Submitted:
12 May 2023
Posted:
15 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
2.1. Mosquitoes Collection and Taxonomic Identification
2.2. Mosquitoes maceration
2.3. RNA extraction
2.4. Real-Time Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
2.5. Nucleotide Sequencing
2.6. Bioinformatic Analysis
3. Results
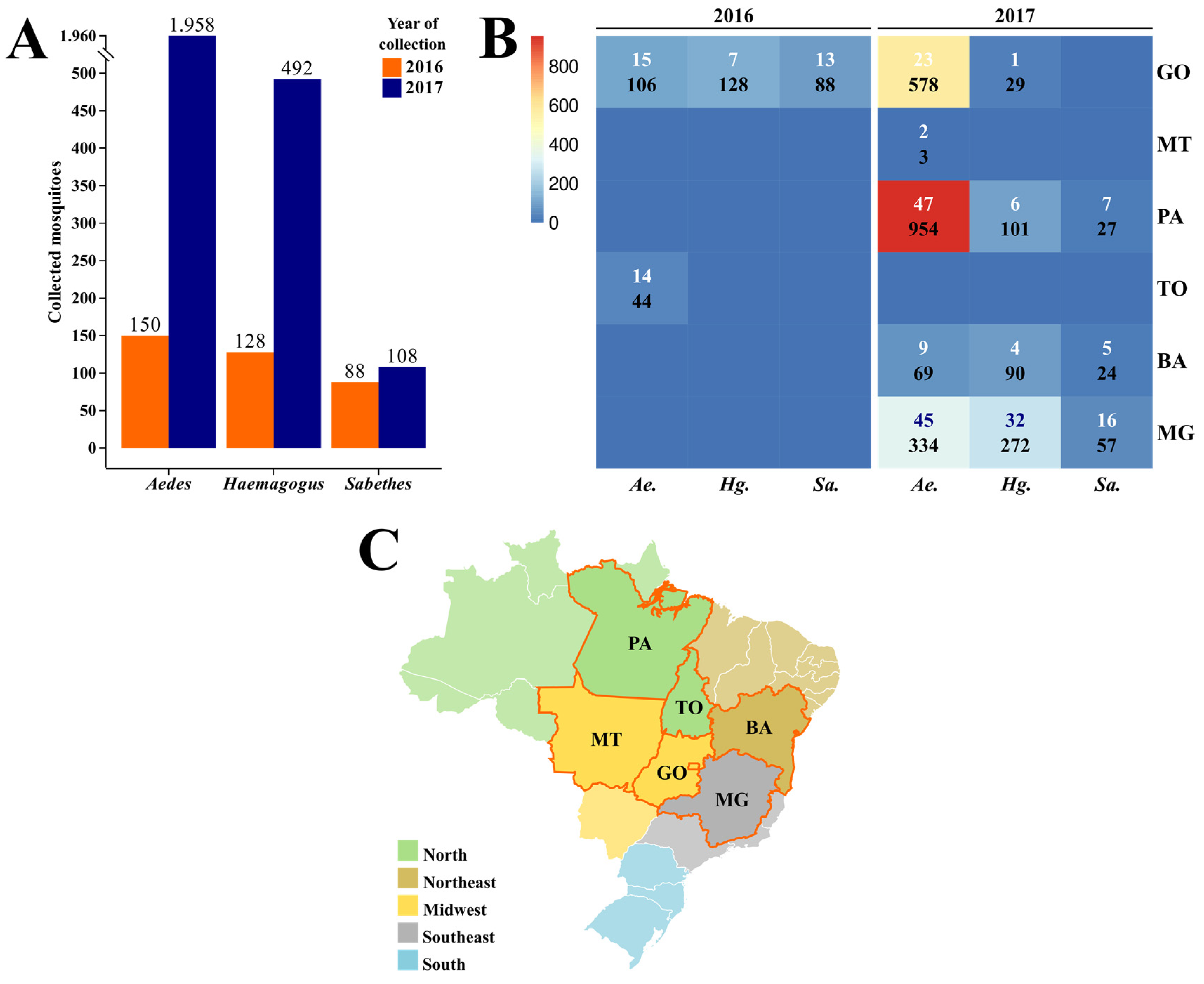
3.1. Collection and Taxonomic Identification
3.2. RT-qPCR Detection
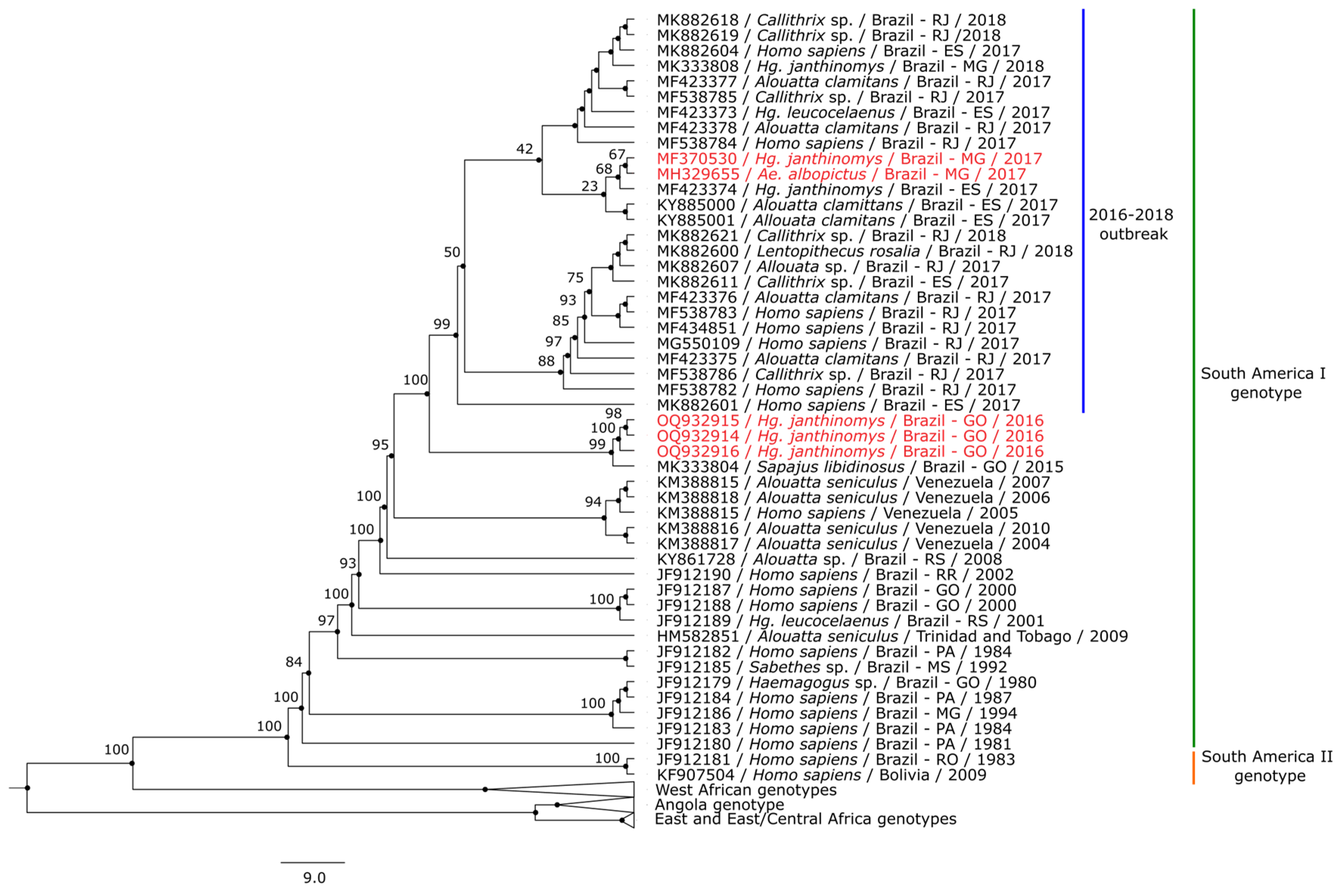
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Committee on Taxonomy of Viruses. Available online: https://ictv.global/taxonomy (accessed on 09 April 2023).
- Vasconcelos, P.F.C. Febre amarela. Rev. Soc. Bras. Med. Trop. 2003, 36, 275–293. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P.; Vasconcelos, P.F.C. Yellow fever. J. Clin. Virol. 2015, 64, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Possas, C.; Lourenço-de-Oliveira, R.; Tauil, P.L.; Pinheiro, F.P.; Pissinatti, A.; Cunha, R.V.; Freire, M.; Martins, R.M.; Homma, A. Yellow fever outbreak in Brazil: the puzzle of rapid viral spread and challenges for immunization. Mem. Inst. Oswaldo Cruz 2018, 113, e180278. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Mendonça, M.C.L.; Fonseca, V.; Mares-Guia, M. A.; Fabri, A.; Xavier, J.; Jesus, J. G; Gräf, T; Rodrigues, C.D.S.; Santos, C.C.; et al. Yellow Fever Virus Reemergence and Spread in Southeast Brazil, 2016-2019. J. Virol. 2019, 94, e01623–19. [Google Scholar] [CrossRef]
- Silva, N.I.O.; Sacchetto, L.; Rezende, I.M.; Trindade, G.S.; LaBeaud, A.D.; de Thoisy, B.; Drumond, B.P. Recent sylvatic yellow fever virus transmission in Brazil: the news from an old disease. Virol. J. 2020, 17, 9. [Google Scholar] [CrossRef]
- Brazil. Ministry of Health. Guia de vigilância em saúde, 5th ed.; Ministry of Health: Brasilia, Brazil, 2022; pp. 623–649. ISBN 978-65-5993-102-6. [Google Scholar]
- Brazil. Ministry of Health. Guia de vigilância de epizootias em primatas não humanos e entomologia aplicada à vigilância da febre amarela., 2nd ed.; Ministry of Health: Brasilia, Brazil, 2017; ISBN 978-85-334-2102-8. [Google Scholar]
- Câmara, D.C.P.; Codeço, C.T.; Ayllón, T.; Nobre, A.A.; Azevedo, R.C.; Ferreira, D.F.; Pinel, C.S.; Rocha, G.P.; Honório, N.A. Entomological Surveillance of Aedes Mosquitoes: Comparison of Different Collection Methods in an Endemic Area in RIO de Janeiro, Brazil. Trop. Med. Infect. Dis. 2022, 7, 114. [Google Scholar] [CrossRef]
- Abreu, F.V.S.; Ribeiro, I.P.; Ferreira-de-Brito, A.; Santos, A.A.C.; Miranda, R.M.; Bonelly, I.S.; Neves, M.S.A.S.; Bersot, M.I.; Santos, T.P.; Gomes, M.Q.; et al. Haemagogus leucocelaenus and Haemagogus janthinomys are the primary vectors in the major yellow fever outbreak in Brazil, 2016-2018. Emerg. Microbes Infect. 2019, 8, 218–231. [Google Scholar] [CrossRef]
- Couto-Lima, D.; Madec, Y.; Bersot, M.I.; Campos, S.S.; Motta, M.A.; Santos, F.B.; Vazeille, M.; Vasconcelos, P.F.C.; Lourenço-de-Oliveira, R.; Failloux, A.B. Potential risk of re-emergence of urban transmission of Yellow Fever virus in Brazil facilitated by competent Aedes populations. Sci. Rep. 2017, 7, 4848. [Google Scholar] [CrossRef]
- Santos, T.P.; Roiz, D.; Abreu, F.V.S.; Luz, S.L.B.; Santalucia, M.; Jiolle, D.; Neves, M.S.A.S.; Simard, F.; Lourenço-de-Oliveira, R.; Paupy, C. Potential of Aedes albopictus as a bridge vector for enzootic pathogens at the urban-forest interface in Brazil. Emerg. Microbes Infect. 2018, 7, 191. [Google Scholar] [CrossRef]
- Lane, J. Neotropical Culicidae, Volume 1; Edusp: São Paulo, Brazil, 1953. [Google Scholar]
- Lane, J. Neotropical Culicidae, Volume 2; Edusp: São Paulo, Brazil, 1953. [Google Scholar]
- Forratini, O.P. Entomologia médica. Culicini: Culex, Aedes e Psorophora. Volume 2; Edusp: São Paulo, Brazil, 1965. [Google Scholar]
- Forratini, O.P. Entomologia médica. Culicini: Haemagogus, Mansonia, Culiseta, Sabethini, Toxorhynchitini, Arboviroses, Filariose bancroftiana, Genética. Volume 3; Edusp: São Paulo, Brazil, 1965. [Google Scholar]
- Forratini, O.P. Culicidologia médica: identificação, biologia, epidemiologia; Edusp: São Paulo, Brazil, 2002; ISBN 85-314-0699-4. [Google Scholar]
- Consoli, R.A.G.B.; Lourenço-de-Oliveira, R. Principais mosquitos de importâcia sanitaria no Brasil, 1st ed.; Editora Fiocruz: Rio de Janeiro, Brazil, 1994; ISBN 85-85676-03-5. [Google Scholar]
- Domingo, C.; Patel, P.; Yillah, J.; Weidmann, M.; Méndez, J.A.; Nakouné, E.R.; Niedrig, M. Advanced yellow fever virus genome detection in point-of-care facilities and reference laboratories. J. Clin. Microbiol. 2012, 50, 4054–4060. [Google Scholar] [CrossRef]
- Menting, S.; Thai, K.T.D.; Nga, T.T.T.; Phuong, H.L.; Klatser, P.; Wolthers, K.C.; Binh, T.Q.; de Vries, P.J.; Beld, M. Internally Controlled, Generic Real-Time PCR for Quantification and Multiplex Real-Time PCR with Serotype-Specific Probes for Serotyping of Dengue Virus Infections. Adv. Virol. 2011, 2011, 514681. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Pinotti, F.; Zanluca, C.; Fonseca, V.; Nakase, T.; Koishi, A.C.; Tscha, M.; Soares, G.; Dorl, G.G.; Marques, A.E.M.L.; et al. Genomic epidemiology sheds light on the recent spatio-temporal dynamics of yellow fever virus and the spatial corridor that fueled its ongoing emergence in southern Brazil. medRxiv 2023, 23284525. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pharm, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- DNASTAR. Available online: https://www.dnastar.com (accessed on 05 March 2023).
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Figtree. Available online: https://tree.bio.ed.ac.uk/software/figtree/ (accessed on 14 April 2023).
- Inkscape. Available online: https://inkscape.org/release/inkscape-1.1/ (accessed on 14 April 2023).
- Vasconcelos, P.F.; Sperb, A.F.; Monteiro, H.A.; Torres, M.A.; Sousa, M.R.; Vasconcelos, H.B.; Mardini, L.B.; Rodrigues, S.G. Isolations of yellow fever virus from Haemagogus leucocelaenus in Rio Grande do Sul State, Brazil. Trans. R. Soc. Trop. Med. Hyg. 2003, 97, 60–62. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, J.C.; Almeida, M.A.B.; Santos, E.; Fonseca, D.F.; Sallum, M.M.; Noll, C.A.; Monteiro, H.A.O.; Cruz, A.C.R.; Carvalho, V.L.; Pinto, E.V.; et al. Yellow fever virus in Haemagogus leucocelaenus and Aedes serratus mosquitoes, Southern Brazil, 2008. Emerg. Infect. Dis. 2010, 16, 1918–1924. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, M.A.; dos Santos, E.; da Cruz Cardoso, J.; da Fonseca, D.F.; Noll, C.A.; Silveira, V.R.; Maeda, A.Y.; de Souza, R.P.; Kanamura, C.; Brasil, R.A. Yellow fever outbreak affecting Allouata populations in southern Brazil (Rio Grande do Sul State), 2008-2009. Am. J. Primatol. 2012, 74, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Kumm, H.W.; Cerqueira, N.L. The role of Aëdes leucocelaenus in the epidemiology of jungle yellow fever in Brazil. Bull. Ent. Res. 1952, 42, 195–199. [Google Scholar] [CrossRef]
- Vasconcelos, P.F.; Rodrigues, S.G.; Degallier, N.; Moraes, M.A.; da Rosa, J.F.; da Rosa, E.S.; Mondet, B.; Barros, V.L.; da Rosa, A.P. An epidemic of sylvatic yellow fever in the Southeast region of Maranhao State, Brazil, 1993-1994: epidemiologic and entomologic findings. Am. J. Trop. Med. Hyg. 1997, 57, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, P.F.; Costa, Z.G.; Travassos da Rosa, E.S.; Luna, E.; Rodrigues, S.G.; Barros, V.L.; Dias, J.P.; Monteiro, H.A.; Oliva, O.F.; Vasconcelos, H.B.; et al. Epidemic of jungle yellow fever in Brazil, 2000: implications of climatic alterations in disease spread. J. Med. Virol. 2001, 65, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Finlay, C. Yellow fever: its transmission by means of the Culex mosquito. Am. J. Med. Sci. 1986, 92, 395–409. [Google Scholar] [CrossRef]
- Reed, W.; Carrol, J.; Agramonte, A. The etiology of yellow fever. J. Am. Med. Assoc. 1901, 36, 415–440. [Google Scholar] [CrossRef]
- Goldani, L.Z. Yellow fever outbreak in Brazil. Braz. J. Infect. Dis. 2017, 21, 123–124. [Google Scholar] [CrossRef]
- Amraoui, F.; Vazeille, M.; Failloux, A.B. French Aedes albopictus are able to transmit yellow fever virus. Euro Surveill. 2016, 21, 30361. [Google Scholar] [CrossRef]
- Cunha, M.S.; Faria, N.R.; Caleiro, G.S.; Candido, D.S.; Hill, S.C.; Claro, I.M.; da Costa, A.C.; Nogueira, J.S.; Maeda, A.Y.; da silva, F.G.; et al. Genomic evidence of yellow fever virus in Aedes scapularis, southeastern Brazil, 2016. ACTA Trop 2020, 205, 105390. [Google Scholar] [CrossRef] [PubMed]
- Damasceno-Caldeira, R.; Nunes-Neto, J.P.; Aragão, C.F.; Freitas, M.N.O.; Ferreira, M.S.; Castro, P.H.G.d.; Dias, D.D.; Araújo, P.A.d.S.; Brandão, R.C.F.; Nunes, B.T.D.; et al. Vector competence of Aedes albopictus for yellow fever virus: risk of reemergence of urban yellow fever in Brazil. Viruses 2023, 15, 1019. [Google Scholar] [CrossRef] [PubMed]
- Agência Brasil. Available online: https://agenciabrasil.ebc.com.br/geral/noticia/2018-02/pesquisa-detecta-virus-da-febre-amarela-em-novo-tipo-de-mosquito (accessed on 18 April 2023).
- Mitchell, C.J. Mosquito vector competence and arboviruses. In Current topics in vector research; Harris, K.F., Ed.; Praeger: New York, NY, USA, 1983; Volume 1, pp. 63–92. ISBN 0030586372. [Google Scholar]
- Mitchell, C.J. The role of Aedes albopictus as an arbovirus vector. Parassitologia 1995, 37, 109–113. [Google Scholar] [PubMed]
- Kramer, L.D.; Ebel, G.D. Dynamics of flavivirus infection in mosquitoes. Adv. Virus Res. 2003, 60, 187–232. [Google Scholar] [PubMed]
- Bonaldo, M.C.; Gómez, M.M.; Santos, dos Santos, A.C.C.; Abreu, F.V.S.; Ferreira-de-Brito, A.; Miranda, R.M.; Castro, M.G.; Lourenço-de-Oliveira, R. Genome analysis of yellow fever virus of the ongoing outbreak in Brazil revealspolymorphisms. Mem. Inst. Oswaldo Cruz, 2017, 112, 447–451. [Google Scholar] [CrossRef]
- Delatorre, E.; Abreu, F.V.S.; Ribeiro, I.P.; Gómez, M.M.; dos Santos, A.A.C.; Ferreira-de-Brito, A.; Neves, M.S.A.S.; Bonelly, I.; de Miranda, R.M.; Furtado, N.D.; et al. Distinct YFV lineages co-circulated in the Central-Western and Southeastern Brazilian Regions from 2015 to 2018. Front. Microbiol. 2019, 10, 1079. [Google Scholar] [CrossRef]
- Gómez, M.M.; Abreu, F.V.S.; Santos, A.A.C.D.; Mello, I.S.; Santos, M.P.; Ribeiro, I.P.; Ferreira-de-Brito, A.; Miranda, R.M.; Castro, M.G.; Ribeiro, M.S.; et al. Genomic and structural features of the yellow fever virus from the 2016-2017 Brazilian outbreak. J. Gen. Virol. 2018, 99, 536–548. [Google Scholar] [CrossRef]
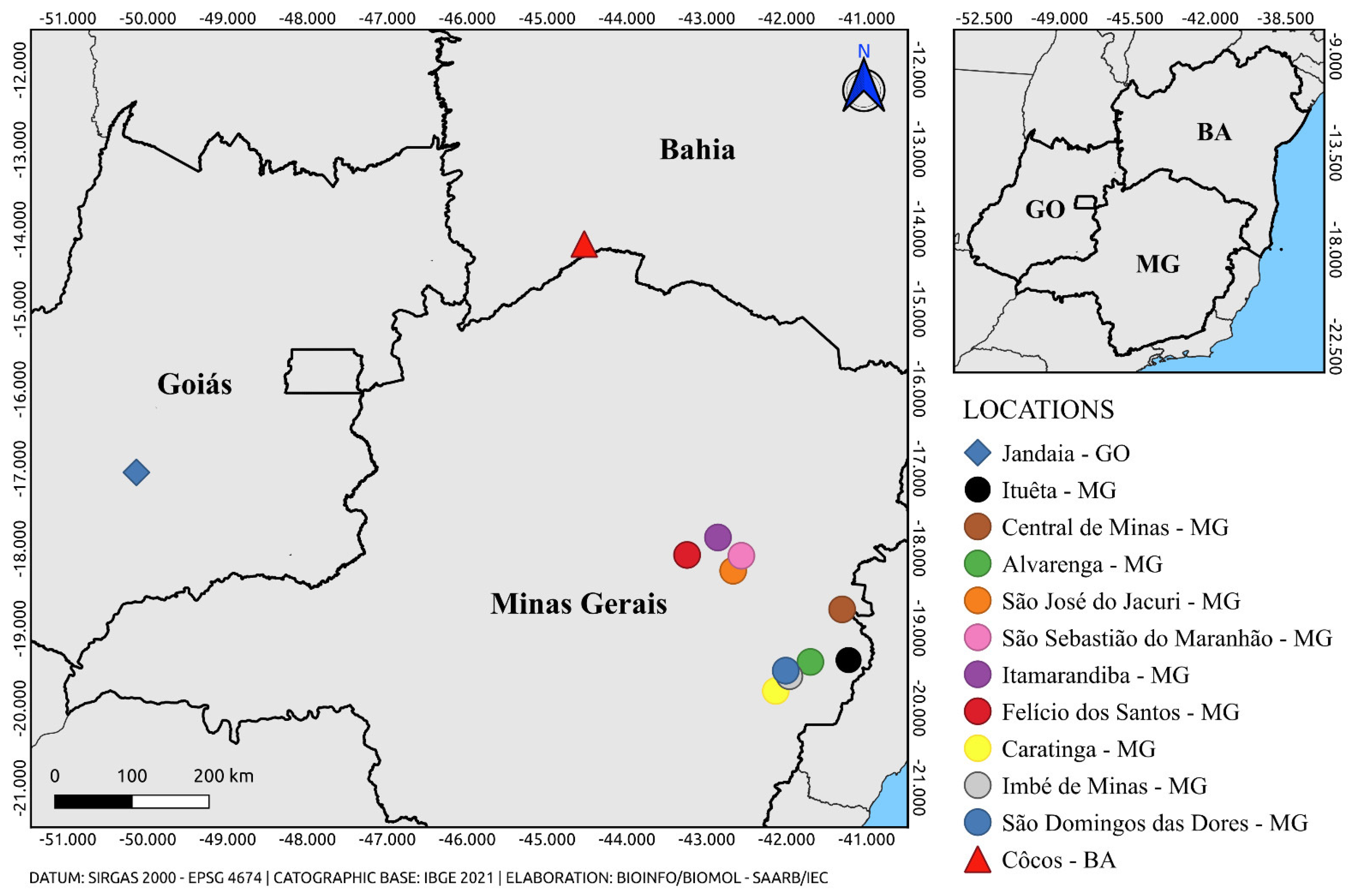
| Sample ID |
Species | Specimens per pool | Location | Collection Date | RT-qPCR Ct value |
|---|---|---|---|---|---|
| AR831906 | Hg. janthinomys | 30 | Jandaia – GO | 24-Sep-2016 | 26.17 |
| AR831907 | Hg. janthinomys | 30 | 24-Sep-2016 | 23.52 | |
| AR831908 | Hg. janthinomys | 28 | 24-Sep-2016 | 21.26 | |
| AR831909 | Hg. janthinomys | 7 | 26-Sep-2016 | 18.27 | |
| AR831914 | Sa. glaucodaemon | 30 | 24-Sep-2016 | 31.27 | |
| AR843690 | Ae. albopictus ♀ | 25 | Ituêta – MG | 14-Jan-2017 | 23.88 |
| AR843692 | Ae. scapularis | 25 | 31.08 | ||
| AR843693 | Ae. argyrothorax | 2 | 33.79 | ||
| AR843713 | Hg. janthinomys | 15 | Central de Minas – MG | 13-Jan-2017 | 32.23 |
| AR843715 | Ae. albopictus ♀ | 4 | Alvarenga – MG | 15-Jan-2017 | 34.28 |
| AR843716 | Ae. argyrothorax | 2 | 32.88 | ||
| AR843717 | Aedes sp. | 1 | 35.55 | ||
| AR843720 | Hg. janthinomys | 25 | 29.08 | ||
| AR843721 | Hg. janthinomys | 25 | 28.44 | ||
| AR843728 | Hg. janthinomys | 19 | São José do Jacuri – MG | 16-Jan-2017 | 32.09 |
| AR843738 | Ae. scapularis | 3 | São Sebastião do Maranhão – MG |
17-Jan-2017 | 24.08 |
| AR843741 | Hg. leucocelaenus | 2 | 24.09 | ||
| AR843745 | Ae. serratus | 1 | Itamarandiba – MG | 18-Jan-2017 | 35.46 |
| AR843765 | Hg. janthinomys | 6 | Felício dos Santos – MG | 20-Jan-2017 | 32.52 |
| AR843771 | Ae. albopictus ♀ | 2 | Caratinga – MG | 10-Jan-2017 | 35.12 |
| AR843772 | Ae. serratus | 2 | 32.16 | ||
| AR843777 | Hg. janthinomys | 1 | 27.60 | ||
| AR843807 | Hg. janthinomys | 4 | Imbé de Minas – MG | 16-Jan-2017 | 34.81 |
| AR843821 | Ae. albopictus ♀ | 1 | São Domingos das Dores – MG |
19-Jan-2017 | 35.61 |
| AR843829 | Ae. albopictus ♀ | 4 | 35.18 | ||
| AR845803 | Hg. janthinomys | 18 | Cocos – BA | 17,18,20-Mar-2017 | 28.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





