1. Introduction
Autism Spectrum Disorder (ASD) is a neurodevelopmental disorder characterized by social communication and interaction challenges, restricted interests, and repetitive behaviors [
1]. Early intervention has been shown to improve developmental outcomes for individuals with ASD, with earlier onset in infancy leading to better outcomes [
2]. In the United States, the prevalence of ASD has been increasing and is currently estimated to affect approximately 1 in 36 children [
3], which represents about 2.3% of the population. This statistic underscores the need for developing early and low-cost ASD indicators that could support earlier, more efficient diagnoses, personalized treatments, and improved quality of life for people with ASD. Furthermore, earlier diagnosis of ASD could lead to the development of even earlier parent-implemented behavioral interventions that support very early communication and social interaction skills [
4].
Widely accepted assessment tools for ASD include the Autism Diagnostic Observation Schedule (ADOS) [
5,
6] and the Autism Diagnostic Interview (ADI) [
5,
7]. These measures offer many advantages. First, when conducted by trained clinicians, they inform reliable diagnoses and allow for a more systematic and reproducible behavioral characterization of participants across studies. Second, ADI and ADOS profiles enable the formation of ASD subgroups based on perceived or confirmed symptoms that may be associated with various clinical trajectories and genetic markers [
5]. While these standardized assessment of ASD requires advanced professional training and a more comprehensive evaluation than many medical conditions.
Physiological recordings can potentially be used to derive reliable diagnostic biomarkers of ASD. One such modality, the electroencephalogram (EEG), is being increasingly investigated as a possible clinical tool for tracking abnormal brain development. For example, one study used spectral and nonlinear features of EEG signals to predict the diagnosis of ASD in infancy [
8]. However, collection of high quality EEG data in infants or young participants is subject to substantial methodological hurdles that are contingent upon the experimental settings and the intended analyses [
9].
Previous research indicates that ASD symptoms could be linked with sympathetic arousal and parasympathetic activity [
10,
11]. Electrodermal activity (EDA) is one of the primary measures for sympathetic arousal. In [
12], the authors found EDA in children with ASD to be significantly correlated with anxiety symptoms. Children with ASD and high anxiety levels demonstrated substantially reduced EDA relative to ASD participants with low anxiety and neurotypical controls [
13]. To the best of our knowledge, no one has studied the use of extracted features from ECG with parasympathetic and sympathetic symptoms in individuals with ASD, despite the potential benefits of such an approach.
In recent decades, some studies have identified features extracted from electrocardiogram (ECG) signals to be associated with ASD [
14,
15]. Among these promising ECG features is heart rate variability (HRV), which has been studied as a possible non-invasive biomarker for ASD. HRV reflects alterations in parasympathetic and sympathetic nervous system activity, and lower HRV in ASD may reflect abnormal neuronal connectivity within the autonomic nervous system (ANS) and between the ANS and other brain regions [
15,
16]. A whole-brain functional magnetic resonance imaging (fMRI) study [
17] of children with ASD and children with developmental language disorders revealed decreased functional connectivity between the frontal and parietal-occipital regions, and between the anterior and posterior insular cortex and the limbic cortex. In addition, due to common anatomical brain regions involved in both autonomic dysfunction and social-emotional dysregulation [
18,
19] measures of autonomic activity constitute a prime candidate for the development of a biomarker and for the study of ASD in general.
Recently, many Machine Learning (ML) approaches have been proposed to improve the early identification of ASD and to study potential ASD biomarkers with MRI [
20,
21,
22]. However, utilizing MRI may not be viable for infants between the ages of 3-6 months. Infants at this age are typically very active and may not be able to stay still for such an extended period, which can result in poor image quality. Earlier contributions used classical machine learning methods (such as with MRI/fMRI [
20,
23,
24,
25,
26]. More recently, deep learning methods have demonstrated a considerable advantage over classical approaches due to their ability to rely on hidden representations of extracted features in MRI/fMRI [
27]. Although an increasing number of studies proposed ML methods for ASD diagnosis using behavioral or psychophysiological data, such as EEG [
28,
29], scant attention has been given to the application of ML to ECG signals for predicting ASD. This study used non-invasive ECG sensors to record infants’ heart activity during interactions with toys and their caregivers. Participants were infants with a typical or elevated likelihood of developing ASD, based on familial history. The ECG signals were pre-processed, and HRV features were extracted and used to train machine learning models. By investigating how predictive these models are, we aim to evaluate the potential of ECG-captured autonomic activity to indicate ASD likelihood.
2. Methods
2.1. Acquisition protocol
A small ECG sensor (Actiwave Cardio; CamNTech) with two electrodes was put on the infants’ chest, and raw signals were recorded. The infants then participated in two experiments: one involving interactions with objects only (OIX) and a second involving interactions with their caregiver (PIX). During the OIX condition, infants were placed on their caregiver’s lap, and five objects were presented sequentially on the table for 1-2 minutes each. The caregiver did not interact with their infant during this time. During the PIX condition, infants were lying on their backs on a table, and caregivers interacted with them without toys for nine minutes. Both experiments were conducted with the same infants. Only OIX or PIX recording is available for some participants, either because of technical issues or because the infant’s fussing required early experiment termination. Recordings from N=70 infants of age 3-6 months were available for this study. Participants were either at elevated familial likelihood for ASD (EL) because they had an older full biological sibling with ASD (n=31) or at a typical likelihood for ASD (TL) given the absence of a family history of ASD in first or second-degree relatives (n=39).
2.2. ECG Preprocessing
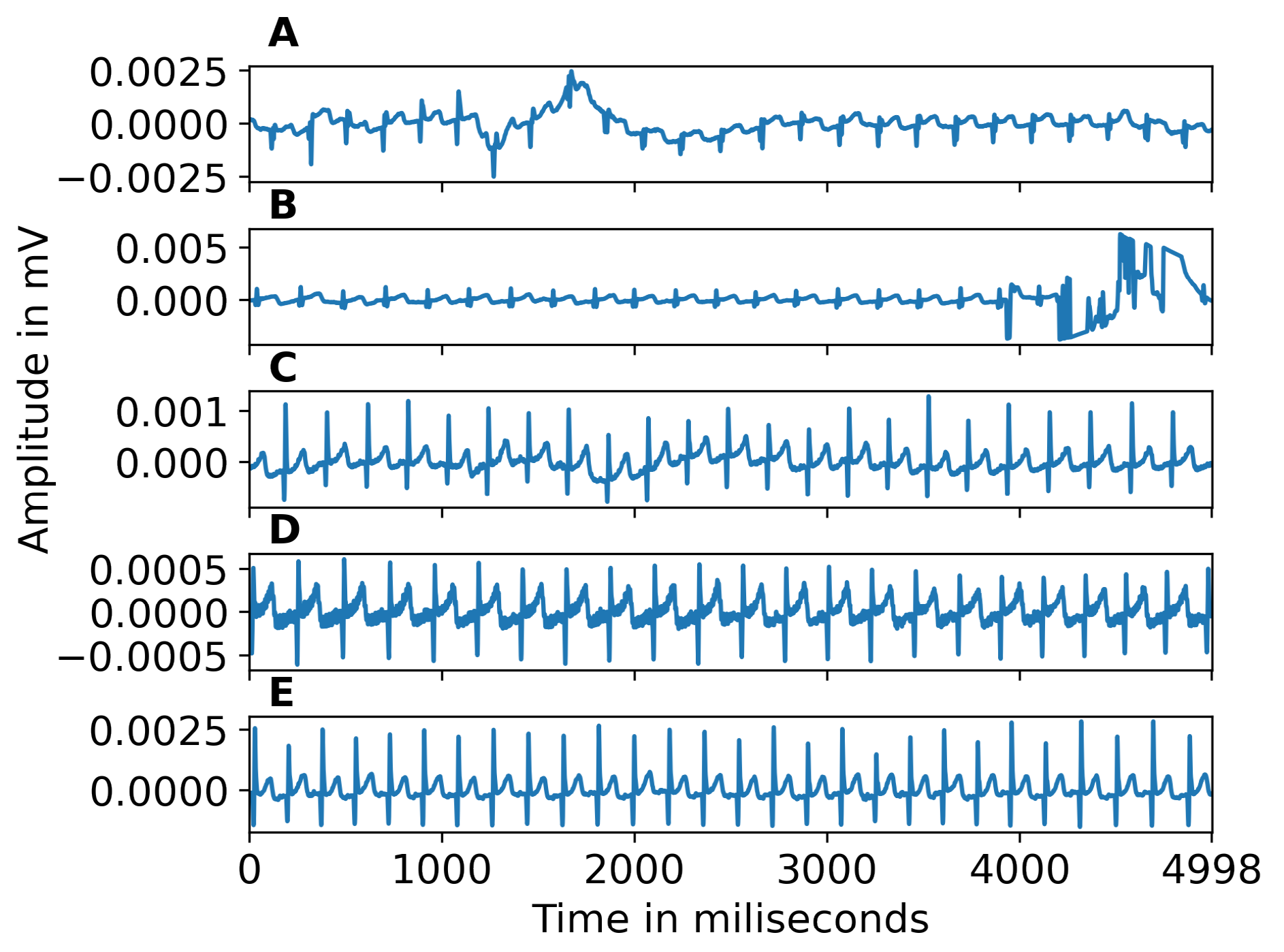
Due to changes in hardware during the data collection, ECG signal collection was done at three different frequencies, i.e., 128 Hz, 512 Hz, and 1024 Hz. Signals were processed at their original frequency. Various artifacts can contaminate ECG, including power line interference, channel noise, electromyographic artifacts, electrode contact noise, and baseline wandering due to body movement and respiration. Since such artifacts have a detrimental impact on automatic classification, we cleaned the ECG signals using the Python HeartPy library [
30]. Missing beats (i.e., heartbeats that were HeartPy failed to detect) were detected by computing a value
X as the rounded value of each interbeat interval divided by the median interbeat interval. We considered every of these integer values
X larger than one as indicative of
missing beats. Beats were then associated with an index starting at zero and incremented by one for every present or missing beat (identified as previously described). We used this index as an independent variable and the beat times as a dependent variable to linearly interpolate the timing of missing beats. To mitigate the impact of missing R peaks, we implemented a threshold. We rejected segments with less than 30 beats or more than 30% of interpolated peaks to ensure enough peaks for reliable feature estimation and to avoid biasing our analysis with recordings that were too noisy for reliable R peak extraction. Examples of noisy segments removed because of high noise are shown in Figure 1(A,B). The ECG signals corrected for baseline wandering is shown in Figure 1(C). Figure 1(D) illustrates power line interference or muscle response, which is corrected during pre-processing. Figure 1(E) gives an example of a clean signal following pre-processing. Figure 2 shows an example of zoomed-in clean R peaks. We performed this pre-processing on different segment lengths (the 30s, 60s, 90s, 120s, and full-length) of ECG data to test the effect of such a segmentation. Since the recording length can impact the success of the inclusion criteria aforementioned, different epochs lengths result in different sample sizes (30s: N=61, 60s: N=54, 90s: N=56, 120s: N=57, full-length: N=51). Figure 4(A) illustrates the automated ECG pre-processing process as explained.
Figure 1.
A-B) Examples of segments excluded from the dataset because they were too noisy. C) Example of baseline wandering. D) Example of power line interference. E) Clean signal, after pre-processing.
Figure 1.
A-B) Examples of segments excluded from the dataset because they were too noisy. C) Example of baseline wandering. D) Example of power line interference. E) Clean signal, after pre-processing.
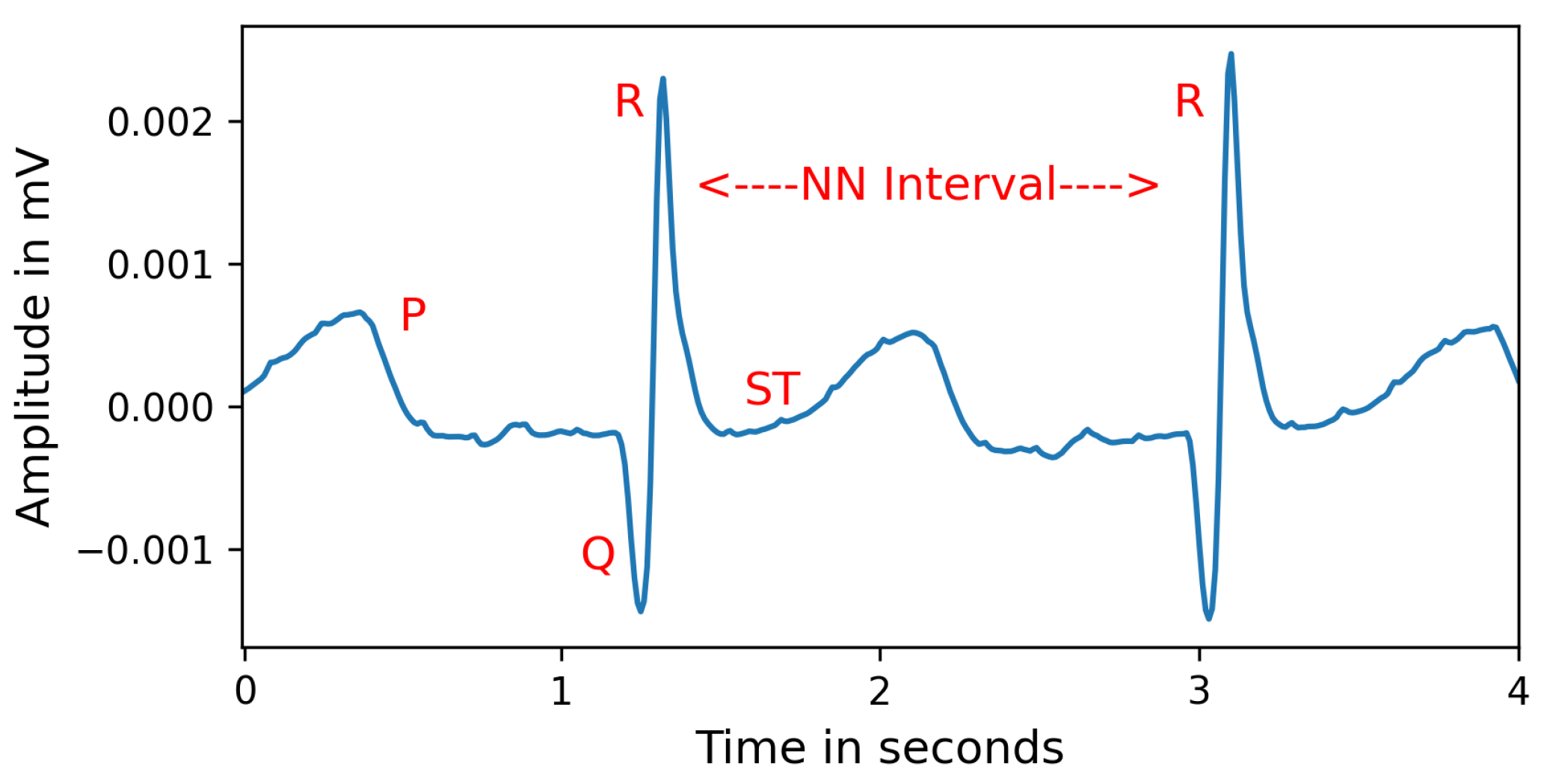
Figure 2.
An example of zoomed-in cleaned ECG beats. NN stands for normal beat intervals, and is similar to “R-R intervals", but highlights that these values represent normal cardiac timing, free from artifacts.
Figure 2.
An example of zoomed-in cleaned ECG beats. NN stands for normal beat intervals, and is similar to “R-R intervals", but highlights that these values represent normal cardiac timing, free from artifacts.
2.3. Feature extraction and Preliminary Analysis
After pre-processing, the process takes the average of the features extracted from each segment within a recording. For instance, if we divided a recording into five 90-second segments, we averaged the features across these five segments. We employed a Linear Mixed Effects Model ("feature condition+Labels+Month") to analyze the impact of a particular feature, while controlling for other variables including age (measured in months), condition (PIX/OIX) and labels (LL/EL). There was not a statistically significant impact of age on the feature values (p’s > 0.1), and so repeated within-subject recordings were aggregated by taking the average of the segment-averaged vectors across the different recording sessions (i.e., at different age). For example, if we have recordings at three, four, and six months for a given participant, each consisting of 90 second averaged vectors, we took the average of these vectors to obtain a single averaged vector for this participant across the three-time points. It is important to note that this process generates a matrix dependent on the segments’ length. For each segment length, we have different sets of subjects (due to pre-processing criteria) and ten HRV features per subject. These features were extracted (Table 1) using the Python neurokit2 library [
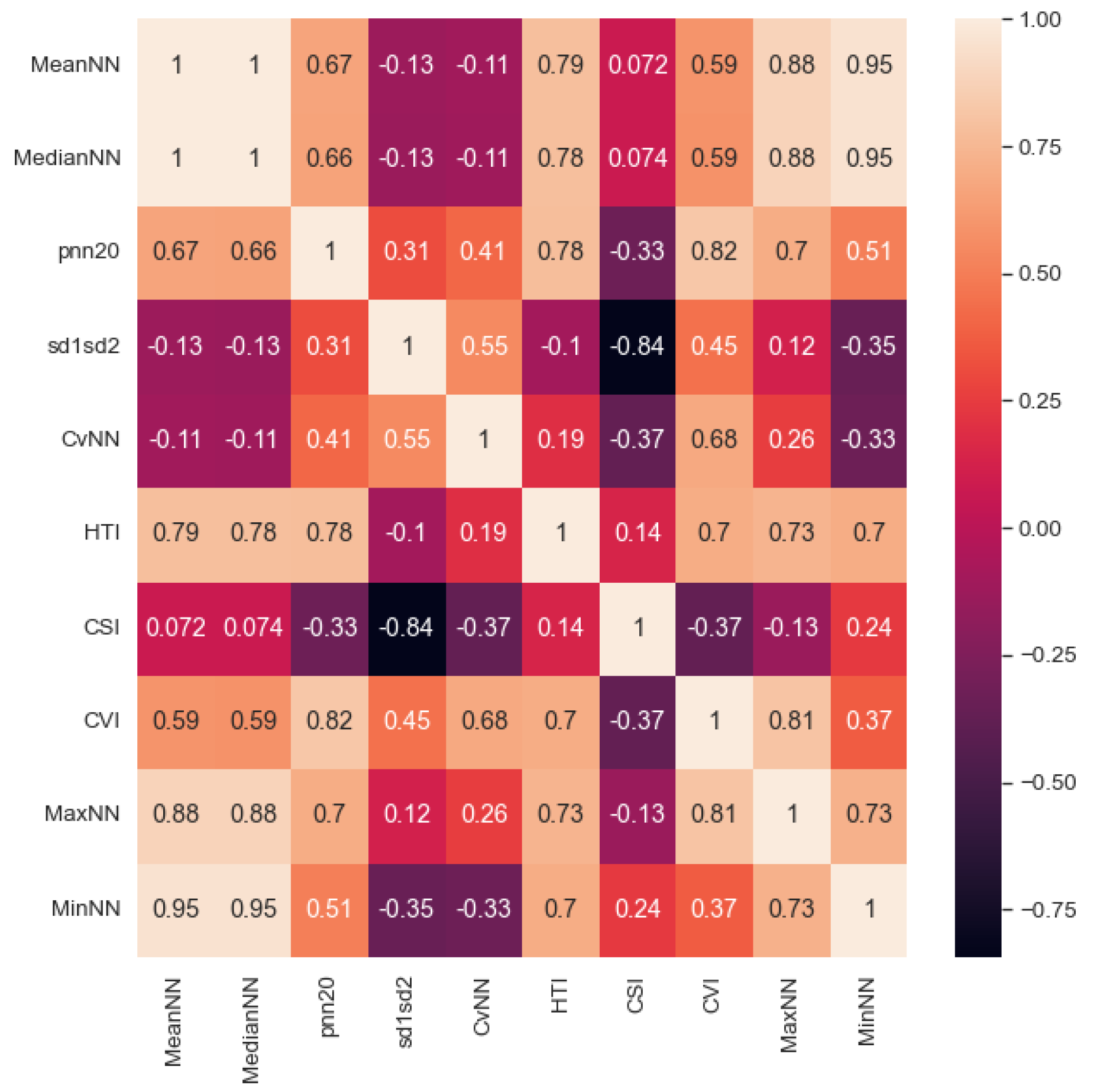
31]. We provided the clean peaks as an array of ECG data points to calculate HRV features (MeanNN, MaxNN, MinNN, MedianNN, pnn20, CVNN, sd1sd2) based on normal beat intervals (NN). NN is the interval between consecutive R peaks (Figure 2). Further, we calculated the Cardiac Sympathetic Index (CSI) and the Cardiovagal Index (CVI) as proxies for sympathetic and parasympathetic activation and their neuronal and cardiac dependencies. To calculate CSI and CVI, the variability of R peaks is observed and transformed into an elliptic distribution by using Lorenz plots [45]. The length of the longitudinal (L) and transverse (T) axes is calculated within each elliptic distribution. We then obtain CV I = log10(L*T) and CSI = L/T. After extracting the features, we calculated the correlation coefficient between each feature (Figure 3) and rejected redundant features if their correlation coefficient was greater than 0.90. The final set of included features was: MedianNN, pnn20, CVNN, sd1sd2, HTI, CSI, and CVI.
Table 1.
Extracted features from ECG Signals after preprocessing step.
Table 1.
Extracted features from ECG Signals after preprocessing step.
| Features |
Feature Description |
| MeanNN |
Mean of the NN [32,33]. |
| MedianNN |
Median of the NN [32,33]. |
| pNN20 |
Proportion of successive NN
with a difference larger than 20 ms.
This measures the relative frequency of
changes in heart rhythm [16]. |
| sd1sd2 |
In Poincaré plot sd1 is standard deviation
perpendicular the line of identity and sd2
is standard deviation along the line of
identity. sd1sd2 is ratio of (sd1/sd2), indicator of the
unpredictability of the NN to measure
autonomic balance when there
is sympathetic activation. [16]. |
| CVNN |
Coefficient of variation of NN,
calculated as standard deviation of NN
divided by mean of NN [34]. |
| HTI |
HRV Triangular Index is the integral of the
NN density histrogram divided by its height [16]. |
| CSI |
The CSI quantifies the sympathetic nervous
system activity indicating increased arousal [35]. |
| CVI |
The CVI quantifies the parasympathetic
nervous system activity [36]. |
| MaxNN |
Max of the NN [32,33] |
| MinNN |
Min of NN [32,33] |
Figure 3.
Heatmap of Correlation between all the HRV features averaged across 90 seconds segment length.
Figure 3.
Heatmap of Correlation between all the HRV features averaged across 90 seconds segment length.
2.4. EL vs. TL Classification Using Machine Learning
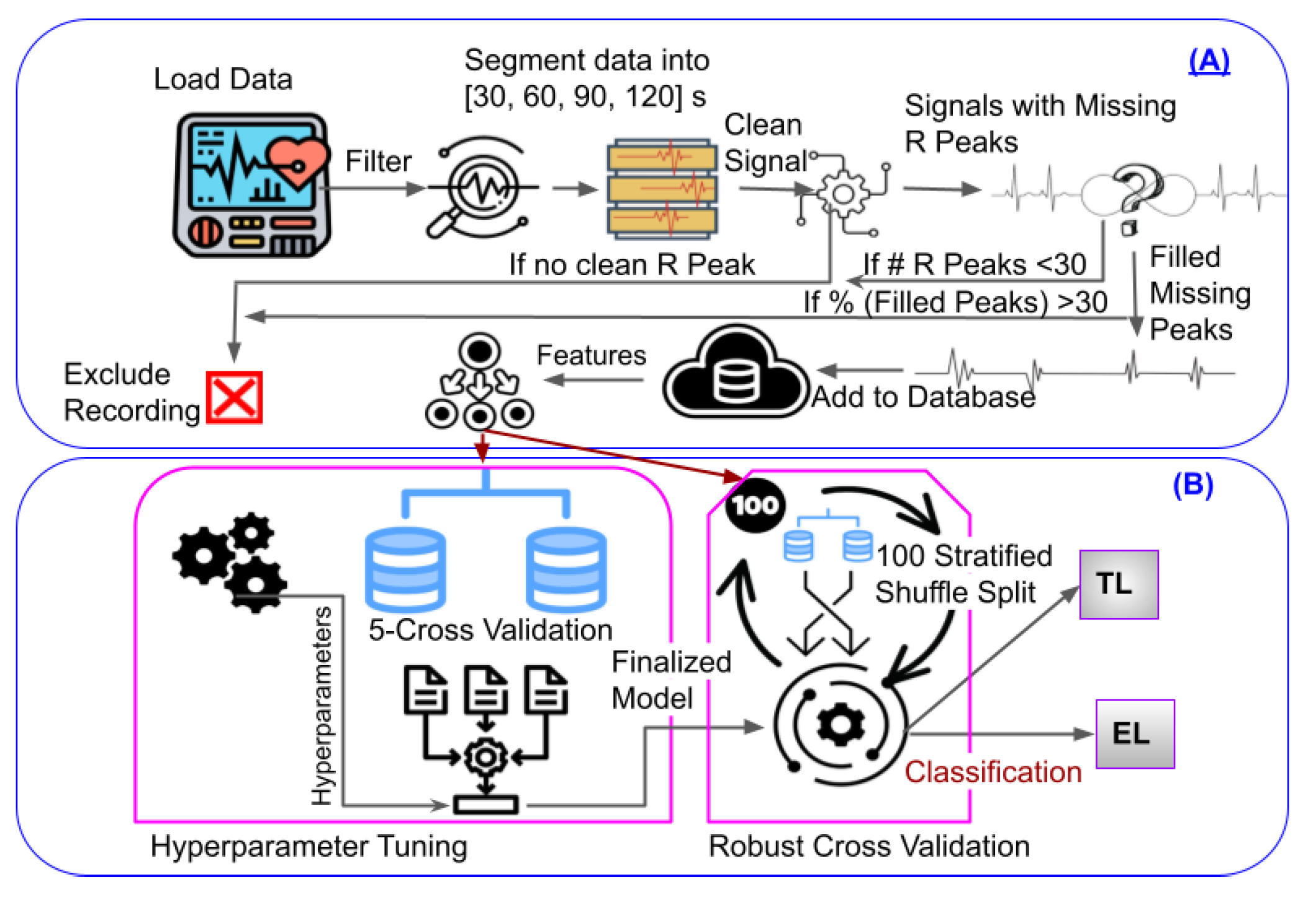
We tested various classifiers (Gradient Boosting (GB), Extra Trees (ET), Random Forest (RF), Ada Boost (AB), Decision Tree (DT), K Nearest Neighbors (KNN), XGBoost (XGB), and Multi-layer Perceptron (MLP)) to classify infants into EL and TL groups. For training and testing, we used an 80/20% split. We used the grid search with a 5-fold cross validation (GS-CV) to systematically evaluate the performance of different combinations of hyperparameters for each model. GS-CV enabled us to identify the optimal set of hyperparameters. Subsequently, we trained the final model using the best set of hyperparameters, as obtained from the GS-CV. For the final evaluation, we used 100 stratified shuffle splits (Figure 4 (B)) to obtain performance distributions rather than point estimates. Finally, we extracted feature importance. The importance of each feature is calculated based on how much they contribute to the model’s predictions. We utilized the permutation importance function in Scikit-learn [
37], which is model agnostic. This function assesses a feature’s significance by randomly changing its values and measuring the model’s resulting decrease in performance. This enables us to gauge a feature’s importance in predicting outcomes since an important feature should lead to a more significant performance decrease than a less important one when its values are randomly shuffled.
Figure 4.
Schematic representation of the automated pipeline we used for ASD classification. A) Pre-processing stage. B) Machine learning and the robust cross-validation used for classification.
Figure 4.
Schematic representation of the automated pipeline we used for ASD classification. A) Pre-processing stage. B) Machine learning and the robust cross-validation used for classification.
2.5. Analysis Pipeline
The ECG sensors were well-tolerated by all infants. Overall, we rejected 12% recordings (9 participants). Figure 4 illustrates our methodology for determining ASD genetic likelihood using ECG signals. It includes automated ECG pre-processing with the following steps: filtering, segmentation of ECG data, pre-processing to eliminate artifacts, extraction of clean R peaks, missing peaks correction, feature extraction, and classification. After pre-processing, the features described in subsection 2.3 were extracted. For each participant, we computed the average of the extracted features from the remaining segments across all age time points. Then we performed ML with GS-CV, as discussed in subsection 2.4.
Table 2.
Mean±Std values of the HRV features extracted averaged across 90 seconds segments of ECG.
Table 2.
Mean±Std values of the HRV features extracted averaged across 90 seconds segments of ECG.
| Feature |
MeanNN |
HTI |
| TL |
571.33±214.77 |
5.64±2.38 |
| EL |
419.47±175.86 |
5.01±2.02 |
| Feature |
MedianNN |
CSI |
| TL |
569.13±213.75 |
3.38±1.23 |
| EL |
417.09±176.22 |
3.56±1.61 |
| Feature |
pnn20 |
CVI |
| TL |
11.7±11.2 |
3.67±0.52 |
| EL |
8.75±11.86 |
3.57±0.58 |
| Feature |
sd1sd2 |
MaxNN |
| TL |
0.35±0.15 |
704.71±313.01 |
| EL |
0.39±0.27 |
520.11±225.89 |
| Feature |
CvNN |
MinNN |
| TL |
0.04±0.02 |
501.48±198.36 |
| EL |
0.06±0.03 |
347.18±149.89 |
3. Results
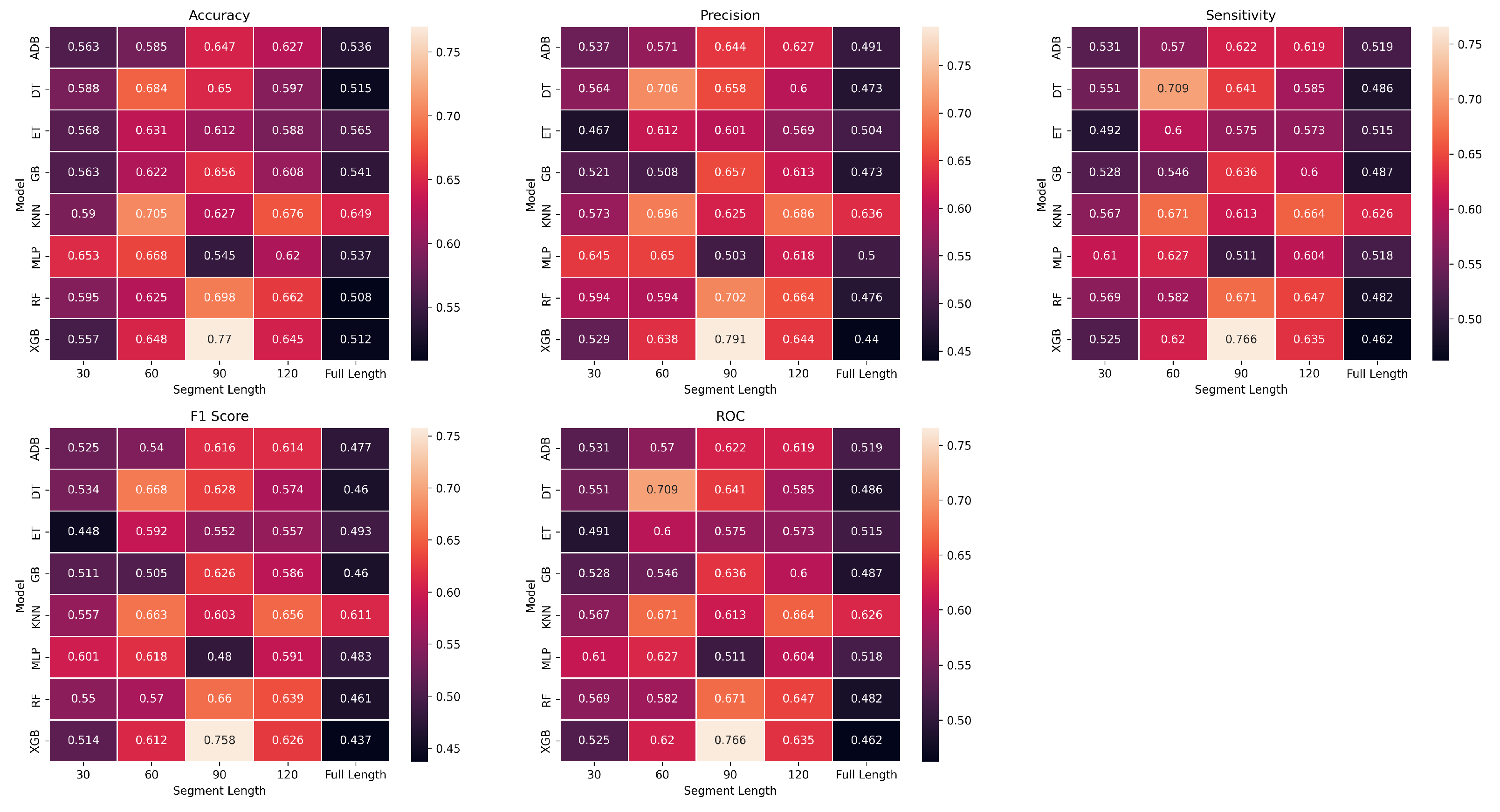
We compared 8 classification algorithms (ADB, DT, ET, GB, KNN, MLP, RF, and XGB) to identify the best predictive model for EL (positive) and TL (negative) groups. The results obtained from these algorithms have been represented using heatmaps in Figure 6. Heatmaps are color-coded to show the level of performance, with brighter colours indicating better performance and darker colours indicating poorer performance. We used accuracy, precision, sensitivity, specificity, and f1score to assess the performances. Sensitivity is defined as the true positive rate (true positives over all positives), while specificity is defined as the true negative rate (true negatives over all negatives). Precision is a metric that measures the accuracy of positive predictions (true positives over the number of predicted positives) in a classification model, and accuracy is the proportion of correctly predicted subjects. The f1-score is the harmonic mean of precision and sensitivity. See [
38] for a complete definition of these metrics. We also calculated the area under the receiver operating characteristic curve (ROC-AUC), a commonly used metric for evaluating the performance of classification models [
39]. It is also less sensitive to class imbalance than accuracy [
40]. Considering the difficulty of classifying EL vs. TL infants based only on ECG, our results using XGB showed good sensitivity (0.76±0.12), f1-score (0.75±0.12), precision (0.79±0. 12), accuracy (0.77±0.12, pvalue = 0.01), and ROC-AUC (0.76±0.12, p-value = 0.02). For comparison, Table 3 also reports the results of two other studies using ML classifiers [
33,
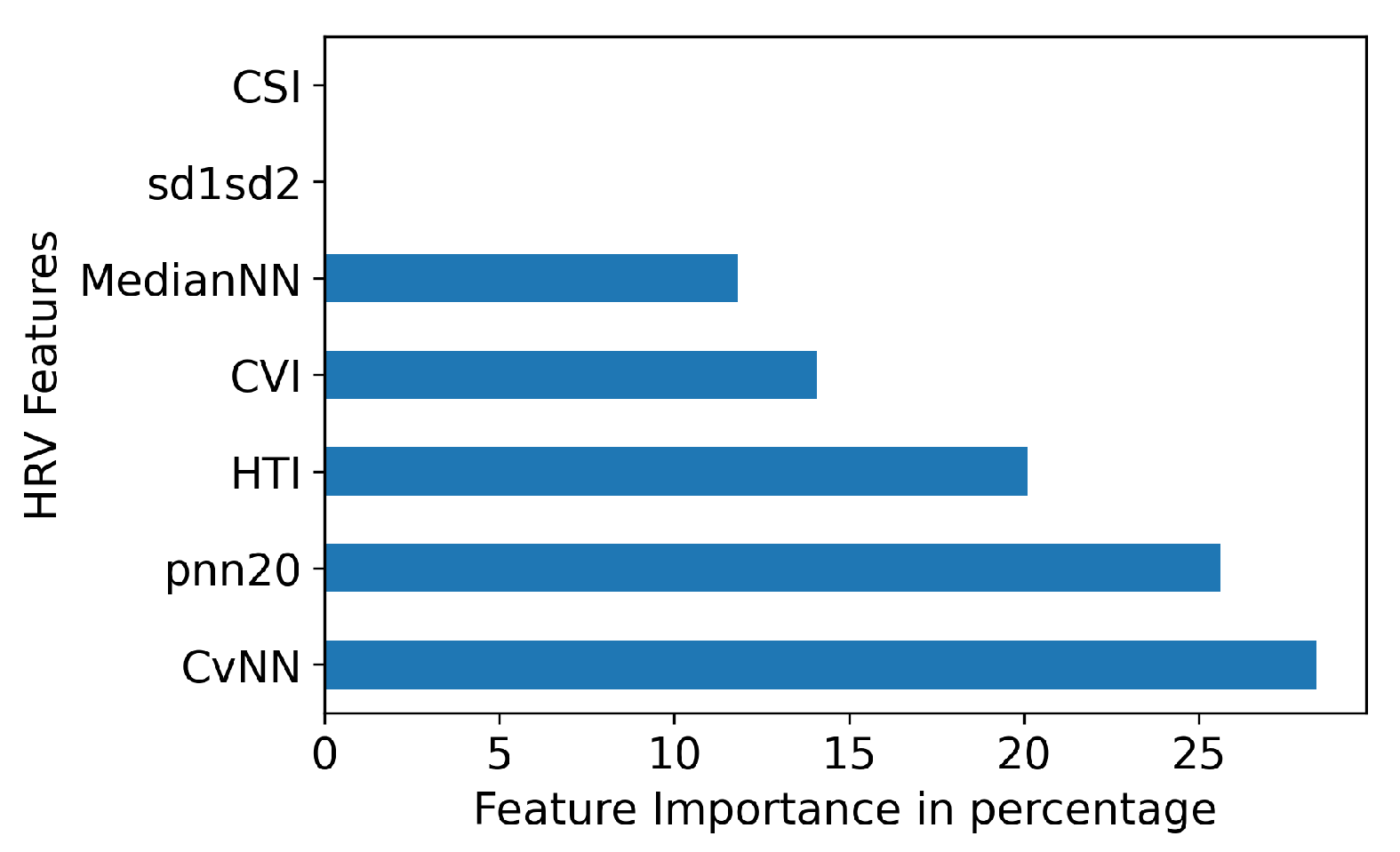
41]. In Figure 5, we identified the top five features based on feature importance from the XGB model. We found that all features, except for CSI and sd1sd2, have a significant contribution to the accuracy of classification.
Figure 5.
HRV Feature importance for XGB.
Figure 5.
HRV Feature importance for XGB.
Figure 6.
Heatmaps represent various metrics, across different segment lengths (x-axis) for ML classifier experiments (y-axis). The heatmap provides a color-coded summary of the performance of each classifier for segment length, with warmer colours indicating poorer results and cooler colours showing better results.
Figure 6.
Heatmaps represent various metrics, across different segment lengths (x-axis) for ML classifier experiments (y-axis). The heatmap provides a color-coded summary of the performance of each classifier for segment length, with warmer colours indicating poorer results and cooler colours showing better results.
Table 3.
Comparison of our proposed approach with results from the literature (mean±std).
Table 3.
Comparison of our proposed approach with results from the literature (mean±std).
| Methodology |
Precision |
Sensitivity |
F1-Score |
ROC-AUC |
Accuracy |
| Ensemble [33] |
- |
- |
- |
- |
0.75 |
| |
|
|
|
|
(avg. accuracy) |
| XGB (their highest |
|
|
|
|
|
| performing model)[41] |
0.57±0.22 |
0.59±0.14 |
- |
0.88±0.12 |
0.59±0.22 |
| XGB with Robust |
|
|
|
|
|
| CV (ours) |
0.79±0. 12 |
0.76±0.12 |
0.75±0.12 |
0.76±0.12 |
0.77±0.12 |
4. Discussion
The primary goal of this study was to determine whether infant ECG signals are useful in classifying ASD likelihood. Statistically, EL infants show elevated rates of ASD diagnosis and elevated ASD symptoms, making this preliminary study a first step in developing biomarkers for early ASD diagnosis and better understanding the relationships between autonomic regulation and ASD. Our approach has high ecological validity considering our naturalistic experimental paradigm (self-initiated, naturalistic interactions with objects and parents) and the wearable, wireless, and non-invasive sensors. In addition, such an approach can be easily scaled up and reach more families by collecting data at home or remotely (i.e., without an on-site experimenter). Traditional ECG pre-processing techniques have several constraints, including their incapability to scale up for remote setups, proneness to human errors, limitations in handling noise, and difficulties in standardization. As a result, there is a growing interest in automated ECG pre-processing algorithms, which have the potential to enhance the accuracy, efficiency, and standardization of ECG signal analysis in clinical and research settings. Our pipeline explained in Figure 4(A). explains automated algorithm which can detect and remove artifacts in the ECG signal, apply filters to remove noise and unwanted components, segment the signal into relevant segments, and improve overall signal quality. By reducing the need for manual inspection and annotation of the signal, automated pre-processing can save time and increase efficiency in ECG signal analysis.
Several studies suggest a significant correlation between social functioning and sympathetic and parasympathetic markers, which can be assessed through HRV, in typical development [
42,
43,
44]. This has led to a growing interest in exploring their association with ASD. By studying HRV features, we may better understand how ANS function is related to the core and associated deficits of ASD and other diagnoses related to neurodevelopmental disorders. In addition, understanding ANS could lead to a clearer understanding of the underlying mechanisms contributing to the development and manifestation of these conditions. Initially, we selected ten features as described in subsection 2.3 for our study. However, we chose to exclude MaxNN, MinNN, and MeanNN because of high correlations between these features and MedianNN, leaving seven features. The median is a measure of central tendency less sensitive to extreme values or outliers than the mean
We aimed to evaluate whether the overall HRV features could separate the EL group from the TD group. Research shows a link between lower HRV and atypical brain functioning in individuals with ASD. Therefore, our study hypothesized that differences in autonomic regulation specific to ASD would result in HRV differences between groups. To achieve this, we calculated two variables, CSI and CVI, associated with various psychological and physical health outcomes, including social behaviour, cognition, and emotional functioning. Our results showed that CSI values were higher and CVI values were lower in EL infants compared to TL infants, indicating lower-paced breathing and a lower heart rate in EL infants. Additionally, EL infants exhibited lower median, pNN20, HTI, and CVNN values than TL infants. These findings suggest a potential relationship between ASD and altered ANS functioning, particularly regarding parasympathetic regulation. These values relate to a previous study that found that children and adults with ASD have lower HRV compared to TL individuals, indicating decreased parasympathetic activity and/or increased sympathetic activity [
45]. This autonomic dysregulation has been associated with various behavioral and physiological problems in individuals with ASD, including social difficulties, anxiety, and gastrointestinal issues. For reference, Table 2 shows the mean±std of HRV features for each group in the 90s segment length.
Our f1-score of 0.75±0.12 suggest that wearable, wireless, and easy-to-use ECG devices hold potential for clinical applications in evaluating ANS activation and screening for ASD likelihood. By comparison, multi-electrodes EEG devices are more sensitive to artifacts, making data collection with infants more complicated [
46]. Other neuroimaging techniques, like functional magnetic resonance imaging, are even less practical due to higher costs and lower accessibility. While ECG signal disruption can occur, denoising and pre-processing methods can help recover some portion of noisy recordings.
Comparing the accuracy of our system with previously reported results on a very similar problem [
33] (i.e., ASD diagnosis vs. ASD likelihood classification) suggests the better performance of our approach. XGBoost is well-suited for managing complex datasets, and when applied to HRV features, it can effectively capture nonlinear relationships and interactions among features. This is achieved by constructing decision trees using gradient boosting, allowing XGBoost to efficiently capture and model complex relationships in the HRV data. Unfortunately, earlier relevant studies, [
33] did not report specificity, sensitivity, precision, or f1-score, limiting the extent to which we can make a fair comparison. Moreover, reporting only accuracy is known to be biased in the case of an unbalanced dataset (i.e., high accuracy can be obtained on an unbalanced dataset even if the classifier performs very poorly for the smaller class). Since reporting only accuracy is generally insufficient to assess the performance of a classifier properly, we also reported on precision, sensitivity, specificity, and f1-score. Further, instead of reporting point estimates, we reported the mean and standard deviation of these measures to allow future studies to test statistical significance of differences in performances. The results reported by the authors [
41] show a higher ROC-AUC than what we found, which seems inconsistent with their reported accuracy. Further, the authors report multi-class classification performance for several ML algorithms using HRV for ASD, conduct problems, depression, and typical development. To our knowledge, these are the only other studies reporting classification results using ECG and HRV with ML for diagnostic applications in ASD. Also, these performances are only partly comparable (e.g., school-aged children were studied in [
41], while we reported on infants aged 3-6 months). The encouraging performances observed in our study support the use of HRV as a potential biomarker for monitoring the effectiveness of interventions for ASD and evaluating the physiological changes that occur in infants with ASD over time.
Some limitations need to be considered when interpreting the results of this study. Firstly, the study analyzed HRV parameters in a moderate sample size. Therefore, corroborating our conclusions in larger samples is essential to ensure the generalizability of the findings. In addition, this study balanced infants with an elevated familial likelihood for ASD and controls, resulting in a small proportion of infants with later ASD diagnoses. Thus, it is worth emphasizing that we did not aim to classify ASD diagnosis but the likelihood of ASD (or risk level).
5. Conclusion and Future Work
ECG recordings are non-invasive, easy-to-use signals that can be leveraged in biomarker research and provide measures (e.g., heart rate variability) that index key neurological systems, including the sympathetic and parasympathetic nervous systems. This study found that we can use HRV features extracted from ECG to predict the familial likelihood of ASD in 3-6-month-old infants. In this study, we only used 10 HRV features. In the future, this study can be expanded along different axes including 1) exploring additional HRV features, including time-domain, frequency-domain, and nonlinear HRV features; 2) replicating these findings in larger samples of infants and children, including those with confirmed ASD diagnoses and those with non-ASD developmental disorders such as language or attention deficit hyperactivity disorder.
Author Contributions
Conceptualization, D.T., C.O’R.; methodology, D.T., C.O’R.; software, D.T.; 504 validation, C.O’R.; formal analysis, D.T., C.O’R.; investigation, D.T.; resources, J.B., A.S; data curation, J.B., 505 D.T.; writing—original draft preparation, D.T., C.O’R.; writing—review and editing, C.O’R., D.T., J.B., A.S., 506 J.B.; visualization, D.T.; supervision, A.S., C.O’R.; project administration, D.T.; funding acquisition, 507 A.S., J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a pilot grant from the Carolina Autism & Neurodevelopment 501 Center at the University of South Carolina (PI: J B.). ECG was collected under a National Institute 502 of Mental Health grant (PI: J.B.; No: K23MH120476)
Institutional Review Board Statement
All study procedures were reviewed and approved by 494 the University of South Carolina Institutional Review Board (project #00081992, approved on 495 10/24/2019).
Informed Consent Statement
Informed consent was obtained from the legal guardian of all subjects 497 involved in the study.
Data Availability Statement
Data are not shared at this time because the data collection is still 499 ongoing.
Conflicts of Interest
The author(s) declare no competing interests.
Abbreviations
The following abbreviations are used in this manuscript:
| ADI |
Autism Diagnostic Interview |
| ADOS |
Autism Diagnostic Observation Schedule |
| ANS |
Autonomic Nervous System |
| ASD |
Autism Spectrum Disorder |
| CVI |
Cardiovagal Index |
| CSI |
Cardiac Sympathetic Index |
| ECG |
Electrocardiogram |
| EDA |
Electrodermal activity |
| EEG |
Electroencephalogram |
| EL |
Elevated Likelihood |
| fMRI |
functional Magnetic Resonance Imaging |
| HRV |
Heart Rate Variability |
| IQR |
Interquartile Range |
| ML |
Machine Learning |
| MRI |
Magnetic Resonance Imaging |
| OIX |
Object Only Intreactions |
| PIX |
Caregiver Only Interactions |
| RR |
Inter-beat Interval |
| TL |
Typical Likelihood |
References
- Carpenter, B., DSM-5; ,, 2015; pp. –. [CrossRef]
- Guthrie, W.; Wetherby, A.M.; Woods, J.; Schatschneider, C.; Holland, R.D.; Morgan, L.; Lord, C.E. The earlier the better: An RCT of treatment timing effects for toddlers on the autism spectrum. Autism 0, 0, 13623613231159153, [https://doi.org/10.1177/13623613231159153]. [CrossRef]
- Autism Statistics and Facts | Autism Speaks. (n.d.). Autism Speaks; 2023. doi:https://www.autismspeaks.org/autism-statistics-asd.
- Bradshaw, J.; Steiner, A.; Gengoux, G.; Koegel, L. Feasibility and Effectiveness of Very Early Intervention for Infants At-Risk for Autism Spectrum Disorder: A Systematic Review. Journal of autism and developmental disorders 2014, 45. [Google Scholar] [CrossRef] [PubMed]
- Frigaux, A.; Evrard, R.; Lighezzolo-Alnot, J. ADI-R and ADOS and the differential diagnosis of autism spectrum disorders: Interests, limits and openings. L Encéphale 2019.
- Hurwitz, S.; Yirmiya, N., Autism Diagnostic Observation Schedule (ADOS) and Its Uses in Research and Practice; 2014; pp. 345–353. [CrossRef]
- Bildt, A.; Sytema, S.; Zander, E.; Bölte, S.; Sturm, H.; Yirmiya, N.; Yaari, M.; Charman, T.; Salomone, E.; LeCouteur, A.; Green, J.; Canal, R.; García-Primo, P.; Daalen, E.; Jonge, M.; Guðmundsdóttir, E.; Jóhannsdóttir, S.; Raleva, M.; Boskovska, M.; Oosterling, I. Autism Diagnostic Interview-Revised (ADI-R) Algorithms for Toddlers and Young Preschoolers: Application in a Non-US Sample of 1,104 Children. Journal of autism and developmental disorders 2015, 45. [Google Scholar] [CrossRef] [PubMed]
- Bosl, W.; Tager-Flusberg, H.; Nelson, C. "EEG" Analytics for Early Detection of Autism Spectrum Disorder: A data-driven approach. Scientific Reports 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Estelle, H.; Mento, G.; Desnous, B.; François, C. Challenges and new perspectives of developmental cognitive EEG studies. NeuroImage 2022, 260, 119508. [Google Scholar] [CrossRef]
- Fenning, R.; Erath, S.; Baker, J.; Messinger, D.; Moffitt, J.; Baucom, B.; Kaeppler, A. Sympathetic-Parasympathetic Interaction and Externalizing Problems in Children with Autism Spectrum Disorder. Autism Research 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Panju, S.; Brian, J.; Dupuis, A.; Anagnostou, E.; Kushki, A. Atypical sympathetic arousal in children with autism spectrum disorder and its association with anxiety symptomatology. Molecular Autism 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Prince, E.; Kim, E.; Wall, C.; Gisin, E.; Goodwin, M.; Schoen Simmons, E.; Chawarska, K.; Shic, F. The relationship between autism symptoms and arousal level in toddlers with autism spectrum disorder, as measured by electrodermal activity. Autism 2016, 21. [Google Scholar] [CrossRef]
- MacNeil, B.; Lopes, V.; Minnes, P. Anxiety in children and adolescents with Autism Spectrum Disorders. Research in Autism Spectrum Disorders 2009, 3, 1–21. [Google Scholar] [CrossRef]
- Alvares, G.; Quintana, D.; Kemp, A.; Zwieten, A.; Balleine, B.; Hickie, I.; Guastella, A. Reduced Heart Rate Variability in Social Anxiety Disorder: Associations with Gender and Symptom Severity. PloS one 2013, 8, e70468. [Google Scholar] [CrossRef]
- Benevides, T.; Lane, S. A Review of Cardiac Autonomic Measures: Considerations for Examination of Physiological Response in Children with Autism Spectrum Disorder. Journal of autism and developmental disorders 2013, 45. [Google Scholar] [CrossRef]
- Eilam-Stock, T.; Xu, P.; Cao, M.; Gu, X.; Van Dam, N.; Anagnostou, E.; Kolevzon, A.; Soorya, L.; Park, Y.; Siller, M.; He, Y.; Hof, P.; Fan, J. Abnormal autonomic and associated brain activities during rest in autism spectrum disorder. Brain : a journal of neurology 2014, 137, 153–71. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Campos, B.; Coan, A.; Pegoraro, L.; Rezende, T.; Obeso, I.; Dalgalarrondo, P.; Dacosta, J.; Dreher, J.C.; Cendes, F. Differences in Cortical Structure and Functional MRI Connectivity in High Functioning Autism. Frontiers in Neurology 2018, 9, 539. [Google Scholar] [CrossRef] [PubMed]
- Bauman, M.; Kemper, T. Neuroanatomic observations of the brain in autism: A review and future directions. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience 2005, 23, 183–7. [Google Scholar] [CrossRef] [PubMed]
- Ecker, C. The neuroanatomy of autism spectrum disorder: An overview of structural neuroimaging findings and their translatability to the clinical setting. Autism 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Pagnozzi, A.; Conti, E.; Calderoni, S.; Fripp, J.; Rose, S. A systematic review of structural MRI biomarkers in Autism Spectrum Disorder: A Machine Learning perspective. International Journal of Developmental Neuroscience 2018, 71. [Google Scholar] [CrossRef] [PubMed]
- Yassin, W.; Nakatani, H.; Zhu, Y.; Kojima, M.; Owada, K.; Kuwabara, H.; Gonoi, W.; Aoki, Y.; Takao, H.; Natsubori, T.; Iwashiro, N.; Kasai, K.; Kano, Y.; Abe, O.; Yamasue, H.; Koike, S. Machine-learning classification using neuroimaging data in schizophrenia, autism, ultra-high risk and first-episode psychosis. Translational Psychiatry 2020, 10, 278. [Google Scholar] [CrossRef] [PubMed]
- Eslami, T.; Almuqhim, F.; Raiker, J.S.; Saeed, F. Machine Learning Methods for Diagnosing Autism Spectrum Disorder and Attention- Deficit/Hyperactivity Disorder Using Functional and Structural MRI: A Survey. Frontiers in Neuroinformatics 2021, 14, 62. [Google Scholar] [CrossRef]
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.; Saeed, F. ASD-DiagNet: A hybrid learning approach for detection of Autism Spectrum Disorder using f"MRI" data. Frontiers in Neuroinformatics 2019. [Google Scholar] [CrossRef]
- Han, Y.; Rizzo, D.; Hanley, J.; Coderre, E.; Prelock, P. Identifying neuroanatomical and behavioral features for autism spectrum disorder diagnosis in children using machine learning. PLOS ONE 2022, 17, e0269773. [Google Scholar] [CrossRef]
- Just, M.; Cherkassky, V.; Buchweitz, A.; Keller, T.; Tom, M. Identifying Autism from Neural Representations of Social Interactions: Neurocognitive Markers of Autism. PloS one 2014, 9, e113879. [Google Scholar] [CrossRef]
- B J, B.; Ashok, G.; Sreekumar, N. Classification of autism based on feature extraction from segmented brain MRI. International Journal of Recent Technology and Engineering 2019, 7, 85–89. [Google Scholar]
- Song, J.W.; Yoon, N.R.; Jang, S.M.; Lee, G.Y.; Kim, B. Neuroimaging-Based Deep Learning in Autism Spectrum Disorder and Attention-Deficit/Hyperactivity Disorder. Journal of the Korean Academy of Child and Adolescent Psychiatry 2020, 31, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Brihadiswaran, G.; Haputhanthri, D.; Gunathilaka, S.; Meedeniya, D.; Jayarathna, S. "EEG"-based processing and classification methodologies for Autism Spectrum Disorder: A Review. Journal of Computer Science 2019, 15, 1161–1183. [Google Scholar] [CrossRef]
- Hyde, K.; Novack, M.; LaHaye, N.; Parlett-Pelleriti, C.; Anden, R.; Dixon, D.; Linstead, E. Applications of Supervised Machine Learning in Autism Spectrum Disorder Research: A Review. Review Journal of Autism and Developmental Disorders 2019, 6. [Google Scholar] [CrossRef]
- van Gent, P.; Farah, H.; Nes, N.; Arem, B. HeartPy: A novel heart rate algorithm for the analysis of noisy signals. Transportation Research Part F: Traffic Psychology and Behaviour 2019, 66, 368–378. [Google Scholar] [CrossRef]
- Makowski, D.; Pham, T.; Lau, Z.J.; Brammer, J.C.; Lespinasse, F.; Pham, H.; Schölzel, C.; Chen, S.H.A. NeuroKit2: A Python toolbox for neurophysiological signal processing. Behavior Research Methods 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- kumar, C.; Mullagiri, M.; Maruthy, K.; Siva Kumar, A.V.; Kuppusamy, M. Association of Heart rate variability measured by RR interval from ECG and pulse to pulse interval from Photoplethysmography. Clinical Epidemiology and Global Health 2021, 10, 100698. [Google Scholar] [CrossRef]
- Anandhi, B.; Selvaraj, J.; Anusuya, I.; Das, H. Time Domain Analysis of Heart Rate Variability Signals in Valence Recognition for Children with Autism Spectrum Disorder (ASD). IRBM 2021. [Google Scholar] [CrossRef]
- Billeci, L.; Tonacci, A.; Narzisi, A.; Manigrasso, Z.; Varanini, M.; Fulceri, F.; Lattarulo, C.; Calderoni, S.; Muratori, F. Heart Rate Variability During a Joint Attention Task in Toddlers With Autism Spectrum Disorders. Frontiers in Physiology 2018, 9, 467. [Google Scholar] [CrossRef]
- Dodo, N.; Hashimoto, R. Autonomic Nervous System Activity During a Speech Task. Frontiers in Neuroscience 2019, 13. [Google Scholar] [CrossRef]
- Low, P.; Tomalia, V.; Park, K.J. Autonomic Function Tests: Some Clinical Applications. Journal of clinical neurology (Seoul, Korea) 2013, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Machine Learning 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Tohka, J.; Gils, M. Evaluation of machine learning algorithms for Health and Wellness applications: a tutorial. Computers in Biology and Medicine 2020. [Google Scholar] [CrossRef] [PubMed]
- Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef]
- He, H.; Ma, Y. Imbalanced Learning: Foundations, Algorithms, and Applications. Imbalanced Learning: Foundations, Algorithms, and Applications 2013, p. page 27. [CrossRef]
- Frasch, M.; Shen, C.; Wu, H.T.; Müller, A.; Neuhaus, E.; Bernier, R.; Kamara, D.; Beauchaine, T. Can a composite heart rate variability biomarker shed new insights about autism spectrum disorder in school-age children? Journal of Autism and Developmental Disorders 2020, 51. [Google Scholar] [CrossRef]
- Doussard-Roosevelt, J.A.; Porges, S.W.; Scanlon, J.W.; Alemi, B.; Scanlon, K.B. Vagal regulation of heart rate in the prediction of developmental outcome for very low birth weight preterm infants. Child development 1997, 68 2, 173–86. [Google Scholar] [CrossRef]
- Patriquin, M.; Lorenzi, J.; Scarpa, A.; Bell, M.A. Developmental trajectories of respiratory sinus arrhythmia: Associations with social responsiveness. Developmental psychobiology 2014, 56. [Google Scholar] [CrossRef]
- Taylor, Z.; Eisenberg, N.; Spinrad, T. Respiratory Sinus Arrhythmia, Effortful Control, and Parenting as Predictors of Children’s Sympathy Across Early Childhood. Developmental psychology 2014, 51. [Google Scholar] [CrossRef]
- Bal, E.; Harden, E.; Lamb, D.; Van Hecke, A.; Denver, J.; Porges, S. Emotion Recognition in Children with Autism Spectrum Disorders: Relations to Eye Gaze and Autonomic State. Journal of Autism and Developmental Disorders 2010, 40, 358–370. [Google Scholar] [CrossRef]
- Bell, M.A.; Cuevas, K. Using EEG to Study Cognitive Development: Issues and Practices. Journal of cognition and development : official journal of the Cognitive Development Society 2012, 13, 281–294. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).