Introduction
In our Faculty, Microbial Physiology is a mandatory course for undergraduate students for the Bachelor’s degree in Biotechnology. This course involves 6 weeks of practices in the lab, during a two-month period with one lab practice per week, meaning that they take, at most, six practice classes during the course. An extensive list of concepts must be acquired by the students throughout the Microbial Physiology course, including regulation of microbial physiology, synthesis and degradation of macromolecules, metabolic pathways, among others. Other concepts (e.g., microbial growth, nutrition, and antibiosis) and skills (e.g., aseptic techniques, preparation of culture media, seeding and incubation) are acquired in General Microbiology course in the previous semester.
The global pandemic situation that we have gone through in the last 2 and a half years, has led us to rethink our traditional laboratory practices. In our Faculty, the coming back to the in-person classes occurred in the middle of the semester. With less weeks for face-to-face teaching, teachers needed to achieve the study goals without lowering the education quality. For students, they had to acquire, in a short time, the corresponding skills related to their degree.
Employing Pseudomonas aeruginosa strain PAO1, we propose a series of lab practices in which students, in a short period of time, learn different concepts of microbial physiology in an interrelated way:
Pseudomonas (P.) aeruginosa is a well know Gram-negative strain; an opportunistic pathogen widely spread in soil and aquatic environments (Qin et al. 2022). In P. aeruginosa PAO1, several virulence factors, including extracellular enzymes, production of biofilm, toxins, secondary metabolites, and siderophores, are regulated by its Quorum Sensing (QS) system (Chadha, Harjai, and Chhibber 2022).
QS is a well know cell density-dependent intercellular communication system that allows individual cells to act as a community. QS mechanisms are based on the production, release, and detection of signal molecules called autoinducers (Fuqua and Greenberg 2002). Similar to several other Alphaproteobacteria, Betaproteobacteria and Gammaproteobacteria, in P. aeruginosa, these bioactive molecules are N-acyl homoserine lactones (AHLs) (Chadha, Harjai, and Chhibber 2022). To date, three QS systems intimately linked among them and arranged in a hierarchical regulatory cascade are known in P. aeruginosa. The autoinducer synthase LasI and RhlI produces AHL molecules as autoinducers: N-oxododecanoyl-L-homoserine lactone (3OC12-HSL) and N-butyryl-L-homoserine lactone (C4-HSL), respectively. When these autoinducers reach a critical threshold level by accumulation due to bacterial population growth, they interact with their receptors LasR and RhlR, respectively, which are also transcriptional activators (Chadha, Harjai, and Chhibber 2022). LasR and RhlR stimulates the lasI and rhlI expression, creating autoinduction loops. A third QS system that relies on the production of 2-heptyl-3-hydroxy-4-quinolone (Pseudomonas quinolone signal or PQS) is interconnected with the LasI-LasR and RhlI-RhlR systems.
In nature, microorganisms interfere with the QS systems of competitors mainly through the enzymatic inactivation of the QS signals, or the production of metabolites that impede the normal functioning of the signal receptors, a phenomenon known as Quorum Quenching (QQ) (Grandclément et al. 2016). In the lab, the cell-to-cell communication can be blocked with easy and fast experiments to understand the functioning of the QS system. In the approach chosen for the present work, a aiiA gene encoding the AiiA lactonase from Bacillus sp. A24, an AHL-degrading enzyme, is expressed under a constitutive plac promoter in P. aeruginosa PAO1, reducing the accumulation of autoinducers and the expression of several traits (Reimmann et al. 2002). The easiest way to expose the QQ phenomenon is evidencing with biosensor strains the decrease in the AHL levels. These strains have a genetic deficiency on signal production, but detect and respond to exogenous signals by means of a functional receptor coupled to a reporter gene that produces an observable phenotype when specific QS signals are present (Miller and Gilmore 2020; Steindler and Venturi 2007). Chromobacterium (C.) subtsugae CV026 and P. putida F117 (pKR-C12) are the biosensors used in these practical lab works. C. subtsugae CV026 produce violacein, a purple pigment, in presence of AHLs with acyl side chains from C4 to C8 in length (McClean et al. 1997). P. putida F117 (pKR-C12) produce green fluorescent protein in presence of AHLs with acyl side chains from C10 to C14 in length (Riedel et al. 2001).
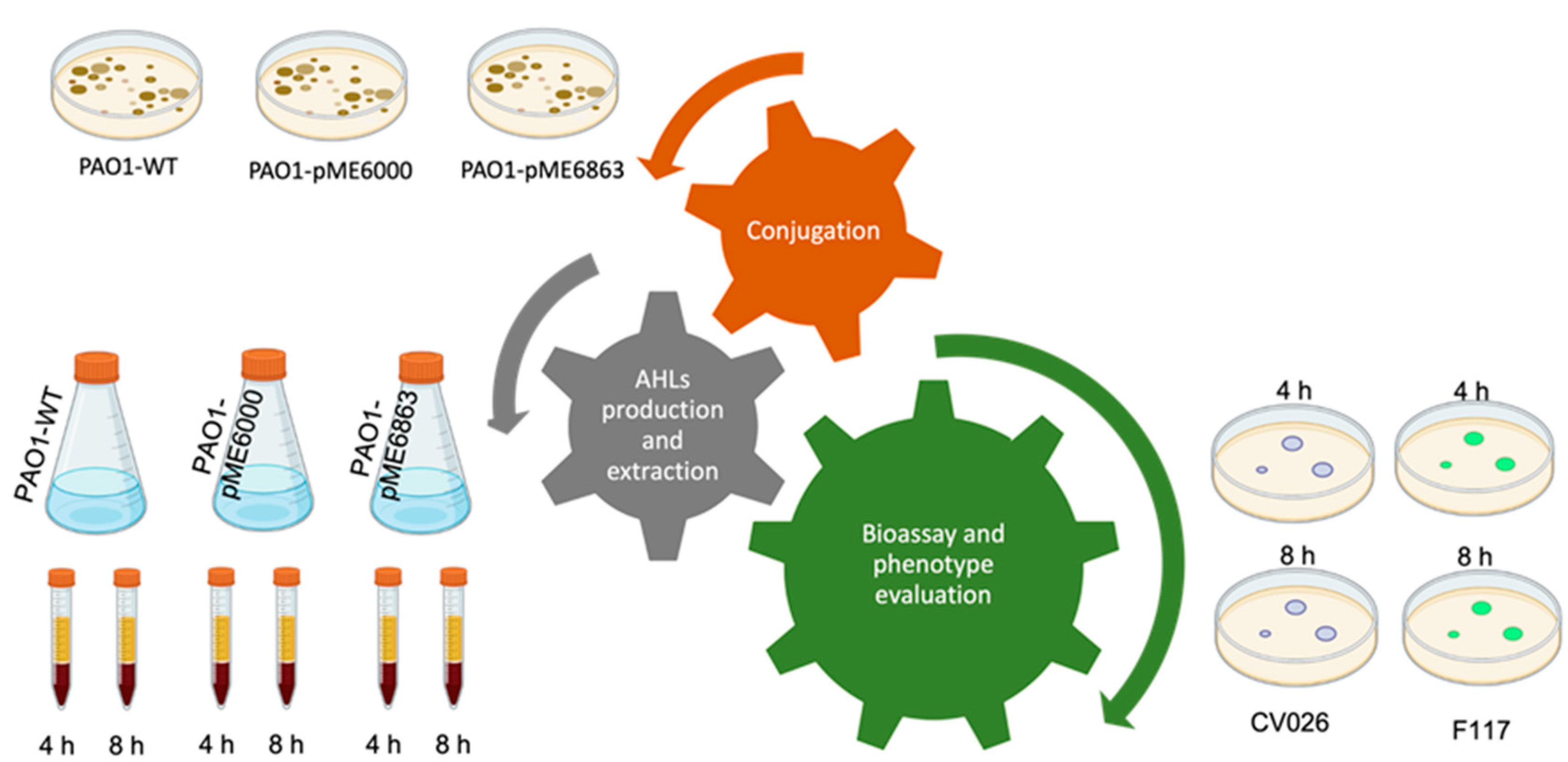
The experimental workflow presented in this work is detailed in
Figure 1. Each individual assay is thought of as a piece of a puzzle that fits together giving a single, complete, overall, and multi-focused approach for the lab classes.
Materials and Methods
Equipment
The experimental procedures require the use of laminar flow chambers, micropipettes, centrifuges, vortex, transilluminator, spectrophotometers, optical microscope, a rotary shaker, a rotary evaporator and a 37 ºC lab stove.
Biosafety
Even when these lab practice are not particularly risky, all students must wear appropriate lab clothes and protective equipment including gloves and glasses for handling Ps. aeruginosa PAO1, considering that this strain is a Biosafety Level 2 organism.
Strains and culture conditions
Bacterial strains used in this work are listed in
Table 1. The culture media used for the assays are Luria Bertani (LB) broth composed of 10 g L
-1 tryptone, 5 g L
-1 yeast extract, 10 g L
-1 NaCl; and nutrient yeast broth (NYB) composed of peptone 10 g L
-1; NaCl 5 g L
-1; meat extract 10 g L
-1; yeast extract 5 g L
-1. The composition of Cetrimide agar utilized for selective isolation of transconjugants is gelatin peptone 20 g L
-1, MgCl 1.4 g L
-1, K
2SO
4 10 g L
-1, centrimide 0.3 g L
-1, glycerol 10 mL L
-1. When required, culture media are solidified with agar 15 g L
-1 and/or tetracycline (Tc) 125 µg ml
-1 sterilized by filtration through sterile 0,22 µ membrane filters.
P. aeruginosa PAO1 and the transconjugants are grown at 37 ºC and 180 rpm in an orbital shaker. Biosensor strains,
Chromobacterium subtsugae CV026 and
P. putida F117 (pKR-C12) are grown at 30 ºC and 180 rpm in an orbital shaker.
Attenuation of Quorum Sensing activity
A Quorum Quenching (QQ) strategy is utilized to achieve the attenuation of P. aeruginosa PAO1 QS activity. Plasmid pME6863 contains the Bacillus sp. A24 aiiA AHL-lactonase gene cloned in the pME6000 control vector (Reimmann et al. 2002). AiiA lactonase degrades the AHLs leading to the attenuation of the QS activity. Both pME6863 and pME6000 are conjugated from E. coli S17-1 into P. aeruginosa PAO1 by biparental mating, in two independent assays, as follows. Overnight cultures of P. aeruginosa PAO1, E. coli S17-1 (pME6000) and E. coli S17-1 (pME6863) on LB agar are utilized for the conjugation. For each conjugation, an inoculation loop of P. aeruginosa PAO1 is mixed with two loops of E. coli S17-1 (pME6000) or E. coli S17-1 (pME6863). The mixtures are prepared as small spots of 0.5-1 cm diameter on top of a LB agar plate. Conjugations are performed at 30 ºC for 24 h. The cell pellets are aseptically withdrawn and suspended in physiological solution. Transconjugants are selected after these dilutions on cetrimide agar plates supplemented with Tc. Cetrimide is a quaternary ammonium compound that inhibits E. coli strains; tetracycline allows the growth of only P. aeruginosa transconjugants. Isolated colonies are finally streaked on fresh cetrimide agar plates with Tc.
AHLs production and extraction
P. aeruginosa PAO1, P. aeruginosa PAO1 (pME6000) and P. aeruginosa PAO1 (pME6863) are independently pre-cultured for 18 h in NYB broth. These precultures are utilized to inoculate 125 mL flasks containing 20 mL of fresh culture media without antibiotics at an initial OD600nm=0,1. After 4h and 8h, samples of 5 mL are withdrawn for AHLs extraction following the protocol described elsewhere (Shaw et al. 1997). Briefly, one volume of ethyl acetate acidified with 0.1 % acetic acid is mixed with one volume of cell-free supernatants. The samples are then vigorously extracted in a vortex for 5 min. After resting, the organic phase is removed, and a second extraction is performed from the aqueous phase. Both organic extractions are combined, and the solvent is evaporated to dryness in a rotary evaporator at 35-40 °C. Finally, the extracts are solubilized in 10 µL acetonitrile.
Bioassays
Biosensor strains C. subtsugae CV026 and P. putida F117 (pKR-C12) lack their own AHL production, but respond to exogenous AHLs through the production of violacein and green fluorescent protein, respectively. 10 mL of melted LB soft (0.6 %) agar are independently seeded with overnight cultures of each biosensor strain, and then poured on top of a layer of LB 1.5 % agar. 2 µL of the organic extracts are spotted on top of the soft layer, and the solvent is left to dry spontaneously. Plates are incubated at 30 ºC for 18-24 h. A positive result with C. subtsugae CV026 is evidenced by the presence of violet spots, which can be observed at the naked eye without the requirement of specific equipment. A positive result with P. putida F117 (pKR-C12) is evidenced by green fluorescent spots that observed with a UV transilluminator or a blue light transilluminator.
Evaluation of Quorum Sensing-regulated phenotypes
In all assays, P. aeruginosa PAO1, P. aeruginosa (pME6000) and P. aeruginosa (pME6863) are grown overnight in NYB broth and then 5 µL are used to inoculate the corresponding plates. All incubations are carried out at 37 ºC for 24 h. Skim milk agar medium is used for the evaluation of protease production (Ohman, Cryz, and Iglewski 1980). A positive proteolytic activity is observed as clear halos around colonies due to degradation casein protein. Interestingly, although King B medium is recommended for the evaluation of siderophore production, this phenotype can also be observed in skim milk agar (see below). Hemolytic activity is visualized on blood agar plates. A positive hemolytic activity is observed as clear halos around the colonies. In this medium, differences in the morphology of the colonies between the strains also can be observed.
Implementation
The practice is proposed in three days during two weeks (
Table 2). While the first week is planned for
P. aeruginosa conjugations, the second week is used for the analysis of AHL production and the characterization of the phenotypes regulated by QS in the transconjugants. In addition,
Table 2 shows the material required for each assay and the theoretical concepts to be learned that open the discussion and encourage critical thinking in the students. The class design is based on a typical lab seance in our Faculty that is organized in 4 hours. At the beginning, an appropriate introductory explanation is provided by the teacher before starting the lab practice. Then, students carry out the experimental part. Finally, the analysis and discussion of the results obtained is carried out with the aim that students reinforce the concepts learned during the class.
Results and Discussion
First week
First day, Petri dishes containing sterile LB agar medium and overnight cultures of bacterial strains are available for conjugations assays (see
Table 2). After a review about how to work under aseptic techniques, students are organized in working groups, as required, and conjugations with
E. coli S17-1 strains carrying pME6000 or pME6863 are performed as described before. To note, functions for transfer helper are integrated in the chromosome of
E. coli S17-1, which allows a biparental mating. Students also prepare cetrimide agar supplemented with Tc solution required for the selection of transconjugants. Next day, students aseptically remove the bacterial growth from the plates where the conjugations were performed, and serial dilutions in sterile physiological solution are seeded on cetrimide agar plates supplemented with Tc. The plates are incubated at 37 °C for 24 hours. As negative controls,
P. aeruginosa PAO1,
E. coli S17-1 (pME6000) and
E. coli S17-1 (pME6863) should also be plated on cetrimide agar supplemented with Tc. The next day, isolated colonies from biparental mating can be obtained. No colonies should be observed in the negative controls. In this point, a proper explanation about the importance of the composition of the culture medium and of the complementation with Tc in the selection of transconjugants is critical. The teacher can also exemplify a hypothetical situation where no antibiotic or a non-selective medium (e.g., LB medium) is used in the selection of transconjugants. To note, cetrimide is the selective component that allows the growth of only
P. aeruginosa, due to its surfactant activity. Finally, students select a plate with isolated colonies to continue the assays the following week.
Second week
Previous to the lab session, liquid cultures of each strain are prepared as described above and, after 4h and 8h of incubation, samples are withdrawn, which can be stored frozen until the organic extractions. First day, AHLs are extracted from supernatants with acidified ethyl acetate, and the extracts are concentrated to dryness. The teacher supports this experience explaining the higher affinity of AHLs molecules for the organic phase recovered during the extraction process. To be utilized the following day, students also prepare cultures of
P. aeruginosa PAO1,
P. aeruginosa PAO1 (pME6000) and
P. aeruginosa PAO1 (pME6863) for protease and hemolytic activity assays, as well as cultures of
C. subtsugae CV026 and
P. putida F117 (pKR-C12) for bioassays. On the second day, students analyze the concentrated extracts using the two biosensor strains. Plates for protease and hemolytic activities are also seeded using overnight cultures of
P. aeruginosa PAO1,
P. aeruginosa PAO1 (pME6000) and
P. aeruginosa PAO1 (pME6863). Ten µL of each culture are seeded as equidistant spots on skim milk agar plates and blood agar plates. Finally, on the third day, students examine and discuss the results found in the bioassays (
Figure 2 and
Figure 3) and for the phenotypic properties (
Figure 4 and 5).
In the bioassays, the production of short- and long-chain AHLs can be distinguished, using
C. subtsugae CV026 and
P. putida F117 (pKR-C12) biosensor strains, respectively (see more details in
Table 1). Students can also observe the effect of the QQ strategy in
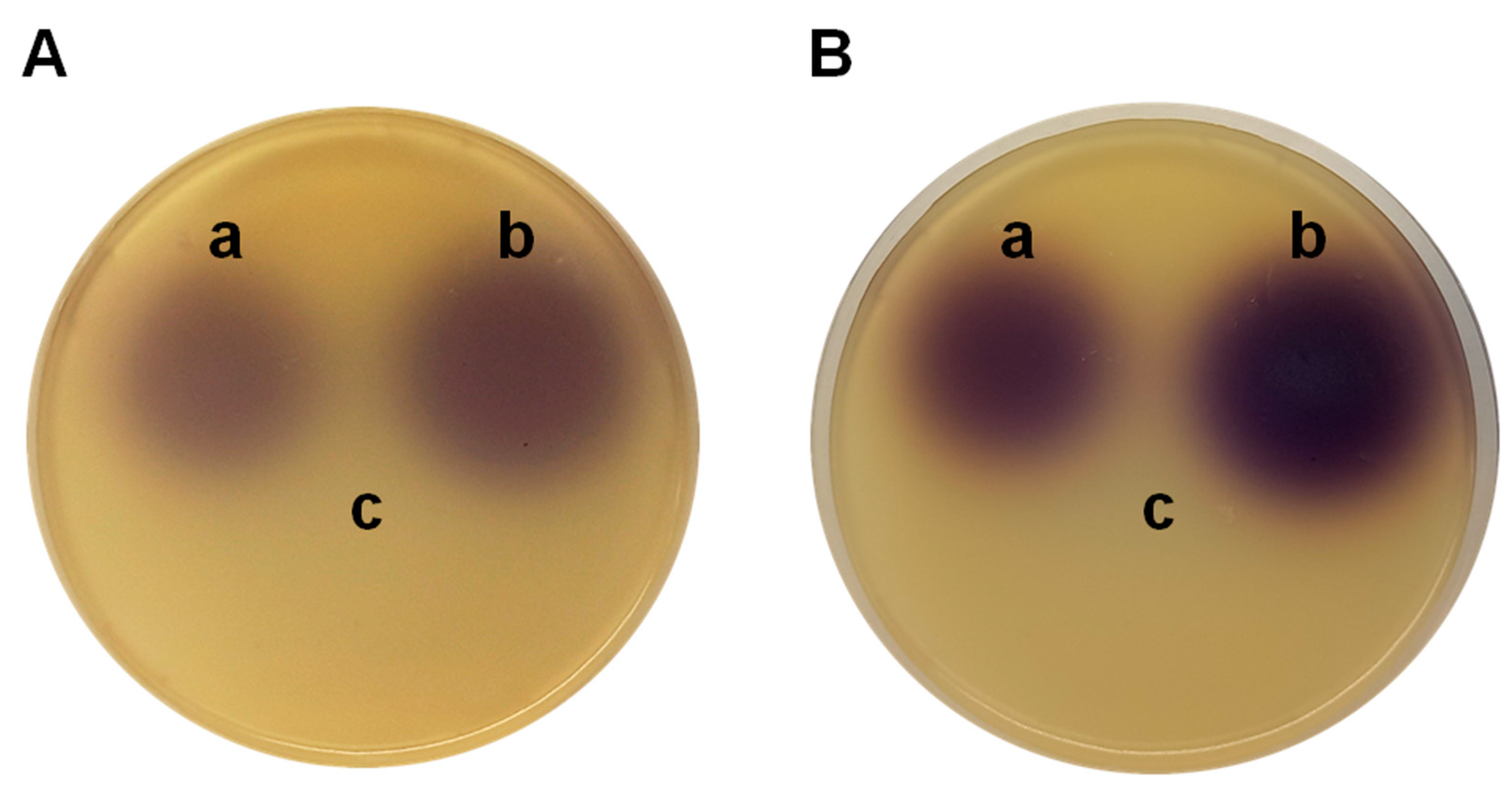
P. aeruginosa PAO1 (pME6863). The lactonase enzyme, coded in the pME6863 vector, degrades the synthesized AHLs; then, a smaller or barely detectable spots in the bioassays is expected. It should be noted that the synthesis of AHLs is not blocked, but their accumulation is prevented by the enzymatic degradation. The violet spots at the top of the plates in Figures 2A (samples obtained after 4 h of incubation), corresponding to
P. aeruginosa PAO1 (left) and
P. aeruginosa PAO1 (pME6000) (right), can be attributed to the induction of violacein production in
C. subtsugae CV026 by C4-HSL. In the lower part of the plate, where the extract obtained from the
P. aeruginosa PAO1 (pME6863) culture was seeded, the pigment production is not detected due to degradation of the QS signal molecules.
Figure 2B shows the results with samples obtained after 8 h of incubation. In this case, no differences are observed with respect to the extracts obtained after 4 h of incubation, which contrasts when
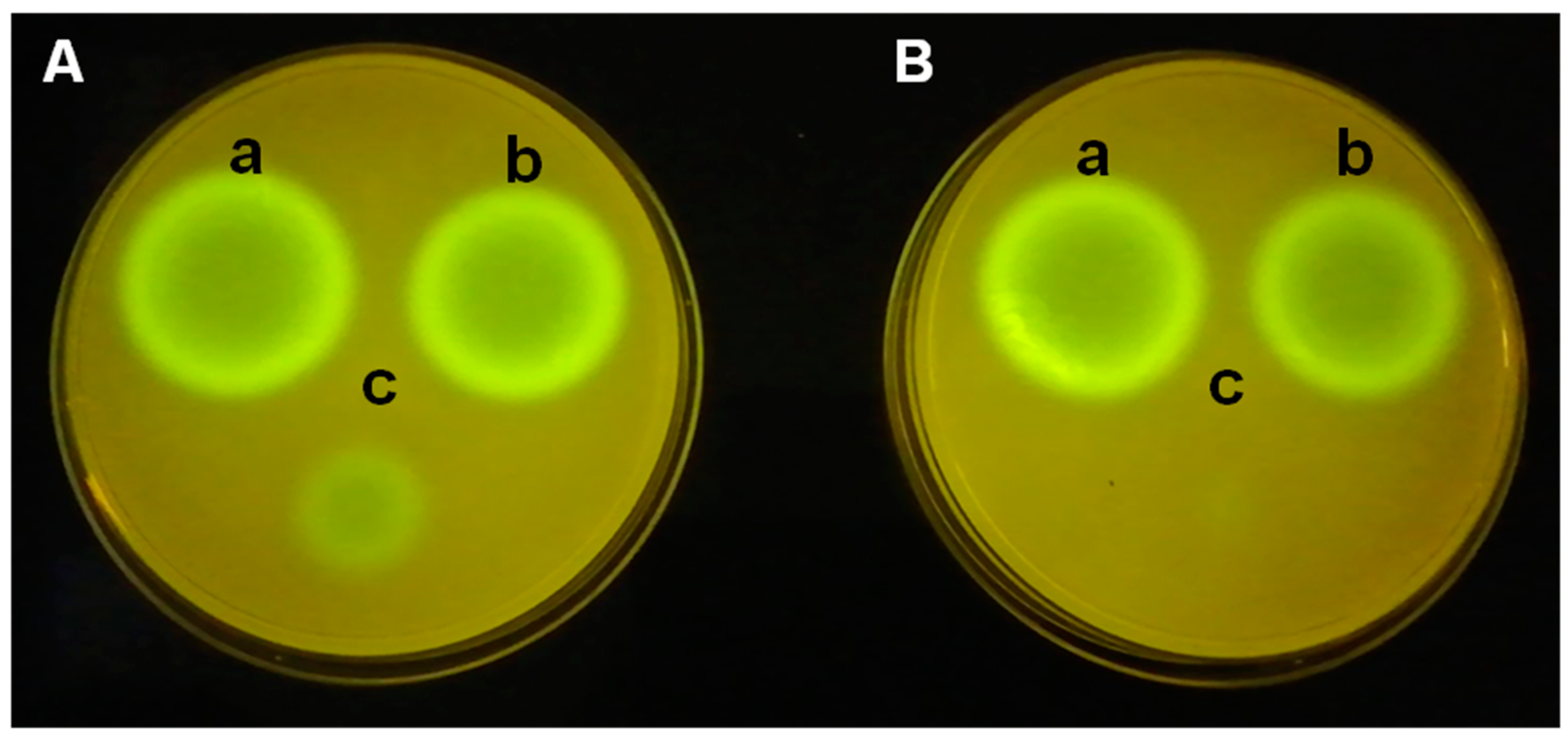
P. putida F117 (pKR-C12) biosensor strains is used. In this biosensor, 3OC12-HSL induce the production of green fluorescent protein (
Figure 3). The analysis of the extracts of both
P. aeruginosa PAO1 (left) and
P. aeruginosa PAO1 (pME6000) (right) exhibits similar size of fluorescence spots. The small spot, corresponding to
P. aeruginosa PAO1 (pME6863), is due to the lower levels of 3OC12-HSL related to the AHL degradation after 4 h of incubation (
Figure 3A), and that is not detectable after 8 h of incubation (
Figure 3B). The differences observed between the biosensor strains and the incubation time are related not only to the different levels of the AHLs but also to the different sensitivity to the QS signals.
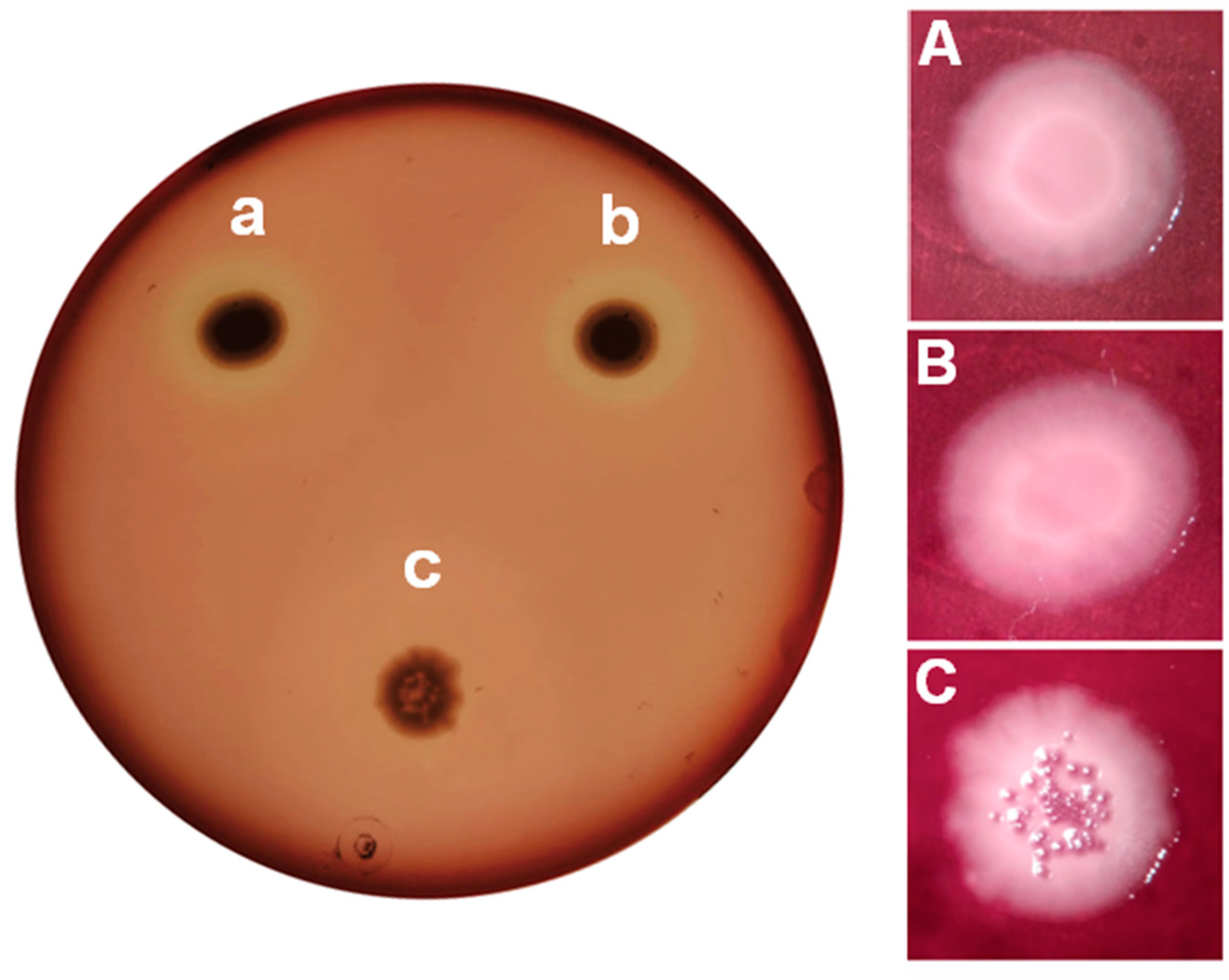
Figure 4 shows the results obtained for protease activity. As expected from the bioassays, similar behaviors with
P. aeruginosa PAO1 and
P. aeruginosa PAO1 (pME6000) are observed. Both strains show positive protease activity evidenced by the presence of a clear halo around the colonies due to the casein degradation. Among other, several genes coding for extracellular proteases are activated by QS in
P. aeruginosa PAO1. LasA protease and LasB elastase are the best characterized and the ones that give the name to the LasI-LasR QS system (Simanek and Paczkowski 2022; Schuster et al. 2003). Thought both Las and Rhl QS systems upregulate the expression of the corresponding
lasA and
lasB coding genes, the former is more prevalent than the latter. In addition, the green color of the colonies can be attributed to the siderophores production. Siderophores are organic iron chelators, which also gives the typical green colour to the
Pseudomonas culture.
P. aeruginosa produces two siderophores in iron-limited media: pyoverdine and pyochelin. Its production can also be evidenced in skim milk agar (
Figure 4).
pvd and
pch genes, which are involved in pyoverdine and pyochelin, are upregulated by LasI-LasR (Stintzi et al. 1998). In
P. aeruginosa PAO1 (pME6863), with an attenuated QS system, both protease activity and siderophore production are affected.
A similar situation is observed on blood agar plates (
Figure 5A) where positive hemolytic activities are detected for
P. aeruginosa PAO1 and
P. aeruginosa PAO1 (pME6000), but not for
P. aeruginosa PAO1 (pME6863). Bacterial phospholipases aide in the hemolysis, though this activity is mainly related to the production of the surfactant rhamnolipids. Expression of
rhlA and
rhlB, which code for enzymes involved in the rhamnolipid synthesis, is positively regulated by the RhlI-RhlR QS system (Ochsner and Reiser 1995).
Differences in the morphology of the colonies can also be observed on blood agar plates (
Figure 5 B, C and D). Bacterial colonies are static biofilms formed in the interphase between the culture medium and the air (Merritt, Kadouri, and O’Toole 2005). A large set of genes are involved in biofilm formation in
P. aeruginosa PAO1 (Waite et al. 2005). Among them,
alg genes involved in the production of the exopolysaccharide alginate, and the aforementioned
rhlA and
rhlB are upregulated by the RhlI-RhlR QS system (Ochsner and Reiser 1995).
Finally, the teacher may introduce students to the biotechnological applications derived from the use of QQ strategies for blocking the cell-to-cell communication systems. Reference materials including scientific articles can support this discussion (Chen et al. 2018; Romero, Acuna, and Otero 2012). Students are assessed through a final report covering the entire practical, which also contributes to the scientific training of how to formulate the hypothesis and objectives of a work.
Final considerations
In the present work, P. aeruginosa PAO1 is used as a model strain because of its QS system involved in the regulation of several readily demonstrable traits, including proteases and surfactants. These properties can be directly observed by students in a simple and practical laboratory class. It has to be noted that the general concepts explored in these experiments are not specific to PAO1. Other bacteria with well-known QS systems can also be used, considering that the methodology should be adapted accordingly. However, in our opinion P. aeruginosa PAO1 is an excellent choice due to the availability of protocols and the facility for the observation and interpretation of the results. These laboratory experiences allow, in a short time, to familiarize the students with several concepts and techniques, ranging from the regulation of genetic expression to the evidence of several physiological traits.
Acknowledgments
This work was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, PIP 2015-0946, PIP 2021-2436, PUE 22920160100012CO), Agencia Nacional de Promoción Científica y Tecnológica (PICT 2016 Nº 0532; PICT 2019 Nº 03336), and Secretaría de Ciencia, Arte e Innovación Tecnológica from the Universidad Nacional de Tucumán (PIUNT D609). We thank Patricia Peralta (PROIMI-CONICET) for her valuable support in the preparation of photos and images.
References
- Chadha, Jatin, Kusum Harjai, and Sanjay Chhibber. 2022. “Revisiting the Virulence Hallmarks of Pseudomonas Aeruginosa: A Chronicle through the Perspective of Quorum Sensing.” Environmental Microbiology 24 (6): 2630–2656. [CrossRef]
- Chen, Xin, Likun Zhang, Mingxiang Zhang, Huayu Liu, Panrui Lu, and Kejiang Lin. 2018. “Quorum Sensing Inhibitors: A Patent Review (2014–2018).” Expert Opinion on Therapeutic Patents 28 (12): 849–865. [CrossRef]
- Fuqua, C, and E P Greenberg. 2002. “Listening in on Bacteria: Acyl-Homoserine Lactone Signalling.” Nature Reviews. Molecular Cell Biology 3 (9): 685–695. [CrossRef]
- Grandclément, Catherine, Mélanie Tannières, Solange Moréra, Yves Dessaux, and Denis Faure. 2016. “Quorum Quenching: Role in Nature and Applied Developments.” FEMS Microbiology Reviews 40 (1): 86–116. [CrossRef]
- McClean, K, M Winson, L Fish, A Taylor, S Chhabra, Camara M, M Daykin, et al. 1997. “Quorum Sensing and Chromobacterium Violaceum: Exploitation of Violacein Production and Inhibition for the Detection of N-Acylhomoserine Lactones.” Microbiology 143: 3703–3711.
- Merritt, J. H., D. E. Kadouri, and G. A. O’Toole. 2005. “Growing and Analyzing Static Biofilms.” In Current Protocols in Microbiology, 1B.1.1-1B.1.18.
- Miller, Craig, and Jordon Gilmore. 2020. “Detection of Quorum-Sensing Molecules for Pathogenic Molecules Using Cell-Based and Cell-Free Biosensors.” Antibiotics 9 (5): 259. [CrossRef]
- Ochsner, U A, and J Reiser. 1995. “Autoinducer-Mediated Regulation of Rhamnolipid Biosurfactant Synthesis in Pseudomonas Aeruginosa.” Proceedings of the National Academy of Sciences 92 (14): 6424–6428. [CrossRef]
- Ohman, D E, S J Cryz, and B H Iglewski. 1980. “Isolation and Characterization of Pseudomonas Aeruginosa PAO Mutant That Produces Altered Elastase.” Journal of Bacteriology 142 (3): 836–842. [CrossRef]
- Qin, Shugang, Wen Xiao, Chuanmin Zhou, Qinqin Pu, Xin Deng, Lefu Lan, Haihua Liang, Xiangrong Song, and Min Wu. 2022. “Pseudomonas Aeruginosa: Pathogenesis, Virulence Factors, Antibiotic Resistance, Interaction with Host, Technology Advances and Emerging Therapeutics.” Signal Transduction and Targeted Therapy 7 (1): 199. [CrossRef]
- Reimmann, Cornelia, Nathalie Ginet, Laurent Michel, Christoph Keel, Patrick Michaux, Viji Krishnapillai, Marcello Zala, et al. 2002. “Genetically Programmed Autoinducer Destruction Reduces Virulence Gene Expression and Swarming Motility in Pseudomonas Aeruginosa PAO1.” Microbiology 148 (4): 923–932.
- Riedel, Kathrin, Morten Hentzer, Otto Geisenberger, Birgit Huber, Anette Steidle, Hong Wu, Niels Høiby, Michael Givskov, Søren Molin, and Leo Eberl. 2001. “N-Acylhomoserine-Lactone-Mediated Communication between Pseudomonas Aeruginosa and Burkholderia Cepacia in Mixed Biofilms.” Microbiology 147 (12): 3249–3262.
- Romero, Manuel, Laura Acuna, and Ana Otero. 2012. “Patents on Quorum Quenching: Interfering with Bacterial Communication as a Strategy to Fight Infections.” Recent Patents on Biotechnology 6 (1): 2–12. [CrossRef]
- Schuster, Martin, C Phoebe Lostroh, Tomoo Ogi, and E P Greenberg. 2003. “Identification, Timing, and Signal Specificity of Pseudomonas Aeruginosa Quorum-Controlled Genes: A Transcriptome Analysis.” Journal of Bacteriology 185 (7): 2066–2079. [CrossRef]
- Shaw, P D, G Ping, S L Daly, C Cha, J E Cronan, K L Rinehart, and S K Farrand. 1997. “Detecting and Characterizing N-Acyl-Homoserine Lactone Signal Molecules by Thin-Layer Chromatography.” Proceedings of the National Academy of Sciences of the United States of America 94 (12): 6036–6041. [CrossRef]
- Simanek, Kayla A, and Jon E Paczkowski. 2022. “Resistance Is Not Futile: The Role of Quorum Sensing Plasticity in Pseudomonas Aeruginosa Infections and Its Link to Intrinsic Mechanisms of Antibiotic Resistance.” Microorganisms 10 (6): 1247. [CrossRef]
- Steindler, Laura, and Vittorio Venturi. 2007. “Detection of Quorum-Sensing N-Acyl Homoserine Lactone Signal Molecules by Bacterial Biosensors.” FEMS Microbiology Letters 266 (1): 1–9. [CrossRef]
- Stintzi, A, K Evans, J M Meyer, and K Poole. 1998. “Quorum-Sensing and Siderophore Biosynthesis in Pseudomonas Aeruginosa: LasR/LasI Mutants Exhibit Reduced Pyoverdine Biosynthesis.” FEMS Microbiology Letters 166 (2): 341–345. [CrossRef]
- Waite, Richard D, Anastasia Papakonstantinopoulou, Eddie Littler, and Michael A Curtis. 2005. “Transcriptome Analysis of Pseudomonas Aeruginosa Growth: Comparison of Gene Expression in Planktonic Cultures and Developing and Mature Biofilms.” Journal of Bacteriology 187 (18): 6571–6576. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).