1. Introduction
Metastasis is the leading cause of death from cancer, the second leading cause of death worldwide, accounting for 8.8 million deaths [
1]. Metastasis begins to release from primary tumors, which invade adjacent tissue, and angiogenesis to form blood vessels to form new ones to nourish and maintain the activity of these cells [
2]. Once there is an invasion of neighboring tissues, intravasation into the circulation may occur, reaching the interior of blood or lymphatic vessels, which disseminates throughout the body and will allow these cells to survive the physical stress of the environment [
3]. Currently, detecting circulating tumor cells (CTCs) in blood has become a front-line technology that can screen cancer noninvasively, diagnose cancer early, monitor tumor progression, and evaluate therapy responses [
5,
6]. The diagnosis of tumors is usually made through punctures and biopsies, where a small sample of the patient’s tumor is taken during a surgical procedure [
7]. These cells are analyzed by specialists and classified according to tissue and cellular morphology by the presence of already-established markers that allow the correct identification of the tumor. However, the traditional method of tissue removal via biopsy is a technique that requires highly trained personnel and high cost as it involves patient hospitalization, is painful, and is time-consuming. It still has some limitations, such as (1) tumor heterogeneity; (2) follow-up, because the difficulty in taking biopsies can compromise the quality of the analysis. However, developing non-invasive methods to detect and monitor tumors remains a significant challenge in oncology.
It is worth mentioning that detecting circulating tumor cells (CTCs) poses a significant challenge due to their rarity. Typically, only one to a few CTCs are present in 1 ml of a patient’s blood, which contains about 10 million white blood cells and 5 billion red blood cells [
7]. Therefore, there is an urgent need for high-efficiency and high-purity methods to detect CTCs in blood samples, which can aid in faster decision-making and a better understanding of the type of tumor the patient has. An alternative detecting approach based on antibodies [
8,
9,
10], aptamers [
11,
12,
13,
14], and ct-DNA [
15,
16] has been a breakthrough in the isolation of circulating tumor cells (CTCs) from patient blood. Many methodologies have received approval from the U.S. Food and Drug Administration (FDA) for clinical use, providing efficient and specific methods for capturing CTCs [
14,
15,
16]. However, despite their effectiveness, some techniques lack interaction between cells and the ligand-coated surface due to the large surface-to-volume ratio and short diffusion distance. As such, further advancements are needed to minimize these limitations and enhance the accuracy of CTC isolation methods.
The isolation and detection of circulating tumor cells (CTCs) has been made possible through a variety of techniques, including immunomagnetic beads [
18], size-based filtration systems [
19], and microfluidic devices [
20,
21]. Of these techniques, microfluidics have emerged as a promising approach due to their ability to handle small blood volumes and provide high sensitivity and specificity. Detecting CTCs using microfluidics involves passing a small blood sample through a device designed to capture and identify CTCs, which may use antibody-coated surfaces or microfluidic structures that mimic blood vessels to isolate and trap CTCs. Once captured, CTCs can be analyzed using various methods such as imaging, genetic analysis, or drug sensitivity testing, providing valuable insights into the type and progression of cancer and the potential effectiveness of specific treatments [
21].
Later, using nanoparticles in conjunction with microfluidics has greatly improved the sensitivity and specificity of CTCs detection [
22,
23,
24]. Nanoparticles offer a unique platform for biomarker detection due to their easy synthesis, surface chemistry, biocompatibility, and remarkable structure. Here, we have investigated using gold nanoparticles on a lateral filter array (LFA) to capture and release cancer cells from whole blood. Gold nanoparticles were functionalized with anti-EpCAM antibodies to coat a microfluidic device, allowing for the specific capture of CTCs that express EpCAM (Epithelial Cell Adhesion Molecule). This approach represents a significant advancement in CTC detection, enabling more accurate and reliable cancer diagnosis.
2. Experimental part
2.1. Reagents and materials
The following reagents were purchased from Sigma-Aldrich (St. Louise, MO, USA) and used as received: Sylgard 184, ethanol, Dulbecco’s phosphate buffered saline (DPBS) with calcium chloride and magnesium chloride, 11-mercaptoundecanoic acid (MUA), 12-mercaptododecanoic acid, N-hydroxysuccinimide (NHS) ester, Avidin, gold nanoparticle 5 nm, bovine serum albumin (BSA), N-[3-(trimethoxysilyl)propyl]ethylenediamine (AEAPTMS), and Tween 20. Anti-EpCam (Anti-Human CD326) was purchased from eBioscience, San Diego, CA.
2.2. Fabrication of the LFA Device
The LFA device was fabricated using a previously described method [
25]. The channel pattern of the LFA device was sketched using AutoCAD. LFA consists of a 1-in. × 3-in. glass slide and a poly(dimethylsiloxane) (PDMS) layer containing four microchannels with a pattern on the upper surface. Based on the silicon master, a PDMS substrate was fabricated using soft lithography. Thoroughly mixed liquid state PDMS (base/curing agent = 10:1) was cast on the silicon master. The PDMS -loaded aluminum foil bowl was put in a vacuum chamber to remove bubbles from PDMS. The aluminum foil bowl was cured at 65°C for at least 4 hours. PDMS was polymerized and formed a transparent elastic substrate after curing it in the oven. The PDMS was then peeled off from the silicon master, trimmed to fit a microscope slide, and punched holes at the inlet and outlet. U.V. Ozone bonded the glass microscope slide and PDMS substrate for 5 minutes. The image of the filters are shown in
Figure S1A, and device is shown in
Figure S1b.
2.3. Functionalization of the LFA device
To prepare the device for CTCs capture, the LFA underwent a series of chemical modifications. Initially, the LFA was treated with 1 wt% solution of AEAPTMS in ethanol for one hour at room temperature and subsequently washed with ethanol. Then, a 0.01 wt% ethanol solution of NHS-functionalized gold nanoparticles (AuNPs) was applied to the device and incubated for 30 minutes at room temperature. The device was rinsed with ethanol to facilitate amide formation, which binds the AuNPs to the aminated surface. Finally, the channels were filled with a 20 µg/mL avidin solution in DPBS and left to incubate for 30 minutes, completing the preparation of the device for CTC capture.
Then 200 µl of DPBS was introduced to wash away excess avidin in the LFA device. Then LFA device was functionalized with anti-EpCAM. A biotinylated anti-EpCAM solution of 70 µL with a concentration of 10 µg/mL was introduced to the LFA device and incubated for 30 minutes. Before each experiment, the LFA device was passivated with a DPBS buffer containing 1% BSA (bovine serum albumin), followed by rinsing with DPBS to prevent nonspecific capture of normal blood cells.
2.4. Cell Culture
The L3.6PL and BxPC-3 pancreatic cancer cell lines were acquired from the American Type Culture Collection (ATCC, Manassas, VA). Following the recommended protocols, the cancer cell lines were expanded and used when they reached 85% confluence. These cells were cultured in DMEM medium (ATCC) supplemented with 10% fetal bovine serum (FBS; GIBCO of Thermo Fisher) and 100 units/mL penicillin-streptomycin (Cellgro, Manassas, VA) under incubation conditions of 37 °C with 5% CO2. The CCRF-CEM cells purchased from the ATCC were cultured in RPMI 1640 medium (ATCC) with 10% FBS and 100 units/mL penicillin-streptomycin. Before preparing the cell samples, the culture medium was removed from the flask, and DPBS was added to rinse the flask and remove any impurities. Trypsin EDTA (GIBCO, Fisher Scientific) of 2 mL, a concentration of 0.25%, was introduced and incubated for 10 minutes to detach the cells from the flask. The growth medium of 6 mL was added to the flask to neutralize the cells. The detached cells were then rinsed with DPBS 2 times to remove impurities. Finally, the cells were resuspended in 1 mL DPBS. CCRF-CEM cells were floating cells for cell sample preparation. The cells were withdrawn from the flask, rinsed with DPBS 2 times, and resuspended in 1 mL of DPBS. Following the manufacturer’s instruction, these cells were stained with Vybrant dyes (Thermo Fisher Scientific, NH). The dyed cells were then rinsed with DPBS before spiking into either DPBS buffer or blood samples. The infusion flow rate of the sample into the device varied from 0.5 ml/h to 2.0 µl/s. After the sample infusion, the device was washed by infusing 250 µl of DPBS.
2.5. Imaging Fluorescence
Fluorescence signals of tumor cells captured in the device were collected using an Olympus IX71 fluorescence microscope (Olympus America, PA) equipped with a scientific-grade CCD camera (Hamamatsu C4742-80-12AG). For devices functionalized with antibodies, the captured cells were first trypsinized with one channel volume of 0.25% trypsin-EDTA for 10 minutes, followed by pumping DPBS from the outlet at 2.0 µL/s. The viability of released L3.6PLpl cells was determined by staining with 4% Trypan blue (Fisher Scientific, NH) following the manufacturer’s instruction.
2.6. Blood Processing
Clinical Samples Blood samples were collected from patients with metastatic colorectal cancers at the U.F. Health Cancer Center. According to the protocol approved by the U.F. institutional review board (IRB), all specimens were processed within three h after the blood draw. Before the clinical test, two LFA devices were functionalized with anti-EpCAM and passivated with 1% BSA solution, as discussed above. The clinical samples were diluted two times (blood: DPBS=1:1) and infused into the antibody-functionalized device at 1.5 µL/s. A total of 8 mL of diluted clinical samples were injected into two LFA devices in parallel (i.e., 4 mL for each device). After the infusion process, each device was washed with DPBS for impurity removal. Next, 50 µL of a 4% paraformaldehyde (PFA) solution was added to the device and incubated for 10 minutes for cell fixation. The device was washed with DPBS before adding 50 µL of a 0.2% Triton X-100 solution for 10 minutes to permeabilize the cell membrane. After washing again, a mixture of 10 µg/mL anti-CD45-PE, 10 µg/mL anti-cytokeratin-FITC, and 500 nM DAPI was introduced to the device for staining and incubated for 30 minutes. The device was then rewashed and mounted onto the fluorescence microscope stage for CTC enumeration. DAPI+, CD45-, and CK+ cells are counted as CTCs. Other cells, such as white blood cells (DAP+, CD45+, C.K.-, bought at Sigma-Aldrich MA), red blood cells (DAPI-), or others (e.g., triple positive), are excluded.
3. Results and discussion
To detect CTCs, we developed a microfluidic laminar flow device (LFA) and evaluated its capture performance with and without nanoparticles. We optimized the flow rate to determine the ideal conditions for capturing cells. To test the device’s efficacy, we infused approximately 1000 fluorescence-labeled L3.6PL cells into the LFA.
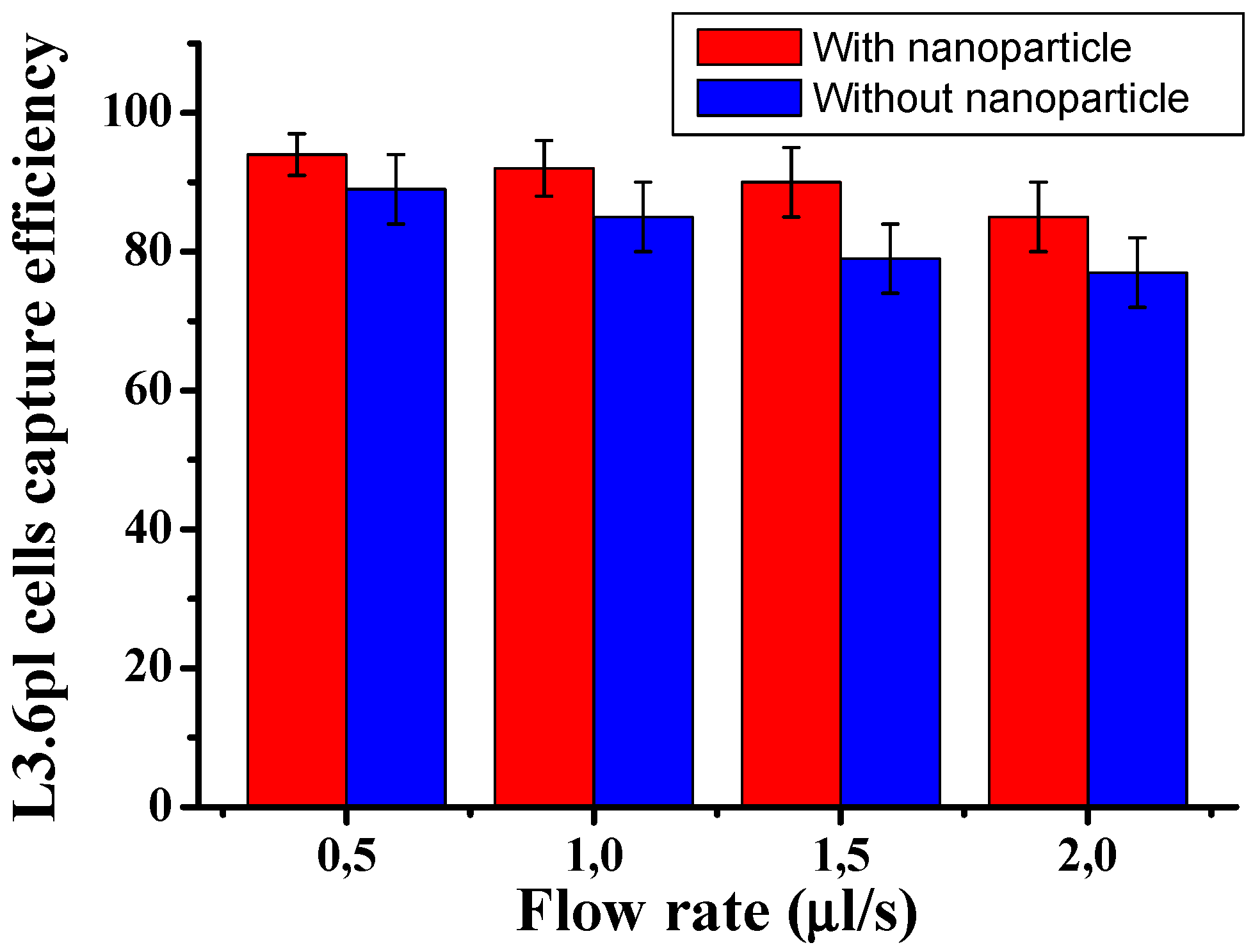
Figure 1 shows the cell capture ratio under different flow rates. The capture efficiency was optimized by varying the flow rate and comparing the capture performance in the presence and absence of nanoparticles. The results showed that the capture efficiency could reach over 94±3.8% at a flow rate of 0.5 µL/s when nanoparticles were present. However, as the flow rate was increased from 0.5 to 1.5 µL/s, the capture efficiency decreased from 94% to 90%. The difference is statistically not significant at a low flow rate CTCs. Also,
Figure 1 shows that using AuNP-conjugated antibodies significantly enhanced the capture of target cells compared to without nanoparticles. The capture rate of CTCs in the microfluidic system was calculated using the formula: Cf/Cs x 100%, where Cf is the number of cancer cells counted under a fluorescence microscope, and Cs is the total number of cancer cells spiked into the DPBS buffer. These results suggest that the LFA device functionalized with nanoparticles can achieve highly efficient and specific capture of CTCs.
Figure 1.
Cells captured in the LFA device under two conditions: with and without gold nanoparticle coated. L3.6PL cells were captured in the LFA device under different flow rates. Error bars show a range (n=3).
Figure 1.
Cells captured in the LFA device under two conditions: with and without gold nanoparticle coated. L3.6PL cells were captured in the LFA device under different flow rates. Error bars show a range (n=3).
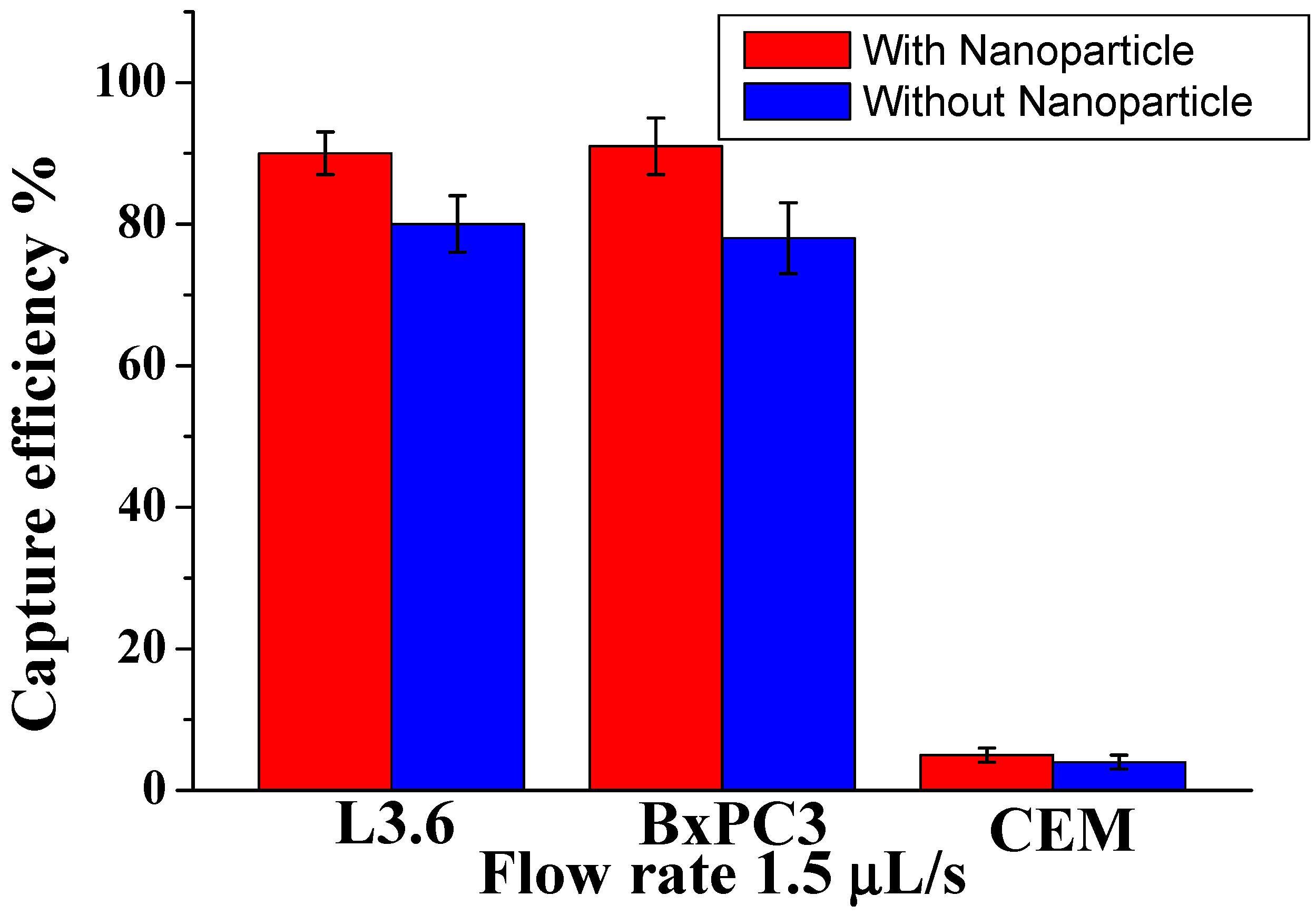
To demonstrate the efficiency among different types of cells with different sizes, we studied a mixture of pancreatic cancer cell lines: target L3.6PL cells, BxPC3 (EpCAM+), and control CCRF-CEM cells (EpCAM−). As shown in
Figure 2, for L3.6PL cells (diameter 16±3 µm), the capture efficiency was 91±4.5%. at 1.5 µL/s; for BxPC3 cells (diammeter 16.0±2 µm) the corresponding capture efficiency was around 88±3.5%, while most control CEM cells (diammeter 13.0±2 µm) were removed by washing (capture of only 6.0±2%). The possible reason is that the CCRF-CEM cells do not express EpCAM, and their sizes are slightly smaller than other cells tested. These results demonstrate that our device, conjugated with nanoparticles and Anti-Epcam, can efficiently isolate CTCs at low numbers by decreasing surface contact and increasing the binding affinity efficiency. Also, our methodology increased the surface roughness compared with the plain surface, allowing local topographic interactions between the aptamer-coated and nanoparticle.
Figure 2.
Comparison of capture efficiency using specific and nonspecific cells at an optimal flow rate of 1.5 µL/s. Error bars show a range (n=3).
Figure 2.
Comparison of capture efficiency using specific and nonspecific cells at an optimal flow rate of 1.5 µL/s. Error bars show a range (n=3).
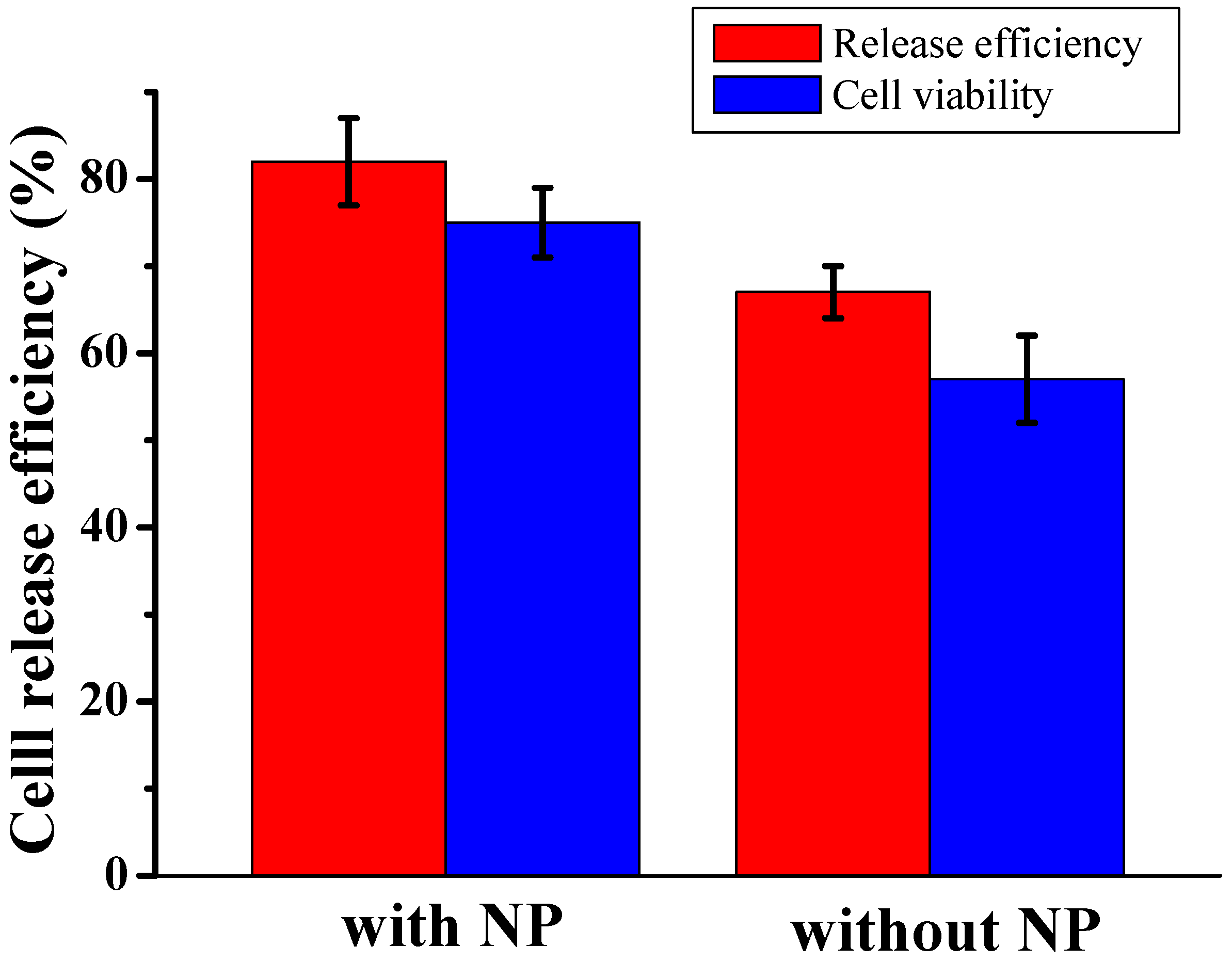
The release of captured cells, both in the presence and absence of nanoparticles, was achieved through trypsinization. The efficiency of release was measured as the ratio of the number of cells released to the number of cells captured.
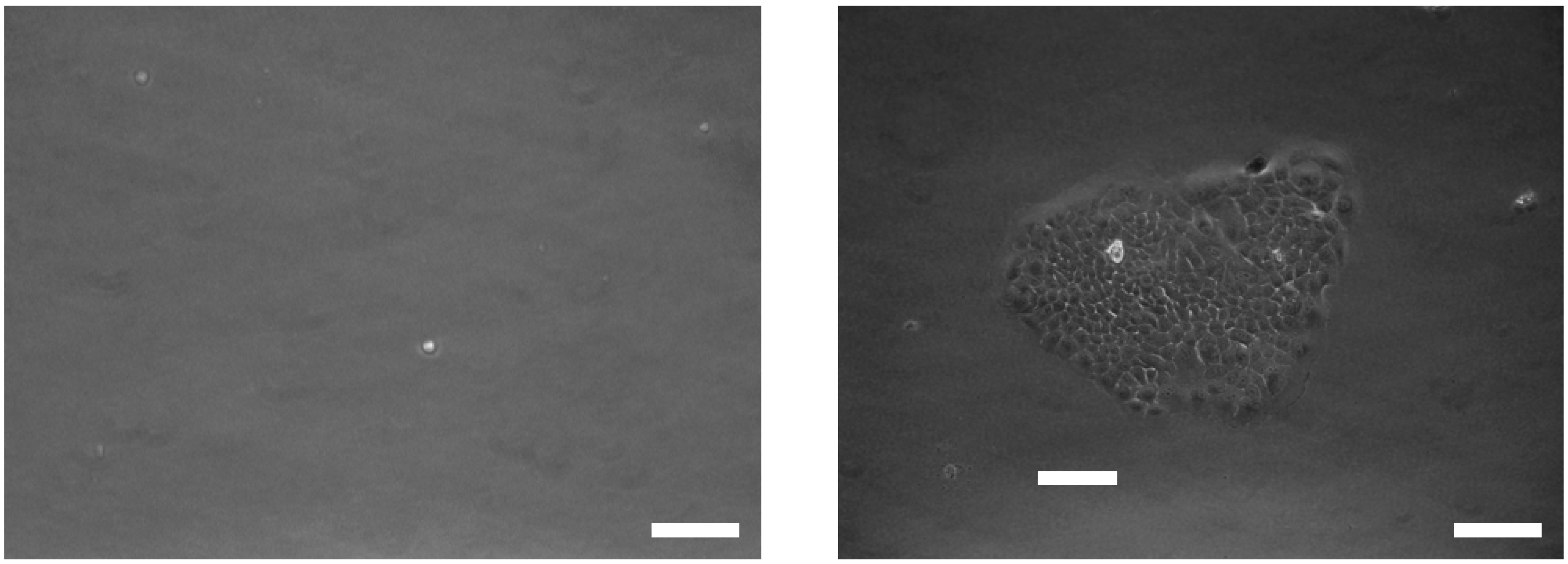
Figure 3 shows the release efficiency achieved by flowing trypsin (1 mg/mL) for 30 minutes and the viability of the recovered CTCs. In the presence of nanoparticles, 80±7% of the captured cells remained viable after the capture and release process, making them suitable for subsequent cellular analysis. In contrast, the device without nanoparticles showed a lower release efficiency and cell viability, with a decrease of approximately 20%. These improvements were strongly correlated with the inclusion of the nanoparticle into the microfluidic chip. As expected, our results show slightly higher cell viability with the nanoparticles, oscillating between 87 and 93% with control and released cells. The released L3.6PL cells were cultured in a complete growth medium after capture and release, and
Figure 4 shows the cell culture at different time points after release.
Figure 3.
Release efficiency and cell viability of recovered L3.6PL cell in DPBS between presence and absence of nanoparticle, at flow rates 1.5 μL/s.
Figure 3.
Release efficiency and cell viability of recovered L3.6PL cell in DPBS between presence and absence of nanoparticle, at flow rates 1.5 μL/s.
Figure 4.
Bright-field and fluorescent images of released cells. Images of released cells, two days (left) and seven days (right) are postrelease (scale bar 10 μm).
Figure 4.
Bright-field and fluorescent images of released cells. Images of released cells, two days (left) and seven days (right) are postrelease (scale bar 10 μm).
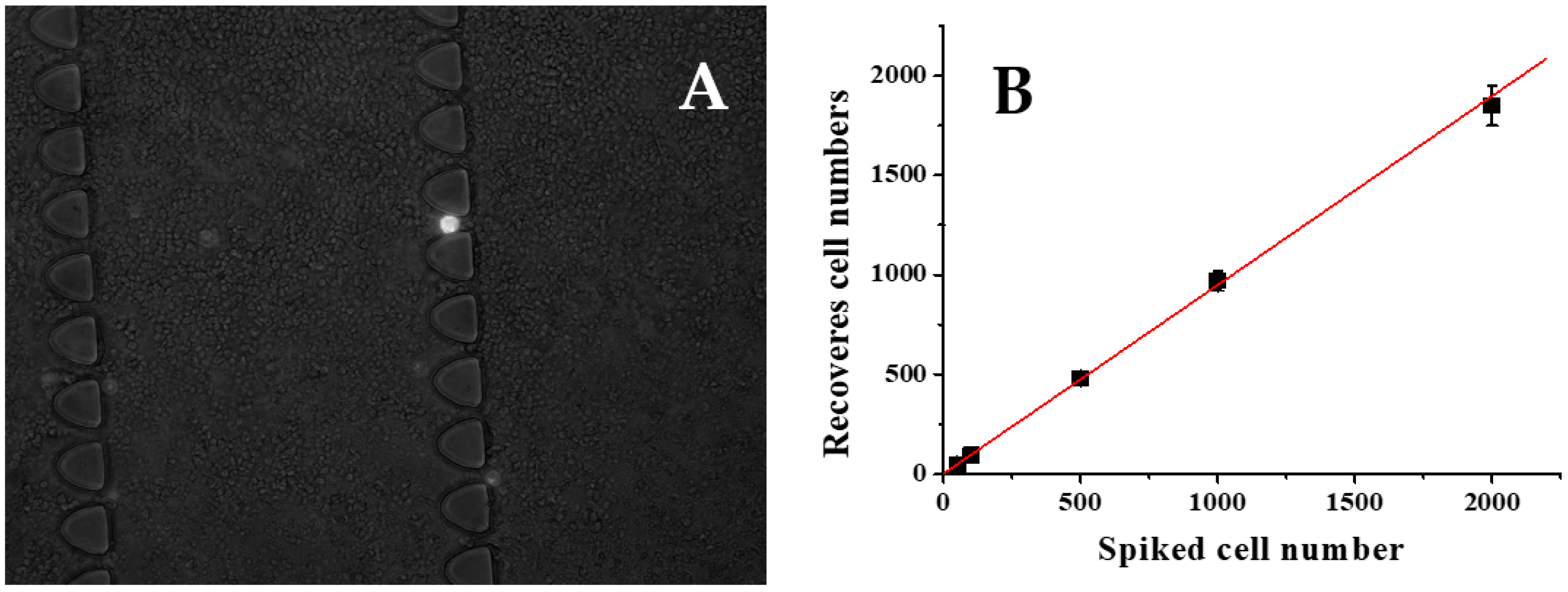
To assess the effectiveness of our CTC capture system in whole human blood, we conducted experiments using blood samples spiked with varying concentrations of L3.6PL cells under optimal conditions. Specifically, 2 mL of whole blood was spiked with L3.6PL cells at concentrations of 50, 100, 500, 1000, and 2000 cells per mL, and the samples were processed using the microfluidic device with a flow rate of 1.5 µL/s. Our results, presented in
Figure 5B, demonstrate a strong linear correlation (R2 = 0.998, n = 3) between the number of spiked cells and the number of cells captured, with an average capture efficiency of 90±3.5%. These findings suggest that our system can effectively capture CTCs from whole blood, even at low concentrations, and has the potential for use in clinical applications.
Figure 5.
The capture of target cells from diluted blood. Different L3.6pl cells are spiked in mL of 2-time diluted healthy whole blood and infused into the functionalized LFA device. A) Comparison of spiked L3.6pl cells and captured L3.6pl cells. B) Captured L3.6pl cells (white) and nonspecific captured white blood cells (gray).
Figure 5.
The capture of target cells from diluted blood. Different L3.6pl cells are spiked in mL of 2-time diluted healthy whole blood and infused into the functionalized LFA device. A) Comparison of spiked L3.6pl cells and captured L3.6pl cells. B) Captured L3.6pl cells (white) and nonspecific captured white blood cells (gray).
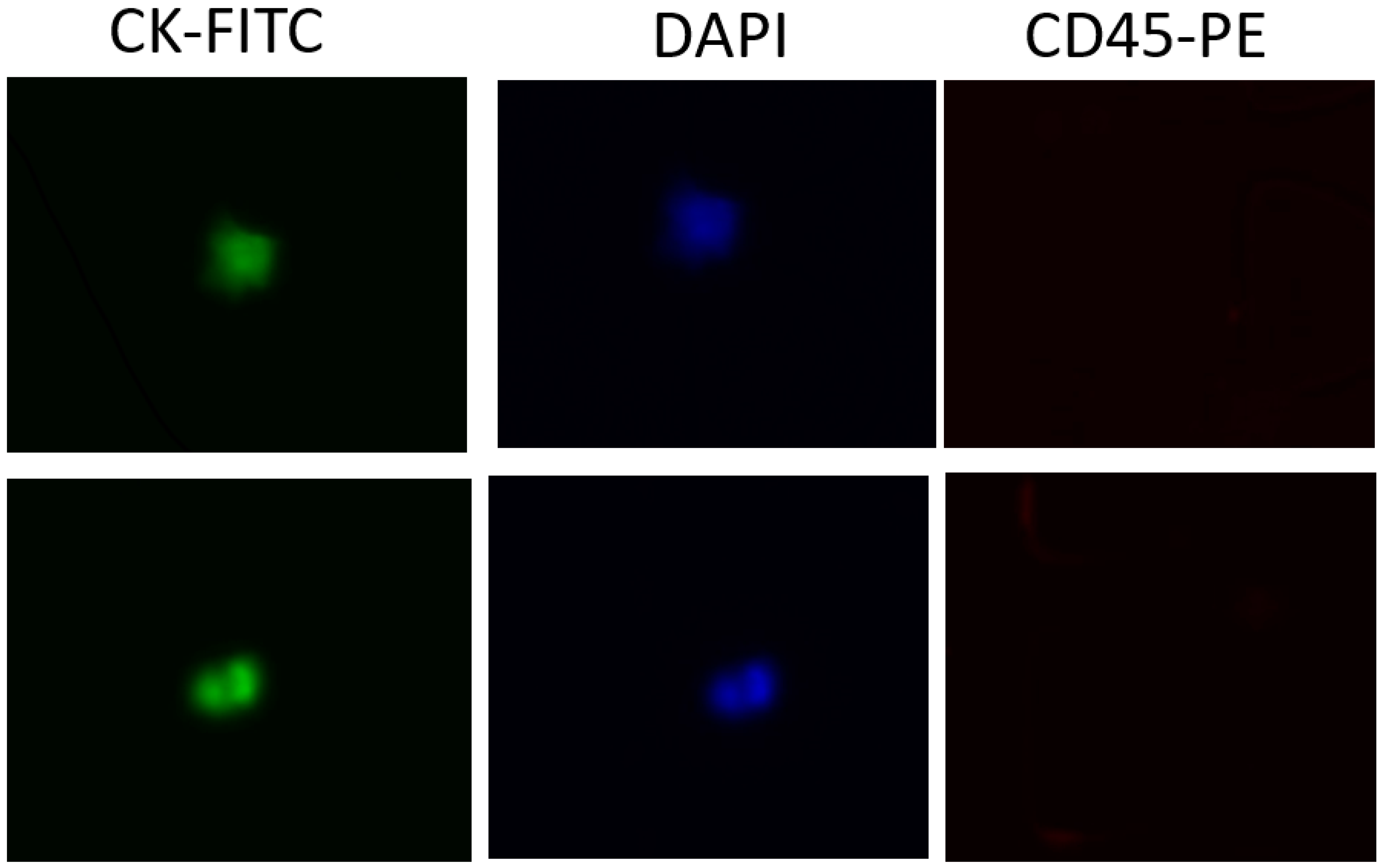
We evaluated the clinical utility of our methodology using blood samples obtained from a small group of patients with metastatic pancreatic cancer. Half of a blood sample (4 mL) was diluted with an equal volume of DPBS and then processed in the antibody-functionalized LFA device to reduce the effect of viscosity variation among patients. This dilution also helped reduce blood viscosity. The nucleated cell sample was then introduced to the LFA device at a flow rate of 1.5 µl/s. After a DPBS washing step, captured cells were fixed with 4% paraformaldehyde (PFA) for 10 minutes and then permeabilized with 0.2% Triton X-100 for 10 minutes, followed by permeabilized with 0.2% Triton X-100 for 10 minutes. Then, a mixture of labeling dye containing 10% of 10 µg/ml FITC anti-cytokeratin, 10% of 10 µg/ml P.E. anti-CD45 and 80% of 500 nM DAPI was introduced into the LFA device and incubated for 30 min. After DPBS washing, CTCs were enumerated under an optical microscope. To eliminate false-positive signals, we only considered cytokeratin-positive, CD45-negative, DAPI -positive (CK+/CD45-/DAPI+) cells as CTCs (
Figure 6). Any other labeling formats were considered false positive signals or cell debris. The average CTCs detected in our device was 3-8 CTCs/mL.
Figure 6.
Sample images of captured CTCs. A) CK-FITC channel image B) DAPI channel image. C) CD45-PE channel image.
Figure 6.
Sample images of captured CTCs. A) CK-FITC channel image B) DAPI channel image. C) CD45-PE channel image.