Submitted:
10 May 2023
Posted:
11 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3.1. Virus-containing materials, serum and ticks
3.2. RNA isolation
3.3. Preparation of the standard RNA samples
3.3. Preparation of porcine embryo kidney total RNA
3.4. Reverse transcription and qPCR
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lindenbach, B.D.; Thiel, H.-J.; Rice, C.M. Flaviviridae: The Viruses and Their Replication. In Fields Virology, 5th Edition.; Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven Publishers: Philadelphia, 2007; pp. 1101–1151; ISBN 0781760607. [Google Scholar]
- Rossi, S.L.; Ross, T.M.; Evans, J.D. West Nile virus. Clin. Lab. Med. 2010, 30, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Whitehorn, J.; Simmons, C.P. The pathogenesis of dengue. Vaccine 2011, 29, 7221–8. [Google Scholar] [CrossRef] [PubMed]
- Wikan, N.; Smith, D.R. Zika virus: History of a newly emerging arbovirus. Lancet Infect. Dis. 2016, 16, e119–e126. [Google Scholar] [CrossRef] [PubMed]
- Ruzek, D.; Av, T.; Borde, J.; Chrdle, A.; Eyer, L.; Karganova, G.; Kholodilov, I.; Kozlovskaya, L.; Matveev, A.; Miller, A.D.; Osolodkin, D.I.; Överby, A.K.; Tikunova, N.; Tkachev, S.; Zajkowska, J. Tick-borne encephalitis in Europe and Russia : Review of pathogenesis, clinical features, therapy, and vaccines. Antiviral Res. 2019, 164, 23–51. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Shi, M.; Tian, J.; Lin, X.; Kang, Y.; Chen, L.; Qin, X.; Xu, J.; Holmes, E.C.; Zhang, Y. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 2015, 4, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.; Tian, J.; Chen, L.; Chen, X.; Li, C.; Qin, X.; Li, J.; Cao, J.; Eden, J.; Buchmann, J.; Wang, W.; Xu, J.; Holmes, E.C.; Zhang, Y. Redefining the invertebrate RNA virosphere. Nature 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, S.; Ka, S.; Zirkel, F.; Donath, A.; Petersen, M.; Liu, S.; Zhou, X.; Drosten, C.; Misof, B.; Junglen, S. Viromics of extant insect orders unveil the evolution of the flavi-like superfamily. Virus 2021, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.-C.; Shi, M.; Tian, J.-H.; Lin, X.-D.; Gao, D.-Y.; He, J.-R.; Wang, J.-B.; Li, C.-X.; Kang, Y.-J.; Yu, B.; Zhou, D.-J.; Xu, J.; Plyusnin, A.; Holmes, E.C.; Zhang, Y.-Z. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl. Acad. Sci. 2014, 111, 6744–6749. [Google Scholar] [CrossRef]
- Gao, X.; Qin, B.; Li, Z.; Wang, M.; Cui, S. Crystal structure of the NS3-like helicase from Alongshan virus. IUCrJ. 2020, 7, 375–382. [Google Scholar] [CrossRef]
- Domains, M.; Garry, C.E.; Garry, R.F. Proteomics Computational Analyses Suggest That the Envelope Glycoproteins of Segmented Jingmen Flavi-Like Viruses are Class II Viral Fusion Proteins. Viruses 2020, 12. [Google Scholar]
- Kholodilov, I.S.; Belova, O.A.; Morozkin, E.S.; Litov, A.G.; Ivannikova, A.Y.; Makenov, M.T.; Shchetinin, A.M.; Aibulatov, S.V.; Bazarova, G.K.; Bell-sakyi, L.; Bespyatova, L.A.; Bugmyrin, S.V.; Chernetsov, N.; Chernokhaeva, L.L.; Gmyl, L.V.; Khaisarova, A.N.; Khalin, A.V.; Klimentov, A.S.; Kovalchuk, I.V.; Luchinina, S.V.; Medvedev, S.G.; Nafeev, A.A.; Oorzhak, N.D.; Panjukova, E.V.; Polienko, A.E.; Purmak, K.A.; Romanenko, E.N.; Rozhdestvenskiy, E.N.; Saryglar, A.A.; Shamsutdinov, A.F.; Karganova, G.G. Geographical and Tick-Dependent Distribution of Flavi-Like Alongshan and Yanggou Tick Viruses in Russia. Viruses 2021, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ladner, J.T.; Wiley, M.R.; Beitzel, B.; Kramer, L.D.; Tesh, R.B.; Palacios, G.; Ladner, J.T.; Wiley, M.R.; Beitzel, B.; Auguste, A.J.; Ii, A.P.D.; Lindquist, M.E.; Sibley, S.D.; Kota, K.P.; Fetterer, D.; Eastwood, G.; Kimmel, D.; Prieto, K.; Guzman, H.; Aliota, M.T.; Reyes, D.; Brueggemann, E.E.; John, L.S.; Hyeroba, D.; Lauck, M. A Multicomponent Animal Virus Isolated from Mosquitoes. Cell Host Microbe 2016, 20, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Kholodilov, I.S.; Litov, A.G.; Klimentov, A.S.; Belova, O.A.; Polienko, A.E.; Nikitin, N.A.; Shchetinin, A.M.; Ivannikova, A.Y.; Bell-sakyi, L.; Yakovlev, A.S.; Bugmyrin, S.V.; Bespyatova, L.A.; Gmyl, L.V.; Luchinina, S.V.; Gmyl, A.P.; Gushchin, V.A.; Karganova, G.G. Isolation and Characterisation of Alongshan Virus in Russia. Viruses 2020, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-D.; Wang, B.; Wei, F.; Han, S.-Z.; Zhang, L.; Yang, Z.-T.; Yan, Y.; Lv, X.-L.; Li, L.; Wang, S.-C.; Song, M.-X.; Zhang, H.-J.; Huang, S.-J.; Chen, J.; Huang, F.-Q.; Li, S.; Liu, H.-H.; Hong, J.; Jin, Y.-L.; Wang, W.; Zhou, J.-Y.; Liu, Q. A New Segmented Virus Associated with Human Febrile Illness in China. N. Engl. J. Med. 2019, 380, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, D.; Kuwata, R.; Kimura, T.; Shimoda, H.; Fujita, R.; Faizah, A.N.; Kai, I.; Matsumura, R.; Kuroda, Y.; Watanabe, S.; Kuniyoshi, S.; Yamauchi, T.; Watanabe, M.; Higa, Y.; Hayashi, T.; Shinomiya, H.; Maeda, K.; Kasai, S.; Sawabe, K.; Isawa, H. Detection of Jingmenviruses in Japan with Evidence of Vertical Transmission in Ticks. Viruses 2021, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.; Vasilakis, N.; Tian, J.; Li, C.; Chen, L.; Eastwood, G.; Diao, X.; Chen, M.-H.; Chen, X.; Qin, X.-C.; Widen, S.G.; Wood, T.G.; Tesh, R.B.; Xu, J.; Holmes, E.C.; Zhanga, Y.-Z. Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses. J. Virol. 2016, 90, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Villa, E.C.; Maruyama, S.R.; de Miranda-Santos, I.K.F.; Palacios, G.; Ladner, J.T. Complete Coding Genome Sequence for Mogiana Tick Virus, a Jingmenvirus Isolated from Ticks in Brazil. Genome Announc. 2017, 5, 17–18. [Google Scholar] [CrossRef]
- Kuivanen, S.; Levanov, L.; Kareinen, L.; Sironen, T.; Jääskeläinen, A.J.; Plyusnin, I.; Zakham, F. Detection of novel tick-borne pathogen, Alongshan virus, in Ixodes ricinus ticks, south-eastern Finland. Eurosurveillance 2019, 24. [Google Scholar] [CrossRef]
- Stefanie, Stegmüller; Fraefel, C.; Kubacki, J. 20. Stefanie Stegmüller; Fraefel, C.; Kubacki, J. Genome Sequence of Alongshan Virus from Ixodes ricinus Ticks Collected in Switzerland. Microbiol. Resour. Announc. 2023, 2022–2023. [Google Scholar]
- Jia, N.; Liu, H.B.; Ni, X.B.; Bell-Sakyi, L.; Zheng, Y.C.; Song, J.L.; Li, J.; Jiang, B.G.; Wang, Q.; Sun, Y.; Wei, R.; Yuan, T.T.; Xia, L.Y.; Chu, Y.L.; Wei, W.; Li, L.F.; Ye, J.L.; Lv, Q.Y.; Cui, X.M.; Guan, Y.; Tong, Y.G.; Jiang, J.F.; Lam, T.T.Y.; Cao, W.C. Emergence of human infection with Jingmen tick virus in China: A retrospective study. EBioMedicine 2019, 43, 317–324. [Google Scholar] [CrossRef]
- Colmant, A.M.G.; Charrel, R.N.; Coutard, B. Jingmenviruses : Ubiquitous, understudied, segmented flavi-like viruses. Front. Microbiol. 2022, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-D.; Wang, W.; Wang, N.N.; Qiu, K.; Zhang, X.; Tana, G.; Liu, Q. Prevalence of the emerging novel Alongshan virus infection in sheep and cattle in Inner Mongolia, northeastern China. Parasit. Vectors 2019, 12, 1–7. [Google Scholar] [CrossRef]
- Ebert, C.L.; Söder, L.; Kubinski, M.; Glanz, J.; Gregersen, E.; Dümmer, K.; Grund, D.; Wöhler, A.; Könenkamp, L.; Liebig, K.; Knoll, S.; Hellhammer, F.; Topp, A.; Becher, P.; Springer, A.; Strube, C.; Nagel-kohl, U.; Nordhoff, M.; Steffen, I.; Bauer, B.U.; Ganter, M.; Feige, K.; Becker, S.C.; Boelke, M. Detection and Characterization of Alongshan Virus in Ticks and Tick Saliva from Lower Saxony, Germany with Serological Evidence for Viral Transmission to Game and Domestic Animals. Microorganisms 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Temmam, S.; Bigot, T.; Chrétien, D.; Gondard, M.; Pérot, P.; Pommelet, V.; Dufour, E.; Petres, S.; Devillers, E.; Hoem, T.; Pinarello, V.; Hul, V.; Vongphayloth, K.; Hertz, J.C.; Loiseau, I.; Dumarest, M.; Duong, V.; Vayssier-Taussat, M.; Grandadam, M.; Albina, E.; Dussart, P.; Moutailler, S.; Cappelle, J.; Brey, P.T.; Eloit, M. Insights into the Host Range, Genetic Diversity, and Geographical Distribution of Jingmenviruses. mSphere 2019, 4, e00645–19. [Google Scholar] [CrossRef] [PubMed]
- Kholodilov, I.S.; Belova, O.A.; Ivannikova, A.Y.; Gadzhikurbanov, M.N.; Makenov, M.T.; Yakovlev, A.S.; Polienko, A.E.; Dereventsova, A.V.; Litov, A.G.; Gmyl, L.V.; Okhezin, E.V.; Luchinina, S.V.; Klimentov, A.S.; Karganova, G.G. Distribution and Characterisation of Tick-Borne Flavi-, Flavi-like, and Phenuiviruses in the Chelyabinsk Region of Russia. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Yu, Z.-M.; Chen, J.-T.; Qin, J.; Guo, J.-J.; Li, K.; Xu, Q.-Y.; Wang, W.; Lu, M.; Qin, X.-C.; Zhang, Y.-Z. Identification and characterization of Jingmen tick virus in rodents from Xinjiang, China. Infect. Genet. Evol. 2020, 84, 1–7. [Google Scholar] [CrossRef]
- Cai, X.; Cai, X.; Xu, Y.; Shao, Y.; Fu, L.; Men, X.; Zhu, Y. Virome analysis of ticks and tick-borne viruses in Heilongjiang and Jilin. Virus Res. 2023, 323, 199006. [Google Scholar] [CrossRef]
- Bell-Sakyi, L. Continuous cell lines from the tick Hyalomma anatolicum anatolicum. J. Parasitol. 1991, 77, 1006–1008. [Google Scholar] [CrossRef]
- Bell-Sakyi, L.; Zweygarth, E.; Blouin, E.F.; Gould, E.A.; Jongejan, F. Tick cell lines: tools for tick and tick-borne disease research. Trends Parasitol. 2007, 23, 450–457. [Google Scholar] [CrossRef]
- Maikova, G.B.; Chernokhaeva, L.L.; Rogova, Y.V.; Kozlovskaya, L.I.; Kholodilov, I.S.; Romanenko, V.V.; Esyunina, M.S.; Ankudinova, A.A.; Kilyachina, A.S.; Vorovitch, M.F.; Karganova, G.G. Ability of inactivated vaccines based on far-eastern tick-borne encephalitis virus strains to induce humoral immune response in originally seropositive and seronegative recipients. J. Med. Virol. 2019, 91, 190–200. [Google Scholar] [CrossRef]
- Bugmyrin, S.V.; Romanova, L.Y.; Belova, O.A.; Kholodilov, I.S.; Bespyatova, L.A.; Chernokhaeva, L.L.; Gmyl, L.V.; Klimentov, A.S.; Ivannikova, A.Y.; Polienko, A.E.; Yakovlev, A.S.; Ieshko, E.P.; Gmyl, A.P.; Karganova, G.G. Pathogens in Ixodes persulcatus and Ixodes ricinus ticks ( Acari, Ixodidae ) in Karelia ( Russia ). Ticks Tick. Borne. Dis. 2022, 13, 102045. [Google Scholar] [CrossRef] [PubMed]
- Kholodilov, I.; Belova, O.; Burenkova, L.; Korotkov, Y.; Romanova, L.; Morozova, L.; Kudriavtsev, V.; Gmyl, L.; Belyaletdinova, I.; Chumakov, A.; Chumakova, N.; Dargyn, O.; Galatsevich, N.; Gmyl, A.; Mikhailov, M.; Oorzhak, N.; Polienko, A.; Saryglar, A.; Volok, V.; Yakovlev, A.; Karganova, G. Ixodid ticks and tick-borne encephalitis virus prevalence in the South Asian part of Russia (Republic of Tuva). Ticks Tick. Borne. Dis. 2019, 10, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Scaramozzino, N.; Crance, J.; Jouan, A.; Briel, D.A.D.E. Comparison of Flavivirus Universal Primer Pairs and Development of a Rapid, Highly Sensitive Heminested Reverse Transcription-PCR Assay for Detection of Flaviviruses Targeted to a Conserved Region of the NS5 Gene Sequences. 2001, 39, 1922–1927. [Google Scholar] [CrossRef] [PubMed]
- Saksida, A.; Jakopin, N.; Jelovšek, M.; Knap, N.; Fajs, L.; Lusa, L.; Lotri, S.; Bogovi, P.; Arnež, M.; Strle, F.; Avšič-županc, T. Virus RNA Load in Patients with Tick-borne Encephalitis, Slovenia. Emerg. Infect. Dis. 2018, 24, 1315–1323. [Google Scholar] [CrossRef]
- Zou, S.; Foster, G.A.; Dodd, R.Y.; Petersen, L.R.; Stramer, S.L. West Nile Fever Characteristics among Viremic Persons Identified through Blood Donor Screening. J. Infect. Dis. 2010, 20855, 1354–1361. [Google Scholar] [CrossRef]
- Brown, J.A.; Factor, D.L.; Tkachenko, N.; Templeton, S.M.; Crall, N.D.; Pape, W.J.; Bauer, M.J.; Ambruso, D.R.; Dickey, W.C.; Marfin, A.A. West Nile Viremic Blood Donors and Risk Factors for Subsequent West Nile Fever. VECTOR-BORNE ZOONOTIC Dis. 2007, 7, 479–488. [Google Scholar] [CrossRef]
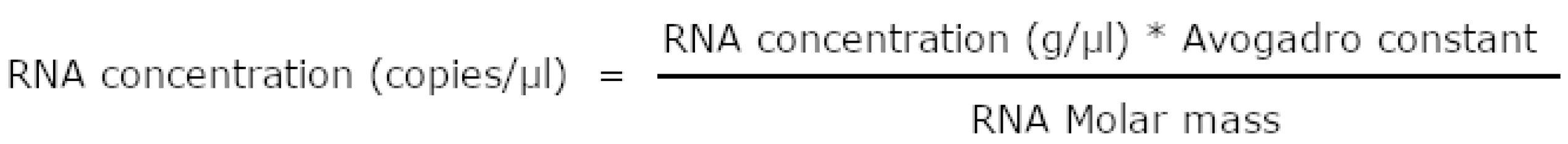
| Virus | Strain | Virus amount, PFU/ml | Virus Origin |
|---|---|---|---|
| ALSV | Miass519 | + 1 | HAE/CTVM8 2 cells |
| ALSV | Miass527 | + | IRE/CTVM19 3 cells |
| YGTV | Plast-T22438 | + | HAE/CTVM8 cells |
| YGTV | Bredy-T22181 | + | HAE/CTVM8 cells |
| TBEV | Sofjin | 6.7 | Mouse brain |
| TBEV | EK-328 | 7.1 | PEK 4 cells |
| TBEV | Absettarov | 9.5 | Mouse brain |
| OHFV | Nikitina | 7.6 | PEK cells |
| WNV | SHUA-3 | 7.7 | Vero cells |
| LIV | S1 | 6.2 | Mouse brain |
| POWV | Pow-24 | 7.6 | PEK cells |
| JEV | Gagar | 8.1 | PEK cells |
| Dengue-4 | Cambodja | + | Mouse brain |
| KEMV | KEM-21/10 | 7.1 | PEK cells |
| Tick species | Number of Ticks in a Pool |
Collection site | Collection Date |
|---|---|---|---|
| Ixodes ricinus | 6 ♂ | Russia, Kaliningrad Region | 2017 |
| Ixodes ricinus | 5 ♀ | Russia, Kaliningrad Region | 2017 |
| Ixodes persulatus | 5 ♀ | Russia, Primorsky Territory | 2021 |
| Dermacentor retuculatus | 3 ♀ | Russia, Chelyabinsk Region | 2015 |
| Dermacentor marginatus | 4 ♀ | Russia, Chelyabinsk Region | 2015 |
| Haemaphysalis conccina | 2 ♂ | Russia, Primorsky Territory | 2021 |
| Haemaphysalis japonica | 2 ♀ | Russia, Primorsky Territory | 2021 |
| Oligonucleotide | Sequence |
|---|---|
| BHT7_Miass_VP1a_F3 | 5′-ATGACTGGATCCTAATACGACTCACTATAGGCTTGTAAAGCTAGCGACTGGA-3′ |
| Miass_gly_2R | 5′-AAAGCCTCATGGACGGTCTG-3′ |
| Oligonucleotide | Sequence | Location |
|---|---|---|
| Miass_gly_3F | 5`-TGGATCAGCTCACACCACAC-3` | VP1a |
| Miass_gly_3R | 5`-TCACCGTCACAGTGGAATGG-3` | VP1a |
| Miass_gly3_PROBE | (FAM)-TTGCGACCCCGTTGTCGTCG-(BHQ-1) | VP1a |
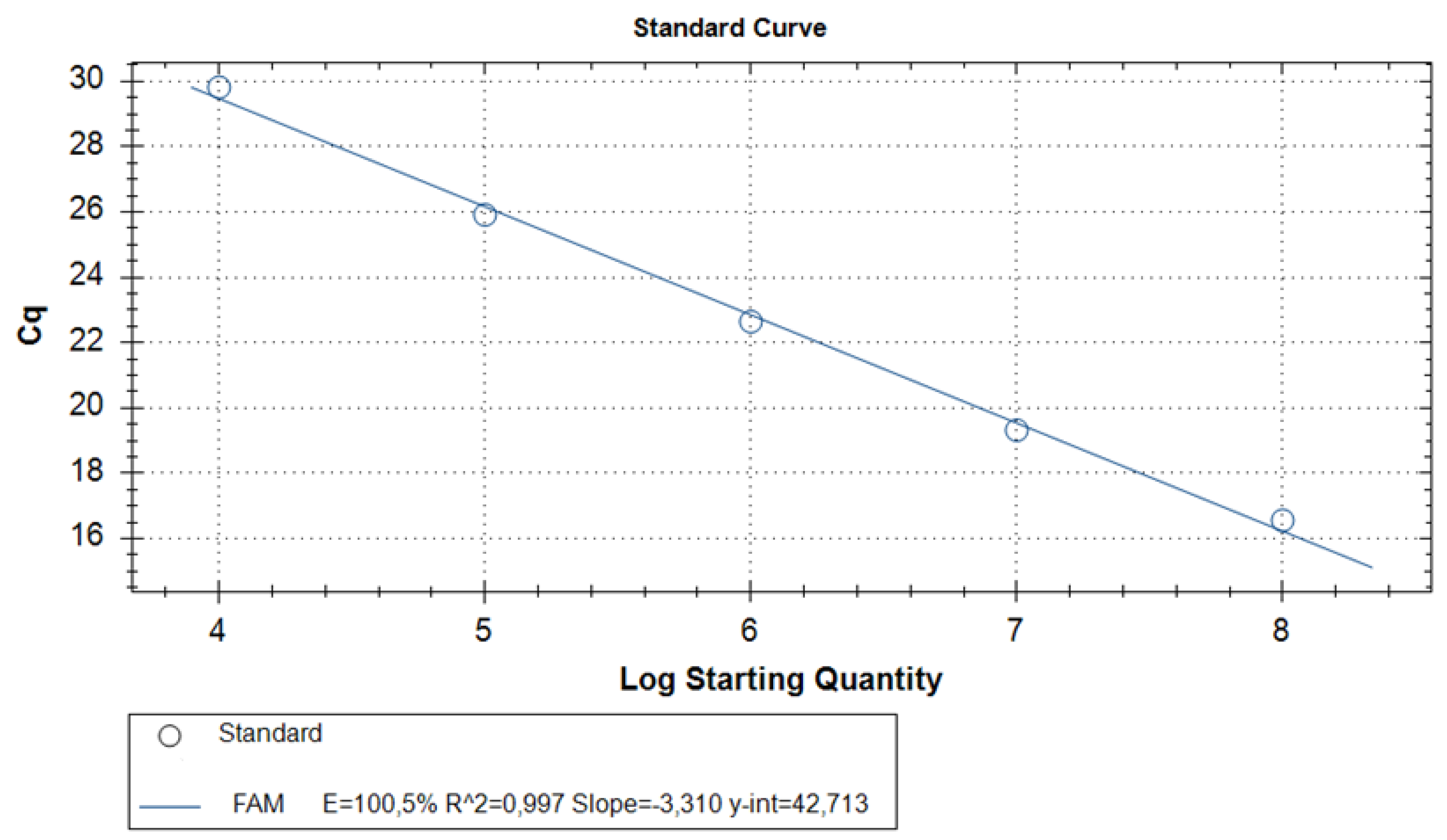
| Probe | Cq | Quantity | Detection result |
|---|---|---|---|
| 108 Standard RNA | 16.4±0.9 * | 108 | - |
| 107 Standard RNA | 19.0±0.6 * | 107 | - |
| 106 Standard RNA | 22.1±1.2 * | 106 | - |
| 105 Standard RNA | 25.5±1* | 105 | - |
| 104 Standard RNA | 29.7±0.9* | 104 | - |
| ALSV strain Miass519 | 15.8 | 9.7*107 | positive |
| ALSV strain Miass527 | 7.4 | 3.6*1010 | positive |
| Human serum, spiked with ALSV strain Miass519 | 18.5 | 1.7*107 | positive |
| Sheep serum, spiked with ALSV strain Miass519 | 19 | 1.2*107 | positive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





