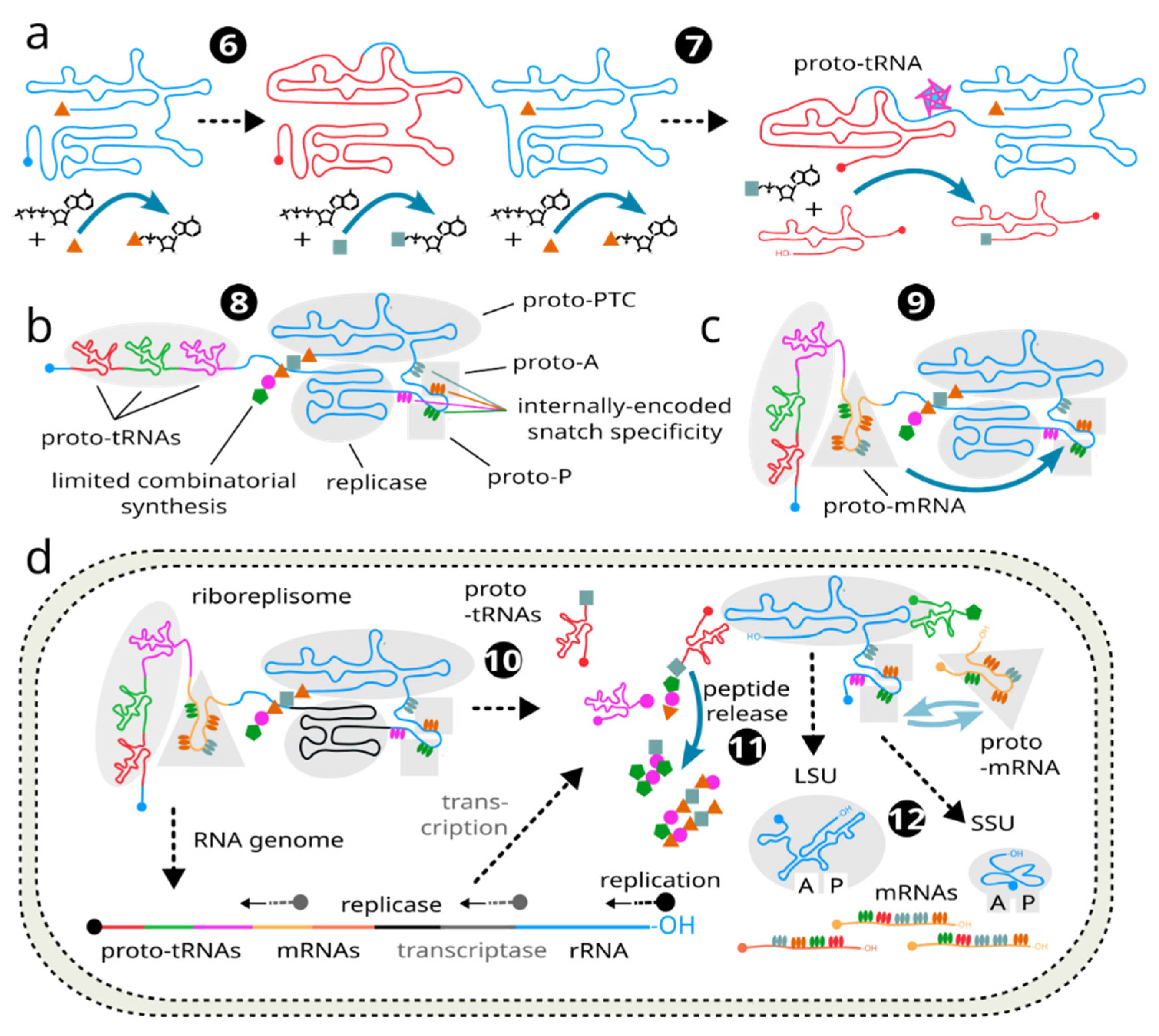
(6) At some point between (3) and (5) a duplication or multiplication event occurs as an “error” of replication (
e.g. slippage with subsequent production of a covalently-linked another copy of riboreplisome) (
Figure 3a; left), or riboreplisome is
ab initio a multiplicated arrangement as one of the possibilities discussed in the “
Early link of replication with the mechanisms of replication enhancement” and
Figure 1). This greatly speeds up the evolution by relaxing the selection criteria and allowing “wild” alterations without loss of “viability”, in a similarity to the currently-observed evolution principles where a new function acquisition often starts with gene duplication or multiplication(Walsh, 1995). Now each riboreplisome has an opportunity to charge multiple aminoacyl-AMPs (or their analogues) independently and possibly on demand, and can give rise to the sequence of different proto-tRNAs.
(7) The “auxiliary” riboreplisome copies evolve very rapidly to lose the now-redundant RNA replication capacity, and only retain the 3’ end aminoacylation and amino-acid-charging activities (
Figure 3a; right). They gradually evolve towards proto-tRNAs that initially integrated ARS function and functioned as self-aminoacylating ribozymes.
(8) Based on the facilitated evolution permitted by “gene” multiplication, riboreplisome develops and expands the internally-encoded codon repertoire to enable more and more sophisticated use of proto-tRNAs and synthesis of more complex peptides. As these are now also subject to genetic encoding and thus natural selection, co-evolution of the code base and peptides is initiated at this point (
Figure 3b).
(9) A major limitation remains in the ribosomal coding capacity which can be altered only genetically between generations. This limitation can be lifted by a mechanism somewhat similar to the proto-tRNA generation, where different fragments of the riboreplisome, most likely initially occurring due to multiplication and initial unguided fragment generation, substitute for the internally-coded “message” (
Figure 3c).
(10) Critically, sometime at this point the riboreplisome system develops abilities for diffusion restriction and some form of physical containment. The proto-tRNAs may begin to evolve accurateself-cleavage and maturation functions at this point, driven by collective advantage of the aminoacyl-proto-tRNA availability. Thus, some of the copies of riboreplisome begin to be “sacrificed” for the proto-tRNA production, as well as proto-mRNA production, and the own 3’ end modification is replaced with transfer to one of the bound proto-tRNAs, completing the A- and P-site arrangement (
Figure 3d). It is conceivable at this point that some of the proto-tRNA and proto-mRNA code is being included in the “negative” riboreplisome strand, and thus the separation of gene expression from replication may begin by an excessive or preferential synthesis of the “negative” strand. Mechanisms develop to control the level of “expression” or replication.
(11) As a variety of peptides can now be dynamically encoded, and their detachment into the environment can enable dynamic re-purposing and “mode switching” of the riboreplisome even between generations. This was achieved by releasing the “old” peptide and synthesising new one in its place (
Figure 3d). As keeping the energy-expensive peptides with certain function around the genetic material for longer could be extremely advantageous, the proto-mRNA code develops to further facilitate encapsulation and coacervation of riboreplisome to take full advantage of its products. We think this event could have been the major driver of compartmentalisation and emergence of the cell-based life.
(12) As compartmentalisation becomes definitively possible, proto-ribosome part of the riboreplisome is now made permanently programmable by removing (cutting out in the maturation process) its integrated mRNA bits and splitting it into SSU and LSU rRNA as the result. Unprocessed riboreplisome becomes a “genome” and loses its proto-ribosomal identity. The riboreplisome now has completed its evolution cycle and is transformed to an RNA genome and system of genetic code, its decoding, and gene expression (
Figure 3d). This system is interconnected sufficiently to be “channelled” towards the development of the current ribosome, such as its function vitally depends on the protein products it produces. For example, these protein products were required for compartmentalisation and cell-like behaviour.
The subsequent development occurs by well-theorised or evidenced events, including code expansion, translation factor introduction and replacement of ribozyme-based tRNA pre-charging with the aminoacyl-synthetatse based chemistry. Self-cleavage of the RNA genome copies is gradually displaced by gene-selective copying of proto-m-, t- and rRNA as a more refined mechanism. This, and the appearance of more stable cell-like structures, eventually enables the emergence of a special RNA “mutation” in the form of DNA for information storage, which is transcribed into RNA by demand. It is indeed notable that some phylogenetic investigations indicate the possible emergence of DNA after the separation of the major cellular branches of life(Forterre, 2015).
Discussion
To summarise, despite being relictual to the extreme, the catalytic RNA core of the ribosomal decoding centre carries through some of the designs of the original universal RNA machine, the riboreplisome. Until we could discover life based on a different driving principle of information transformation, ribosome remains the closest structure resembling early “life” and can be considered as the unifying molecule of the living. Until proven otherwise, it should be questioned if life inevitably depends on the existence of the ribosome or any similarly organised RNA. Critically, riboreplisome concept provides an uninterrupted link between the worlds of chemical and biological evolution and a pathway for the emergence of the ribosome.
Several riboreplisome features that arise from the purely theoretical considerations bear a remarkable resemblance to the facts and evidence we derive from the current ribosomal, genetic code and cellular organisation. A stunningly simple observation is that the contemporary ribosome is predominantly transcribed as a single pre-rRNA unit (except 5S rRNA in eukaryotes), which also contains tRNA(s) in place of the 5.8S rRNA and after 23S and 5S rRNAs in eubacteria(Stoppel and Meurer, 2012; Henras et al., 2015). Ribosome production is still tightly linked even in higher eukaryotes through the ribosome biogenesis “regulon”(Brown et al., 2008). In addition to the tethered rRNA and subunits, a naturally-occurring transfer messenger (tm)RNA evidences a next possibility of consolidation towards a singular RNA of the riboreplisome(Withey and Friedman, 2003; Bessho et al., 2007; Neubauer et al., 2012; Macé and Gillet, 2016). There are suggestions that rRNA, at least SSU rRNA, appeared by concatemerisation of proto-tRNA molecules(de Farias et al., 2021), a process that would result in features similar to the proposed here multiplication of the riboreplisome aminoacylation site. This is further supported by the view that rRNA sequences may be linked to the tRNA sequences, including tracing the PTC and SSU rRNA similarities with a consensus tRNA(Farias et al., 2014; de Farias et al., 2021). There is some indication that fragments resembling tRNAs and ORFs of key peptides such as ribosomal proteins, polymerases, ligases etc. can be found or even reconstructed within the rRNA itself, which probably is not an absolute evidence due to the immense evolutionary distances involved, but a factual indication of some possibilities that these sequences have had a “common ancestor”(Root-Bernstein and Root-Bernstein, 2015, 2019; de Farias et al., 2021). A converging indication is presented by the “circular code” hypothesis which postulates a reduced early code that excluded circular permutations and homopolymeric codons, potentially resulting in some tolerance to frameshifts(Dila et al., 2019). Circular code results in self-complementary sequences and can be located in the structural rRNA regions(Dila et al., 2019). These considerations coupled with experiments confirming a possibility of relatively simple synthetic self-replicating systems, such as translation-coupeled RNA replication, provide a plausible path towards a riboreplisome(Mizuuchi et al., 2015; Root-Bernstein and Root-Bernstein, 2015).
In regard to the larger-scale dynamics of the ribosome (and riboreplisome), initial ribosomal operation can be explained through the interaction of minihelixes with tRNA-like function with the original PTC-like structure(Tamura, 2011). Indeed, there is evidence that the CCA stem and sequence are the more ancient parts of the tRNAs, with the anticodon loop being a separate module that has signs of a later evolution(Schimmel et al., 1993; Maizels and Weiner, 1994; Schimmel and Henderson, 1994; Noller, 1999; Weiner and Maizels, 1999; Di Giulio, 2009; Fox, 2010). Ligation and hybridisation were proposed as the possible evolutionary mechanisms in further increasing complexity and diversity of tRNAs. An interesting proposal could be made that the earliest regions of the SSU RNA were located closest to the parts of the repliribiosome defining RNA-dependent RNA polymerase (the riboreplisome replicase), or initially were that replicase. The evolutionary-older h44 and h28 of the SSU (bacterial nomenclature) were proposed to also be linked to the inter-subunit (inter-domain in the context of riboreplisome) movement that eventually becomes the ratcheting decoding motion(Fox et al., 2015). These regions and mostly codon:anticodon interactions with tRNA and mRNA could promoted early proto-mrNA interaction and translation initiation, with the more specialised Shine-Dalgarno sequence evolving later as a result of transcriptome-level selection(Saito et al., 2020). We know that base-complementarity interactions for translation initiation can be discarded, as exemplified by Bacteroidetes(Accetto and Avguštin, 2011), which represent a “living relic” of eukaryote-type translation initiation emergence(Shirokikh and Preiss, 2018; Shirokikh et al., 2019). The amino acid diversity can then be sequentially developed and further segregated by, for example, a mechanism similar to the proposed cofactor-based synthesis of new aminoacids at the codon-anticodon minihelix(Martínez Giménez and Tabares Seisdedos, 2022).
Much evidence points towards the tRNA-delivering GTPase (EF-Tu/aeEF1) evolving earlier and before than translocation-promoting GTPase (EF-G/aeEF2), once again suggesting that riboreplisome could initially have just a single full site, the A-site(Hartman and Smith, 2010). Appearance of a capability to bind a complex ligand (tRNA) first in a non-templated manner and then in conjunction with a guiding template (mRNA) resulted in rRNA-driven translocation first, as we know from the factor-free translation cycle(Gavrilova et al., 1976), and then in the adoption on molecular mimicry-based displacement of tRNA and “powered” translocation with EF-G. Investigations using EF-Tu resurrected to different ancestral age indicate the likely less accurate but more promiscuous and permissive early factor variants, that were surprisingly still active with the present-day ribosome(De Tarafder et al., 2021).
Because here it is proposed that the initial activity of riboreplisome was self-aminoacylation, which can be regarded as RNA modification activity, it is interesting to discuss the hypermodification of ribosomal RNA. It has been long noted that tRNAs and rRNA have some of the highest content of the modified nucleotides(Decatur and Fournier, 2002; Roundtree et al., 2017; Sloan et al., 2017). Quite notably, rRNA modifications are important for the optimal performance of the ribosome and are mostly deposited during rRNA maturation and in the regions inner to the ribosomal structure, nearby to the rRNA functional centres(Decatur and Fournier, 2002; Sloan et al., 2017). This phenomena can be seen as an alternative to creating chemical diversity (compared to the proteins), that has been developing in parallel with the evolution of the ribosome or even riboreplisome, and also paralleled peptide and protein evolution. Perhaps, the fact that these modifications are preserved throughout time is unsurprising until the rRNA catalytic core of the ribosome remains relatively protein-free. Thus, rRNA modifications can be seen as landmarks or “scars” of the rRNA evolution and can represent an overall vector of RNA systems perpetuating long enough to accumulate functional improvements through chemical modification of nucleotides. In this regard it is interesting to speculate about the RNA modification machinery, that may have evolved from ribozymes to specific modification “writer” proteins, following the sequential expansion of the rRNA with new sequences and performing its “patching”. With the co-evolution of the rRNA modifications we still can expect a link to Lamarkist ribosome evogenesis, whereby ectopic upregulation of certain RNA modification enzymes that favoured enhanced function or robustness(Sloan et al., 2017) gave rise to their eventual hard-wiring as a separate, and then specialised gene in the genome. The likeliness of this process is verified by recent experiments using unmodified SSU rRNA, where after just 15 cycles of evolution it was possible to obtain a non-modified derivative with a high relative ribosome activity(Murase et al., 2018). Facilitated by the multiplicity of ribosomal genes, active selection, adjustment and refinement of the ribosome is continuously happening, with heterogeneous ribosome being one of the examples how function of such a critical component can be altered without compromising its activity(Barna, 2017). RNA modifying enzymes thus may carry some of the most ancient protein code functions, if this primordial code can still be extracted from them.
What could have been the aboriginal function of the proto-ribosomal component of the riboreplisome? Here, we need to abstract from the current diversity of molecular interactions seen in complex live systems and imagine the extremely chemically diverse but functionally limited environment of the riboreplisome at its emergence. Any gain of function and function selectivity in these circumstances would carry an enormous benefit, which, coupled with an inheritance-driven selection, could boost the emergence of life. The riboreplisome concept offers inheritance. In fact, inheritance of features that in any way stimulates propagation would be the most important function of the riboreplisome. These features could be primarily of two types: (1) direct enhancement of replication and (2) preservation, stability and “survival”. We do not consider indirect replication enhancement at this point, such as facilitation of production of the replication intermediates, as the the early riboreplisome capacity would not allow complex peptides, and means for diffusion restriction were unlikely available, cancelling any advantages of “local” replication intermediate synthesis. Direct enhancement of replication could have been through facilitating unspecific (later – specific) binding to and retention of the interacting replication partner, including through formation of molecular condensates, and enhancement of the ribozyme activity. For example, by providing an additional functional group, such as positively charged amino acid residue. Preservation, stability and “survival” could have been through blocking access to the riboreplisome’s own 3’ end to avoid inclusion into adverse polymerisation reactions and, perhaps, any other terminal transfer/exchange reactions or exonuclease (ribozyme-based) activity that could be conceived as early “antibiotic” or “predatory” activity. Of these, “parasitic” continued extension of the 3’ end was perhaps the most immediate and greater threat, given the self-replication properties of the riboreplisome, and should be named as the most likely initial feature. The riboreplisome’s stabilisation could also be important for physically and chemically adverse circumstances, such as temperature, pH and metal ion presence. Indeed, recently it was demonstrated that hydrophobic cationic peptides can substantially modulate, enhance and organise structural and catalytic RNA, and accelerate RNA polymerase rybozyme(Li et al., 2022). A follow-up hypothesis can be that the aboriginal proteome has been composed of the peptides that functionally improved riboreplisone function and were also its constituents upon the emergence of the peptide release mechanism. This way, the initial genetic selection could have occurred even in a cell-free form, whereby more successful peptides and proteins remained associated with their genetic carrier simply through direct binding, as it is experimentally implemented in the ribosome display technique(Hanes and Plückthun, 1997; Hoffmüller and Schneider-Mergener, 1998; Lipovsek and Plückthun, 2004).
While there are obviously deep reasons for the homogeneity and persistence of the genetic code over time, some of which are perhaps rooted in fundamental properties of the available molecules, it has been prominently demonstrated in many synthetic biology experiments that the code can be altered and expanded, also expanding the functionality of the ribosome(Hayashi et al., 2010; Neumann et al., 2010). The riboreplisome hypothesis offers an uninterrupted path of a sequential sophistication of the genetic code, through the ease of alteration of the proto-tRNAs and their expansion into new codons. This classical evolution process involves selecting “genes” by advantageous features and occurs as plausible, given there is a relative freedom for the generation of the new features and these innovations do not easily break the entire system. In this case, there were, inadvertently, all the different versions of the genetic code – however, these versions seem to have been out-competed by the current variant. Likely, there are deep thermodynamic, kinetic and combinatorial reasons for the success of the current code as it was suggested(Grosjean and Westhof, 2016; Westhof et al., 2022), and in the paradigm of the historic ribosomal organisation there really is no better variant, which would be sufficient to explain the observed universality of the genetic code. Interestingly, from this perspective the code does never appear as in any way randomly-occurred coincidence of interactions, but is always a sequential sophistication from a very simple initial state. We thus cannot exclude early diverse forms of even cellular life that used alternative code variants, but became fully extinct due to the code sub-optimality.
Consistent with the proposed functional expansion of the RNA world by the proteome(Noller, 2004) – a process that still continues – ribosome, in some respects, is a minitiaurisation attempt in progress. Some minimal limits of the current ribosomal structures can be assessed by deriving minimal protein and rRNA orthologues set across all branches of life(Mushegian, 2005). In an analogy to the current race to upgrade lithographic process, the larger building blocks (RNA) are being replaced buy the smaller and more diverse building blocks (proteins)(Bernhardt and Tate, 2015). It remains to be determined if it is possible to fully replace the older “RNA technology” while preserving all of its fuctions(Bernhardt and Tate, 2015). This replacement has certainly occurred nearly completely in RNA and DNA replication (priming still requires DNA or RNA pieces), lokely owing to the lesser enzymatic complexity of the replication processes. Mitochondrial ribosome appears as most evolved, potentially resulting from a relaxed selection due to nuclear-encoded substitution of function and thus a more rapid pace of improvement(O’Brien, 2002; Sloan et al., 2014; Bernhardt and Tate, 2015). Indeed, at least 19 new ribosomal proteins were added (with some rRNA replacement) to the mitochondrial ribosome compared to the common ancestor since the endosymbiosis establishment time(Desmond et al., 2011). A ribosomal protein “promiscuity” observed in different archaeal species whereby S24e, L14e and L8e could bind different rRNA structures(Armache et al., 2013), could also explain an “easy” evolutionary path for rRNA function replacement or augmentation via sequential modification of a ribosomal protein copy. A fascinating observation can be that while evolution of the ribosome as a performant decoding ribo/enzyme shows evidence of “polymer transition” in the replacement of nucleic acids with proteins, its function as part of the gene expression control machinery clearly and paradoxically evidences the inverse trend, with ribosomes of organisms of arguably more complex gene expression patterns having longer rRNA(Bowman et al., 2015; Simsek et al., 2017). We think this might be, once again, a result of the fact that rRNA provides a genetic and functional binding matrix and continuity of the function development, preventing excessive “channelling” of the system to a state of an irreversible perfection. Essentially, the riboreplisome was, and ribosome being its current most close approximation is, an irreplaceable evolutionary vehicle, a catalytic replication-enhancing machine that is sufficiently crude and imperfect in its design not to be easily broken, and also a genetic system that is forgiving, robust and modular.
Intriguingly, ribosomes and “replication” have already been linked through a “duplicator RNA” hypothesis, in which short tRNA-like homodimers could assist in duplication of the original RNA template by bringing in new nucleotide triplets through a codon:anticodon-like interactions between the original and duplicated strands(Noller, 2012). A difficult part of this hypothesis is the presence of a relatively sophisticated template RNA and multiple components that require additional production, and the absence of “holding matrix” that would allow coherent co-evolution of these components. Remarkably, the riboreplisome hypothesis presented here resolves these difficulties and assumes just two non-Darwinian steps (
Figure 2a; red asterisks), namely the formation of RNA-dependent RNA polymerase replicase and the acquisition of said replicase a 3’ end-modifying function. The first step has been fully proven and the second step has been partially confirmed by generating RNA ribozymes that are capable of 3’ end operations, but are not replicases.
To conclude, in a conversation where we consider what could be the smallest biological entity that can be deemed as “live”, the riboreplisome offers many, possibly all, necessary answers. The riboreplisome hypothesis posits a possibility of a much earlier onset of Darvinian evolution compared to the RNA:protein mutual or parallel evolution pathways(Bowman et al., 2015). The riboreplisome explains a seamless, sequential and genetic sophistication of programmed features based on progressively more complex encoding, without requiring strict (or any) compartmentalisation arrangements for the genetic material and its products. At the same time, with the onset of the peptide release mechanisms, the riboreplisome also provides a driving force towards the selection of compartmentalised genetic entities and the emergence of cellular organisation. The riboreplisome explains the origins of all three main and universal types of RNA, the rRNA, tRNA and mRNA, and also provides a basis to the development of RNA processing and maturation, and eventually transcription functions. Perhaps, the riboreplisome was still present in its more evolved form in early cell-based life up until more controlled mechanisms of cell division enabled possibility of information storage function in the form of DNA. While we unlikely to recover the aboriginal riboreplisome, we will likely to continue to discover its relics scattered across the biological RNA. “Reverse engineering” a riboreplisome can nonetheless be possible and would help to provide novel synthetic biology opportunities in self-contained translation/replication, which could be built using completely “parallel” genetic code, and also would be a first step in replicating the emergence of life.
Funding
This work was supported by Australian Government, National Health and Medical Research Council (NHMRC): Investigator Grant GNT1175388, and the Bootes Foundation.
Acknowledgements
The author is very grateful for the critical scientific discussions with all members of the Division of Genome Sciences and Cancer of The John Curtin School of Medical Research, The Australian National University.
References
- Accetto, T. , and Avguštin, G. (2011). Inability of Prevotella bryantii to Form a Functional Shine-Dalgarno Interaction Reflects Unique Evolution of Ribosome Binding Sites in Bacteroidetes. PLOS ONE 6, e22914. [CrossRef]
- Agmon, I. (2009). The Dimeric Proto-Ribosome: Structural Details and Possible Implications on the Origin of Life. International Journal of Molecular Sciences 10, 2921–2934. [CrossRef]
- Agmon, I. (2012). “The Dimeric Proto-Ribosome Within the Modern Ribosome,” in Genesis - In The Beginning: Precursors of Life, Chemical Models and Early Biological Evolution Cellular Origin, Life in Extreme Habitats and Astrobiology., ed. J. Seckbach (Dordrecht: Springer Netherlands), 653–668. [CrossRef]
- Agmon, I. , Bashan, A., and Yonath, A. (2006). On Ribosome Conservation and Evolution. Israel Journal of Ecology & Evolution 52, 359–374. [CrossRef]
- Agmon, I. , Bashan, A., Zarivach, R., and Yonath, A. (2005). Symmetry at the active site of the ribosome: structural and functional implications. Biol Chem 386, 833–844. [CrossRef]
- Agmon, I. , Davidovich, C., Bashan, A., and Yonath, A. (2009). Identification of the prebiotic translation apparatus within the contemporary ribosome. Nat Prec, 1–1. [CrossRef]
- Aleksashin, N. A. , Leppik, M., Hockenberry, A. J., Klepacki, D., Vázquez-Laslop, N., Jewett, M. C., et al. (2019). Assembly and functionality of the ribosome with tethered subunits. Nat Commun 10, 930. Nat Commun. [CrossRef]
- Alksne, L. E. , Anthony, R. A., Liebman, S. W., and Warner, J. R. (1993). An accuracy center in the ribosome conserved over 2 billion years. Proceedings of the National Academy of Sciences 90, 9538–9541. [CrossRef]
- Andras, P. , and Andras, C. (2005). The origins of life -- the “protein interaction world” hypothesis: protein interactions were the first form of self-reproducing life and nucleic acids evolved later as memory molecules. Med Hypotheses 64, 678–688. [CrossRef]
- Armache, J.-P. , Anger, A. M., Márquez, V., Franckenberg, S., Fröhlich, T., Villa, E., et al. (2013). Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution. Nucleic Acids Research 41, 1284–1293. [CrossRef]
- Attwater, J. , Raguram, A., Morgunov, A. S., Gianni, E., and Holliger, P. (2018). Ribozyme-catalysed RNA synthesis using triplet building blocks. eLife 7, e35255. [CrossRef]
- Ban, N. , Nissen, P., Hansen, J., Moore, P. B., and Steitz, T. A. (2000). The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science 289, 905–920. [CrossRef]
- Barna, M. (2017). Specialized Ribosomes: A New Frontier in Gene Regulation, Organismal Biology, & Evolution. The FASEB Journal 31, 387.1-387.1. [CrossRef]
- Bartel, D. P. , and Szostak, J. W. (1993). Isolation of New Ribozymes from a Large Pool of Random Sequences. Science 261, 1411–1418. [CrossRef]
- Belousoff, M. J. , Davidovich, C., Zimmerman, E., Caspi, Y., Wekselman, I., Rozenszajn, L., et al. (2010). Ancient machinery embedded in the contemporary ribosome. Biochemical Society Transactions 38, 422–427. [CrossRef]
- Bernhardt, H. S. , and Tate, W. P. (2015). A Ribosome Without RNA. Frontiers in Ecology and Evolution 3. Available online: https://www.frontiersin.org/article/10.3389/fevo.2015.00129 (accessed on 20 June 2022).
- Bessho, Y. , Shibata, R., Sekine, S., Murayama, K., Higashijima, K., Hori-Takemoto, C., et al. (2007). Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proceedings of the National Academy of Sciences 104, 8293–8298. [CrossRef]
- Beuning, P. J. , and Musier-Forsyth, K. (1999). Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers 52, 1–28. [CrossRef]
- Bogorad, L. (1975). Evolution of Organelles and Eukaryotic Genomes. Science 188, 891–898. [CrossRef]
- Bokov, K. , and Steinberg, S. V. (2009). A hierarchical model for evolution of 23S ribosomal RNA. Nature 457, 977–980. [CrossRef]
- Bose, T. , Fridkin, G., Davidovich, C., Krupkin, M., Dinger, N., Falkovich, A. H., et al. (2022). Origin of life: protoribosome forms peptide bonds and links RNA and protein dominated worlds. Nucleic Acids Research, gkac052. [CrossRef]
- Bowman, J. C. , Hud, N. V., and Williams, L. D. (2015). The Ribosome Challenge to the RNA World. J Mol Evol 80, 143–161. [CrossRef]
- Brandman, R. , Brandman, Y., and Pande, V. S. (2012). Sequence Coevolution between RNA and Protein Characterized by Mutual Information between Residue Triplets. PLOS ONE 7, e30022. [CrossRef]
- Brown, S. J. , Cole, M. D., and Erives, A. J. (2008). Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics 9, 442. [CrossRef]
- Buckle, A. M. , and Buckle, M. (2019). Ribosome Evolution and Structural Capacitance. Frontiers in Molecular Biosciences 6. Available online: https://www.frontiersin.org/article/10.3389/fmolb.2019.00123 (accessed on 20 June 2022).
- Caetano-Anollés, D. , and Caetano-Anollés, G. (2017). Commentary: History of the ribosome and the origin of translation. Frontiers in Molecular Biosciences 3. Available online: https://www.frontiersin.org/article/10.3389/fmolb.2016.00087 (accessed on 20 June 2022).
- Caetano-Anollés, G. (2002). Tracing the evolution of RNA structure in ribosomes. Nucleic Acids Research 30, 2575–2587. [CrossRef]
- Canceill, D. , Viguera, E., and Ehrlich, S. D. (1999). Replication Slippage of Different DNA Polymerases Is Inversely Related to Their Strand Displacement Efficiency*. Journal of Biological Chemistry 274, 27481–27490. [CrossRef]
- Castelvecchi, D. (2022). Origin of life theory involving RNA–protein hybrid gets new support. Nature 605, 409–409. [CrossRef]
- Cech, T. R. (2012). The RNA Worlds in Context. Cold Spring Harb Perspect Biol 4, a006742. [CrossRef]
- Clark, C. G. (1987). On the evolution of ribosomal RNA. J Mol Evol 25, 343–350. [CrossRef]
- Crick, F. H. , Brenner, S., Klug, A., and Pieczenik, G. (1976). A speculation on the origin of protein synthesis. Orig Life 7, 389–397. [CrossRef]
- Crick, F. H. C. (1968). The origin of the genetic code. Journal of Molecular Biology 38, 367–379. [CrossRef]
- Davidovich, C. , Belousoff, M., Wekselman, I., Shapira, T., Krupkin, M., Zimmerman, E., et al. (2010). The Proto-Ribosome: An Ancient Nano-machine for Peptide Bond Formation. Israel Journal of Chemistry 50, 29–35. [CrossRef]
- Davis, J. H. , Tan, Y. Z., Carragher, B., Potter, C. S., Lyumkis, D., and Williamson, J. R. (2016). Modular Assembly of the Bacterial Large Ribosomal Subunit. Cell 167, 1610-1622.e15. [CrossRef]
- de Farias, S. T. , Rêgo, T. G., and José, M. V. (2021). Origin of the 16S Ribosomal Molecule from Ancestor tRNAs. J Mol Evol 89, 249–256. [CrossRef]
- De Tarafder, A. , Parajuli, N. P., Majumdar, S., Kaçar, B., and Sanyal, S. (2021). Kinetic Analysis Suggests Evolution of Ribosome Specificity in Modern Elongation Factor-Tus from “Generalist” Ancestors. Molecular Biology and Evolution 38, 3436–3444. [CrossRef]
- Decatur, W. A. , and Fournier, M. J. (2002). rRNA modifications and ribosome function. Trends in Biochemical Sciences 27, 344–351. [CrossRef]
- Delihas, N. (2006). The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA World. Third Edition. Cold Spring Harbor Monograph Series, Volume 43. Edited by Raymond F Gesteland, Thomas R Cech and, John F Atkins. The Quarterly Review of Biology 81, 272–273. [CrossRef]
- Desmond, E. , Brochier-Armanet, C., Forterre, P., and Gribaldo, S. (2011). On the last common ancestor and early evolution of eukaryotes: reconstructing the history of mitochondrial ribosomes. Research in Microbiology 162, 53–70. [CrossRef]
- Di Giulio, M. (2009). A Comparison Among the Models Proposed to Explain the Origin of the tRNA Molecule: A Synthesis. J Mol Evol 69, 1–9. [CrossRef]
- Dila, G. , Ripp, R., Mayer, C., Poch, O., Michel, C. J., and Thompson, J. D. (2019). Circular code motifs in the ribosome: a missing link in the evolution of translation? RNA 25, 1714–1730. [CrossRef]
- Dong, X. , Doerfel, L. K., Sheng, K., Rabuck-Gibbons, J. N., Popova, A. M., Lyumkis, D., et al. (2023). Near-physiological in vitro assembly of 50S ribosomes involves parallel pathways. Nucleic Acids Research, gkad082. [CrossRef]
- Eigen, M. , and Schuster, P. (1979). The Hypercycle. Berlin, Heidelberg: Springer. [CrossRef]
- Ekland, E. H. , and Bartel, D. P. (1996). RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 382, 373–376. [CrossRef]
- Ekland, E. H. , Szostak, J. W., and Bartel, D. P. (1995). Structurally Complex and Highly Active RNA Ligases Derived from Random RNA Sequences. Science 269, 364–370. [CrossRef]
- Farias, S. T. , Rêgo, T. G., and José, M. V. (2014). Origin and evolution of the Peptidyl Transferase Center from proto-tRNAs. FEBS Open Bio 4, 175–178. [CrossRef]
- Farías-Rico, J. A. , and Mourra-Díaz, C. M. (2022). A Short Tale of the Origin of Proteins and Ribosome Evolution. Microorganisms 10, 2115. [CrossRef]
- Forterre, P. (2015). The universal tree of life: an update. Frontiers in Microbiology 6. Available online: https://www.frontiersin.org/article/10.3389/fmicb.2015.00717 (accessed on 20 June 2022).
- Fournier, G. P. , and Gogarten, J. P. (2010). Rooting the Ribosomal Tree of Life. Molecular Biology and Evolution 27, 1792–1801. [CrossRef]
- Fox, G. E. (2010). Origin and Evolution of the Ribosome. Cold Spring Harb Perspect Biol 2, a003483. [CrossRef]
- Fox, G. E. , Paci, M., Tran, Q., Petrov, A. S., and Williams, L. D. (2015). Ribosome dynamics and the evolutionary history of ribosomes. in Instruments, Methods, and Missions for Astrobiology XVII (SPIE), 88–92. [CrossRef]
- Garrett, R. (1999). Mechanics of the ribosome. Nature 400, 811–812. [CrossRef]
- Gavrilova, L. P. , Kostiashkina, O. E., Koteliansky, V. E., Rutkevitch, N. M., and Spirin, A. S. (1976). Factor-free (“Non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. Journal of Molecular Biology 101, 537–552. [CrossRef]
- Gilbert, W. (1986). Origin of life: The RNA world. Nature 319, 618–618. [CrossRef]
- Gomez, M. A. R. , and Ibba, M. (2020). Aminoacyl-tRNA synthetases. RNA 26, 910–936. [CrossRef]
- Grosjean, H. , and Westhof, E. (2016). An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res 44, 8020–8040. [CrossRef]
- Hanes, J. , and Plückthun, A. (1997). In vitro selection and evolution of functional proteins by using ribosome display. Proceedings of the National Academy of Sciences 94, 4937–4942. [CrossRef]
- Harish, A. , and Caetano-Anollés, G. (2012). Ribosomal History Reveals Origins of Modern Protein Synthesis. PLOS ONE 7, e32776. [CrossRef]
- Hartman, H. , and Smith, T. F. (2010). GTPases and the origin of the ribosome. Biology Direct 5, 36. [CrossRef]
- Hartman, H. , and Smith, T. F. (2014). The Evolution of the Ribosome and the Genetic Code. Life 4, 227–249. [CrossRef]
- Hayashi, G. , Goto, Y., and Suga, H. (2010). Ribosome Evolution for Two Artificial Amino Acids in E. coli. Chemistry & Biology 17, 320–321. [CrossRef]
- Henras, A. K. , Plisson-Chastang, C., O’Donohue, M.-F., Chakraborty, A., and Gleizes, P.-E. (2015). An overview of pre-ribosomal RNA processing in eukaryotes. WIREs RNA 6, 225–242. [CrossRef]
- Higgs, P. G. , and Lehman, N. (2015). The RNA World: molecular cooperation at the origins of life. Nat Rev Genet 16, 7–17. [CrossRef]
- Hoffmüller, U. , and Schneider-Mergener, J. (1998). In Vitro Evolution and Selection of Proteins: Ribosome Display for Larger Libraries. Angewandte Chemie International Edition 37, 3241–3243. [CrossRef]
- Hsiao, C. , Mohan, S., Kalahar, B. K., and Williams, L. D. (2009). Peeling the Onion: Ribosomes Are Ancient Molecular Fossils. Molecular Biology and Evolution 26, 2415–2425. [CrossRef]
- Hsiao, C. , and Williams, L. D. (2009). A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Res 37, 3134–3142. [CrossRef]
- Ibba, M. , and Söll, D. (2000). Aminoacyl-tRNA Synthesis. Annual Review of Biochemistry 69, 617–650. [CrossRef]
- Ichihashi, N. , and Yomo, T. (2016). Constructive Approaches for Understanding the Origin of Self-Replication and Evolution. Life 6, 26. [CrossRef]
- Ikehara, K. (2014). [GADV]-Protein World Hypothesis on the Origin of Life. Orig Life Evol Biosph 44, 299–302. [CrossRef]
- Ishikawa, H. (1977). Evolution of ribosomal RNA. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry 58, 1–7. [CrossRef]
- Jacob, F. (1977). Evolution and Tinkering. Science 196, 1161–1166. [CrossRef]
- Johnston, W. K. , Unrau, P. J., Lawrence, M. S., Glasner, M. E., and Bartel, D. P. (2001). RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325. [CrossRef]
- Joyce, G. F. (1989). RNA evolution and the origins of life. Nature 338, 217–224. [CrossRef]
- Jukes, T. H. (1967). Indications of an evolutionary pathway in the amino acid code. Biochemical and Biophysical Research Communications 27, 573–578. [CrossRef]
- Kim, K. M. , and Caetano-Anollés, G. (2011). The proteomic complexity and rise of the primordial ancestor of diversified life. BMC Evol Biol 11, 140. [CrossRef]
- Kim, K. M. , and Caetano-Anollés, G. (2012). The evolutionary history of protein fold families and proteomes confirms that the archaeal ancestor is more ancient than the ancestors of other superkingdoms. BMC Evol Biol 12, 13. [CrossRef]
- Krupkin, M. , Matzov, D., Tang, H., Metz, M., Kalaora, R., Belousoff, M. J., et al. (2011). A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philosophical Transactions of the Royal Society B: Biological Sciences 366, 2972–2978. [CrossRef]
- Kurland, C. G. (2010). The RNA dreamtime: modern cells feature proteins that might have supported a prebiotic polypeptide world but nothing indicates that RNA world ever was. Bioessays 32, 866–871. [CrossRef]
- Lake, J. A. (1983). “Ribosome Evolution: The Structural Bases of Protein Synthesis in Archaebacteria, Eubacteria, and Eukaryotes,” in Progress in Nucleic Acid Research and Molecular Biology, eds. W. E. Cohn and K. Moldave (Academic Press), 163–194. [CrossRef]
- Lake, J. A. , Henderson, E., Clark, M. W., and Matheson, A. T. (1982). Mapping evolution with ribosome structure: intralineage constancy and interlineage variation. Proceedings of the National Academy of Sciences 79, 5948–5952. [CrossRef]
- Lazcano, A. , and Miller, S. L. (1996). The Origin and Early Evolution of Life: Prebiotic Chemistry, the Pre-RNA World, and Time. Cell 85, 793–798. [CrossRef]
- Lee, N. , Bessho, Y., Wei, K., Szostak, J. W., and Suga, H. (2000). Ribozyme-catalyzed tRNA aminoacylation. Nat Struct Mol Biol 7, 28–33. [CrossRef]
- Li, P. , Holliger, P., and Tagami, S. (2022). Hydrophobic-cationic peptides modulate RNA polymerase ribozyme activity by accretion. Nat Commun 13, 3050. [CrossRef]
- Lipovsek, D. , and Plückthun, A. (2004). In-vitro protein evolution by ribosome display and mRNA display. Journal of Immunological Methods 290, 51–67. [CrossRef]
- Loening, U. E. (1968). Molecular weights of ribosomal RNA in relation to evolution. Journal of Molecular Biology 38, 355–365. [CrossRef]
- Lohse, P. A. , and Szostak, J. W. (1996). Ribozyme-catalysed amino-acid transfer reactions. Nature 381, 442–444. [CrossRef]
- Lüking, A. , Stahl, U., and Schmidt, U. (1998). The Protein Family of RNA Helicases. Critical Reviews in Biochemistry and Molecular Biology 33, 259–296. [CrossRef]
- Macé, K. , and Gillet, R. (2016). Origins of tmRNA: the missing link in the birth of protein synthesis? Nucleic Acids Research 44, 8041–8051. [CrossRef]
- Maizels, N. , and Weiner, A. M. (1994). Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc Natl Acad Sci U S A 91, 6729–6734.
- Martínez Giménez, J. A. , and Tabares Seisdedos, R. (2022). A Cofactor-Based Mechanism for the Origin of the Genetic Code. Orig Life Evol Biosph 52, 149–163. [CrossRef]
- Mattick, J. , and Amaral, P. (2022). RNA, the Epicenter of Genetic Information. Boca Raton: CRC Press. [CrossRef]
- Melnikov, S. , Manakongtreecheep, K., and Söll, D. (2018). Revising the Structural Diversity of Ribosomal Proteins Across the Three Domains of Life. Molecular Biology and Evolution 35, 1588–1598. [CrossRef]
- Mizuuchi, R. , Ichihashi, N., Usui, K., Kazuta, Y., and Yomo, T. (2015). Adaptive Evolution of an Artificial RNA Genome to a Reduced Ribosome Environment. ACS Synth. Biol. 4, 292–298. [CrossRef]
- Moore, P. B. , and Steitz, T. A. (2002). The involvement of RNA in ribosome function. Nature 418, 229–235. [CrossRef]
- Müller, F. , Escobar, L., Xu, F., Węgrzyn, E., Nainytė, M., Amatov, T., et al. (2022). A prebiotically plausible scenario of an RNA–peptide world. Nature 605, 279–284. [CrossRef]
- Murakami, H. , Saito, H., and Suga, H. (2003). A Versatile tRNA Aminoacylation Catalyst Based on RNA. Chemistry & Biology 10, 655–662. [CrossRef]
- Murase, Y. , Nakanishi, H., Tsuji, G., Sunami, T., and Ichihashi, N. (2018). In Vitro Evolution of Unmodified 16S rRNA for Simple Ribosome Reconstitution. ACS Synth. Biol. 7, 576–583. [CrossRef]
- Mushegian, A. (2005). Protein content of minimal and ancestral ribosome. RNA 11, 1400–1406. [CrossRef]
- Natsidis, P. , Schiffer, P. H., Salvador-Martínez, I., and Telford, M. J. (2019). Computational discovery of hidden breaks in 28S ribosomal RNAs across eukaryotes and consequences for RNA Integrity Numbers. Sci Rep 9, 19477. [CrossRef]
- Nazar, R. N. , Yaguchi, M., and Willick, G. E. (1982). The 5S RNA – protein complex from yeast: a model for the evolution and structure of the eukaryotic ribosome. Can. J. Biochem. 60, 490–496. [CrossRef]
- Neubauer, C. , Gillet, R., Kelley, A. C., and Ramakrishnan, V. (2012). Decoding in the Absence of a Codon by tmRNA and SmpB in the Ribosome. Science 335, 1366–1369. [CrossRef]
- Neumann, H. , Wang, K., Davis, L., Garcia-Alai, M., and Chin, J. W. (2010). Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444. [CrossRef]
- Nierhaus, K. H. , Brimacombe, R., Nowotny, V., Pon, C. L., Rheinberger, H. J., Wittmann-Liebold, B., et al. (1987). New Aspects of Structure, Assembly, Evolution, and Function of Ribosomes. Cold Spring Harb Symp Quant Biol 52, 665–674. [CrossRef]
- Noller, H. F. (1999). 8 On the Origin of the Ribosome: Coevolution of Subdomains of tRNA and rRNA. Cold Spring Harbor Monograph Archive 37, 197–219. [CrossRef]
- Noller, H. F. (2004). The driving force for molecular evolution of translation. RNA 10, 1833–1837. [CrossRef]
- Noller, H. F. (2012). Evolution of Protein Synthesis from an RNA World. Cold Spring Harb Perspect Biol 4, a003681. [CrossRef]
- O’Brien, T. W. (2002). Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene 286, 73–79. [CrossRef]
- Oparin, A. I. (1953). Origin of Life. 2 edition. New York, NY: Dover Publications Inc.
- Parker, M. D. , and Karbstein, K. (2023). Quality control ensures fidelity in ribosome assembly and cellular health. Journal of Cell Biology 222, e202209115. [CrossRef]
- Petrov, A. S. , Bernier, C. R., Hsiao, C., Norris, A. M., Kovacs, N. A., Waterbury, C. C., et al. (2014). Evolution of the ribosome at atomic resolution. Proceedings of the National Academy of Sciences 111, 10251–10256. [CrossRef]
- Petrov, A. S. , Gulen, B., Norris, A. M., Kovacs, N. A., Bernier, C. R., Lanier, K. A., et al. (2015). History of the ribosome and the origin of translation. Proceedings of the National Academy of Sciences 112, 15396–15401. [CrossRef]
- Pflug, H. D. (1986). Morphological and chemical record of the organic particles in precambrian sediments. Systematic and Applied Microbiology 7, 184–189. [CrossRef]
- Pietromonaco, S. F. , Hessler, R. A., and O’Brien, T. W. (1986). Evolution of proteins in mammalian cytoplasmic and mitochondrial ribosomes. J Mol Evol 24, 110–117. [CrossRef]
- Polacek, N. , and Mankin, A. S. (2005). The Ribosomal Peptidyl Transferase Center: Structure, Function, Evolution, Inhibition. Critical Reviews in Biochemistry and Molecular Biology 40, 285–311. [CrossRef]
- Radakovic, A. , DasGupta, S., Wright, T. H., Aitken, H. R. M., and Szostak, J. W. (2022). Nonenzymatic assembly of active chimeric ribozymes from aminoacylated RNA oligonucleotides. Proceedings of the National Academy of Sciences 119, e2116840119. [CrossRef]
- Radford, F. , Elliott, S. D., Schepartz, A., and Isaacs, F. J. (2022). Targeted editing and evolution of engineered ribosomes in vivo by filtered editing. Nat Commun 13, 180. [CrossRef]
- Rajkowitsch, L. , Chen, D., Stampfl, S., Semrad, K., Waldsich, C., Mayer, O., et al. (2007). RNA Chaperones, RNA Annealers and RNA Helicases. RNA Biology 4, 118–130. [CrossRef]
- Ramakrishnan, V. (2009). The Ribosome: Some Hard Facts about Its Structure and Hot Air about Its Evolution. Cold Spring Harb Symp Quant Biol 74, 25–33. [CrossRef]
- Ramakrishnan, V. , and White, S. W. (1998). Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends in Biochemical Sciences 23, 208–212. [CrossRef]
- Roberts, E. , Sethi, A., Montoya, J., Woese, C. R., and Luthey-Schulten, Z. (2008). Molecular signatures of ribosomal evolution. Proceedings of the National Academy of Sciences 105, 13953–13958. [CrossRef]
- Root-Bernstein, M. , and Root-Bernstein, R. (2015). The ribosome as a missing link in the evolution of life. Journal of Theoretical Biology 367, 130–158. [CrossRef]
- Root-Bernstein, R. , and Root-Bernstein, M. (2019). The Ribosome as a Missing Link in Prebiotic Evolution III: Over-Representation of tRNA- and rRNA-Like Sequences and Plieofunctionality of Ribosome-Related Molecules Argues for the Evolution of Primitive Genomes from Ribosomal RNA Modules. International Journal of Molecular Sciences 20, 140. [CrossRef]
- Roundtree, I. A. , Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200. [CrossRef]
- Saito, H. , Kourouklis, D., and Suga, H. (2001). An in vitro evolved precursor tRNA with aminoacylation activity. The EMBO Journal 20, 1797–1806. [CrossRef]
- Saito, K. , Green, R., and Buskirk, A. R. (2020). Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing. eLife 9, e55002. [CrossRef]
- Schimmel, P. (1987). AMINOACYL tRNA SYNTHETASES: GENERAL SCHEME OF STRUCTURE-FUNCTION RELATIONSHIPS IN THE POLYPEPTIDES AND RECOGNITION OF TRANSFER RNAS. Annu. Rev. Biochem. 56, 125–158. [CrossRef]
- Schimmel, P. , Giegé, R., Moras, D., and Yokoyama, S. (1993). An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci U S A 90, 8763–8768. [CrossRef]
- Schimmel, P. , and Henderson, B. (1994). Possible role of aminoacyl-RNA complexes in noncoded peptide synthesis and origin of coded synthesis. Proc Natl Acad Sci U S A 91, 11283–11286.
- Schmied, W. H. , Tnimov, Z., Uttamapinant, C., Rae, C. D., Fried, S. D., and Chin, J. W. (2018). Controlling orthogonal ribosome subunit interactions enables evolution of new function. Nature 564, 444–448. [CrossRef]
- Schopf, J. W. , and Packer, B. M. (1987). Early Archean (3.3-billion to 3.5-billion-year-old) microfossils from Warrawoona Group, Australia. Science 237, 70–73. [CrossRef]
- Shajani, Z. , Sykes, M. T., and Williamson, J. R. (2011). Assembly of bacterial ribosomes. Annu Rev Biochem 80, 501–526. [CrossRef]
- Shirokikh, N. E. , Dutikova, Y. S., Staroverova, M. A., Hannan, R. D., and Preiss, T. (2019). Migration of Small Ribosomal Subunits on the 5′ Untranslated Regions of Capped Messenger RNA. International Journal of Molecular Sciences 20, 4464. [CrossRef]
- Shirokikh, N. E. , and Preiss, T. (2018). Translation initiation by cap-dependent ribosome recruitment: Recent insights and open questions. WIREs RNA 9, e1473. [CrossRef]
- Simsek, D. , Tiu, G. C., Flynn, R. A., Byeon, G. W., Leppek, K., Xu, A. F., et al. (2017). The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 169, 1051-1065.e18. [CrossRef]
- Sloan, D. B. , Triant, D. A., Wu, M., and Taylor, D. R. (2014). Cytonuclear Interactions and Relaxed Selection Accelerate Sequence Evolution in Organelle Ribosomes. Molecular Biology and Evolution 31, 673–682. [CrossRef]
- Sloan, K. E. , Warda, A. S., Sharma, S., Entian, K.-D., Lafontaine, D. L. J., and Bohnsack, M. T. (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biology 14, 1138–1152. [CrossRef]
- Smith, T. F. , Lee, J. C., Gutell, R. R., and Hartman, H. (2008). The origin and evolution of the ribosome. Biology Direct 3, 16. [CrossRef]
- Steitz, T. A. , and Moore, P. B. (2003). RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends in Biochemical Sciences 28, 411–418. [CrossRef]
- Stoppel, R. , and Meurer, J. (2012). The cutting crew – ribonucleases are key players in the control of plastid gene expression. Journal of Experimental Botany 63, 1663–1673. [CrossRef]
- Sun, F.-J. , and Caetano-Anollés, G. (2010). The ancient history of the structure of ribonuclease P and the early origins of Archaea. BMC Bioinformatics 11, 153. [CrossRef]
- Tagami, S. , and Li, P. (2023). The origin of life: RNA and protein co-evolution on the ancient Earth. Dev Growth Differ. [CrossRef]
- Tamura, K. (2011). Ribosome evolution: Emergence of peptide synthesis machinery. J Biosci 36, 921–928. [CrossRef]
- Taylor, D. J. , Devkota, B., Huang, A. D., Topf, M., Narayanan, E., Sali, A., et al. (2009). Comprehensive Molecular Structure of the Eukaryotic Ribosome. Structure 17, 1591. [CrossRef]
- Tjhung, K. F. , Shokhirev, M. N., Horning, D. P., and Joyce, G. F. (2020). An RNA polymerase ribozyme that synthesizes its own ancestor. Proceedings of the National Academy of Sciences 117, 2906–2913. [CrossRef]
- Trifonov, E. N. (2000). Consensus temporal order of amino acids and evolution of the triplet code. Gene 261, 139–151. [CrossRef]
- Van Treeck, B. , and Parker, R. (2018). Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell 174, 791–802. [CrossRef]
- Walker, S. I. , Packard, N., and Cody, G. D. (2017). Re-conceptualizing the origins of life. Philos Trans A Math Phys Eng Sci 375, 20160337. [CrossRef]
- Walsh, J. B. (1995). How often do duplicated genes evolve new functions? Genetics 139, 421–428. [CrossRef]
- Weiner, A. M. , and Maizels, N. (1999). The genomic tag hypothesis: modern viruses as molecular fossils of ancient strategies for genomic replication, and clues regarding the origin of protein synthesis. Biol Bull 196, 327–328; discussion 329-330. [CrossRef]
- Westhof, E. , Thornlow, B., Chan, P. P., and Lowe, T. M. (2022). Eukaryotic tRNA sequences present conserved and amino acid-specific structural signatures. Nucleic Acids Research 50, 4100–4112. [CrossRef]
- Wimberly, B. T. , Brodersen, D. E., Clemons, W. M., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., et al. (2000). Structure of the 30S ribosomal subunit. Nature 407, 327–339. [CrossRef]
- Withey, J. H. , and Friedman, D. I. (2003). A Salvage Pathway for Protein Synthesis: tmRNA and Trans-Translation. Annual Review of Microbiology 57, 101–123. [CrossRef]
- Wittmann, H. G. (1980). “Structure and Evolution of Ribosomes,” in Chemical Recognition in Biology Molecular Biology, Biochemistry and Biophysics., eds. F. Chapeville and A.-L. Haenni (Berlin, Heidelberg: Springer), 376–397. [CrossRef]
- Wittmann-Liebold, B. (1986). “Ribosomal Proteins: Their Structure and Evolution,” in Structure, Function, and Genetics of Ribosomes Springer Series in Molecular Biology., eds. B. Hardesty and G. Kramer (New York, NY: Springer), 326–361. [CrossRef]
- Wochner, A. , Attwater, J., Coulson, A., and Holliger, P. (2011). Ribozyme-Catalyzed Transcription of an Active Ribozyme. Science 332, 209–212. [CrossRef]
- Woese, C. (1970). Molecular Mechanics of Translation : a Reciprocating Ratchet Mechanism. Nature 226, 817–820. [CrossRef]
- Woese, C. R. (1965). On the evolution of the genetic code. Proc Natl Acad Sci U S A 54, 1546–1552.
- Woese, C. R. , Dugre, D. H., Dugre, S. A., Kondo, M., and Saxinger, W. C. (1966). On the Fundamental Nature and Evolution of the Genetic Code. Cold Spring Harb Symp Quant Biol 31, 723–736. [CrossRef]
- Zhang, H. , Li, D., Zhao, X., Pan, S., Wu, X., Peng, S., et al. (2020). Relatively semi-conservative replication and a folded slippage model for short tandem repeats. BMC Genomics. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).