1. Introduction
Intestinal parasitic infections have grave consequences on public health globally, especially in developing and underdeveloped countries. According to the World Health Organization, infectious and parasitic diseases are responsible for infecting approximately 24% of the world population and preventive chemotherapy provided to 836 million children globally in 2020 [
1]. Most parasitic infections lead to diarrhea, malnutrition, anemia and other symptoms, mostly in children. More than 100 species of intestinal parasites replicated daily after hatching at a rate of 200,000 eggs per day. Manual light microscopy is the gold standard for the diagnosing of the parasite diseases [
2,
3]. However, microscopy-based parasite species recognition and quantification is complicated, labor-intensive, and time-consuming, requiring a microscope and experts [
4,
5] and is also challenging for human experts to examine microscopic images because of the variation and uncertainties in morphological features like shape, staining color and density of parasite species.
Previously, the automation of intestinal protozoa analysis was restricted due to unavailability of automatic recognition algorithms for microorganisms under the microscope. In recent years, deep learning architectures based on convolutional neural networks (CNNs) have revolutionized the performance of many machine learning tasks, such as medical image identification and classification [
6,
7,
8]. Compared to traditional machine learning methods, deep learning based architectures directly extract features from images and enhance the validation and accuracy of detection [
9,
10]. Application of Plasmodium (malarial parasite) in blood using deep learning architectures achieved detection performance close to human expert [
11,
12,
13]. Amal H. A. et al. developed a deep learning based model by conducting experiments on 13,750 parasitized and 13,7500 non-parasitic samples [
14]. They estimated that the accuracy rate achieved by the deep convolutional neural network model was 97% in recognizing the samples. In a recent study by Z. Jing et al. [
15], a novel approach for fecal cell detection was presented using the Faster R-CNN model combined with the powerful Resnet-152 convolutional neural network architecture. The proposed algorithm demonstrated impressive accuracy, achieving an average precision of 84% on a dataset of 40,560 fecal images. Furthermore, the detection time per sample was significantly reduced to only 723ms, enabling rapid analysis and diagnosis of fecal specimens in clinical settings.
Here, we address the issue of image pre-processing and augmentation for the rapid automatic detection and classification in raw parasite images. We have adopted a multi-steps routine for data preparation, neural network configuration, model training and for prediction analysis. We applied the You Only Look Once (YOLO) CNN based model for object detection [
15,
16,
17], specifically the YOLOv5 architecture for detection and classification of samples in dataset. Yolo is a cutting-edge, real-time object detector, and Yolov5 builds on Yolov1-Yolov4. It has consistently outperformed on two official object detection datasets: Pascal VOC (visual object classes) [
18] and Microsoft COCO (common objects in context) [
19].
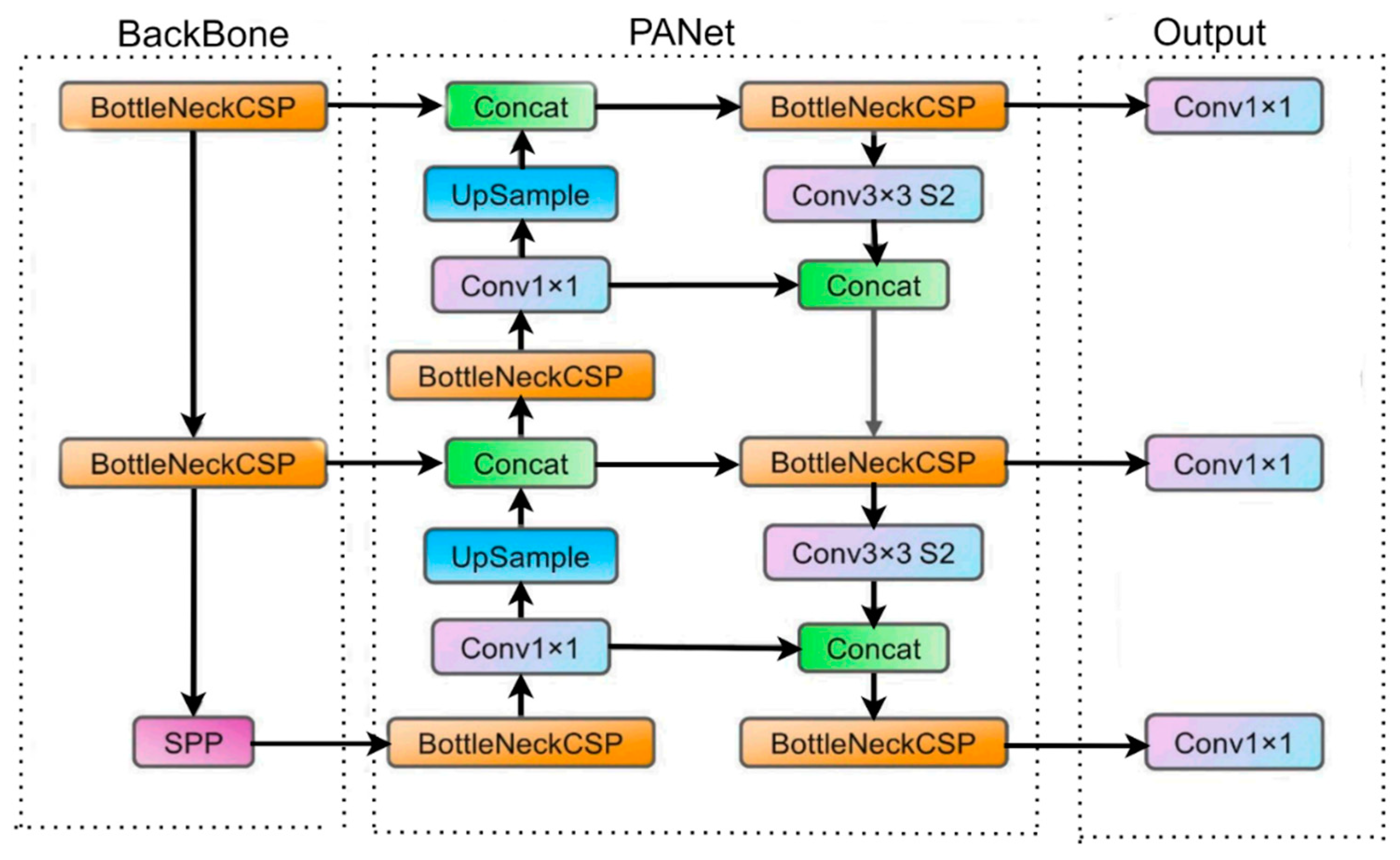
Figure 1 depicts the YOLOv5 network architecture. We have utilized YOLOv5 in this experiment as our initial learner for three reasons. First, YOLOv5 came with CSPDarknet, the backbone of Darknet, including cross stage partial network (CSPNet) [
20] into it. CSPNet minimizes the model's parameters and FLOPS (floating-point operations per second) while ensuring inference speed and accuracy while also reducing the model's size by integrating gradient changes into the feature map and resolving issues with repeated gradient information in large-scale backbones. The task of detecting intestinal parasites required speed and accuracy for detection, and the efficiency of inference on edge devices with limited resources is also derived by the compact model size. The YOLOv5 have used the path aggregation network (PANet) [
21] to improve information flow in its neck. PANet's utilised a novel feature pyramid network (FPN) structure with an improved bottom-up path greatly enhances the transmission of low-level features. Furthermore, adaptive feature pooling is used to link feature grid at all level and permit essential information from each feature level to extend to the next subnetwork. PANet approach improves the utilization of precise localization signals in down layers, which increases the object's location accuracy. Third, the YOLO layer, which serves as the powerhouse of Yolov5, generates feature maps in three different sizes (18×18, 36×36, and 72×72), giving the model the ability to handle objects of varying sizes, from small to medium to large. In a similar way, the growth of a forest fire typically progresses from a small-scale ground fire to a medium-sized trunk fire, and eventually to a large-scale canopy fire. By utilizing multi-scale detection, the Yolov5 model is able to accurately track these size changes, allowing for faster and more effective fire suppression efforts.
We utilized data augmentation techniques on the source parasite images to generate more regularization and divergence in training dataset. We attempt to optimize the model by applying additional dataset by using augmentation/transformation to enhance the results. We are able to accurately detect parasites from a separate test set of microscopic images by using trained YOLOv5 algorithms.
2. Related work
Previous efforts in computational diagnosis of intestinal parasitic infections have primarily focused on improving detection accuracy through hand-engineered feature extraction techniques, which require specialized skills and experts [
28]. However, recent developments in pre-trained deep learning models have the potential to revolutionize medical image analysis, especially in scenarios where data are limited and heterogeneous [
29]. These models [
30,
31,
32] can serve as feature extractors for a new model trained on a dataset with fewer images, improving performance in different tasks of computer vision. Recent studies have applied pre-trained deep learning models to the classification and detection tasks of differentiating infectec parasitized from microscopic images, achieving classification and detection accuracies above 95% and outperforming traditional machine learning algorithms [
33]. However, most of these studies have focused on detection tasks of intestinal parasite images, which are less sensitive and may miss cyst or eggs of parasites due to morphological features. Therefore, there is a need to scale up the approach of transfer learning for object detection in intestinal parasitic infections, which could provide more insights and considerably improve parasite detection.
In recent years, the You Only Look Once (YOLO) model for object detection has gained significant attention and has been continuously developed. YOLO directly detects multiple objects by predicting multiple bounding boxes and class probabilities, making it a popular one-stage detection model [
15,
17,
22]. With subsequent improvements in accuracy and computational performance, YOLO-based architectures such as YOLOv4 [
23], YOLOv5 [
24], and YOLOv8 [
25] have been successfully applied in various research works. Although YOLOv8 is the latest version, there is still ongoing debate about the comparisons between the YOLO versions. While YOLOv5 has higher precision and speed than other state-of-the-art algorithms like Faster R-CNN [
26] and SSD [
27], YOLOv5 has higher accuracy and detection speed compared to YOLOv4 and YOLOv8. However, due to the complexity of the YOLOv8 network, a more simplified version of YOLOv5 has been designed to maximize detection speed and improve computational efficiency. This model has been applied in various applications, including the detection of pine wilt disease, trash, and electronic components. In our research, we focus on the YOLOv5 because of its faster detection results and memory usage with low-end GPU devices. We propose using this model for the automatic recognition of six common classes of protozoan cysts and helminthic eggs in parasitic products of stool examination, which is suitable for remote areas with limited laboratory testing capabilities. By utilizing the YOLOv5 model, we aim to improve the detection and classification of protozoan cysts and helminthic eggs for more effective and prompt treatment.
3. Materials and Methods
3.1. Dataset Collection
The intestinal parasites dataset used in this research report contain 10× magnification microscopic images with resolution of 416×416 pixels. Five different types of intestinal parasites cysts or eggs collectes: hookworm eggs, Hymenolepsis nana, Taenia, Ascaris Lumbricoides and Fasciolopsis Buski. These images obtained from Mulago Referral Hospital located in Uganda and the IEEE Dataport [
34,
35]. The intestinal parasites images were annotated using the open-source data annotation graphical user interface (GUI) tool “Roboflow” ((
https://app.roboflow.com/).
Figure 2 depicts microscopic stool images captured using smartphones. The test performance of the model can be affected by the small number of images available for training. Therefore, to reduce the effect of overfitting after model training, data augmentation techniques was used which include vertical and rotational augmentation.
The images were resized to 416x416 for the YOLOv5 algorithms. Finally, the dataset split into training, validation and testing in the ration 70%, 20% and 10% respectively.
3.2. Data preparation
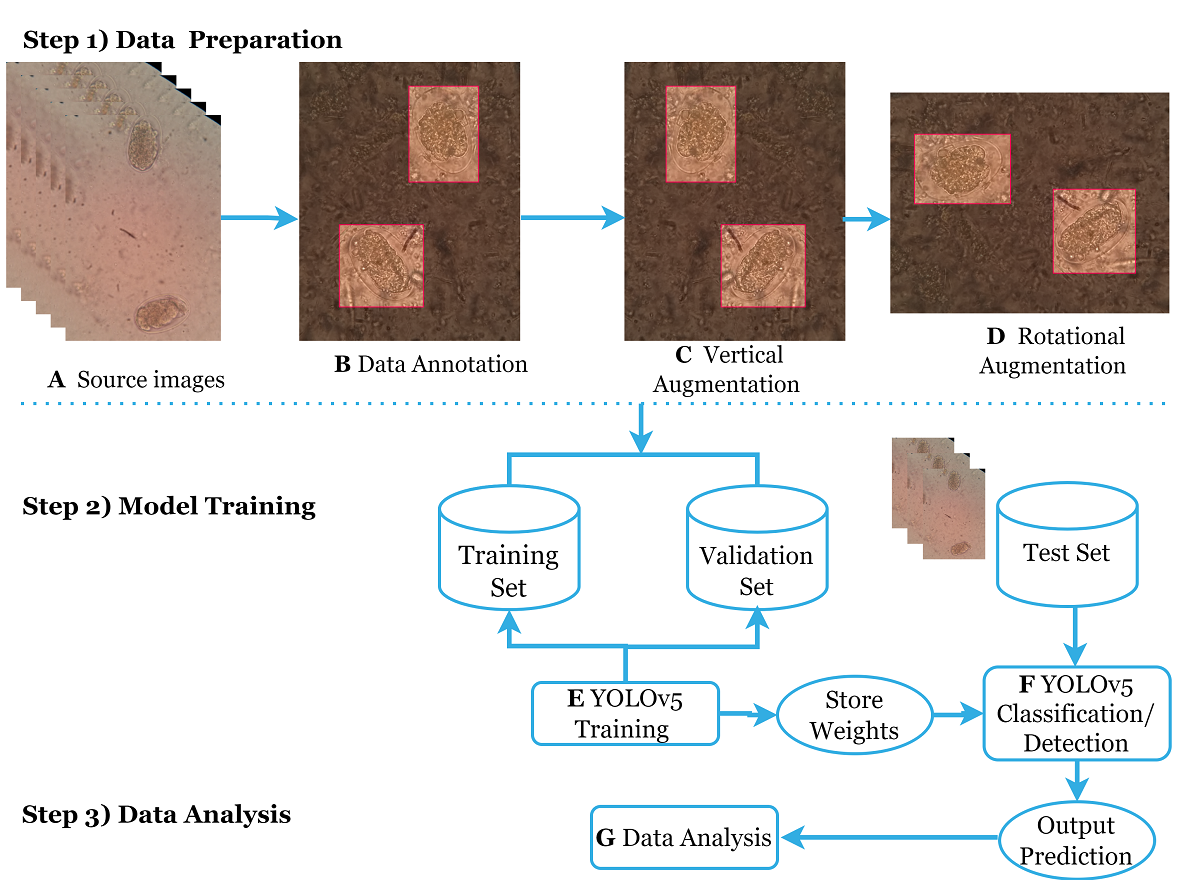
Figure 3 presents a comprehensive overview of the innovative approach adopted to detect parasite images, which involves several crucial stages such as data preparation, annotation, augmentation, training, evaluation, and data analysis. To annotate the parasite images, we utilized the state-of-the-art data annotation tool "Roboflow," which offered a user-friendly graphical user interface (GUI) to draw bounding boxes around the positive and negative images of intestinal parasites accurately. Each bounding box encapsulated the entire parasite species depicted in the image, and the image annotation was saved in YOLO format along with a corresponding text file. To generate a more regularized training dataset, we employed the commonly used technique of data augmentation, which involved subjecting each source image to several data augmentation transformations. The resulting dataset collection contained a vast number of unique images, which were used as the training dataset. We then utilized the test dataset, which only contained images of unannotated parasites, to evaluate the performance of our model. With such an innovative approach, we are confident that our method will pave the way for accurate and efficient parasite detection, bringing us one step closer to eradicating parasitic infections.
3.3. Deep-learning: YOLOv5 training and prediction.
In a recent study, we employed advanced machine learning techniques, specifically YOLOv5 algorithms, to achieve unparalleled accuracy and efficiency in detecting parasites. The use of these cutting-edge algorithms allowed us to extract the best weights from our massive dataset of 2,393 parasite images with varying resolutions. To optimize our results, we experimented with various YOLOv5 algorithm variants and trained our models using both YOLOv5s and YOLOv5l object identification models.
Our decision to work with these particular models was based on their ease of implementation on freely available cloud computing platforms such as Google Colaboratory, making them accessible to a broader audience. Additionally, the YOLOv5 models offer a wide range of pre-written model architectures, each with its own specific speed and accuracy based on the given conditions. We trained our models using a dataset of approximately 950 MB, and the training process took just one hour on a Tesla T4 GPU.The learned weights and biases of our models were automatically saved into a PyTorch weights file, streamlining the process of reusing and improving upon our results. As depicted in
Figure 3, this study demonstrates the potential for these advanced techniques to revolutionize parasite detection, improving healthcare outcomes worldwide and potentially saving countless lives. The ability to detect parasites with such accuracy and efficiency is critical in regions with limited access to healthcare resources and could significantly impact disease prevention and treatment efforts.
The YOLOv5 object recognition version was cloned from the Ultralytics YOLOv5 catalogue [
36]. The YOLOv5s and YOLOv5l models were trained using the train.py function for 100 epochs with an image size of 416x416 and batch size 16. The models were used for both the classification and detection tasks. Both methods were trained with only the default hyper-parameters. On the test dataset, these trained weights were then used to make predictions. A 416x416 image was used for testing using the detect.py function. Utilizing hardware from the Google Colaboratory cloud computing service, models were trained and tested.
In conclusion, this groundbreaking study highlights the power of advanced machine learning techniques in addressing real-world challenges. By utilizing state-of-the-art algorithms and leveraging cloud computing platforms, we can enhance our ability to detect and combat diseases more effectively. This study is a promising step towards transforming healthcare outcomes globally and advancing the field of machine learning.
3.4. Experimental platform.
The experiment was executed on Google Colab System as the base environment with a Nividia K80/T4 GPU at 0.82GHz/1.59GHz GPU memory clock and GPU memory of 12GB/16GB. This experiment was carried out with models built on PyTorch [
37], which provides libraries for building the main architecture of a deep learning model. The training of the models was done using the open-source library Pytorch.
Figure 4 represents a detailed flowchart of the dataset, training and detection process of the YOLOv5 model used in this experiment.
3.5. Statistical analyses.
To determine the strength of the conducted experiments on the trained YOLOv5 algorithms, Precision, Recall, F1-score and mAP measures are used for evaluation. The computation methods are provided in equations 1-4.
Here, TP, TN, FN, and FP are abbreviations for True Positive (correct detections), True Negative, False Positive (incorrect detections), and False Negative (missed detections). The F1-score metric determines the trade-off between Recall and Precision to exhibit the comprehensive performance of the trained model, defined in equation (3). Average Precision-AP was used to demonstrate the overall performance of the models at various confidence levels, as shown below:
where
is the measured Precision at Recall
.
4. Experiment Results
In our pursuit to advance parasite detection, we've gathered a diverse dataset of 5,883 images featuring various parasites, including hookworm eggs, Hymenolepis nana, Taenia, Ascaris Lumbricoides, and Fasciolopsis. Our collection spans across continents, with images acquired from the Mulago Referral Hospital in Uganda and the IEEE Dataport.
With this dataset, we developed a novel detection algorithm using YOLOv5, with each image in the development dataset meticulously annotated with vibrant rectangular boxes as the ground truth. To further improve the robustness of our model, we applied data augmentation and split our dataset into training and testing sets. As we trained our models, we experimented with various image resolutions and settled on 416x416 pixels to achieve optimal performance.
The YOLOv5 algorithms detect parasites in the test dataset by achieving good detection results. The Precision, Recall, F1-score and AP of the detected parasites were calculated and compared with YOLOv5s and YOLOv5l models. SGD optimizer was used, and the training lasted for 100 epochs completed in 1.04 hours and 0.918 hours for YOLOv5l and YOLOv5s, respectively. Additionally, we used the YOLOv5 feature to augment training images by cropping images, dynamic range, and changing scale during the training process. YOLOv5 applied image space and color space augmentations in the training dataset to present an original image + 3 random images each time an image is loaded for training.
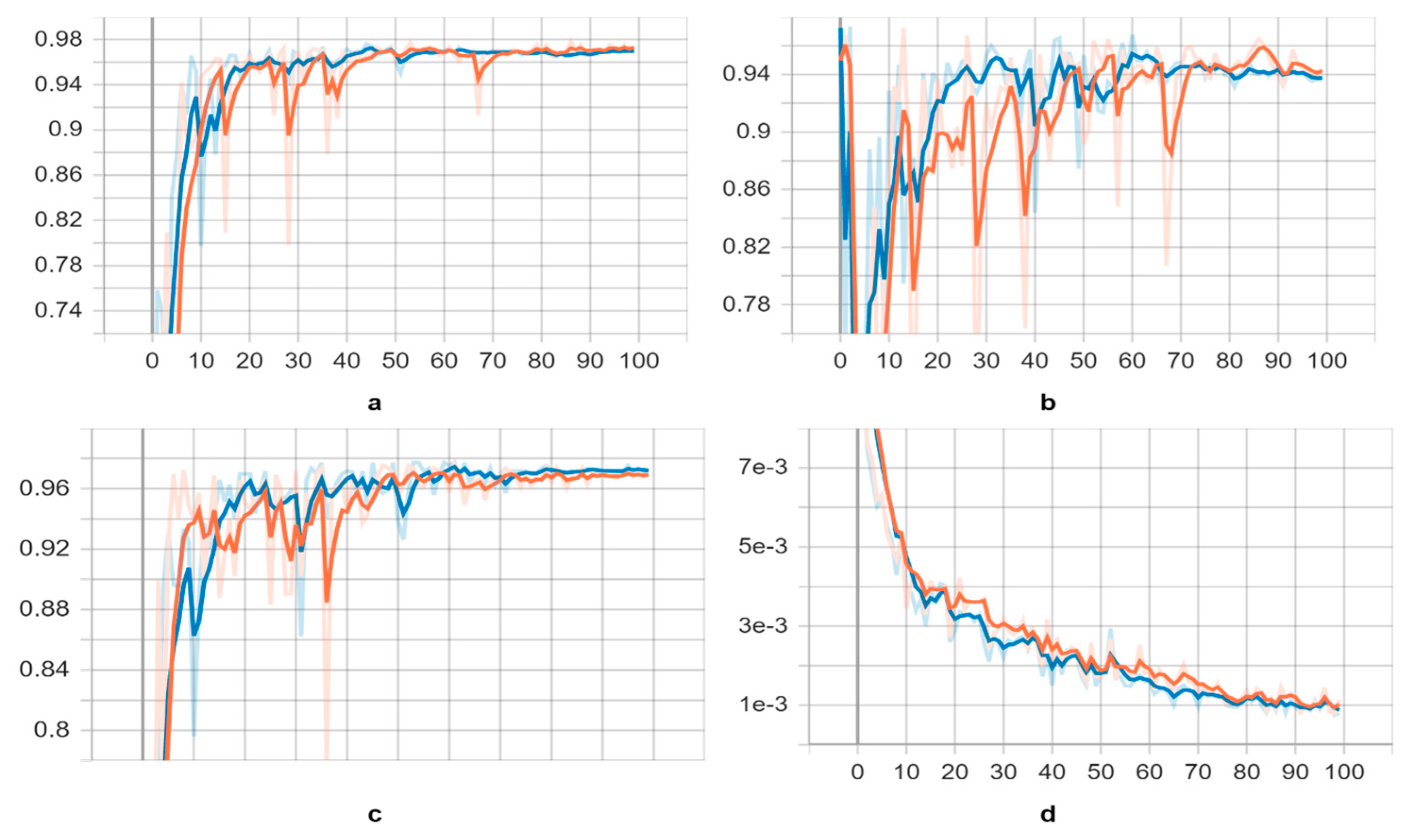
Initially, we trained the “s” and “l” variants of the YOLOv5 model on training datasets. As the best indicative of the overall training stage, we generally selected the weights with the best available scores. The score of trained neural networks dramatically increased in the 20th and 24th epochs on both trained neural networks and the corresponding loss fall down (see
Figure 5). These are the points at which neural networks began to identify parasite species. However, when we extended both training processes to 100 epochs, the validation scores began to fall gradually after the 90th epoch, and the trained neural networks were never able to recognize parasites on a meaningful level.
After applying pre-trained weights to both YOLOv5 models for validation, we obtained results that are presented in
Table 1 and visualized in
Figure 5. The trained neural networks showed better recognition of parasite species, with slightly higher average mAP (0.5), mAP (0.5:0.95), recall, and precision scores, as shown in
Table 2. In
Figure 6, we provide examples of parasite species detection by YOLOv5l models, which demonstrate their impressive performance, but also significant distortion of bounding boxes around the detected species. It is worth noting that the evaluated performance varies depending on the method used. Although the YOLOv5l model exhibits higher accuracy, it also has more layers and parameters, making it slower than the YOLOv5s model, as shown in
Table 3. Overall, our findings suggest that the YOLOv5 models are effective in detecting parasite species and provide valuable insights for future research in this field.
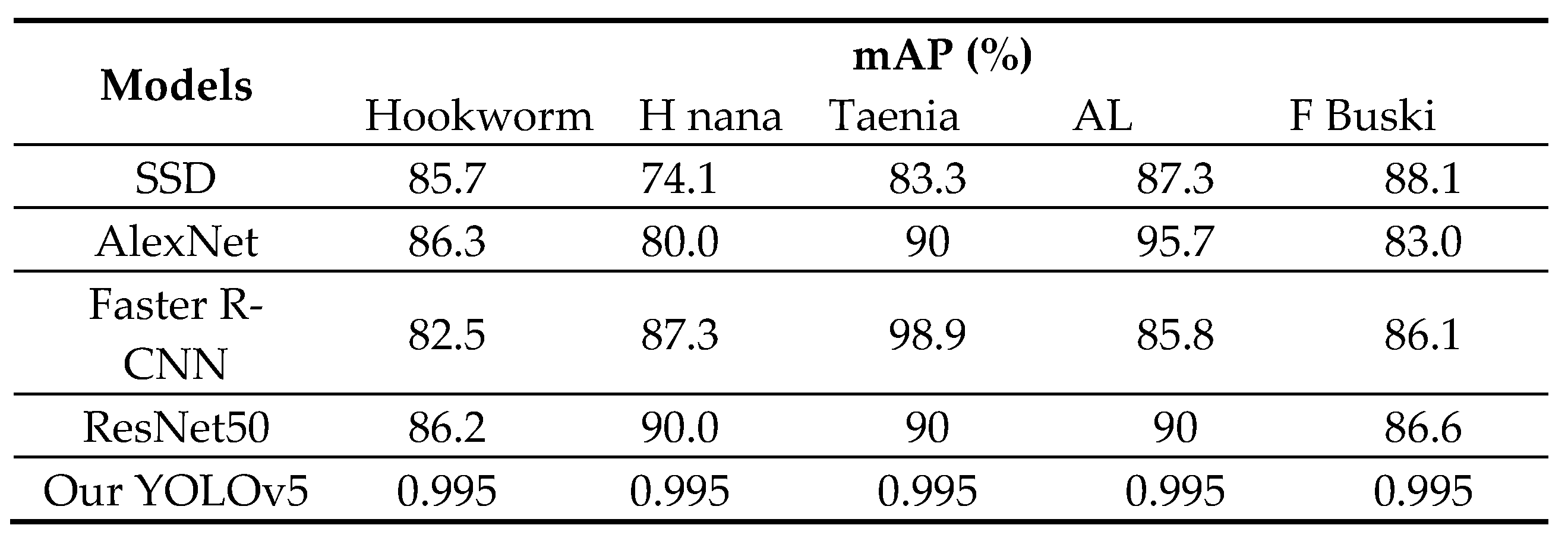
4.1. Comparison with object detection models
We evaluated the performance of our proposed methods against four state-of-the-art object detection models: Single shot detector (SSD) [
18], Faster R-CNN [
19], AlexNet, and ResNet50. While SSD is a lightweight network that detects multiple objects in a single shot, Faster R-CNN requires two shots for object detection - one to identify regions of interest (ROI) and another to detect the object in each ROI using CNNs. We used VGG-16 and ResNet50 as the backbones for SSD and Faster R-CNN, respectively.
Table 4 summarizes the precision results of our methods compared to these four models. Interestingly, our proposed YOLOv5 model outperforms Faster R-CNN with the best average precision. OurYOLOv5 model also outperforms SSD despite having a smaller architecture and fewer parameters. The sliding window technique employed in our method works well for parasite egg detection as it allows us to fix the size of each patch. Unlike natural images containing objects with various shapes and aspect ratios, parasite eggs have a consistent aspect ratio. Thus, the adaptive size of a bounding box offered by SSD and Faster R-CNN does not improve parasite egg detection.
5. Discussion
This research paper explores the use of advanced deep learning techniques and cloud computing platforms to enhance parasite detection. The study utilized a diverse dataset of 5,883 images featuring various parasites, including hookworm eggs, Hymenolepis nana, Taenia, Ascaris Lumbricoides, and Fasciolopsis, acquired from the Mulago Referral Hospital in Uganda and the IEEE Dataport. The researchers developed a novel detection algorithm using YOLOv5 and meticulously annotated each image in the development dataset with vibrant rectangular boxes as the ground truth. The dataset was split into training and testing sets, and data augmentation was applied to improve the robustness of the model. The YOLOv5 algorithms achieved good detection results, with Precision, Recall, F1-score, and AP calculated and compared with YOLOv5s and YOLOv5l models. Image and color space augmentations were applied in the training dataset to present an original image + 3 random images each time an image is loaded for training.
The study trained the “s” and “l” variants of the YOLOv5 model on the training datasets, and the best weights were selected based on the overall training stage. The score of the trained neural networks dramatically increased in the 20th and 24th epochs on both trained neural networks, and the corresponding loss fell down, indicating that neural networks began to identify parasite species. However, when the training process was extended to 100 epochs, the validation scores began to fall gradually after the 90th epoch, and the trained neural networks were never able to recognize parasites on a meaningful level.
After applying pre-trained weights to both YOLOv5 models for validation, the trained neural networks showed better recognition of parasite species, with slightly higher average mAP (0.5), mAP (0.5:0.95), recall, and precision scores. The YOLOv5l model exhibited higher accuracy but was slower than the YOLOv5s model, with more layers and parameters. The researchers provided examples of parasite species detection by YOLOv5l models, demonstrating their impressive performance but also significant distortion of bounding boxes around the detected species. Overall, the study's findings suggest that the YOLOv5 models are effective in detecting parasite species and provide valuable insights for future research in this field. The research highlights the power of advanced machine learning techniques in addressing real-world challenges and improving disease detection and combat.
6. Conclusion
In this study, we aimed to optimize the performance of two state-of-the-art deep-learning architectures, YOLOv5s and YOLOv5l, for object detection. Specifically, we evaluated the precision, recall, F1-score, and mean average precision of the algorithms on a dataset of 5,883 parasite microscopic images with input image resolutions of 416x416. Our findings showed that YOLOv5l outperformed YOLOv5s in terms of overall accuracy. However, both algorithms demonstrated excellent performance in recognizing five types of parasites and accurately locating them within images. Notably, the YOLOv5 algorithms were significantly faster than Faser-RCNN [
38] and ResNet50 [
33], which requires selective search and, as a result, considerable running time. Interestingly, the mAP performance for the Hookworm , Taenia and Fasciolopsis Buski parasite was the best, with a score of 0.99. We believe that this is due to the clear characteristics of hookworms and the large number of hookworms in the collected dataset, which enabled effective data enhancement during training. The mAP values for Ascaris Lumbricoides were also fine 0.92, for the YOLOv5 algorithms. However, due to the small dataset size and class imbalance, the training model suffered from some degree of overfitting. Despite this limitation, our proposed deep learning model for intestinal parasite detection achieved the highest mAP and demonstrated rapid detection and location of hookworms, taenia, fasciolopsis buski, ascaris lumbricoides and hymenolepis nana with an mAP of approximately 97% and a detection time of 8.5 ms per image. Future work should aim to expand the dataset size and include hymenolepis nana in the analysis to address the overfitting issue when dealing with small dataset sizes. Overall, our study contributes to the development of effective deep learning models for object detection in the field of parasitology.
Author Contributions
S. A. Abdullah and K. Satish constructed the concept; A. Tasleem, S. A Abdullah performed the data preprocessing and image analysis; and K. Satish proofread and ensured the general quality of manuscript
Data Availability
The datasets used and/or analysed during the current experiment for training and testing purpose are available from the corresponding author on reasonable request. All data annotation and augmentation tools are provided by Roboflow on link (https:
//app.roboflow.com/). The “You Only Look Once” version 5 (YOLOv5) object detection architecture written in Python by Ultralytics, is freely available under a GPL-3.0 license (
https://github.com/ultralytics/yolov5). Two Jupyter notebooks that are interactive implementations of the YOLOv5 methods for training and detection are provided with this publication. All pre-processed data used in this experiment can be made available upon reasonable request.
Acknowledgments
We express our thanks to laboratory experts from Mulago Referral Hospital located in Uganda and the IEEE dataport who collected the parasitic species used in this study
Competing Interests
The authors declare no competing interests.
Additional Information
Correspondence and requests for materials should be addressed to K. Satish
References
- O. Dyer, “African malaria deaths set to dwarf covid-19 fatalities as pandemic hits control efforts, WHO warns,” BMJ, vol. 371, p. m4711, Dec. 2020. [CrossRef]
- P. Berzosa et al., “Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea 11 Medical and Health Sciences 1108 Medical Microbiology,” Malar. J., vol. 17, no. 1, pp. 1–12, Sep. 2018. [CrossRef]
- P. Dahal, B. Khanal, K. Rai, V. Kattel, S. Yadav, and N. R. Bhattarai, “Challenges in Laboratory Diagnosis of Malaria in a Low-Resource Country at Tertiary Care in Eastern Nepal: A Comparative Study of Conventional vs. Molecular Methodologies,” J. Trop. Med., vol. 2021, 2021. [CrossRef]
- S. Li, A. Li, D. A. Molina Lara, J. E. Gómez Marín, M. Juhas, and Y. Zhang, “Transfer Learning for Toxoplasma gondii Recognition,” mSystems, vol. 5, no. 1, pp. 1–12, 2020. [CrossRef]
- N. Tangpukdee, C. Duangdee, P. Wilairatana, and S. Krudsood, “Malaria Diagnosis: A Brief Review,” Korean J. Parasitol., vol. 47, no. 2, pp. 93–102, 2009. [CrossRef]
- P. Rani, S. Kotwal, J. Manhas, V. Sharma, and S. Sharma, Machine Learning and Deep Learning Based Computational Approaches in Automatic Microorganisms Image Recognition: Methodologies, Challenges, and Developments. Springer Netherlands, 2021.
- A. Esteva et al., “A guide to deep learning in healthcare,” Nat. Med. 2019 251, vol. 25, no. 1, pp. 24–29, Jan. 2019. [CrossRef]
- S. Kumar, T. Arif, A. S. Alotaibi, M. B. Malik, and J. Manhas, “Advances Towards Automatic Detection and Classification of Parasites Microscopic Images Using Deep Convolutional Neural Network : Methods , Models and Research Directions,” Arch. Comput. Methods Eng., 2022. [CrossRef]
- M. Salvi, U. R. Acharya, F. Molinari, and K. M. Meiburger, “The impact of pre- and post-image processing techniques on deep learning frameworks: A comprehensive review for digital pathology image analysis,” Comput. Biol. Med., vol. 128, p. 104129, Jan. 2021. [CrossRef]
- Y. Pratama, Y. Fujimura, T. Funatomi, and Y. Mukaigawa, “PARASITIC EGG DETECTION AND CLASSIFICATION BY UTILIZING THE YOLO ALGORITHM WITH DEEP LATENT SPACE IMAGE RESTORATION AND GRABCUT AUGMENTATION Graduate School of Science and Technology , Nara Institute of Science and Technology ( NAIST ),” pp. 4311–4315, 2022.
- G. P. Gopakumar, M. Swetha, G. Sai Siva, and G. R. K. Sai Subrahmanyam, “Convolutional neural network-based malaria diagnosis from focus stack of blood smear images acquired using custom-built slide scanner,” J. Biophotonics, vol. 11, no. 3, p. e201700003, Mar. 2018. [CrossRef]
- L. Rosado, J. M. C. da Costa, D. Elias, and J. S. Cardoso, “Mobile-Based Analysis of Malaria-Infected Thin Blood Smears: Automated Species and Life Cycle Stage Determination,” Sensors 2017, Vol. 17, Page 2167, vol. 17, no. 10, p. 2167, Sep. 2017. [CrossRef]
- G. Díaz, F. A. González, and E. Romero, “A semi-automatic method for quantification and classification of erythrocytes infected with malaria parasites in microscopic images,” J. Biomed. Inform., vol. 42, no. 2, pp. 296–307, Apr. 2009. [CrossRef]
- A. H. Alharbi, C. V. Aravinda, M. Lin, B. Ashwini, M. Y. Jabarulla, and M. A. Shah, “Detection of Peripheral Malarial Parasites in Blood Smears Using Deep Learning Models,” Comput. Intell. Neurosci., vol. 2022, 2022. [CrossRef]
- A. Bochkovskiy, C.-Y. Wang, and H.-Y. M. Liao, “YOLOv4: Optimal Speed and Accuracy of Object Detection,” Apr. 2020. [CrossRef]
- J. Redmon, S. Divvala, R. Girshick, and A. Farhadi, “You Only Look Once: Unified, Real-Time Object Detection,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2016-December, pp. 779–788, Jun. 2015. [CrossRef]
- J. Redmon and A. Farhadi, “YOLO9000: Better, faster, stronger,” Proc. - 30th IEEE Conf. Comput. Vis. Pattern Recognition, CVPR 2017, vol. 2017-January, pp. 6517–6525, Nov. 2017. [CrossRef]
- M. Everingham, S. M. A. Eslami, L. Van Gool, C. K. I. Williams, J. Winn, and A. Zisserman, “The PASCAL Visual Object Classes Challenge: A Retrospective,” Int. J. Comput. Vis., vol. 111, no. 1, pp. 98–136, Jan. 2015. [CrossRef]
- T. Y. Lin et al., “Microsoft COCO: Common Objects in Context,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 8693 LNCS, no. PART 5, pp. 740–755, May 2014. [CrossRef]
- C.-Y. Wang, H.-Y. M. Liao, I.-H. Yeh, Y.-H. Wu, P.-Y. Chen, and J.-W. Hsieh, “CSPNet: A New Backbone that can Enhance Learning Capability of CNN,” Nov. 2019. [CrossRef]
- K. Wang, J. H. Liew, Y. Zou, D. Zhou, and J. Feng, “PANet: Few-Shot Image Semantic Segmentation with Prototype Alignment,” Proc. IEEE Int. Conf. Comput. Vis., vol. 2019-October, pp. 9196–9205, Aug. 2019. [CrossRef]
- G. Jocher et al., “ultralytics/yolov5: v7.0 - YOLOv5 SOTA Realtime Instance Segmentation,” Nov. 2022. [CrossRef]
- S. J. Ji, Q. H. Ling, and F. Han, “An improved algorithm for small object detection based on YOLO v4 and multi-scale contextual information,” Comput. Electr. Eng., vol. 105, p. 108490, Jan. 2023. [CrossRef]
- D. Wan, R. Lu, S. Wang, S. Shen, T. Xu, and X. Lang, “YOLO-HR: Improved YOLOv5 for Object Detection in High-Resolution Optical Remote Sensing Images,” Remote Sens. 2023, Vol. 15, Page 614, vol. 15, no. 3, p. 614, Jan. 2023. [CrossRef]
- J. R. Terven and D. M. Cordova-Esparaza, “A Comprehensive Review of YOLO: From YOLOv1 to YOLOv8 and Beyond,” Apr. 2023, Accessed: Apr. 28, 2023. [Online]. Available: https://arxiv.org/abs/2304.00501v1.
- D. Osaku, C. F. Cuba, C. T. N. Suzuki, J. F. Gomes, and A. X. Falcão, “Automated diagnosis of intestinal parasites: A new hybrid approach and its benefits,” Comput. Biol. Med., vol. 123, pp. 1–18, 2020. [CrossRef]
- R. Nakasi, E. Mwebaze, A. Zawedde, J. Tusubira, B. Akera, and G. Maiga, “A new approach for microscopic diagnosis of malaria parasites in thick blood smears using pre-trained deep learning models,” SN Appl. Sci., vol. 2, no. 7, pp. 1–7, 2020. [CrossRef]
- H. A. Nugroho, S. A. Akbar, and E. E. H. Murhandarwati, “Feature extraction and classification for detection malaria parasites in thin blood smear,” ICITACEE 2015 - 2nd Int. Conf. Inf. Technol. Comput. Electr. Eng. Green Technol. Strength. Inf. Technol. Electr. Comput. Eng. Implementation, Proc., vol. 1, no. c, pp. 197–201, 2016. [CrossRef]
- S. Li, Z. Du, X. Meng, and Y. Zhang, “Multi-stage malaria parasite recognition by deep learning,” Gigascience, vol. 10, no. 6, pp. 1–11, 2021. [CrossRef]
- F. Yang, H. Yu, K. Silamut, R. J. Maude, S. Jaeger, and S. Antani, “Parasite Detection in Thick Blood Smears Based on Customized Faster-RCNN on Smartphones,” Proc. - Appl. Imag. Pattern Recognit. Work., vol. 2019-Octob, pp. 0–3, 2019. [CrossRef]
- C. Zhang et al., “Deep learning for microscopic examination of protozoan parasites,” Comput. Struct. Biotechnol. J., vol. 20, pp. 1036–1043, Jan. 2022. [CrossRef]
- R. Nakasi, E. Mwebaze, A. Zawedde, J. Tusubira, B. Akera, and G. Maiga, “A new approach for microscopic diagnosis of malaria parasites in thick blood smears using pre-trained deep learning models,” SN Appl. Sci., vol. 2, no. 7, pp. 1–7, Jul. 2020. [CrossRef]
- T. Suwannaphong, S. Chavana, S. Tongsom, and D. Palasuwan, “Parasitic Egg Detection and Classification in Low-cost Microscopic Images using Transfer Learning.”.
- J. A. Quinn, R. Nakasi, P. K. B. Mugagga, P. Byanyima, W. Lubega, and A. Andama, “Deep Convolutional Neural Networks for Microscopy-Based Point of Care Diagnostics,” pp. 1–12, 2016, [Online]. Available: http://arxiv.org/abs/1608.02989.
- “Parasitic Egg Detection and Classification in Microscopic Images | IEEE DataPort.” https://ieee-dataport.org/competitions/parasitic-egg-detection-and-classification-microscopic-images (accessed May 19, 2022).
- “GitHub - ultralytics/yolov5: YOLOv5 🚀 in PyTorch > ONNX > CoreML > TFLite.” https://github.com/ultralytics/yolov5 (accessed Jan. 25, 2023).
- A. Paszke et al., “Automatic differentiation in PyTorch.” Oct. 28, 2017.
- N. Q. Viet, “Parasite worm egg automatic detection in microscopy stool image based on Faster R-CNN,” pp. 197–202, 2019.
Figure 1.
The Yolov5 network architecture. It consists of three parts: (a) BackBone (CSPDarknet), (b) Neck (PANet), and (c) Output (YOLO Layer). The data are first input to Backbone for feature extraction, and then applied to PANet for feature fusion. Finally, Yolo Layer outputs detection results.
Figure 1.
The Yolov5 network architecture. It consists of three parts: (a) BackBone (CSPDarknet), (b) Neck (PANet), and (c) Output (YOLO Layer). The data are first input to Backbone for feature extraction, and then applied to PANet for feature fusion. Finally, Yolo Layer outputs detection results.
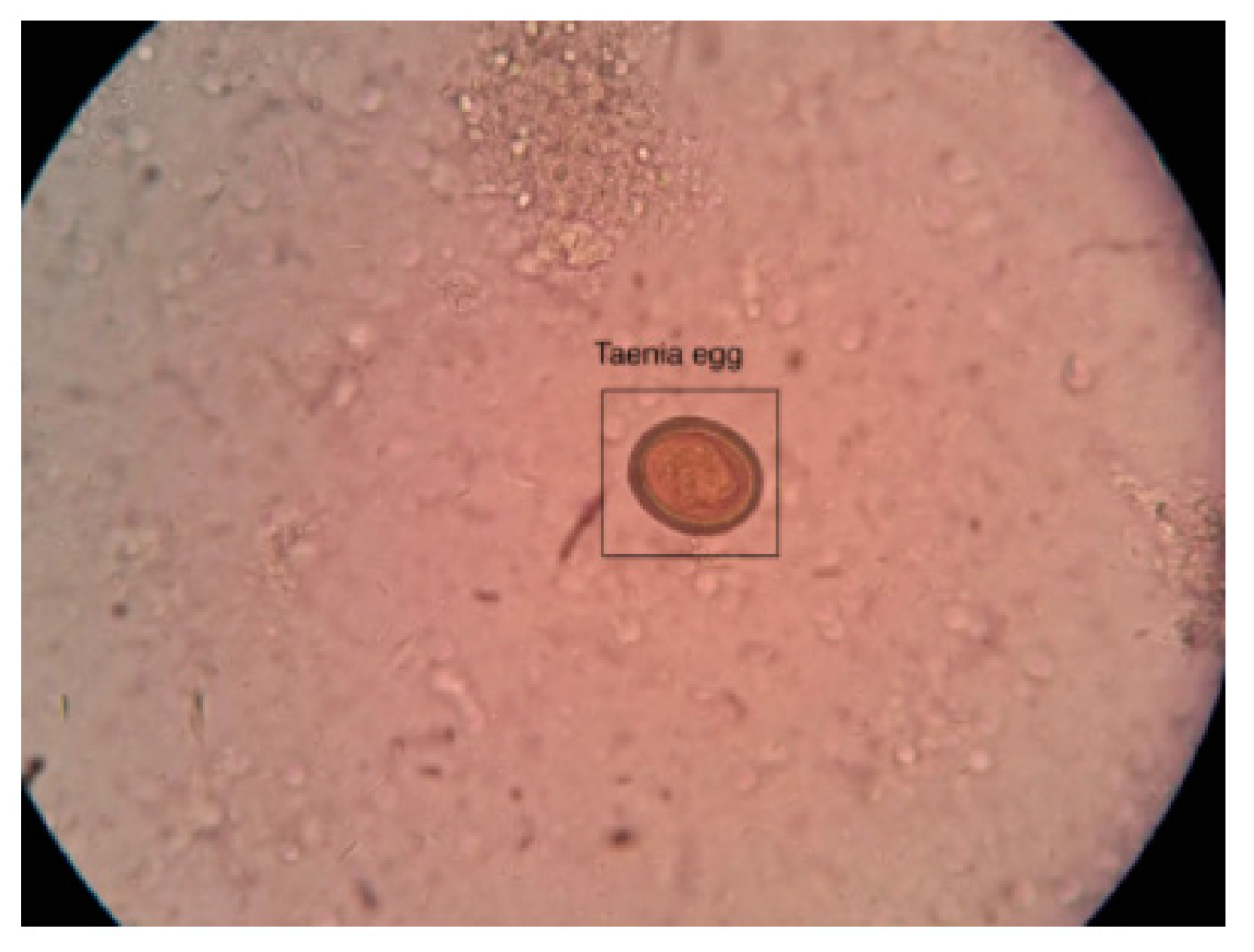
Figure 2.
Microscopic stool sample image captured with a smartphone.
Figure 2.
Microscopic stool sample image captured with a smartphone.
Figure 3.
Flow chart of data preparation and analysis. Depict various steps that configure and utilize YOLOv5s and YOLOv5l models. Step 1 begins with the collection of parasite images, which are then annotated and subjected to data augmentation techniques such as vertical and rotational methods. Step 2 involves training the YOLOv5 models using both training and validation datasets, which are then fit with PyTorch weights files to make predictions on the test dataset. The YOLOv5 detection and classification function, applied with these trained weights, is then used to make output predictions. Finally, in Step 3, we employ advanced data analysis techniques to measure the models' performance. Our innovative flow chart demonstrates the exciting possibilities of combining advanced technology and data analysis in revolutionizing parasite detection and improving global healthcare outcomes.
Figure 3.
Flow chart of data preparation and analysis. Depict various steps that configure and utilize YOLOv5s and YOLOv5l models. Step 1 begins with the collection of parasite images, which are then annotated and subjected to data augmentation techniques such as vertical and rotational methods. Step 2 involves training the YOLOv5 models using both training and validation datasets, which are then fit with PyTorch weights files to make predictions on the test dataset. The YOLOv5 detection and classification function, applied with these trained weights, is then used to make output predictions. Finally, in Step 3, we employ advanced data analysis techniques to measure the models' performance. Our innovative flow chart demonstrates the exciting possibilities of combining advanced technology and data analysis in revolutionizing parasite detection and improving global healthcare outcomes.
Figure 4.
Flowchart of YOLOv5 model for dataset, training and detection process.
Figure 4.
Flowchart of YOLOv5 model for dataset, training and detection process.
Figure 5.
Learning curves of the YOLOv5s and YOLOv5l trained models on parasite dataset, yellow and blue respectively. As a metric, we used the a) mean Average Precision (mAP) (0.5) score, b) precision score, c) recall score and d) class loss score.
Figure 5.
Learning curves of the YOLOv5s and YOLOv5l trained models on parasite dataset, yellow and blue respectively. As a metric, we used the a) mean Average Precision (mAP) (0.5) score, b) precision score, c) recall score and d) class loss score.
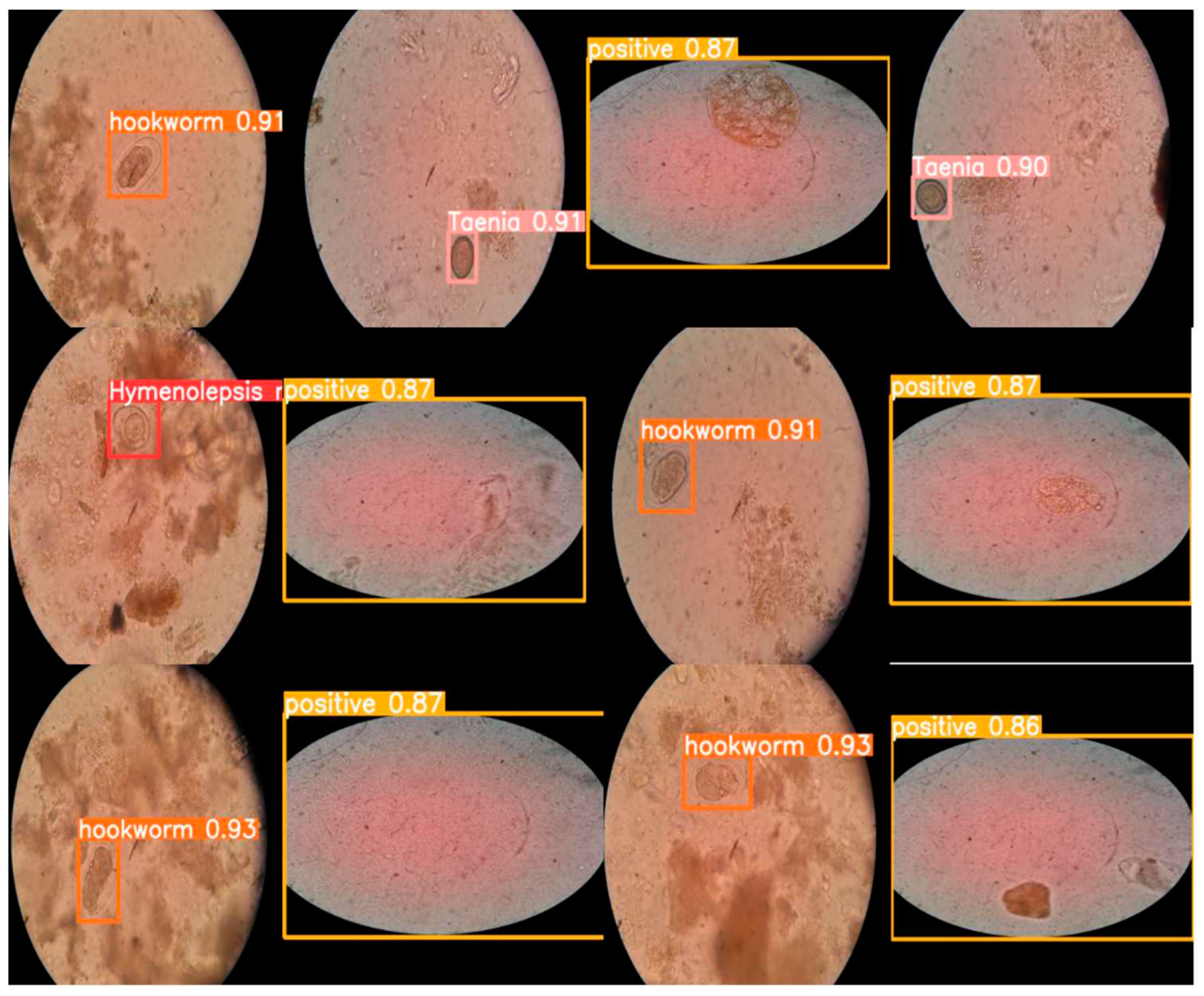
Figure 6.
Image shows several examples of object detections performed by YOLOv5l neural networks.
Figure 6.
Image shows several examples of object detections performed by YOLOv5l neural networks.
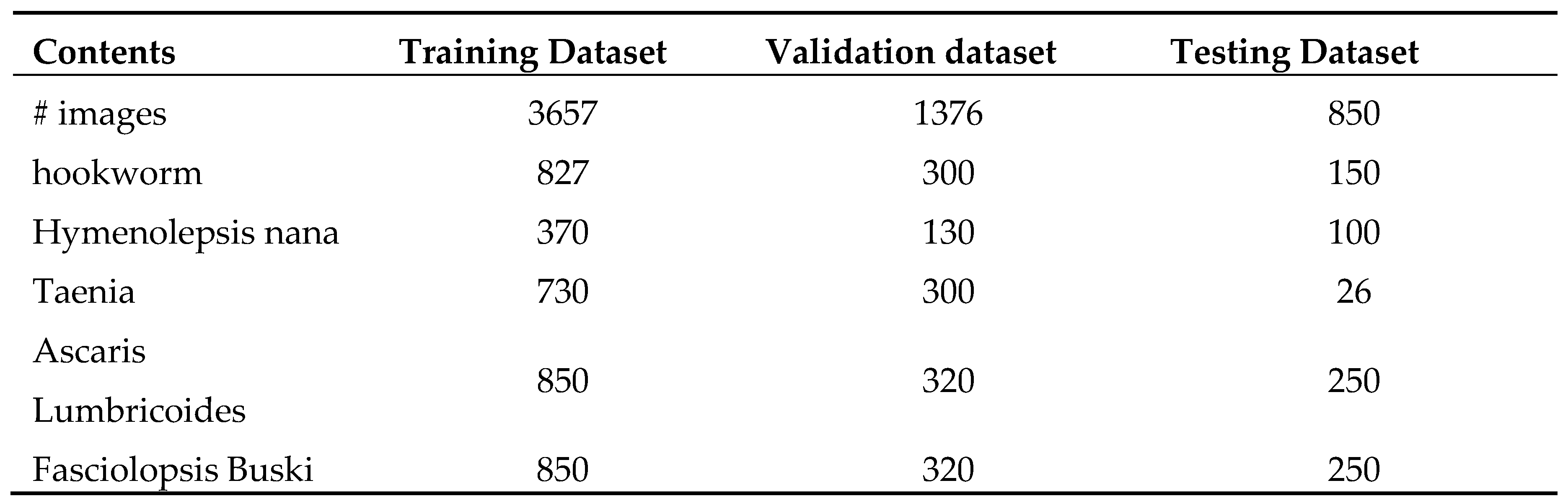
Table 1.
Overview of the intestinal parasites dataset obtained after applying data augmentation techniques. The dataset is divided into a training set (70%), validation set (20%) and testing set (10%).
Table 1.
Overview of the intestinal parasites dataset obtained after applying data augmentation techniques. The dataset is divided into a training set (70%), validation set (20%) and testing set (10%).
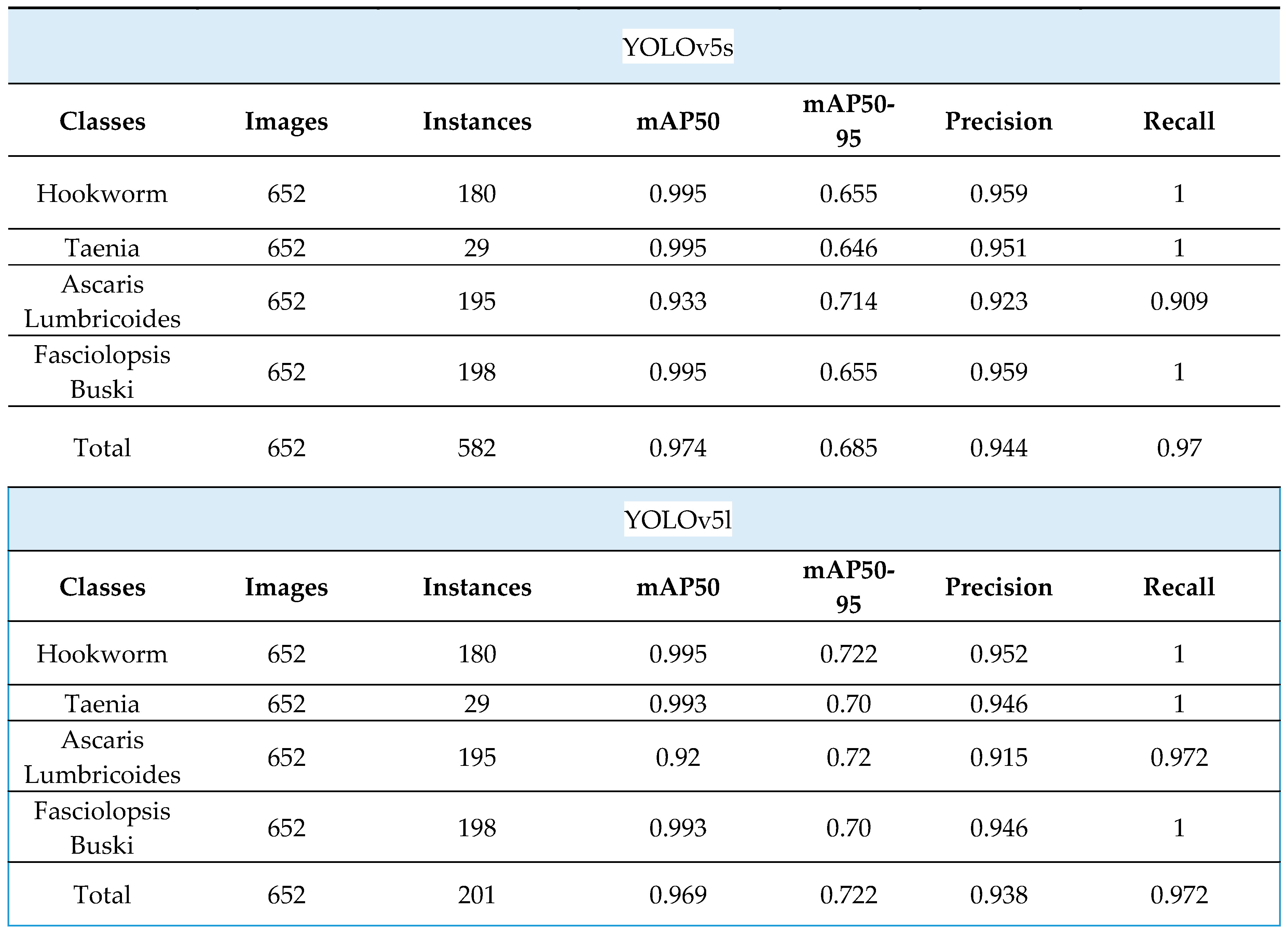
Table 2.
Comparison of the YOLOv5s and YOLOv5l neural networks trained, by using transfer learning and pre-trained on trained weights, on validation datasets.
Table 2.
Comparison of the YOLOv5s and YOLOv5l neural networks trained, by using transfer learning and pre-trained on trained weights, on validation datasets.
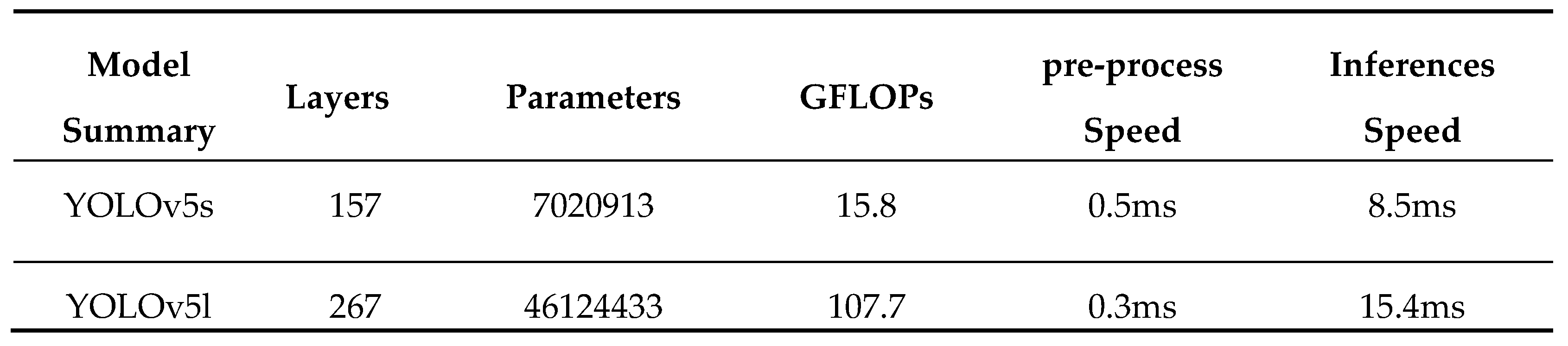
Table 3.
Parasites detection speed on YOLOv5 models.
Table 3.
Parasites detection speed on YOLOv5 models.
Table 4.
Comparison with object detection models.
Table 4.
Comparison with object detection models.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).