Submitted:
12 May 2023
Posted:
16 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
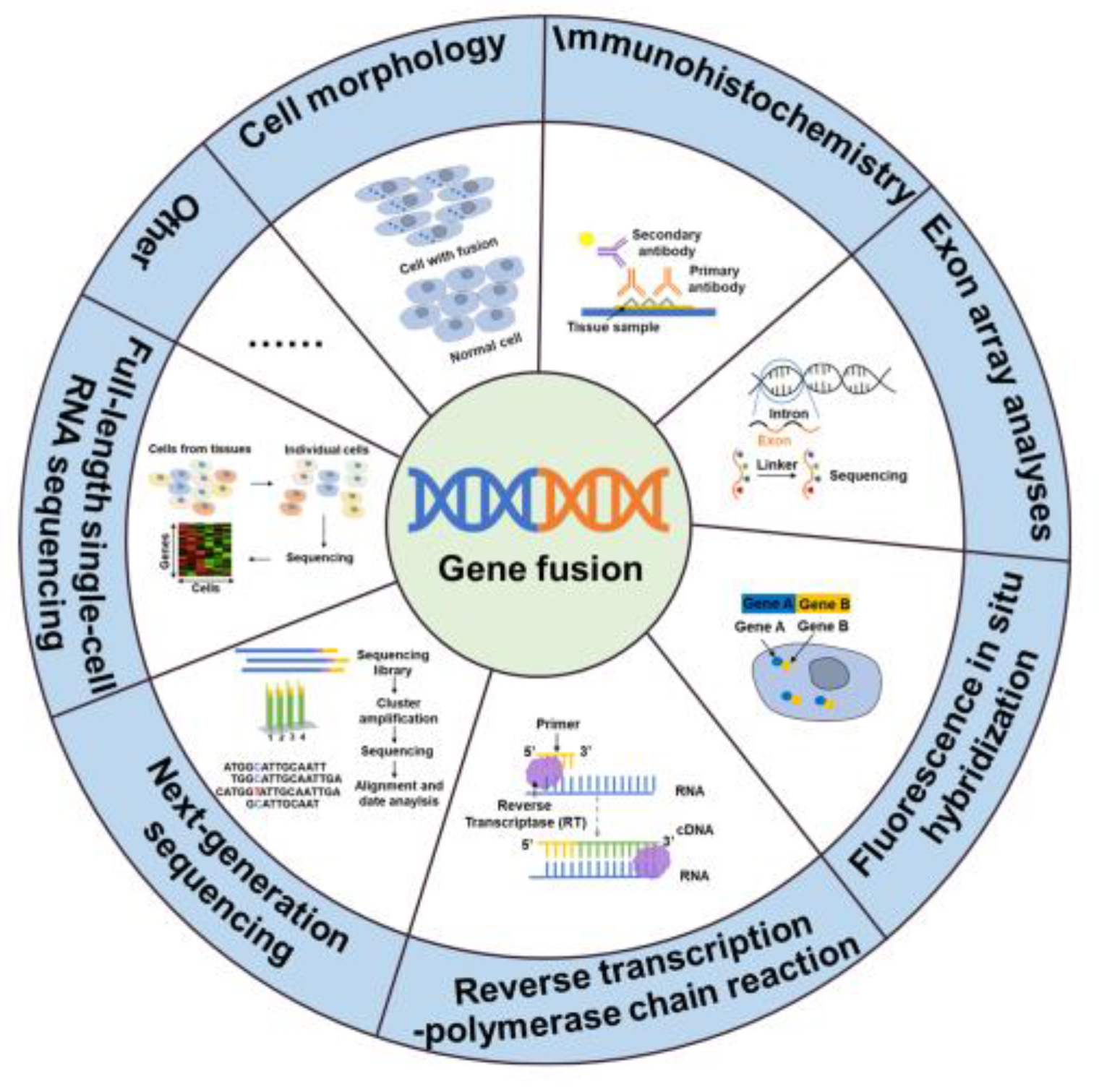
2. Fusion genes in diseases and the detection of fusion genes
3. The development of inhibitors targeted kinase fusion genes
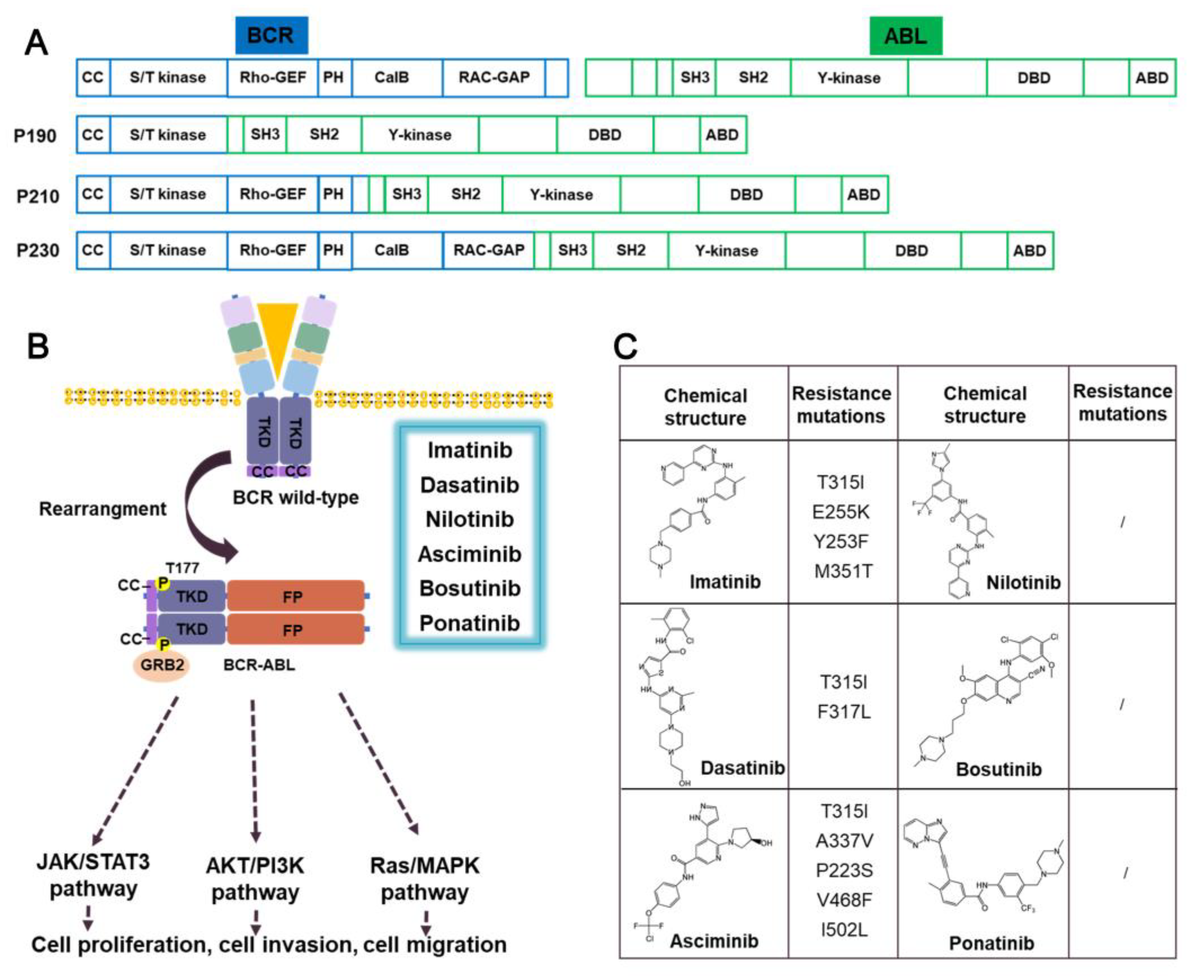
3.1. ABL-fusion inhibitors
3.1.1. Imatinib
3.1.2. Dasatinib
3.1.3. Nilotinib
3.1.4. Asciminib
3.1.5. Prospect
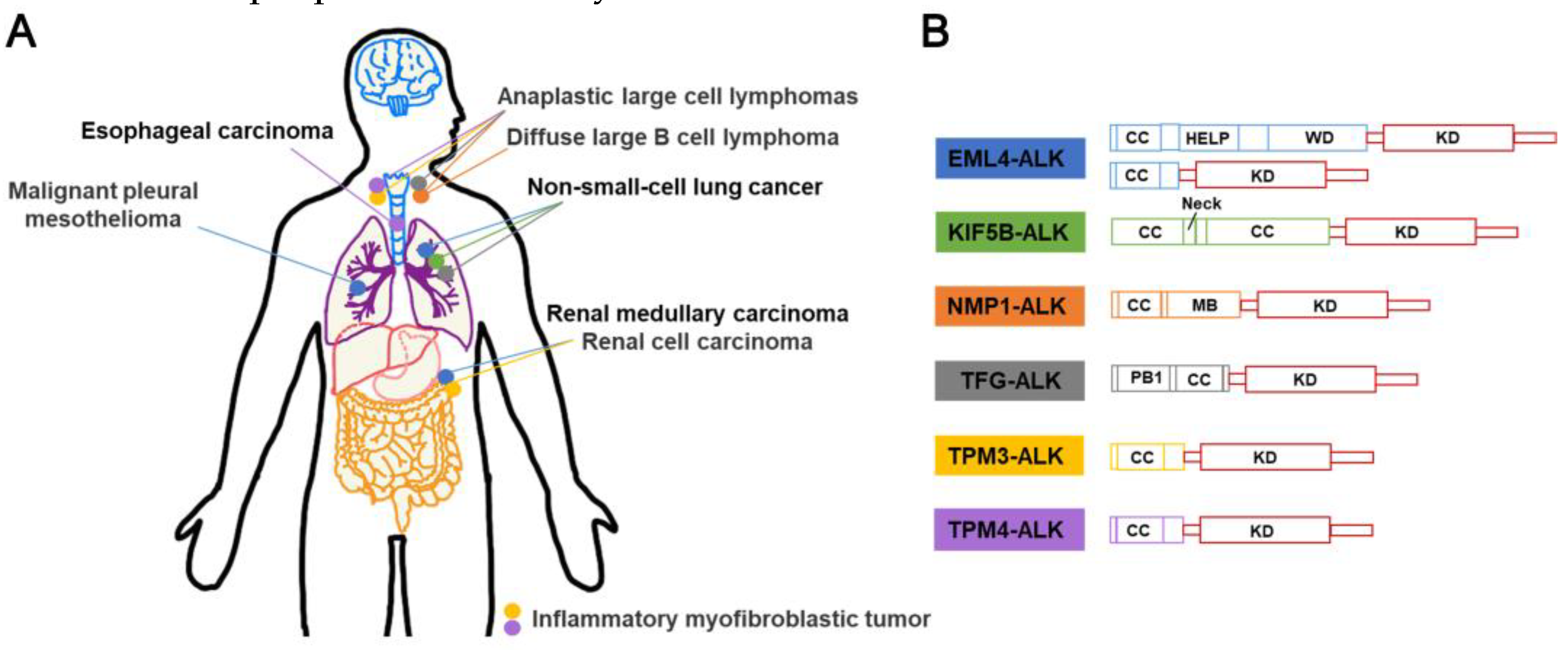
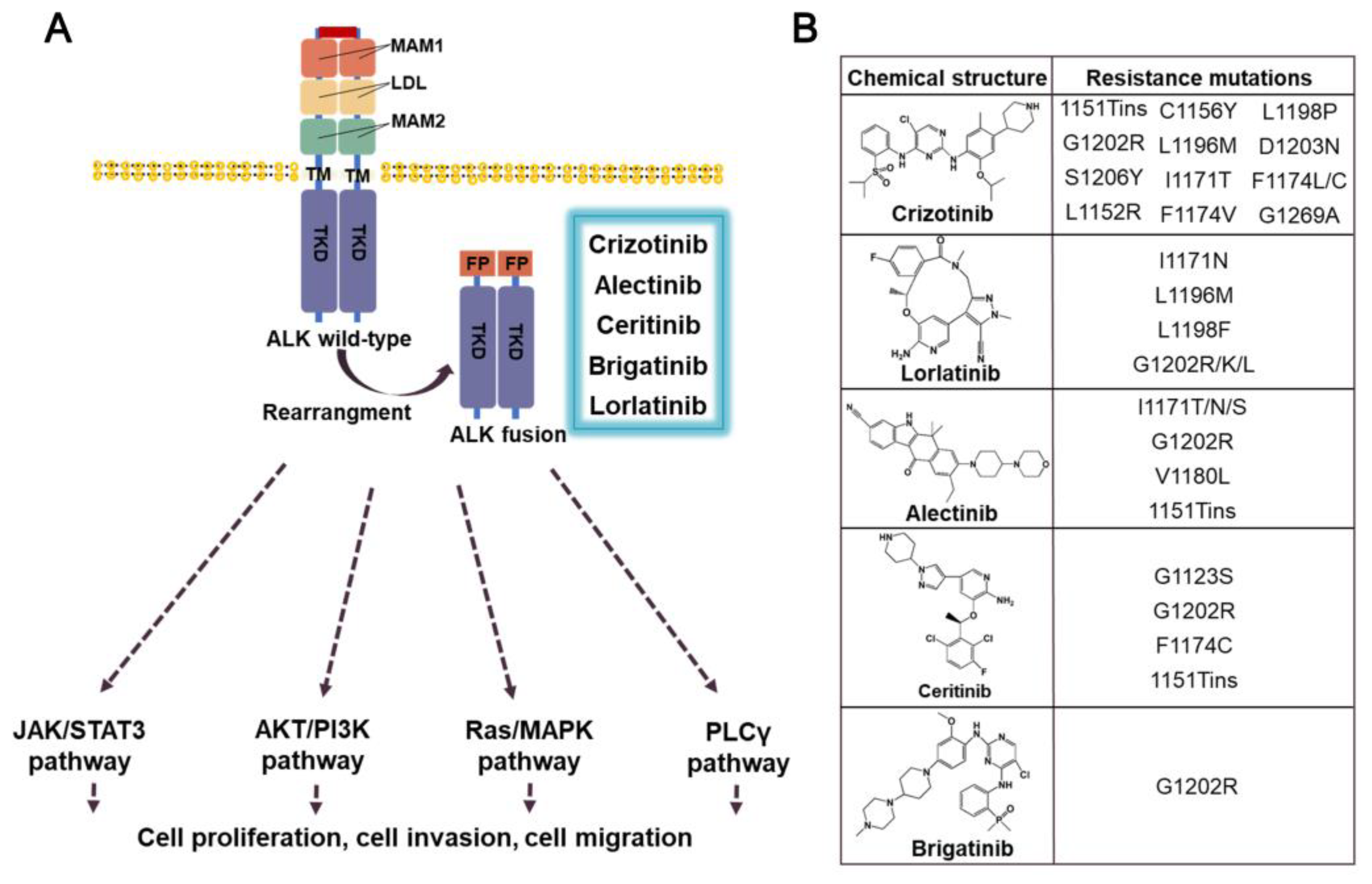
3.2. ALK-fusion inhibitors
3.2.1. Crizotinib
3.2.2. Ceritinib
3.2.3. Alectinib
3.2.4. Brigatinib
3.2.5. Prospect
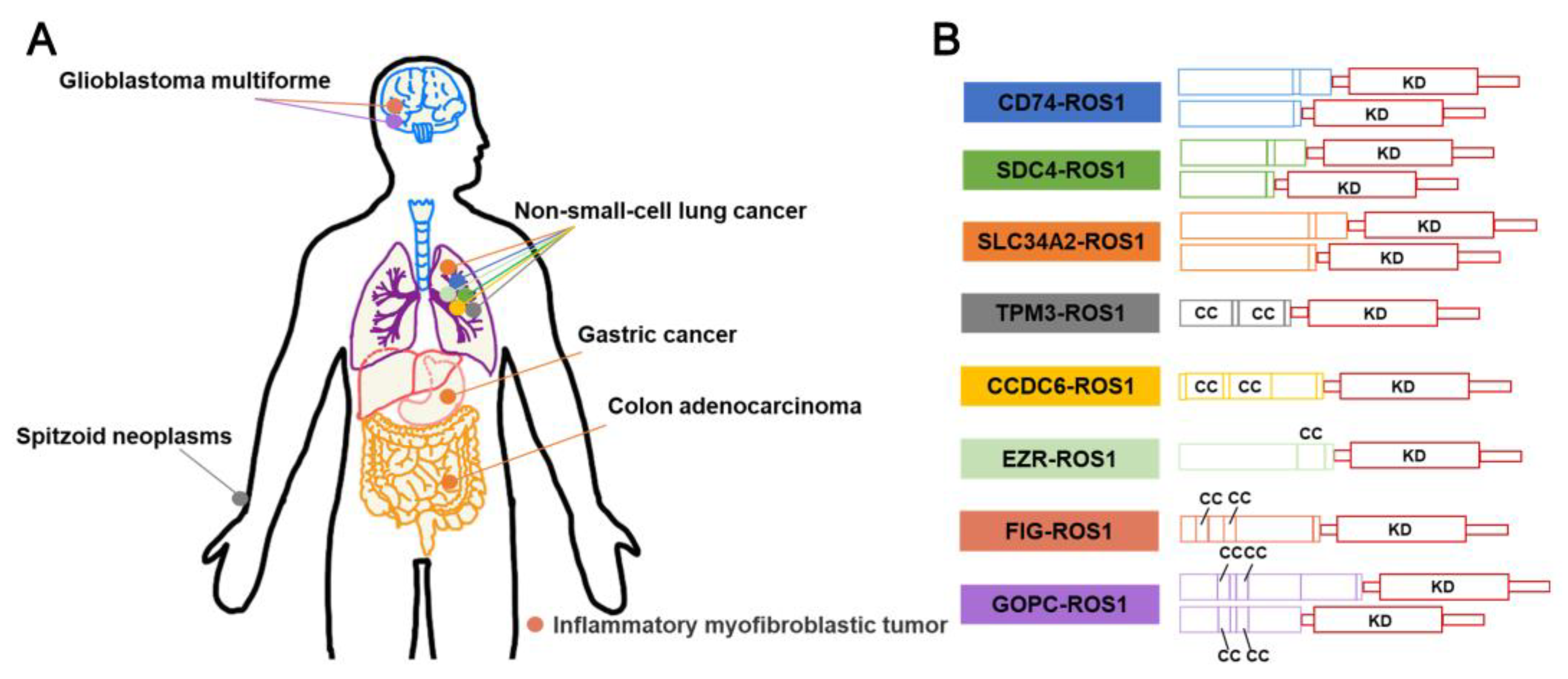
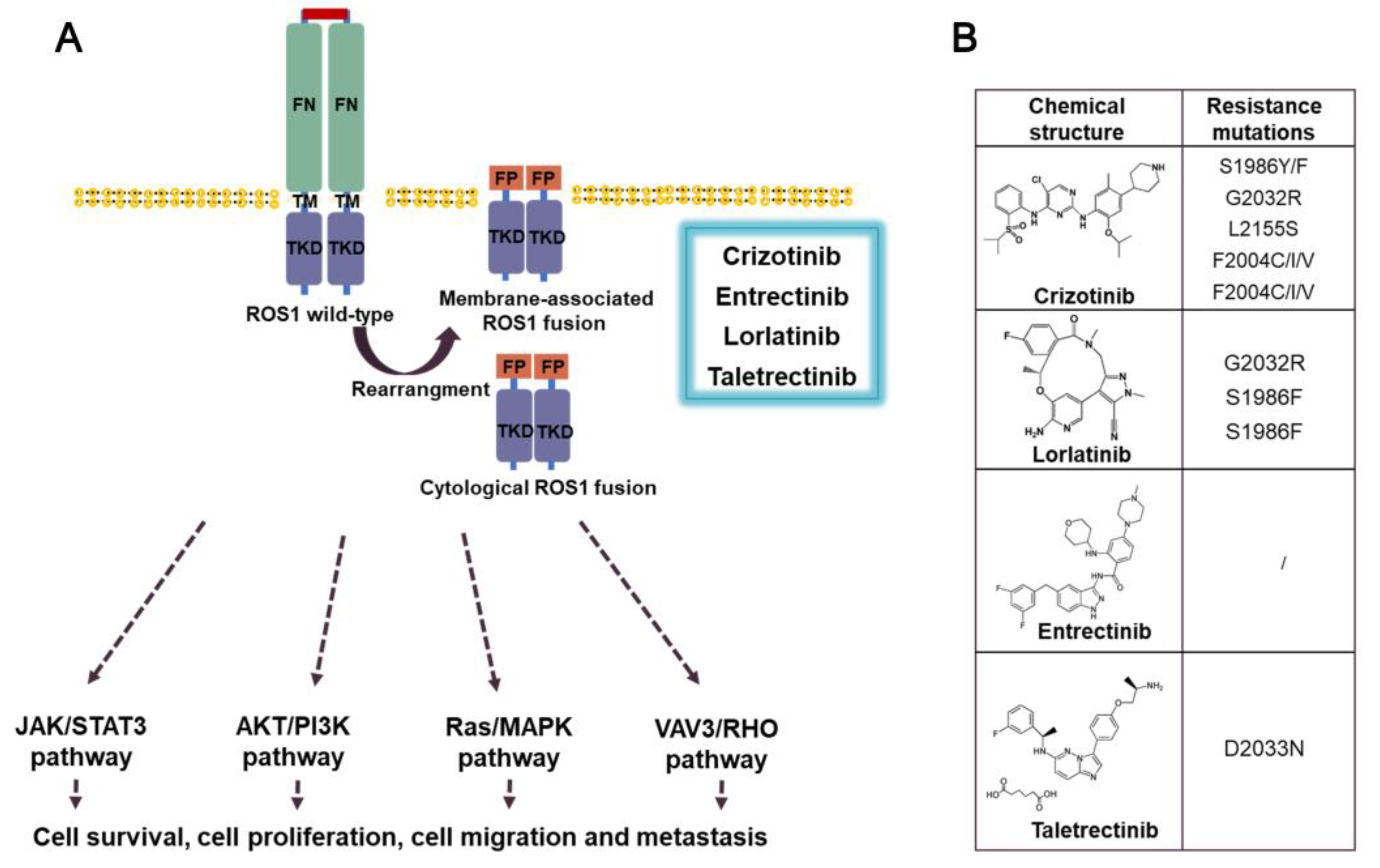
3.3. ROS1 fusion inhibitors
3.3.1. Crizotinib
3.3.2. Entrectinib
3.3.3. Prospect
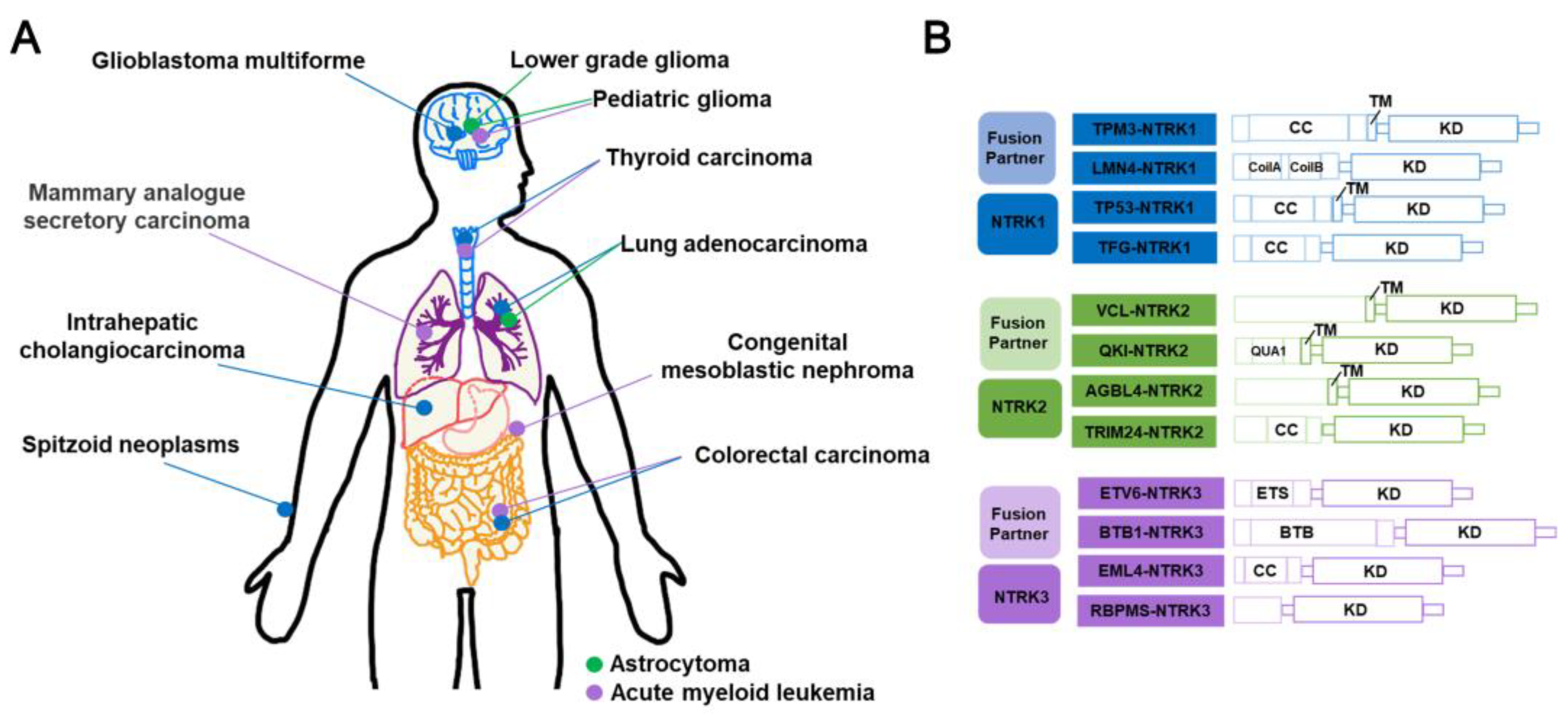
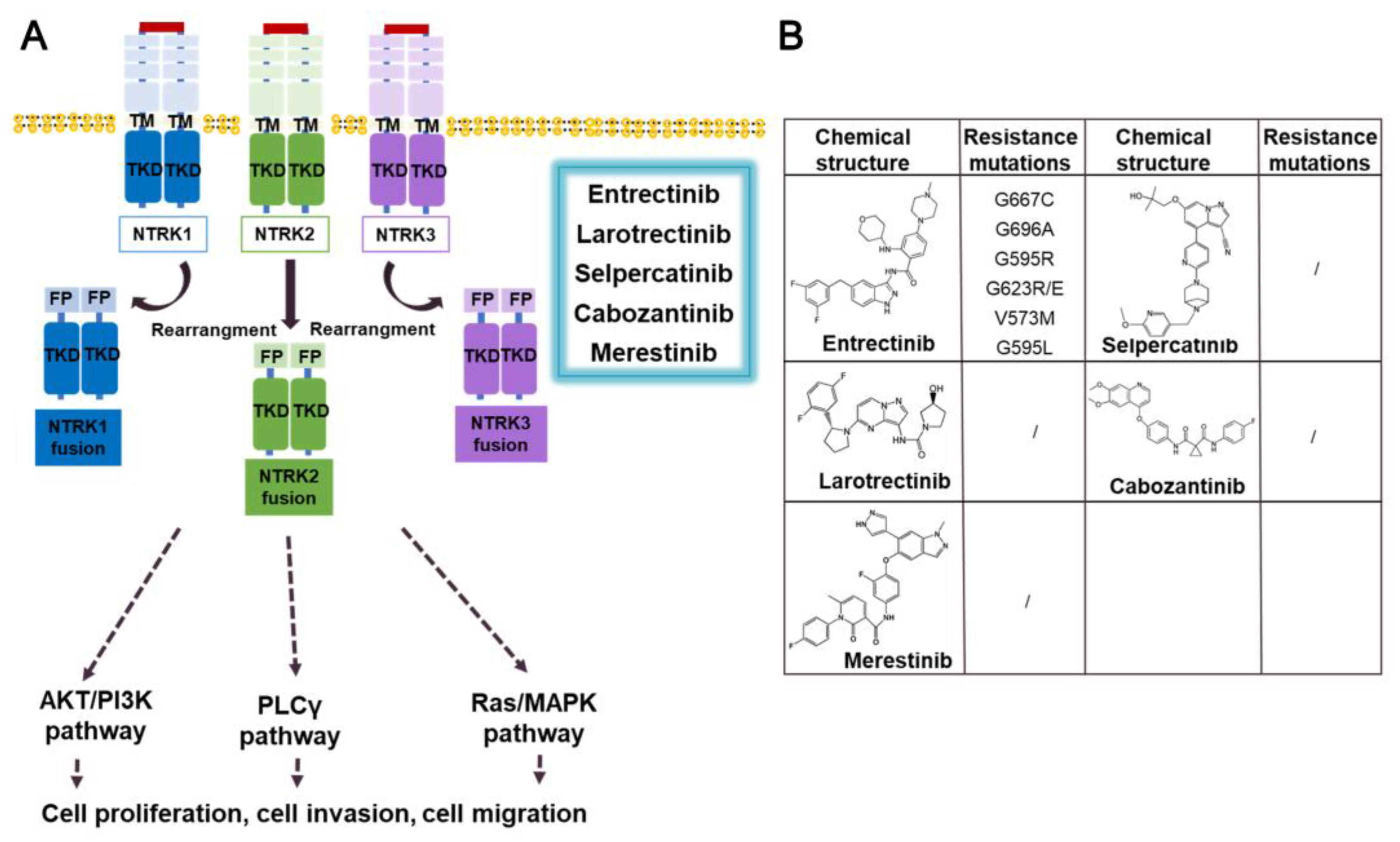
3.4. NTRK fusion inhibitors
3.4.1. Entrectinib
3.4.2. Larotrectinib
3.4.3. Prospect
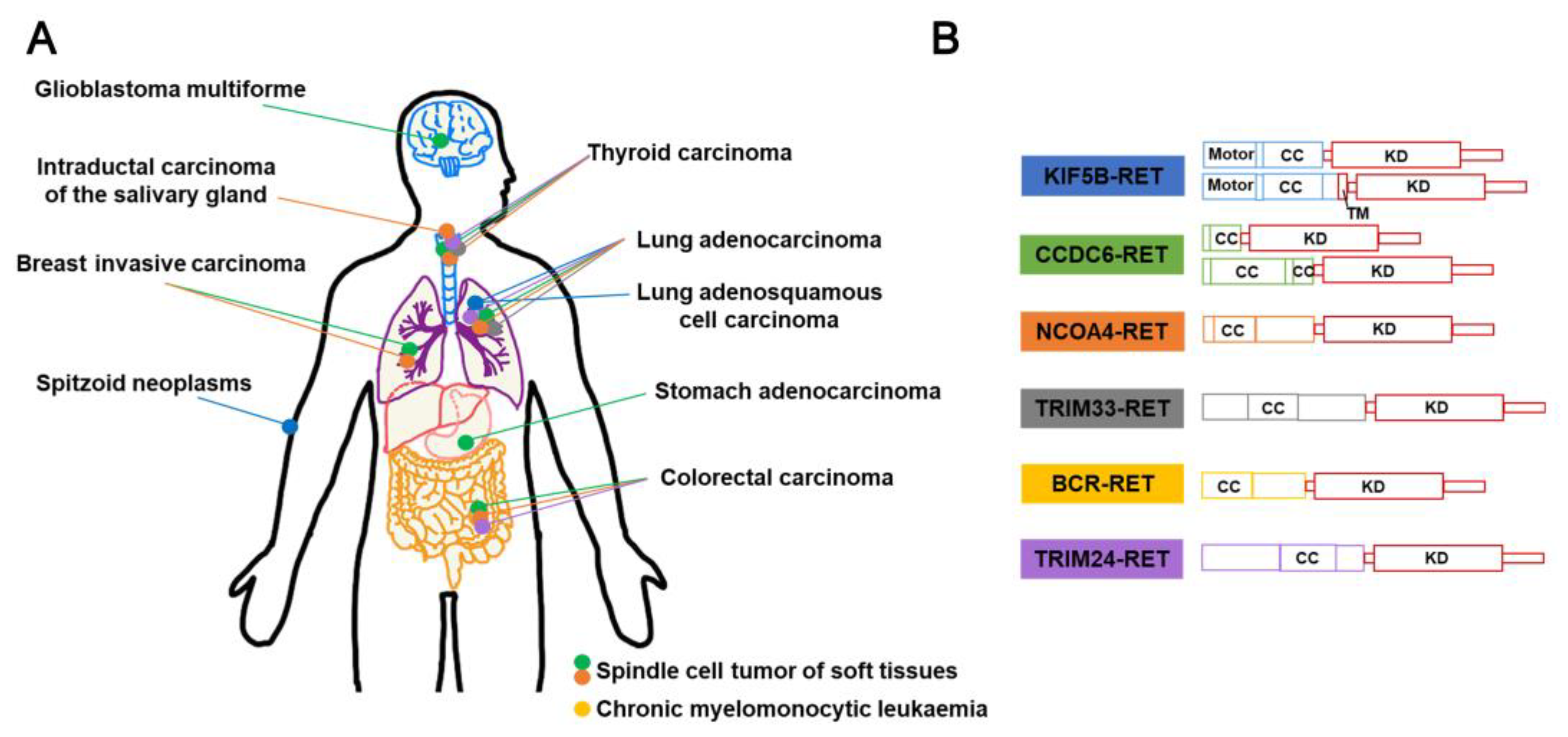
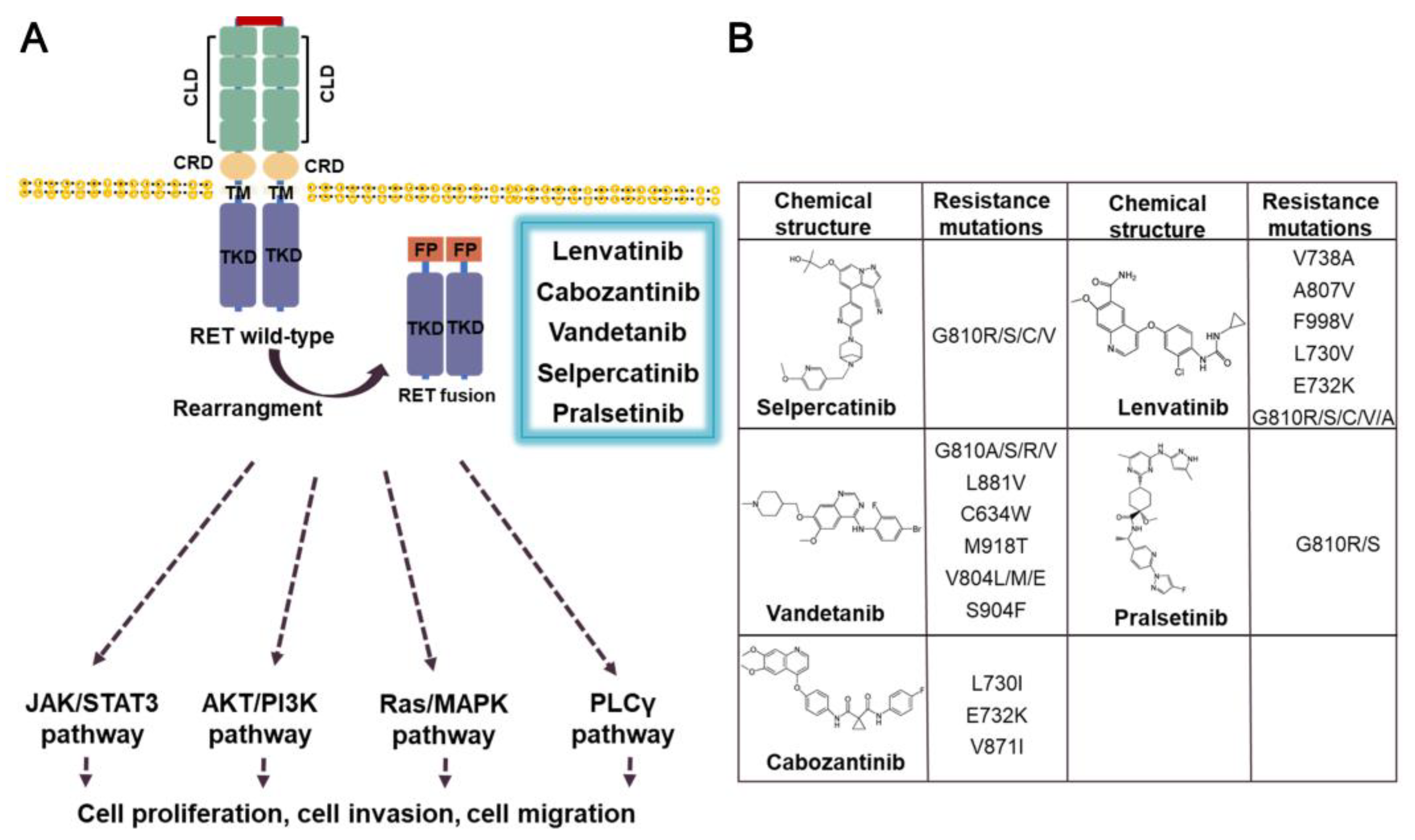
3.5. RET fusion inhibitors
3.5.1. Lenvatinib
3.5.2. Cabozantinib
3.5.3. Vandetanib
3.5.4. Selpercatinib
3.5.5. Pralsetinib
3.5.6. Prospect
3.6. Others
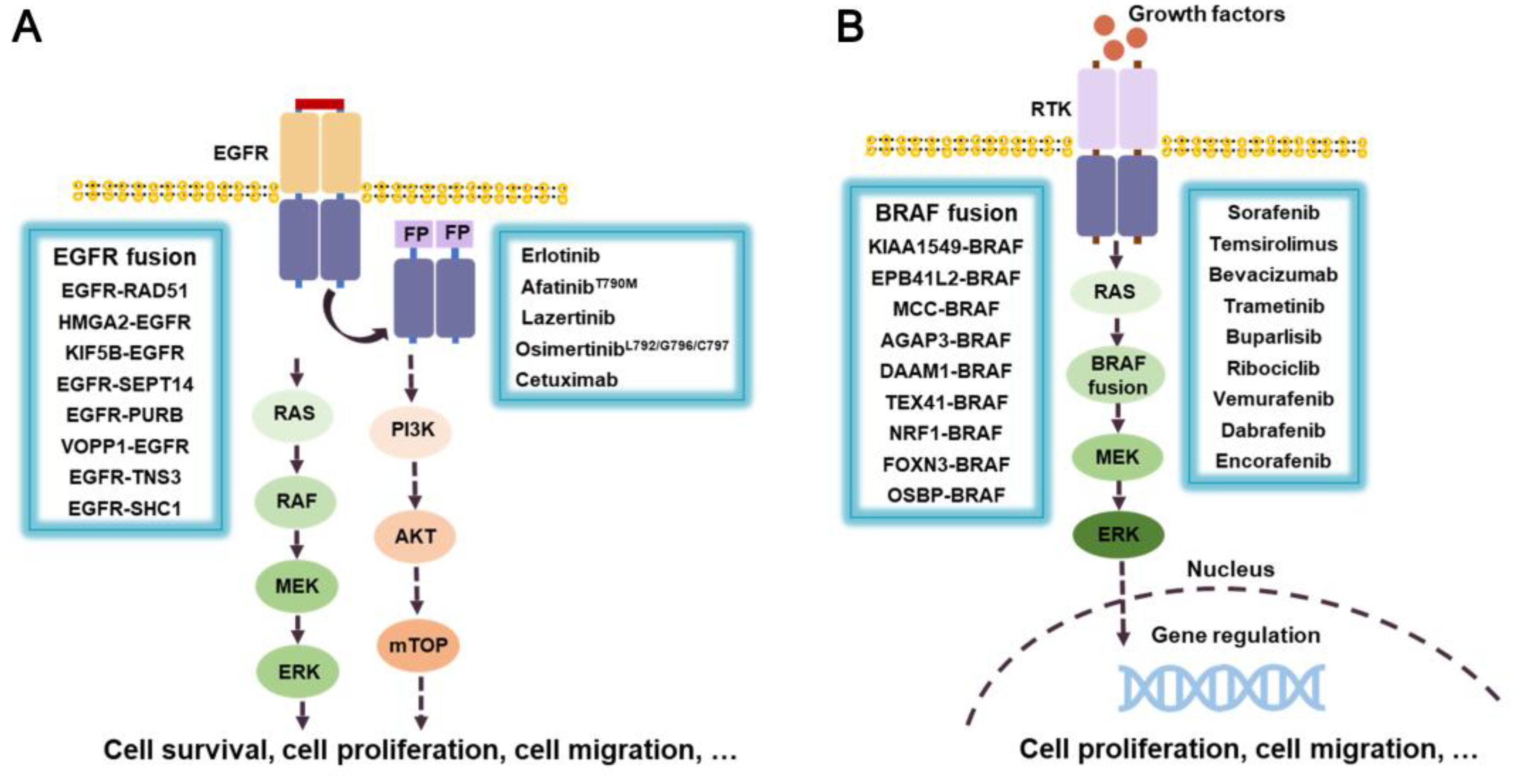
3.6.1. EGFR fusion inhibitors
3.6.2. RAF/BRAF-fusion inhibitors
4. The development of inhibitors targeted non-kinase fusion genes
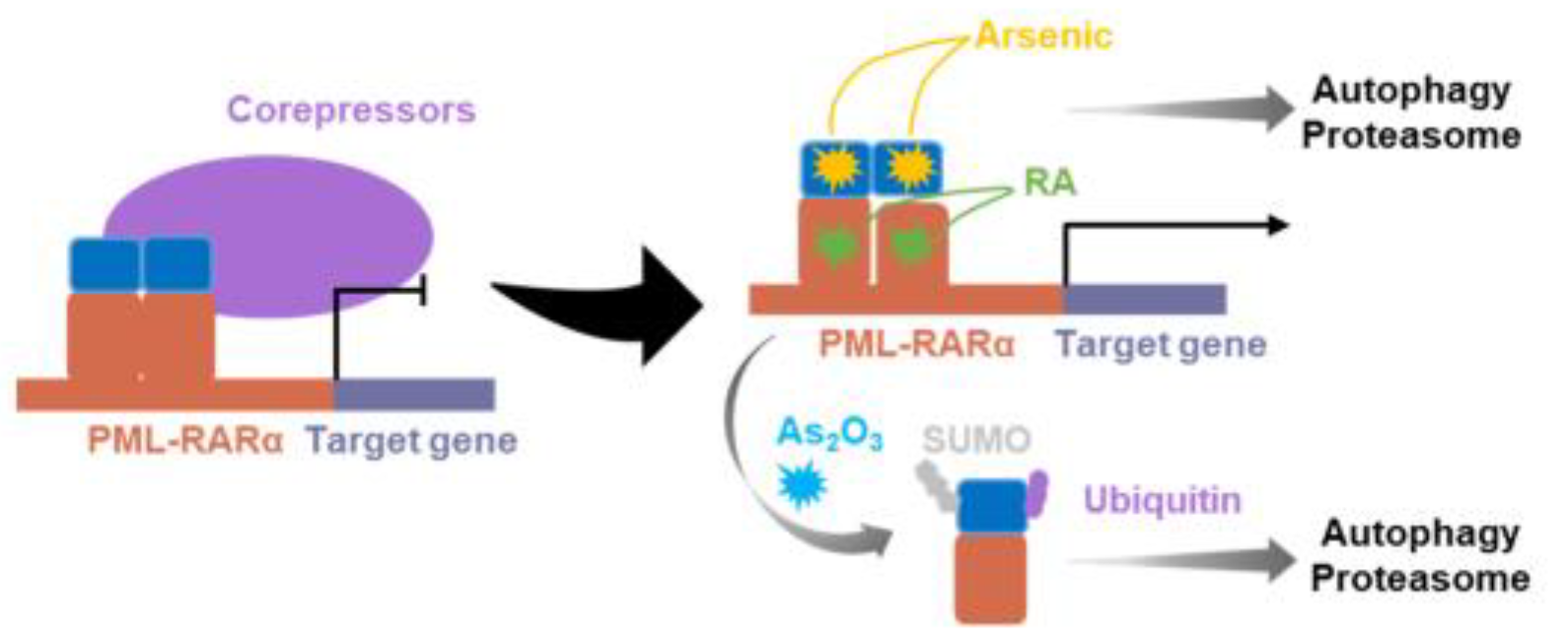
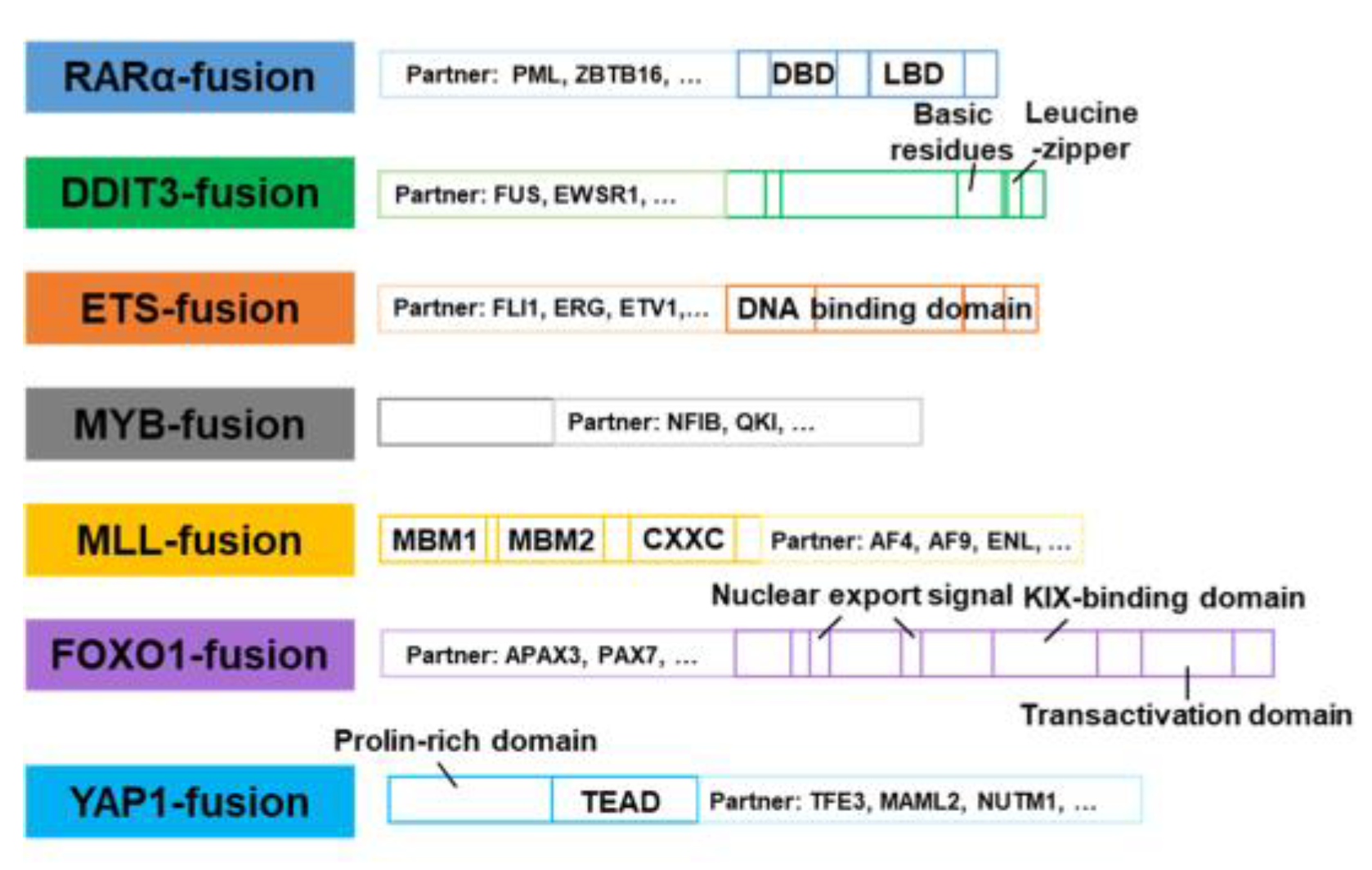
4.1. RARα-fusion inhibitors
4.2. DDIT3-fusion inhibitors
4.3. ETS-fusion inhibitors
4.4. MYB-fusion inhibitors
4.5. MLL-fusion inhibitors
4.6. FOXO1-fusion inhibitors
4.7. YAP1-fusion inhibitors
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Roberts, N.D.; Wala, J.A.; Shapira, O.; Schumacher, S.E.; Kumar, K.; Khurana, E.; Waszak, S.; Korbel, J.O.; Haber, J.E.; Imielinski, M.; Weischenfeldt, J.; Beroukhim, R.; Campbell, P.J. Patterns of somatic structural variation in human cancer genomes. Nature 2020, 578, 112–121. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; Boyault, S.; Burkhardt, B.; Butler, A.P.; Caldas, C.; Davies, H.R.; Desmedt, C.; Eils, R.; Eyfjörd, J.E.; Foekens, J.A.; Greaves, M.; Hosoda, F.; Hutter, B.; Ilicic, T.; Imbeaud, S.; Imielinski, M.; Jäger, N.; Jones, D.T.; Jones, D.; Knappskog, S.; Kool, M.; Lakhani, S.R.; López-Otín, C.; Martin, S.; Munshi, N.C.; Nakamura, H.; Northcott, P.A.; Pajic, M.; Papaemmanuil, E.; Paradiso, A.; Pearson, J.V.; Puente, X.S.; Raine, K.; Ramakrishna, M.; Richardson, A.L.; Richter, J.; Rosenstiel, P.; Schlesner, M.; Schumacher, T.N.; Span, P.N.; Teague, J.W.; Totoki, Y.; Tutt, A.N.; Valdés-Mas, R.; van Buuren, M.M.; van 't Veer, L.; Vincent-Salomon, A.; Waddell, N.; Yates, L.R.; Zucman-Rossi, J.; Futreal, P.A.; McDermott, U.; Lichter, P.; Meyerson, M.; Grimmond, S.M.; Siebert, R.; Campo, E.; Shibata, T.; Pfister, S.M.; Campbell, P.J.; Stratton, M.R. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Mitelman, F.; Johansson, B.; Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 2007, 7, 233–245. [Google Scholar] [CrossRef]
- Panagopoulos, I.; Heim, S. Interstitial deletions generating fusion genes. Cancer Genomics Proteomics 2021, 18, 167–196. [Google Scholar] [CrossRef]
- Pederzoli, F.; Bandini, M.; Marandino, L.; Ali, S.M.; Madison, R.; Chung, J.; Ross, J.S.; Necchi, A. Targetable gene fusions and aberrations in genitourinary oncology. Nat. Rev. Urol. 2020, 17, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.D. A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia identified by Quinacrine Fluorescence and Giemsa Staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef]
- Heydt, C.; Wölwer, C.B.; Velazquez Camacho, O.; Wagener-Ryczek, S.; Pappesch, R.; Siemanowski, J.; Rehker, J.; Haller, F.; Agaimy, A.; Worm, K.; Herold, T.; Pfarr, N.; Weichert, W.; Kirchner, T.; Jung, A.; Kumbrink, J.; Goering, W.; Esposito, I.; Buettner, R.; Hillmer, A.M.; Merkelbach-Bruse, S. Detection of gene fusions using targeted next-generation sequencing: a comparative evaluation. BMC Med. Genomics 2021, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Klijn, C.; Durinck, S.; Stawiski, E.W.; Haverty, P.M.; Jiang, Z.; Liu, H.; Degenhardt, J.; Mayba, O.; Gnad, F.; Liu, J.; Pau, G.; Reeder, J.; Cao, Y.; Mukhyala, K.; Selvaraj, S.K.; Yu, M.; Zynda, G.J.; Brauer, M.J.; Wu, T.D.; Gentleman, R.C.; Manning, G.; Yauch, R.L.; Bourgon, R.; Stokoe, D.; Modrusan, Z.; Neve, R.M.; de Sauvage, F.J.; Settleman, J.; Seshagiri, S.; Zhang, Z. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015, 33, 306–312. [Google Scholar] [CrossRef]
- Cilloni, D.; Saglio, G. Molecular pathways: BCR-ABL. Clin. Cancer Res. 2012, 18, 930–937. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lalazar, G.; Houlihan, S.L.; Tschaharganeh, D.F.; Baslan, T.; Chen, C.C.; Requena, D.; Tian, S.; Bosbach, B.; Wilkinson, J.E.; Simon, S.M.; Lowe, S.W. DNAJB1-PRKACA fusion kinase interacts with β-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 13076–13084. [Google Scholar] [CrossRef]
- Asai, N.; Murakami, H.; Iwashita, T.; Takahashi, M. A mutation at tyrosine 1062 in MEN2A-Ret and MEN2B-Ret impairs their transforming activity and association with shc adaptor proteins. J. Biol. Chem. 1996, 271, 17644–17649. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Yu, Y.P.; Tao, J.; Liu, S.; Tseng, G.; Nalesnik, M.; Hamilton, R.; Bhargava, R.; Nelson, J.B.; Pennathur, A.; Monga, S.P.; Luketich, J.D.; Michalopoulos, G.K.; Luo, J.H. MAN2A1-FER fusion gene is expressed by human liver and other tumor types and has oncogenic activity in mice. Gastroenterology 2017, 153, 1120–1132. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Wang, X.; Song, H.; Zhang, Y.; Zhang, R.; Li, S.; Jin, W.; Chen, S.; Fang, H.; Chen, Z.; Wang, K. A PML/RARα direct target atlas redefines transcriptional deregulation in acute promyelocytic leukemia. Blood 2021, 137, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Zullow, H.J.; Sankar, A.; Ingram, D.R.; Samé Guerra, D.D.; D'Avino, A.R.; Collings, C.K.; Lazcano, R.; Wang, W.L.; Liang, Y.; Qi, J.; Lazar, A.J.; Kadoch, C. The FUS::DDIT3 fusion oncoprotein inhibits BAF complex targeting and activity in myxoid liposarcoma. Mol. Cell 2022, 82, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- Capdeville, R.; Silberman, S. Imatinib: a targeted clinical drug development. Semin. Hematol. 2003, 40, 15–20. [Google Scholar] [CrossRef]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.J.; Tran-Dubé, M.; Shen, H.; Nambu, M.; Kung, P.P.; Pairish, M.; Jia, L.; Meng, J.; Funk, L.; Botrous, I.; McTigue, M.; Grodsky, N.; Ryan, K.; Padrique, E.; Alton, G.; Timofeevski, S.; Yamazaki, S.; Li, Q.; Zou, H.; Christensen, J.; Mroczkowski, B.; Bender, S.; Kania, R.S.; Edwards, M.P. Structure based drug design of Crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J. Med. Chem. 2011, 54, 6342–6363. [Google Scholar] [CrossRef]
- Lallemand-Breitenbach, V.; Zhu, J.; Chen, Z.; de Thé, H. Curing APL through PML/RARA degradation by As2O3. Trends Mol. Med. 2012, 18, 36–42. [Google Scholar] [CrossRef]
- Schram, A.M.; Chang, M.T.; Jonsson, P.; Drilon, A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat. Rev. Clin. Oncol. 2017, 14, 735–748. [Google Scholar] [CrossRef]
- Hahn, Y.; Bera, T.K.; Gehlhaus, K.; Kirsch, I.R.; Pastan, I.H.; Lee, B. Finding fusion genes resulting from chromosome rearrangement by analyzing the expressed sequence databases. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 13257–13261. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; Lee, C.; Montie, J.E.; Shah, R.B.; Pienta, K.J.; Rubin, M.A.; Chinnaiyan, A.M. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Laxman, B.; Dhanasekaran, S.M.; Helgeson, B.E.; Cao, X.; Morris, D.S.; Menon, A.; Jing, X.; Cao, Q.; Han, B.; Yu, J.; Wang, L.; Montie, J.E.; Rubin, M.A.; Pienta, K.J.; Roulston, D.; Shah, R.B.; Varambally, S.; Mehra, R.; Chinnaiyan, A.M. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 2007, 448, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; Bando, M.; Ohno, S.; Ishikawa, Y.; Aburatani, H.; Niki, T.; Sohara, Y.; Sugiyama, Y.; Mano, H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef]
- Koivunen, J.P.; Mermel, C.; Zejnullahu, K.; Murphy, C.; Lifshits, E.; Holmes, A.J.; Choi, H.G.; Kim, J.; Chiang, D.; Thomas, R.; Lee, J.; Richards, W.G.; Sugarbaker, D.J.; Ducko, C.; Lindeman, N.; Marcoux, J.P.; Engelman, J.A.; Gray, N.S.; Lee, C.; Meyerson, M.; Jänne, P.A. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 2008, 14, 4275–4283. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, Q.; Tang, M.; Barthel, F.; Amin, S.; Yoshihara, K.; Lang, F.M.; Martinez-Ledesma, E.; Lee, S.H.; Zheng, S.; Verhaak, R.G.W. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 2018, 46, D1144–D1149. [Google Scholar] [CrossRef]
- Amin, S.M.; Haugh, A.M.; Lee, C.Y.; Zhang, B.; Bubley, J.A.; Merkel, E.A.; Verzi, A.E.; Gerami, P. A comparison of morphologic and molecular features of BRAF, ALK, and NTRK1 fusion Spitzoid Neoplasms. Am. J. Pathol. 2017, 41, 491–498. [Google Scholar] [CrossRef]
- De la Fouchardière, A.; Tee, M.K.; Peternel, S.; Valdebran, M.; Pissaloux, D.; Tirode, F.; Busam, K.J.; LeBoit, P.E.; McCalmont, T.H.; Bastian, B.C.; Yeh, I. Fusion partners of NTRK3 affect subcellular localization of the fusion kinase and cytomorphology of melanocytes. Mod. Pathol. 2021, 34, 735–747. [Google Scholar] [CrossRef]
- Batra, U.; Nathany, S.; Sharma, M.; Pasricha, S.; Bansal, A.; Jain, P.; Mehta, A. IHC versus FISH versus NGS to detect ALK gene rearrangement in NSCLC: all questions answered? J. Clin. Pathol. 2022, 75, 405–409. [Google Scholar] [CrossRef]
- Mangolini, A.; Rocca, C.; Bassi, C.; Ippolito, C.; Negrini, M.; Dell'Atti, L.; Lanza, G.; Gafà, R.; Bianchi, N.; Pinton, P.; Aguiari, G. Detection of disease-causing mutations in prostate cancer by NGS sequencing. Cell Biol. Int. 2022, 46, 1047–1061. [Google Scholar] [CrossRef]
- Wagener-Ryczek, S.; Pappesch, R. Targeted RNA-sequencing for the evaluation of gene fusions in lung tumors: current status and future prospects. Expert Rev. Mol. Diagn. 2021, 21, 531–534. [Google Scholar] [CrossRef]
- Jin, Z.; Huang, W.; Shen, N.; Li, J.; Wang, X.; Dong, J.; Park, P.J.; Xi, R. Single-cell gene fusion detection by scFusion. Nat. Commun. 2022, 13, 1084. [Google Scholar] [CrossRef] [PubMed]
- Hantschel, O.; Superti-Furga, G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004, 5, 33–44. [Google Scholar] [CrossRef]
- Carofiglio, F.; Trisciuzzi, D.; Gambacorta, N.; Leonetti, F.; Stefanachi, A.; Nicolotti, O. Bcr-Abl allosteric inhibitors: Where we are and Where we are going to. Molecules 2020, 25, 4210. [Google Scholar] [CrossRef] [PubMed]
- Kargbo, R.B. Breakthrough in degradation of BCR-ABL fusion protein for the treatment of cancer. ACS Medicinal Chem. Lett. 2020, 11, 2359–2360. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.Q.; Gao, F.H. Regulatory molecules and corresponding processes of BCR-ABL protein degradation. J. Cancer 2019, 10, 2488–2500. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Yang, Y.; Gupta, P.; Wang, A.; Zhao, M.; Zhao, Y.; Qu, M.; Ke, Y.; Liu, Y.; Liu, H.M.; Xu, X.; Sun, Y.; Chen, Z.S.; Hu, Z. A small molecule inhibitor, OGP46, is effective against Imatinib-resistant BCR-ABL mutations via the BCR-ABL/JAK-STAT pathway. Mol. Ther. Oncolytics. 2020, 18, 137–148. [Google Scholar] [CrossRef]
- Chandrasekhar, C.; Kumar, P.S.; Sarma, P. Novel mutations in the kinase domain of BCR-ABL gene causing Imatinib resistance in chronic myeloid leukemia patients. Sci. Rep. 2019, 9, 2412. [Google Scholar] [CrossRef]
- Hochhaus, A. Management of Bcr-Abl-positive leukemias with Dasatinib. Expert Rev. Anticancer Ther. 2007, 7, 1529–1536. [Google Scholar] [CrossRef]
- Talpaz, M.; Shah, N.P.; Kantarjian, H.; Donato, N.; Nicoll, J.; Paquette, R.; Cortes, J.; O'Brien, S.; Nicaise, C.; Bleickardt, E.; Blackwood-Chirchir, M.A.; Iyer, V.; Chen, T.T.; Huang, F.; Decillis, A.P.; Sawyers, C.L. Dasatinib in Imatinib-resistant Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 2006, 354, 2531–2541. [Google Scholar] [CrossRef]
- Müller, M.C.; Erben, P.; Schenk, T.; Lauber, S.; Kruth, J.; Hoffmann, J.; Ernst, T.; Lahaye, T.; Metzgeroth, G.; Acevedo, M.; Nicaise, C.; Hehlmann, R.; Hochhaus, A. Response to Dasatinib after Imatinib Failure According to Type of Preexisting BCR-ABL Mutations. Blood 2006, 108, 748. [Google Scholar] [CrossRef]
- Krijanovski, Y.; Donato, N.; Cardama, A.Q.; Cortes, J.; Talpaz, M. Dasatinib resistance in CML patients. Identification of novel BCR-ABL kinase domain mutation. Blood 2007, 110, 1957. [Google Scholar] [CrossRef]
- Makiko, M.; Yoko, N.; Itaru, K.; Satoshi, S.; Hidefumi, H.; Yasuhiko, K.; Toshio, H.; Tatsutoshi, N.; Souichi, A. Dasatinib induces autophagy in mice with Bcr-Abl-positive leukemia. Int. J. Hematol. 2017, 105, 335–340. [Google Scholar]
- Kantarjian, H.; Giles, F.; Wunderle, L.; Bhalla, K.; O'Brien, S.; Wassmann, B.; Tanaka, C.; Manley, P.; Rae, P.; Mietlowski, W.; Bochinski, K.; Hochhaus, A.; Griffin, J.D.; Hoelzer, D.; Albitar, M.; Dugan, M.; Cortes, J.; Alland, L.; Ottmann, O.G. Nilotinib in Imatinib-resistant CML and Philadelphia chromosome-positive ALL. N. Engl. J. Med. 2006, 354, 2542–2551. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.; Mohi, M.G.; Pride, Y.B.; Quinnan, L.R.; Malouf, N.A.; Podar, K.; Gesbert, F.; Iwasaki, H.; Li, S.; Van Etten, R.A.; Gu, H.; Griffin, J.D.; Neel, B.G. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell 2002, 1, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Breccia, M.; Colafigli, G.; Scalzulli, E.; Martelli, M. Asciminib: an investigational agent for the treatment of chronic myeloid leukemia. Expert Opin. Investig. Drugs 2021, 30, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Eide, C.A.; Zabriskie, M.S.; Savage Stevens, S.L.; Antelope, O.; Vellore, N.A.; Than, H.; Schultz, A.R.; Clair, P.; Bowler, A.D.; Pomicter, A.D.; Yan, D.; Senina, A.V.; Qiang, W.; Kelley, T.W.; Szankasi, P.; Heinrich, M.C.; Tyner, J.W.; Rea, D.; Cayuela, J.M.; Kim, D.W.; Tognon, C.E.; O'Hare, T.; Druker, B.J.; Deininger, M.W. Combining the Allosteric Inhibitor Asciminib with Ponatinib Suppresses Emergence of and Restores Efficacy against Highly Resistant BCR-ABL1 Mutants. Cancer Cell 2019, 36, 431–443.e435. [Google Scholar] [CrossRef] [PubMed]
- A. , E.C.; S., Z.M.; L., S.S.; Orlando, A.; A., V.N.; Hein, T.; Anna, R.S.; M., C.P.; D., B.A.; D., P.A.; Dongqing, Y.; V., S.A.; Qiang, W.; W., K.T.; Philippe, S.; C., H.M.; W., T.J.; Delphine, R.; Michel, C.J.; Wook, K.D.; E., T.C.; Thomas, O.H.; J., D.B.; W., D.M. Combining the allosteric ABL1 inhibitor Asciminib (ABL001) with Ponatinib suppresses emergence of and restores efficacy against highly resistant BCR-ABL1 compound mutants. Blood 2019, 134, 431–443.e435. [Google Scholar]
- Schoepfer, J.; Jahnke, W.; Berellini, G.; Buonamici, S.; Cotesta, S.; Cowan-Jacob, S.W.; Dodd, S.; Drueckes, P.; Fabbro, D.; Gabriel, T.; Groell, J.M.; Grotzfeld, R.M.; Hassan, A.Q.; Henry, C.; Iyer, V.; Jones, D.; Lombardo, F.; Loo, A.; Manley, P.W.; Pelle, X.; Rummel, G.; Salem, B.; Warmuth, M.; Wylie, A.A.; Zoller, T.; Marzinzik, A.L.; Furet, P. Discovery of Asciminib (ABL001), an allosteric inhibitor of the tyrosine kinase activity of BCR-ABL1. J. Med. Chem. 2018, 61, 8120–8135. [Google Scholar] [CrossRef]
- Keller, V.A.G.; Brümmendorf, T.H. Novel aspects of therapy with the dual Src and Abl kinase inhibitor Bosutinib in chronic myeloid leukemia. Expert Rev. Anticancer Ther. 2012, 12, 1121–1127. [Google Scholar] [CrossRef]
- Ducray, S.P.; Natarajan, K.; Garland, G.D.; Turner, S.D.; Egger, G. The transcriptional roles of ALK fusion proteins in tumorigenesis. Cancers (Basel) 2019, 11, 1074. [Google Scholar] [CrossRef]
- Wang, D.; Li, D.; Qin, G.; Zhang, W.; Ouyang, J.; Zhang, M.; Xie, L. The structural characterization of tumor fusion genes and proteins. Comput. Math. Methods Med. 2015, 2015, 912742. [Google Scholar] [CrossRef] [PubMed]
- Rikova, K.; Guo, A.; Zeng, Q.; Possemato, A.; Yu, J.; Haack, H.; Nardone, J.; Lee, K.; Reeves, C.; Li, Y.; Hu, Y.; Tan, Z.; Stokes, M.; Sullivan, L.; Mitchell, J.; Wetzel, R.; Macneill, J.; Ren, J.M.; Yuan, J.; Bakalarski, C.E.; Villen, J.; Kornhauser, J.M.; Smith, B.; Li, D.; Zhou, X.; Gygi, S.P.; Gu, T.L.; Polakiewicz, R.D.; Rush, J.; Comb, M.J. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007, 131, 1190–1203. [Google Scholar] [CrossRef]
- Takeuchi, K.; Choi, Y.L.; Togashi, Y.; Soda, M.; Hatano, S.; Inamura, K.; Takada, S.; Ueno, T.; Yamashita, Y.; Satoh, Y.; Okumura, S.; Nakagawa, K.; Ishikawa, Y.; Mano, H. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin. Cancer Res. 2009, 15, 3143–3149. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.W.; Kirstein, M.N.; Valentine, M.B.; Dittmer, K.; Shapiro, D.N.; Look, A.T.; Saltman, D.L. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 1995, 267, 316–317. [Google Scholar] [CrossRef]
- Lamant, L.; Dastugue, N.; Pulford, K.; Delsol, G.; Mariamé, B. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood 1999, 93, 3088–3095. [Google Scholar] [CrossRef]
- Qin, Z.; Sun, H.; Yue, M.; Pan, X.; Chen, L.; Feng, X.; Yan, X.; Zhu, X.; Ji, H. Phase separation of EML4-ALK in firing downstream signaling and promoting lung tumorigenesis. Cell Discov. 2021, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, B.; Palmer, R.H. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat. Rev. Cancer 2013, 13, 685–700. [Google Scholar] [CrossRef]
- Rotow, J.; Bivona, T.G. Understanding and targeting resistance mechanisms in NSCLC. Nat. Rev. Cancer 2017, 17, 637–658. [Google Scholar] [CrossRef]
- Toyokawa, G.; Takenoyama, M.; Taguchi, K.; Toyozawa, R.; Inamasu, E.; Kojo, M.; Shiraishi, Y.; Morodomi, Y.; Takenaka, T.; Hirai, F.; Yamaguchi, M.; Seto, T.; Shimokawa, M.; Ichinose, Y. An extremely rare case of small-cell lung cancer harboring variant 2 of the EML4-ALK fusion gene. Lung Cancer 2013, 81, 487–490. [Google Scholar] [CrossRef]
- Li, T.; Zhang, F.; Wu, Z.; Cui, L.; Zhao, X.; Wang, J.; Hu, Y. PLEKHM2-ALK: A novel fusion in small-cell lung cancer and durable response to ALK inhibitors. Lung Cancer 2020, 139, 146–150. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Crizotinib resistance: implications for therapeutic strategies. Ann. Oncol. 2016, 27 Suppl 3, iii42–iii50. [Google Scholar] [CrossRef]
- Timm, A.; Kolesar, J.M. Crizotinib for the treatment of non-small-cell lung cancer. Am. J. Health-Syst. Pharm. 2013, 70, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.M.; Maher, V.E.; Bijwaard, K.E.; Becker, R.L.; Zhang, L.; Tang, S.W.; Song, P.; Liu, Q.; Marathe, A.; Gehrke, B.; Helms, W.; Hanner, D.; Justice, R.; Pazdur, R.U.S. Food and Drug Administration approval: Crizotinib for treatment of advanced or metastatic non-small cell lung cancer that is anaplastic lymphoma kinase positive. Clin. Cancer Res. 2014, 20, 2029–2034. [Google Scholar] [CrossRef]
- Katayama, R.; Shaw, A.; Khan, T.; Mino-Kenudson, M.; Solomon, B.; Halmos, B.; Jessop, N.; Wain, J.; Yeo, A.; Benes, C.; Drew, L.; Saeh, J.; Crosby, K.; Sequist, L.; Iafrate, A.; Engelman, J. Mechanisms of acquired Crizotinib resistance in ALK-rearranged lung Cancers. Sci. Transl. Med. 2012, 4, 120ra117. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.L.; Soda, M.; Yamashita, Y.; Ueno, T.; Takashima, J.; Nakajima, T.; Yatabe, Y.; Takeuchi, K.; Hamada, T.; Haruta, H.; Ishikawa, Y.; Kimura, H.; Mitsudomi, T.; Tanio, Y.; Mano, H. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 2010, 363, 1734–1739. [Google Scholar] [PubMed]
- Doebele, R.C.; Pilling, A.B.; Aisner, D.L.; Kutateladze, T.G.; Le, A.T.; Weickhardt, A.J.; Kondo, K.L.; Linderman, D.J.; Heasley, L.E.; Franklin, W.A.; Varella-Garcia, M.; Camidge, D.R. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin. Cancer Res. 2012, 18, 1472–1482. [Google Scholar] [CrossRef]
- Sasaki, T.; Koivunen, J.; Ogino, A.; Yanagita, M.; Nikiforow, S.; Zheng, W.; Lathan, C.; Marcoux, J.P.; Du, J.; Okuda, K.; Capelletti, M.; Shimamura, T.; Ercan, D.; Stumpfova, M.; Xiao, Y.; Weremowicz, S.; Butaney, M.; Heon, S.; Wilner, K.; Christensen, J.G.; Eck, M.J.; Wong, K.K.; Lindeman, N.; Gray, N.S.; Rodig, S.J.; Jänne, P.A. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011, 71, 6051–6060. [Google Scholar] [CrossRef]
- Toyokawa, G.; Hirai, F.; Inamasu, E.; Yoshida, T.; Nosaki, K.; Takenaka, T.; Yamaguchi, M.; Seto, T.; Takenoyama, M.; Ichinose, Y. Secondary mutations at I1171 in the ALK gene confer resistance to both Crizotinib and Alectinib. J. Thorac. Oncol. 2014, 9, e86–e87. [Google Scholar] [CrossRef]
- Fang, D.D.; Zhang, B.; Gu, Q.; Lira, M.; Xu, Q.; Sun, H.; Qian, M.; Sheng, W.; Ozeck, M.; Wang, Z.; Zhang, C.; Chen, X.; Chen, K.X.; Li, J.; Chen, S.H.; Christensen, J.; Mao, M.; Chan, C.C. HIP1-ALK, a novel ALK fusion variant that responds to Crizotinib. J. Thorac. Oncol. 2014, 9, 285–294. [Google Scholar] [CrossRef]
- Shan, L.; Jiang, P.; Xu, F.; Zhang, W.; Guo, L.; Wu, J.; Zeng, Y.; Jiao, Y.; Ying, J. BIRC6-ALK, a novel fusion gene in ALK break-apart FISH-negative lung adenocarcinoma, responds to Crizotinib. J. Thorac. Oncol. 2015, 10, e37–e39. [Google Scholar] [CrossRef]
- Marsilje, T.H.; Pei, W.; Chen, B.; Lu, W.; Uno, T.; Jin, Y.; Jiang, T.; Kim, S.; Li, N.; Warmuth, M.; Sarkisova, Y.; Sun, F.; Steffy, A.; Pferdekamper, A.C.; Li, A.G.; Joseph, S.B.; Kim, Y.; Liu, B.; Tuntland, T.; Cui, X.; Gray, N.S.; Steensma, R.; Wan, Y.; Jiang, J.; Chopiuk, G.; Li, J.; Gordon, W.P.; Richmond, W.; Johnson, K.; Chang, J.; Groessl, T.; He, Y.-Q.; Phimister, A.; Aycinena, A.; Lee, C.C.; Bursulaya, B.; Karanewsky, D.S.; Seidel, H.M.; Harris, J.L.; Michellys, P.-Y. Synthesis, structure–activity relationships, and in vivo efficacy of the novel potent and selective anaplastic lymphoma kinase (ALK) inhibitor 5-Chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulfonyl)phenyl)pyrimidine-2,4-diamine (LDK378) currently in Phase 1 and Phase 2 Clinical Trials. J. Med. Chem. 2013, 56, 5675–5690. [Google Scholar]
- Shaw, A.T.; Kim, D.W.; Mehra, R.; Tan, D.S.; Felip, E.; Chow, L.Q.; Camidge, D.R.; Vansteenkiste, J.; Sharma, S.; De Pas, T.; Riely, G.J.; Solomon, B.J.; Wolf, J.; Thomas, M.; Schuler, M.; Liu, G.; Santoro, A.; Lau, Y.Y.; Goldwasser, M.; Boral, A.L.; Engelman, J.A. Ceritinib in ALK-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 370, 1189–1197. [Google Scholar] [CrossRef]
- Latif, M.; Saeed, A.; Kim, S.H. Journey of the ALK-inhibitor CH5424802 to phase II clinical trial. Arch. Pharm. Res. 2013, 36, 1051–1054. [Google Scholar] [CrossRef]
- McKeage, K. Alectinib: A Review of Its Use in Advanced ALK-Rearranged Non-Small Cell Lung Cancer. Drugs 2015, 75, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Zhu, V.W.; Schrock, A.B.; Bosemani, T.; Benn, B.S.; Ali, S.M.; Ou, S.I. Dramatic response to alectinib in a lung cancer patient with a novel VKORC1L1-ALK fusion and an acquired ALK T1151K mutation. Lung Cancer (Auckl) 2018, 9, 111–116. [Google Scholar] [CrossRef]
- Honda, K.; Kadowaki, S.; Kato, K.; Hanai, N.; Hasegawa, Y.; Yatabe, Y.; Muro, K. Durable response to the ALK inhibitor alectinib in inflammatory myofibroblastic tumor of the head and neck with a novel SQSTM1-ALK fusion: A case report. Invest. New Drugs 2019, 37, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H.; Klempner, S.J.; Greenbowe, J.R.; Azada, M.; Schrock, A.B.; Ali, S.M.; Ross, J.S.; Stephens, P.J.; Miller, V.A. Identification of a novel HIP1-ALK fusion variant in Non-Small-Cell Lung Cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to Alectinib. J. Thorac. Oncol. 2014, 9, 1821–1825. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Jiang, Y.; Jiang, W.; Wang, H.; Liu, S.; Shao, Y.; Zhao, W.; Ning, R.; Yu, Q. STRN-ALK fusion in lung adenocarcinoma with excellent response upon Alectinib treatment: A case report and literature review. Onco. Targets Ther. 2020, 13, 12515–12519. [Google Scholar] [CrossRef]
- Pan, Y.; Xiao, W.; Ye, F.; Wang, H.; Shen, Y.; Yu, X.; Han, X.; Chu, Q.; Zhou, C.; Zhang, Z.; Ren, S. Outcomes of switching from Crizotinib to Alectinib in patients with advanced non-small cell lung cancer with anaplastic lymphoma kinase fusion. Ann. Transl. Med. 2021, 9, 1014. [Google Scholar] [CrossRef]
- Ignatius Ou, S.H.; Azada, M.; Hsiang, D.J.; Herman, J.M.; Kain, T.S.; Siwak-Tapp, C.; Casey, C.; He, J.; Ali, S.M.; Klempner, S.J.; Miller, V.A. Next-generation sequencing reveals a novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to Alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on Crizotinib. J. Thorac. Oncol. 2014, 9, 549–553. [Google Scholar]
- Gettinger, S.N.; Bazhenova, L.; Salgia, R.; Langer, C.J.; Gold, K.A.; Rosell, R.; Shaw, A.T.; Weiss, G.J.; Narasimhan, N.I.; Dorer, D.J.; Rivera, V.M.; Clackson, T.; Haluska, F.G.; Camidge, D.R. Updated efficacy and safety of the ALK inhibitor AP26113 in patients (pts) with advanced malignancies, including ALK+ non-small cell lung cancer (NSCLC). J. Clin. Oncol. 2014, 32, 8047. [Google Scholar] [CrossRef]
- Li, Z.; Liu, F.; Wu, S.; Ding, S.; Chen, Y.; Liu, J. Research progress on the drug resistance of ALK kinase inhibitors. Curr. Med. Chem. 2021, 29, 2456–2475. [Google Scholar] [CrossRef] [PubMed]
- Redaelli, S.; Ceccon, M.; Zappa, M.; Sharma, G.G.; Mastini, C.; Mauri, M.; Nigoghossian, M.; Massimino, L.; Cordani, N.; Farina, F.; Piazza, R.; Gambacorti-Passerini, C.; Mologni, L. Lorlatinib treatment elicits multiple on- and off-Target mechanisms of resistance in ALK-driven cancer. Cancer Res. 2018, 78, 6866–6880. [Google Scholar] [CrossRef] [PubMed]
- Siaw, J.T.; Wan, H.; Pfeifer, K.; Rivera, V.M.; Guan, J.; Palmer, R.H.; Hallberg, B. Brigatinib, an anaplastic lymphoma kinase inhibitor, abrogates activity and growth in ALK-positive neuroblastoma cells, Drosophila and mice. Oncotarget 2016, 7, 29011–29022. [Google Scholar] [CrossRef]
- Galkin, A.V.; Melnick, J.S.; Kim, S.; Hood, T.L.; Li, N.; Li, L.; Xia, G.; Steensma, R.; Chopiuk, G.; Jiang, J.; Wan, Y.; Ding, P.; Liu, Y.; Sun, F.; Schultz, P.G.; Gray, N.S.; Warmuth, M. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 270–275. [Google Scholar] [CrossRef]
- El-Deeb, I.M.; Yoo, K.H.; Lee, S.H. ROS receptor tyrosine kinase: a new potential target for anticancer drugs. Med. Res. Rev. 2011, 31, 794–818. [Google Scholar] [CrossRef]
- Park, S.; Ahn, B.C.; Lim, S.W.; Sun, J.M.; Kim, H.R.; Hong, M.H.; Lee, S.H.; Ahn, J.S.; Park, K.; Choi, Y.; Cho, B.C.; Ahn, M.J. Characteristics and outcome of ROS1-positive Non-Small Cell Lung Cancer patients in routine clinical practice. J. Thorac. Oncol. 2018, 13, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol. Res. 2017, 121, 202–212. [Google Scholar] [CrossRef]
- Zeng, L.; Li, Y.; Xiao, L.; Xiong, Y.; Liu, L.; Jiang, W.; Heng, J.; Qu, J.; Yang, N.; Zhang, Y. Crizotinib presented with promising efficacy but for concomitant mutation in next-generation sequencing-identified ROS1-rearranged non-small-cell lung cancer. Onco. Targets Ther. 2018, 11, 6937–6945. [Google Scholar] [CrossRef]
- Davies, K.D.; Doebele, R.C. Molecular pathways: ROS1 fusion proteins in cancer. Clin. Cancer Res. 2013, 19, 4040–4045. [Google Scholar] [CrossRef]
- Rimkunas, V.M.; Crosby, K.E.; Li, D.; Hu, Y.; Kelly, M.E.; Gu, T.L.; Mack, J.S.; Silver, M.R.; Zhou, X.; Haack, H. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin. Cancer Res. 2012, 18, 4449–4457. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Jenkins, C.; Iyer, S.; Schoenfeld, A.; Keddy, C.; Davare, M.A. ROS1-dependent cancers - biology, diagnostics and therapeutics. Nat. Rev. Clin. Oncol. 2021, 18, 35–55. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, H.; de Figueiredo-Pontes, L.L.; Kobayashi, S.; Costa, D.B. Preclinical rationale for use of the clinically available multitargeted tyrosine kinase inhibitor Crizotinib in ROS1-translocated lung cancer. J. Thorac. Oncol. 2012, 7, 1086–1090. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.Y.; Li, Q.; Engstrom, L.D.; West, M.; Appleman, V.; Wong, K.A.; McTigue, M.; Deng, Y.L.; Liu, W.; Brooun, A.; Timofeevski, S.; McDonnell, S.R.; Jiang, P.; Falk, M.D.; Lappin, P.B.; Affolter, T.; Nichols, T.; Hu, W.; Lam, J.; Johnson, T.W.; Smeal, T.; Charest, A.; Fantin, V.R. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking Crizotinib-resistant ROS1 mutations. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 3493–3498. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.J.; Liu, S.V.; McCoach, C.E.; Zhu, V.W.; Tan, A.C.; Yoda, S.; Peterson, J.; Do, A.; Prutisto-Chang, K.; Dagogo-Jack, I.; Sequist, L.V.; Wirth, L.J.; Lennerz, J.K.; Hata, A.N.; Mino-Kenudson, M.; Nardi, V.; Ou, S.I.; Tan, D.S.; Gainor, J.F. Mechanisms of resistance to selective RET tyrosine kinase inhibitors in RET fusion-positive non-small-cell lung cancer. Ann. Oncol. 2020, 31, 1725–1733. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Zhou, Q. Diagnostics, therapeutics and RET inhibitor resistance for RET fusion-positive non-small cell lung cancers and future perspectives. Cancer Treat. Rev. 2021, 96, 102153. [Google Scholar] [CrossRef]
- Rolfo, C.; Ruiz, R.; Giovannetti, E.; Gil-Bazo, I.; Russo, A.; Passiglia, F.; Giallombardo, M.; Peeters, M.; Raez, L. Entrectinib: a potent new TRK, ROS1, and ALK inhibitor. Expert Opin. Investig. Drugs 2015, 24, 1493–1500. [Google Scholar] [CrossRef]
- Ardini, E.; Menichincheri, M.; Banfi, P.; Bosotti, R.; De Ponti, C.; Pulci, R.; Ballinari, D.; Ciomei, M.; Texido, G.; Degrassi, A.; Avanzi, N.; Amboldi, N.; Saccardo, M.B.; Casero, D.; Orsini, P.; Bandiera, T.; Mologni, L.; Anderson, D.; Wei, G.; Harris, J.; Vernier, J.M.; Li, G.; Felder, E.; Donati, D.; Isacchi, A.; Pesenti, E.; Magnaghi, P.; Galvani, A. Entrectinib, a Pan-TRK, ROS1, and ALK inhibitor with activity in multiple molecularly defined cancer indications. Mol. Cancer Ther. 2016, 15, 628–639. [Google Scholar] [CrossRef]
- Keddy, C.; Shinde, P.; Jones, K.; Kaech, S.; Somwar, R.; Shinde, U.; Davare, M.A. Resistance profile and structural modeling of next-generation ROS1 tyrosine kinase inhibitors. Mol. Cancer Ther. 2022, 21, 336–346. [Google Scholar] [CrossRef]
- Facchinetti, F.; Loriot, Y.; Kuo, M.S.; Mahjoubi, L.; Lacroix, L.; Planchard, D.; Besse, B.; Farace, F.; Auger, N.; Remon, J.; Scoazec, J.Y.; Andre, F.; Soria, J.C.; Friboulet, L. Crizotinib-resistant ROS1 mutations reveal a predictive kinase inhibitor sensitivity model for ROS1- and ALK-rearranged lung cancers. Clin. Cancer Res. 2016, 22, 5983–5991. [Google Scholar] [CrossRef]
- Gandara, D.R.; Li, T.; Lara, P.N.; Kelly, K.; Riess, J.W.; Redman, M.W.; Mack, P.C. Acquired resistance to targeted therapies against oncogene-driven non-small-cell lung cancer: approach to subtyping progressive disease and clinical implications. Clin. Lung Cancer 2014, 15, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Doebele, R.C.; Lin, J.J.; Nagasaka, M.; Zhu, V.W.; Gabrail, N.Y.; Bazhenova, L.; Anderson, P.M.; Solomon, B.J.; Dudek, A.Z.; Pippas, A.W.; Shirinian, M.; Baik, C.S.; Stopatschinskaja, S.; Camidge, D.R.; Cho, B.C.; Drilon, A.E. TRIDENT-1: A global, multicenter, open-label Phase II study investigating the activity of repotrectinib in advanced solid tumors harboring ROS1 or NTRK1-3 rearrangements. J. Clin. Oncol. 2020, 38, TPS9637–TPS9637. [Google Scholar] [CrossRef]
- Markham, A. Brigatinib: first global approval. Drugs 2017, 77, 1131–1135. [Google Scholar] [CrossRef] [PubMed]
- Haruko, D.; Seiji, N.; Jun, S.K.; Hiroshi, T.; Yasushi, G.; Kadoaki, O.; Ryo, T.; Masahiro, K.; Toshiaki, T.; Yoshihiro, H.; Masahiro, M.; Takaya, I.; Shingo, M.; Kiyotaka, Y.; Shogo, N.; Koichi, G. Phase II study of Brigatinib in ROS1 positive non-small cell lung cancer (NSCLC) patients previously treated with Crizotinib: Barossa cohort 2. J. Clin. Oncol. 2021, 39, 9040. [Google Scholar]
- Cai, L.; Duan, J.; Qian, L.; Wang, Z.; Wang, S.; Li, S.; Wang, C.; Zhao, J.; Zhang, X.; Bai, H.; Wang, J. ROS1 fusion mediates immunogenicity by upregulation of PD-L1 after the activation of ROS1-SHP2 signaling pathway in Non-Small Cell Lung Cancer. Front. Immunol. 2020, 11, 527750. [Google Scholar] [CrossRef] [PubMed]
- Amatu, A.; Sartore-Bianchi, A.; Siena, S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 2016, 1, e000023–e000023. [Google Scholar]
- Pekova, B.; Sykorova, V.; Mastnikova, K.; Vaclavikova, E.; Moravcova, J.; Vlcek, P.; Lastuvka, P.; Taudy, M.; Katra, R.; Bavor, P.; Kodetova, D.; Chovanec, M.; Drozenova, J.; Astl, J.; Hrabal, P.; Vcelak, J.; Bendlova, B. NTRK fusion genes in thyroid carcinomas: clinicopathological characteristics and their impacts on prognosis. Cancers (Basel) 2021, 13, 1932. [Google Scholar]
- Ni, J.; Xie, S.; Ramkissoon, S.H.; Luu, V.; Sun, Y.; Bandopadhayay, P.; Beroukhim, R.; Roberts, T.M.; Stiles, C.D.; Segal, R.A.; Ligon, K.L.; Hahn, W.C.; Zhao, J.J. Tyrosine receptor kinase B is a drug target in astrocytomas. Neuro. Oncol. 2017, 19, 22–30. [Google Scholar] [CrossRef]
- Osako, T.; Takeuchi, K.; Horii, R.; Iwase, T.; Akiyama, F. Secretory carcinoma of the breast and its histopathological mimics: value of markers for differential diagnosis. Histopathology 2013, 63, 509–519. [Google Scholar] [CrossRef]
- Vaishnavi, A.; Capelletti, M.; Le, A.T.; Kako, S.; Butaney, M.; Ercan, D.; Mahale, S.; Davies, K.D.; Aisner, D.L.; Pilling, A.B.; Berge, E.M.; Kim, J.; Sasaki, H.; Park, S.; Kryukov, G.; Garraway, L.A.; Hammerman, P.S.; Haas, J.; Andrews, S.W.; Lipson, D.; Stephens, P.J.; Miller, V.A.; Varella-Garcia, M.; Jänne, P.A.; Doebele, R.C. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat. Med. 2013, 19, 1469–1472. [Google Scholar] [CrossRef]
- Marcus, L.; Donoghue, M.; Aungst, S.; Myers, C.E.; Helms, W.S.; Shen, G.; Zhao, H.; Stephens, O.; Keegan, P.; Pazdur, R. FDA approval summary: Entrectinib for the treatment of NTRK gene fusion solid tumors. Clin. Cancer Res. 2021, 27, 928–932. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; Besse, B.; Chawla, S.P.; Bazhenova, L.; Krauss, J.C.; Chae, Y.K.; Barve, M.; Garrido-Laguna, I.; Liu, S.V.; Conkling, P.; John, T.; Fakih, M.; Sigal, D.; Loong, H.H.; Buchschacher, G.L., Jr.; Garrido, P.; Nieva, J.; Steuer, C.; Overbeck, T.R.; Bowles, D.W.; Fox, E.; Riehl, T.; Chow-Maneval, E.; Simmons, B.; Cui, N.; Johnson, A.; Eng, S.; Wilson, T.R.; Demetri, G.D. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet. Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Emiliano, C.; Maurizio, S.; Alexander, D. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar]
- Farago, A.F.; Le, L.P.; Zheng, Z.; Muzikansky, A.; Drilon, A.; Patel, M.; Bauer, T.M.; Liu, S.V.; Ou, S.-H.I.; Jackman, D.; Costa, D.B.; Multani, P.S.; Li, G.G.; Hornby, Z.; Chow-Maneval, E.; Luo, D.; Lim, J.E.; Iafrate, A.J.; Shaw, A.T. Durable clinical response to Entrectinib in NTRK1-rearranged Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2015, 10, 1670–1674. [Google Scholar] [CrossRef] [PubMed]
- Sartore-Bianchi, A.; Ardini, E.; Bosotti, R.; Amatu, A.; Valtorta, E.; Somaschini, A.; Raddrizzani, L.; Palmeri, L.; Banfi, P.; Bonazzina, E.; Misale, S.; Marrapese, G.; Leone, A.; Alzani, R.; Luo, D.; Hornby, Z.; Lim, J.; Veronese, S.; Vanzulli, A.; Bardelli, A.; Martignoni, M.; Davite, C.; Galvani, A.; Isacchi, A.; Siena, S. Sensitivity to Entrectinib associated with a novel LMNA-NTRK1 gene fusion in metastatic colorectal cancer. J. Natl. Cancer Inst. 2015, 108, djv306. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Siena, S.; Ou, S.-H.I.; Patel, M.; Ahn, M.J.; Lee, J.; Bauer, T.M.; Farago, A.F.; Wheler, J.J.; Liu, S.V.; Doebele, R.; Giannetta, L.; Cerea, G.; Marrapese, G.; Schirru, M.; Amatu, A.; Bencardino, K.; Palmeri, L.; Sartore-Bianchi, A.; Vanzulli, A.; Cresta, S.; Damian, S.; Duca, M.; Ardini, E.; Li, G.; Christiansen, J.; Kowalski, K.; Johnson, A.D.; Patel, R.; Luo, D.; Chow-Maneval, E.; Hornby, Z.; Multani, P.S.; Shaw, A.T.; De Braud, F.G. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor Entrectinib: combined results from two Phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 2017, 7, 400–409. [Google Scholar] [CrossRef]
- Drilon, A.; Li, G.; Dogan, S.; Gounder, M.; Shen, R.; Arcila, M.; Wang, L.; Hyman, D.M.; Hechtman, J.; Wei, G.; Cam, N.R.; Christiansen, J.; Luo, D.; Maneval, E.C.; Bauer, T.; Patel, M.; Liu, S.V.; Ou, S.H.I.; Farago, A.; Shaw, A.; Shoemaker, R.F.; Lim, J.; Hornby, Z.; Multani, P.; Ladanyi, M.; Berger, M.; Katabi, N.; Ghossein, R.; Ho, A.L. What hides behind the MASC: clinical response and acquired resistance to Entrectinib after ETV6-NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Ann. Oncol. 2016, 27, 920–926. [Google Scholar] [CrossRef]
- Alvarez-Breckenridge, C.; Miller, J.J.; Nayyar, N.; Gill, C.M.; Kaneb, A.; D'Andrea, M.; Le, L.P.; Lee, J.; Cheng, J.; Zheng, Z.; Butler, W.E.; Multani, P.; Chow Maneval, E.; Ha Paek, S.; Toyota, B.D.; Dias-Santagata, D.; Santagata, S.; Romero, J.; Shaw, A.T.; Farago, A.F.; Yip, S.; Cahill, D.P.; Batchelor, T.T.; Iafrate, A.J.; Brastianos, P.K. Clinical and radiographic response following targeting of BCAN-NTRK1 fusion in glioneuronal tumor. NPJ Precis. Oncol. 2017, 1, 5. [Google Scholar] [CrossRef]
- Rogers, C.; Morrissette, J.J.D.; Sussman, R.T. NTRK point mutations and their functional consequences. Cancer Genet. 2022, 262-263, 5–15. [Google Scholar] [CrossRef]
- Kummar, S.; Lassen, U.N. TRK inhibition:A new tumor-agnostic treatment strategy. Target Oncol. 2018, 13, 545–556. [Google Scholar] [CrossRef]
- Doebele, R.C.; Davis, L.E.; Vaishnavi, A.; Le, A.T.; Estrada-Bernal, A.; Keysar, S.; Jimeno, A.; Varella-Garcia, M.; Aisner, D.L.; Li, Y.; Stephens, P.J.; Morosini, D.; Tuch, B.B.; Fernandes, M.; Nanda, N.; Low, J.A. An oncogenic NTRK fusion in a patient with soft-tissue Sarcoma with response to the Tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015, 5, 1049–1057. [Google Scholar] [CrossRef]
- O'Reilly, E.M.; Hechtman, J.F. Tumour response to TRK inhibition in a patient with pancreatic adenocarcinoma harbouring an NTRK gene fusion. Ann. Oncol. 2019, 30, viii36–viii40. [Google Scholar] [CrossRef]
- Liu, F.; Wei, Y.; Zhang, H.; Jiang, J.; Zhang, P.; Chu, Q. NTRK fusion in Non-Small Cell Lung Cancer: diagnosis, therapy, and TRK inhibitor resistance. Front. Oncol. 2022, 12, 864666. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Nagasubramanian, R.; Blake, J.F.; Ku, N.; Tuch, B.B.; Ebata, K.; Smith, S.; Lauriault, V.; Kolakowski, G.R.; Brandhuber, B.J.; Larsen, P.D.; Bouhana, K.S.; Winski, S.L.; Hamor, R.; Wu, W.I.; Parker, A.; Morales, T.H.; Sullivan, F.X.; DeWolf, W.E.; Wollenberg, L.A.; Gordon, P.R.; Douglas-Lindsay, D.N.; Scaltriti, M.; Benayed, R.; Raj, S.; Hanusch, B.; Schram, A.M.; Jonsson, P.; Berger, M.F.; Hechtman, J.F.; Taylor, B.S.; Andrews, S.; Rothenberg, S.M.; Hyman, D.M. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov. 2017, 7, 963–972. [Google Scholar] [CrossRef]
- Yuekun, W.; Piaopiao, L.; Yu, W.; Wenbin, M. NTRK fusions and TRK inhibitors: potential targeted therapies for adult glioblastoma. Front. Oncol. 2020, 10, 593578. [Google Scholar]
- Krishnan, A.; Berthelet, J.; Renaud, E.; Rosigkeit, S.; Distler, U.; Stawiski, E.; Wang, J.; Modrusan, Z.; Fiedler, M.; Bienz, M.; Tenzer, S.; Schad, A.; Roth, W.; Thiede, B.; Seshagiri, S.; Musholt, T.J.; Rajalingam, K. Proteogenomics analysis unveils a TFG-RET gene fusion and druggable targets in papillary thyroid carcinomas. Nat. Commun. 2020, 11, 2056. [Google Scholar] [CrossRef]
- Li, A.Y.; McCusker, M.G.; Russo, A.; Scilla, K.A.; Gittens, A.; Arensmeyer, K.; Mehra, R.; Adamo, V.; Rolfo, C. RET fusions in solid tumors. Cancer Treat. Rev. 2019, 81, 101911. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Ling, F.; Zhang, J.; Xiao, M.; Mao, W. A novel intergenic LSM14A-RET fusion variant in a patient with lung adenocarcinoma. J. Thorac. Oncol. 2020, 15, e52–e53. [Google Scholar] [CrossRef]
- Vodopivec, D.M.; Hu, M.I. RET kinase inhibitors for RET-altered thyroid cancers. Ther. Adv. Med. Oncol. 2022, 14, 17588359221101691. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, T.; Song, X.; Liu, B.; Wei, J. Gene fusion neoantigens: Emerging targets for cancer immunotherapy. Cancer Lett. 2021, 506, 45–54. [Google Scholar] [CrossRef]
- Takamori, S.; Matsubara, T.; Haratake, N.; Toyokawa, G.; Fujishita, T.; Toyozawa, R.; Ito, K.; Yamaguchi, M.; Taguchi, K.; Okamoto, T.; Seto, T. Targeted therapy for RET fusion lung cancer: breakthrough and unresolved issue. Front. Oncol. 2021, 11, 704084. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Kodama, K.; Takase, K.; Sugi, N.H.; Yamamoto, Y.; Iwata, M.; Tsuruoka, A. Antitumor activities of the targeted multi-tyrosine kinase inhibitor lenvatinib (E7080) against RET gene fusion-driven tumor models. Cancer Lett. 2013, 340, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shen, T.; Mooers, B.H.M.; Hilberg, F.; Wu, J. Drug resistance profiles of mutations in the RET kinase domain. Br. J. Pharmacol. 2018, 175, 3504–3515. [Google Scholar] [CrossRef]
- M. , D.L.; Estelamari, R.; Richa, D.; V., I.C. Therapeutic strategies in RET gene rearranged non-small cell lung cancer. J. Hematol. Oncol. 2021, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Terzyan, S.S.; Shen, T.; Liu, X.; Huang, Q.; Teng, P.; Zhou, M.; Hilberg, F.; Cai, J.; Mooers, B.H.M.; Wu, J. Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib. J. Biol. Chem. 2019, 294, 10428–10437. [Google Scholar] [CrossRef]
- Mologni, L.; Redaelli, S.; Morandi, A.; Plaza-Menacho, I.; Gambacorti-Passerini, C. Ponatinib is a potent inhibitor of wild-type and drug-resistant gatekeeper mutant RET kinase. Mol. Cell Endocrinol. 2013, 377, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Thein, K.Z.; Velcheti, V.; Mooers, B.H.M.; Wu, J.; Subbiah, V. Precision therapy for RET-altered cancers with RET inhibitors. Trends Cancer 2021, 7, 1074–1088. [Google Scholar] [CrossRef]
- Drilon, A.; Oxnard, G.R.; Tan, D.S.W.; Loong, H.H.F.; Johnson, M.; Gainor, J.; McCoach, C.E.; Gautschi, O.; Besse, B.; Cho, B.C.; Peled, N.; Weiss, J.; Kim, Y.J.; Ohe, Y.; Nishio, M.; Park, K.; Patel, J.; Seto, T.; Sakamoto, T.; Rosen, E.; Shah, M.H.; Barlesi, F.; Cassier, P.A.; Bazhenova, L.; De Braud, F.; Garralda, E.; Velcheti, V.; Satouchi, M.; Ohashi, K.; Pennell, N.A.; Reckamp, K.L.; Dy, G.K.; Wolf, J.; Solomon, B.; Falchook, G.; Ebata, K.; Nguyen, M.; Nair, B.; Zhu, E.Y.; Yang, L.; Huang, X.; Olek, E.; Rothenberg, S.M.; Goto, K.; Subbiah, V. Efficacy of Selpercatinib in RET fusion-positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 813–824. [Google Scholar] [CrossRef]
- Bradford, D.; Larkins, E.; Mushti, S.L.; Rodriguez, L.; Skinner, A.M.; Helms, W.S.; Price, L.S.L.; Zirkelbach, J.F.; Li, Y.; Liu, J.; Charlab, R.; Turcu, F.R.; Liang, D.; Ghosh, S.; Roscoe, D.; Philip, R.; Zack-Taylor, A.; Tang, S.; Kluetz, P.G.; Beaver, J.A.; Pazdur, R.; Theoret, M.R.; Singh, H. FDA approval summary: Selpercatinib for the treatment of lung and thyroid cancers with RET gene mutations or fusions. Clin. Cancer Res. 2021, 27, 2130–2135. [Google Scholar] [CrossRef]
- Anthony, M. Selpercatinib: First Approval. Drugs 2020, 80, 1119–1124. [Google Scholar]
- Loong, H.; Goto, K.; Park, K.; Ohe, Y.; Nishio, M.; Cho, B.C.; Kim, Y.J.; French, P.; Soldatenkova, V.; Tan, D. FP14.10 efficacy and safety of Selpercatinib (LOXO-292) in East Asian patients with RET fusion-positive NSCLC. J. Thorac. Oncol. 2021, 16, S231–S232. [Google Scholar] [CrossRef]
- Wirth, L.J.; Sherman, E.J.; Robinson, B.; Solomon, B.; Kang, H.; Lorch, J.H.; Worden, F.; Brose, M.S.; Leboulleux, S.; Godbert, Y.; Meurer, M.; Morris, J.; Owonikoko, T.K.; Shao-Weng Tan, D.; Gautschi, O.; Patel, J.; Yang, L.; Kherani, J.; Cabanillas, M.E.; Shah, M.H. O10-3 Selpercatinib (LOXO-292) in patients (pts) with RET-altered thyroid cancer. Ann. Oncol. 2021, 32, S289. [Google Scholar] [CrossRef]
- Ali, F.; Neha, K.; Chauhan, G. Pralsetinib: chemical and therapeutic development with FDA authorization for the management of RET fusion-positive non-small-cell lung cancers. Arch. Pharm. Res. 2022, 45, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Syed, Y.Y. Pralsetinib: A Review in Advanced RET Fusion-Positive NSCLC. Drugs 2022, 82, 811–816. [Google Scholar] [CrossRef]
- Subbiah, V.; Cassier, P.A.; Siena, S.; Garralda, E.; Paz-Ares, L.; Garrido, P.; Nadal, E.; Vuky, J.; Lopes, G.; Kalemkerian, G.P.; Bowles, D.W.; Seetharam, M.; Chang, J.; Zhang, H.; Green, J.; Zalutskaya, A.; Schuler, M.; Fan, Y.; Curigliano, G. Pan-cancer efficacy of pralsetinib in patients with RET fusion-positive solid tumors from the phase 1/2 ARROW trial. Nat. Med. 2022, 28, 1640–1645. [Google Scholar] [CrossRef]
- Kim, J.; Bradford, D.; Larkins, E.; Pai-Scherf, L.H.; Chatterjee, S.; Mishra-Kalyani, P.S.; Wearne, E.; Helms, W.S.; Ayyoub, A.; Bi, Y.; Sun, J.; Charlab, R.; Liu, J.; Zhao, H.; Liang, D.; Ghosh, S.; Philip, R.; Pazdur, R.; Theoret, M.R.; Beaver, J.A.; Singh, H. FDA approval summary: Pralsetinib for the treatment of lung and thyroid cancers with RET gene mutations or fusions. Clin. Cancer Res. 2021, 27, 5452–5456. [Google Scholar] [CrossRef]
- Shabbir, A.; Kojadinovic, A.; Shafiq, T.; Mundi, P.S. Targeting RET alterations in cancer: recent progress and future directions. Crit. Rev. Oncol. Hematol. 2023, 181, 103882. [Google Scholar] [CrossRef]
- Drusbosky, L.M.; Rodriguez, E.; Dawar, R.; Ikpeazu, C.V. Therapeutic strategies in RET gene rearranged non-small cell lung cancer. J. Hematol. Oncol. 2021, 14, 50. [Google Scholar] [CrossRef]
- Di Federico, A.; Filetti, M.; Palladini, A.; Giusti, R.; Piras, M.; De Giglio, A.; Ardizzoni, A.; Gelsomino, F. EGFR-RAD51 gene fusion NSCLC responsiveness to different generation EGFR-TKIs: two cases and review of the literature. Transl. Lung Cancer Res. 2022, 11, 497–503. [Google Scholar] [CrossRef]
- Komuro, A.; Raja, E.; Iwata, C.; Soda, M.; Isogaya, K.; Yuki, K.; Ino, Y.; Morikawa, M.; Todo, T.; Aburatani, H.; Suzuki, H.; Ranjit, M.; Natsume, A.; Mukasa, A.; Saito, N.; Okada, H.; Mano, H.; Miyazono, K.; Koinuma, D. Identification of a novel fusion gene HMGA2-EGFR in glioblastoma. Int. J. Cancer 2018, 142, 1627–1639. [Google Scholar] [CrossRef]
- Wang, X.; Huang, L.; Cai, J.; Liu, A. A novel KIF5B-EGFR fusion variant in Non-Small-Cell Lung Cancer and response to Afatinib: A case report. Onco. Targets Ther. 2021, 14, 3739–3744. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.B.; Chen, X.; Yang, X.; Ye, Y.; Bekaii-Saab, T.; Zheng, Y.; Zhang, Y. A Rare EGFR-SEPT14 Fusion in a Patient with Colorectal Adenocarcinoma Responding to Erlotinib. Oncologist 2020, 25, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Kartik, K.; Jean-Nicolas, G.; Kwang, C.Y.; J, G.F.; J, G.B.; Kyle, G.; Eiki, I.; K, O.T.; Vijay, P.; S, R.S.; K, R.S.; Beth, E.-S.; Tiziana, V.; Andrew, W.; Heidi, C.; Yingjun, Y.; H, S.J.; Jens, M.; Deborah, M.; S, R.J.; J, S.P.; A, M.V.; M, A.S.; M, L.C. EGFR fusions as novel therapeutic targets in lung cancer. Cancer Discov. 2016, 6, 601–611. [Google Scholar]
- Paik, P.K. Something old, Something new, Something borrowed, Something fused: novel EGFR rearrangements in lung adenocarcinomas. Cancer Discov. 2016, 6, 574–575. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Lazertinib: First Approval. Drugs 2021, 81, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Shao, C. KIF5B-EGFR fusion: a novel EGFR mutation in lung adenocarcinoma. Onco. Targets Ther. 2020, 13, 8317–8321. [Google Scholar] [CrossRef] [PubMed]
- Zhu, V.W.; Klempner, S.J.; Ou, S.I. Receptor Tyrosine Kinase Fusions as an Actionable Resistance Mechanism to EGFR TKIs in EGFR-Mutant Non-Small-Cell Lung Cancer. Trends Cancer 2019, 5, 677–692. [Google Scholar] [CrossRef]
- Xia, W.; Weiwei, P.; Zhimin, Z.; Jing, C.; Anwen, L. Emerging a novel VOPP1-EGFR fusion coexistent with T790M as an acquired resistance mechanism to prior Icotinib and sensitive to osimertinib in a patient With EGFR L858R lung adenocarcinoma: a case report. Front. Oncol. 2021, 11, 720819. [Google Scholar]
- Heppner, D.E.; Wittlinger, F.; Beyett, T.S.; Shaurova, T.; Urul, D.A.; Buckley, B.; Pham, C.D.; Schaeffner, I.K.; Yang, B.; Ogboo, B.C.; May, E.W.; Schaefer, E.M.; Eck, M.J.; Laufer, S.A.; Hershberger, P.A. Structural basis for inhibition of mutant EGFR with Lazertinib (YH25448). ACS Med. Chem. Lett. 2022, 13, 1856–1863. [Google Scholar] [CrossRef]
- Jones, D.T.; Kocialkowski, S.; Liu, L.; Pearson, D.M.; Bäcklund, L.M.; Ichimura, K.; Collins, V.P. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008, 68, 8673–8677. [Google Scholar] [CrossRef]
- Dahiya, S.; Yu, J.; Kaul, A.; Leonard, J.R.; Gutmann, D.H. Novel BRAF alteration in a Sporadic Pilocytic Astrocytoma. Case Rep. Med. 2012, 2012, 418672. [Google Scholar] [CrossRef]
- Cin, H.; Meyer, C.; Herr, R.; Janzarik, W.G.; Lambert, S.; Jones, D.T.; Jacob, K.; Benner, A.; Witt, H.; Remke, M.; Bender, S.; Falkenstein, F.; Van Anh, T.N.; Olbrich, H.; von Deimling, A.; Pekrun, A.; Kulozik, A.E.; Gnekow, A.; Scheurlen, W.; Witt, O.; Omran, H.; Jabado, N.; Collins, V.P.; Brummer, T.; Marschalek, R.; Lichter, P.; Korshunov, A.; Pfister, S.M. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011, 121, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Helgager, J.; Lidov, H.G.; Mahadevan, N.R.; Kieran, M.W.; Ligon, K.L.; Alexandrescu, S. A novel GIT2-BRAF fusion in pilocytic astrocytoma. Diagn. Pathol. 2017, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Tomić, T.T.; Olausson, J.; Wilzén, A.; Sabel, M.; Truvé, K.; Sjögren, H.; Dósa, S.; Tisell, M.; Lannering, B.; Enlund, F.; Martinsson, T.; Åman, P.; Abel, F. A new GTF2I-BRAF fusion mediating MAPK pathway activation in pilocytic astrocytoma. PLoS One 2017, 12, e0175638. [Google Scholar] [CrossRef]
- He, H.; Li, W.; Yan, P.; Bundschuh, R.; Killian, J.A.; Labanowska, J.; Brock, P.; Shen, R.; Heerema, N.A.; de la Chapelle, A. Identification of a recurrent LMO7-BRAF fusion in Papillary Thyroid Carcinoma. Thyroid 2018, 28, 748–754. [Google Scholar] [CrossRef]
- Penman, C.L.; Faulkner, C.; Lowis, S.P.; Kurian, K.M. Current understanding of BRAF alterations in diagnosis, prognosis, and therapeutic targeting in pediatric low-grade gliomas. Front. Oncol. 2015, 5, 54. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Westin, S.N.; Wang, K.; Araujo, D.; Wang, W.L.; Miller, V.A.; Ross, J.S.; Stephens, P.J.; Palmer, G.A.; Ali, S.M. Targeted therapy by combined inhibition of the RAF and mTOR kinases in malignant spindle cell neoplasm harboring the KIAA1549-BRAF fusion protein. J. Hematol. Oncol. 2014, 7, 8. [Google Scholar] [CrossRef]
- Kim, H.S.; Jung, M.; Kang, H.N.; Kim, H.; Park, C.W.; Kim, S.M.; Shin, S.J.; Kim, S.H.; Kim, S.G.; Kim, E.K.; Yun, M.R.; Zheng, Z.; Chung, K.Y.; Greenbowe, J.; Ali, S.M.; Kim, T.M.; Cho, B.C. Oncogenic BRAF fusions in mucosal melanomas activate the MAPK pathway and are sensitive to MEK/PI3K inhibition or MEK/CDK4/6 inhibition. Oncogene 2017, 36, 3334–3345. [Google Scholar] [CrossRef]
- Kulkarni, A.; Al-Hraishawi, H.; Simhadri, S.; Hirshfield, K.M.; Chen, S.; Pine, S.; Jeyamohan, C.; Sokol, L.; Ali, S.; Teo, M.L.; White, E.; Rodriguez-Rodriguez, L.; Mehnert, J.M.; Ganesan, S. BRAF fusion as a novel mechanism of acquired resistance to Vemurafenib in BRAFV600E mutant melanoma. Clin. Cancer Res. 2017, 23, 5631–5638. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Infante, J.R.; Daud, A.; Gonzalez, R.; Kefford, R.F.; Sosman, J.; Hamid, O.; Schuchter, L.; Cebon, J.; Ibrahim, N.; Kudchadkar, R.; Burris, H.A., 3rd; Falchook, G.; Algazi, A.; Lewis, K.; Long, G.V.; Puzanov, I.; Lebowitz, P.; Singh, A.; Little, S.; Sun, P.; Allred, A.; Ouellet, D.; Kim, K.B.; Patel, K.; Weber, J. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012, 367, 1694–1703. [Google Scholar] [CrossRef]
- Grimwade, D.; Lo Coco, F. Acute promyelocytic leukemia: a model for the role of molecular diagnosis and residual disease monitoring in directing treatment approach in acute myeloid leukemia. Leukemia 2002, 16, 1959–1973. [Google Scholar] [CrossRef] [PubMed]
- Nowak, D.; Stewart, D.; Koeffler, H.P. Differentiation therapy of leukemia: 3 decades of development. Blood 2009, 113, 3655–3665. [Google Scholar] [CrossRef]
- Sirulnik, A.; Melnick, A.; Zelent, A.; Licht, J.D. Molecular pathogenesis of acute promyelocytic leukaemia and APL variants. Best Pract. Res. Clin. Haematol. 2003, 16, 387–408. [Google Scholar] [CrossRef] [PubMed]
- De Braekeleer, E.; Douet-Guilbert, N.; De Braekeleer, M. RARA fusion genes in acute promyelocytic leukemia: a review. Expert Rev. Hematol. 2014, 7, 347–357. [Google Scholar] [CrossRef]
- Lu, J.; Chew, E.H.; Holmgren, A. Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 12288–12293. [Google Scholar] [CrossRef]
- Huang, M.E.; Ye, Y.C.; Chen, S.R.; Chai, J.R.; Lu, J.X.; Zhoa, L.; Gu, L.J.; Wang, Z.Y. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988, 72, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, P.; Chomienne, C.; Degos, L. Treatment of acute promyelocytic leukaemia. Best Pract. Res. Clin. Haematol. 2001, 14, 153–174. [Google Scholar] [CrossRef]
- Marasca, R.; Zucchini, P.; Galimberti, S.; Leonardi, G.; Vaccari, P.; Donelli, A.; Luppi, M.; Petrini, M.; Torelli, G. Missense mutations in the PML/RARalpha ligand binding domain in ATRA-resistant As(2)O(3) sensitive relapsed acute promyelocytic leukemia. Haematologica 1999, 84, 963–968. [Google Scholar]
- Lengfelder, E.; Hofmann, W.K.; Nowak, D. Impact of arsenic trioxide in the treatment of acute promyelocytic leukemia. Leukemia 2012, 26, 433–442. [Google Scholar] [CrossRef]
- Strehl, S.; König, M.; Boztug, H.; Cooper, B.W.; Suzukawa, K.; Zhang, S.J.; Chen, H.Y.; Attarbaschi, A.; Dworzak, M.N. All-trans retinoic acid and arsenic trioxide resistance of acute promyelocytic leukemia with the variant STAT5B-RARA fusion gene. Leukemia 2013, 27, 1606–1610. [Google Scholar] [CrossRef]
- Mistry, A.R.; Pedersen, E.W.; Solomon, E.; Grimwade, D. The molecular pathogenesis of acute promyelocytic leukaemia: implications for the clinical management of the disease. Blood Rev. 2003, 17, 71–97. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, L.; Naik, I.; Braunstein, Z.; Zhong, J.; Ren, B. Transcription factor C/EBP homologous protein in health and diseases. Front. Immunol. 2017, 8, 1612. [Google Scholar] [CrossRef] [PubMed]
- Zullow, H.J.; Sankar, A.; Ingram, D.R.; Same Guerra, D.D.; D'Avino, A.R.; Collings, C.K.; Lazcano, R.; Wang, W.L.; Liang, Y.; Qi, J.; Lazar, A.J.; Kadoch, C. The FUS::DDIT3 fusion oncoprotein inhibits BAF complex targeting and activity in myxoid liposarcoma. Mol. Cell 2022, 82, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- Dolatabadi, S.; Jonasson, E.; Andersson, L.; Luna Santamaria, M.; Linden, M.; Osterlund, T.; Aman, P.; Stahlberg, A. FUS-DDIT3 fusion oncoprotein expression affects JAK-STAT signaling in Myxoid Liposarcoma. Front. Oncol. 2022, 12, 816894. [Google Scholar] [CrossRef] [PubMed]
- Marcel, T.; Jasmin, M.; Christian, B.; Magdalene, C.; Ilka, I.; Konrad, S.; Sandra, E.; Inga, G.; Bianca, A.; Claudia, R.; Stefan, F.; Susanne, H.; Thomas, S.; Pierre, Å.; Eva, W.; Sebastian, H.; Wolfgang, H. FUS-DDIT3 fusion protein-driven IGF-IR signaling is a therapeutic target in myxoid liposarcoma. Clin. Cancer Res. 2017, 23, 6227–6238. [Google Scholar]
- Svec, D.; Dolatabadi, S.; Thomsen, C.; Cordes, N.; Shannon, M.; Fitzpatrick, P.; Landberg, G.; Aman, P.; Stahlberg, A. Identification of inhibitors regulating cell proliferation and FUS-DDIT3 expression in myxoid liposarcoma using combined DNA, mRNA, and protein analyses. Lab. Invest. 2018, 98, 957–967. [Google Scholar] [CrossRef]
- Schenkl, C.; Schrepper, A.; Heyne, E.; Doenst, T.; Schwarzer, M. The IGF-1R inhibitor NVP-AEW541 causes insulin-independent and reversible cardiac contractile dysfunction. Biomedicines 2022, 10, 2022. [Google Scholar] [CrossRef]
- Gasi Tandefelt, D.; Boormans, J.; Hermans, K.; Trapman, J. ETS fusion genes in prostate cancer. Endocr. Relat. Cancer 2014, 21, R143–R152. [Google Scholar] [CrossRef]
- Feng, F.Y.; Brenner, J.C.; Hussain, M.; Chinnaiyan, A.M. Molecular pathways: targeting ETS gene fusions in cancer. Clin. Cancer Res. 2014, 20, 4442–4448. [Google Scholar] [CrossRef]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate cancer. Nat. Rev. Dis. Primers 2021, 7, 9. [Google Scholar] [CrossRef]
- L. , S.J.; E., R.R.; Yi-Cheng, W.; R., L.A.; A., A.T.; S., G.A.; Richard, A.; E., K.A.; J., H.B.; Xiaotu, M.; I., S.T.; Soheil, M. ETS family transcription factor fusions in childhood AML: distinct expression networks and clinical implications. Blood 2021, 138, 2356. [Google Scholar] [CrossRef]
- Matjasic, A.; Zupan, A.; Bostjancic, E.; Pizem, J.; Popovic, M.; Kolenc, D. A novel PTPRZ1-ETV1 fusion in gliomas. Brain Pathol. 2020, 30, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Choudhary, G.; Heston, W.D.; Klein, E.A.; Almasan, A. Abstract B27: The PARP inhibitor rucaparib radiosensitizes prostate cancer cells, most effectively those that are PTEN-deficient and are expressing ETS gene fusion proteins, which inhibit NHEJ DNA repair. Cancer Res. 2012, 72. [Google Scholar] [CrossRef]
- Qian, C.; Li, D.; Chen, Y. ETS factors in prostate cancer. Cancer Lett. 2022, 530, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Katayoun, J.; Eric, M.; Lin, C.E.Y.; Lauren, O.; Chiara, T.; Eugenio, G.; James, B.; James, F.; Jeffrey, T.; Francesco, B.; J, L.B. TK216, a Novel, small molecule inhibitor of the ETS-Family of transcription factors, displays anti-tumor activity in AML and DLBCL. Blood 2016, 128, 4035. [Google Scholar]
- Ateeq, B.; Vellaichamy, A.; Tomlins, S.A.; Wang, R.; Cao, Q.; Lonigro, R.J.; Pienta, K.J.; Varambally, S. Role of dutasteride in pre-clinical ETS fusion-positive prostate cancer models. Prostate 2012, 72, 1542–1549. [Google Scholar] [CrossRef] [PubMed]
- Faivre, E.J.; Wilcox, D.; Lin, X.; Hessler, P.; Torrent, M.; He, W.; Uziel, T.; Albert, D.H.; McDaniel, K.; Kati, W.; Shen, Y. Exploitation of Castration-resistant Prostate Cancer transcription factor dependencies by the novel BET inhibitor ABBV-075. Mol. Cancer Res. 2017, 15, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Nataraj, N.B.; Sekar, A.; Ghosh, S.; Bornstein, C.; Drago-Garcia, D.; Roth, L.; Romaniello, D.; Marrocco, I.; David, E.; Gilad, Y.; Lauriola, M.; Rotkopf, R.; Kimchi, A.; Haga, Y.; Tsutsumi, Y.; Mirabeau, O.; Surdez, D.; Zinovyev, A.; Delattre, O.; Kovar, H.; Amit, I.; Yarden, Y. ETS proteins bind with glucocorticoid receptors: relevance for treatment of ewing sarcoma. Cell Rep. 2019, 29, 104–117 e104. [Google Scholar] [CrossRef]
- Humtsoe, J.O.; Kim, H.S.; Jones, L.; Cevallos, J.; Boileau, P.; Kuo, F.; Morris, L.G.T.; Ha, P. Development and characterization of MYB-NFIB fusion expression in adenoid cystic carcinoma. Cancers (Basel) 2022, 14, 2263. [Google Scholar] [CrossRef]
- Wagner, V.P.; Bingle, C.D.; Bingle, L. MYB-NFIB fusion transcript in adenoid cystic carcinoma: Current state of knowledge and future directions. Crit. Rev. Oncol. Hematol. 2022, 176, 103745. [Google Scholar] [CrossRef]
- Andersson, M.K.; Aman, P.; Stenman, G. IGF2/IGF1R signaling as a therapeutic target in MYB-positive adenoid cystic carcinomas and other fusion gene-driven tumors. Cells 2019, 8, 913. [Google Scholar] [CrossRef]
- Shirazi, P.T.; Eadie, L.N.; Heatley, S.L.; Page, E.C.; Francois, M.; Hughes, T.P.; Yeung, D.; White, D.L. Exploring the oncogenic and therapeutic target potential of the MYB-TYK2 fusion gene in B-cell acute lymphoblastic leukemia. Cancer Gene Ther. 2022, 29, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.K.; Mangiapane, G.; Nevado, P.T.; Tsakaneli, A.; Carlsson, T.; Corda, G.; Nieddu, V.; Abrahamian, C.; Chayka, O.; Rai, L.; Wick, M.; Kedaigle, A.; Stenman, G.; Sala, A. ATR is a MYB regulated gene and potential therapeutic target in adenoid cystic carcinoma. Oncogenesis 2020, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Hidemasa, M.; Kenichi, Y.; Kana, N.; Yutarou, H.; Moe, H.; Yuri, I.; Yasuhiko, K.; Yusuke, S.; Yuichi, S.; Kenichi, C.; Hiroko, T.; Ai, O.; Yasuhito, N.; June, T.; Hiroo, U.; Nobutaka, K.; Daisuke, T.; Takashi, T.; Akio, T.; Satoru, M.; Manja, M.; Claudia, H.; Seishi, O.; Souichi, A. KRAS mutations frequently coexist with high-risk MLL fusions and are independent adverse prognostic factors in MLL-rearranged acute Myeloid Leukemia. Blood 2020, 136, 28–29. [Google Scholar]
- Huang, W.; Fang, K.; Chen, T.Q.; Zeng, Z.C.; Sun, Y.M.; Han, C.; Sun, L.Y.; Chen, Z.H.; Yang, Q.Q.; Pan, Q.; Luo, X.Q.; Wang, W.T.; Chen, Y.Q. CircRNA circAF4 functions as an oncogene to regulate MLL-AF4 fusion protein expression and inhibit MLL leukemia progression. J. Hematol. Oncol. 2019, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Slany, R.K. MLL fusion proteins and transcriptional control. Biochim. Biophys. Acta, Gene Regul. Mech. 2020, 1863, 194503. [Google Scholar] [CrossRef]
- Takahashi, S.; Yokoyama, A. The molecular functions of common and atypical MLL fusion protein complexes. Biochim. Biophys. Acta, Gene Regul. Mech. 2020, 1863, 194548. [Google Scholar] [CrossRef]
- Marschalek, R. The reciprocal world of MLL fusions: A personal view. Biochim. Biophys. Acta, Gene Regul. Mech. 2020, 1863, 194547. [Google Scholar] [CrossRef]
- Cantilena, S.; Gasparoli, L.; Pal, D.; Heidenreich, O.; Klusmann, J.H.; Martens, J.H.A.; Faille, A.; Warren, A.J.; Karsa, M.; Pandher, R.; Somers, K.; Williams, O.; de Boer, J. Direct targeted therapy for MLL-fusion-driven high-risk acute leukaemias. Clin. Transl. Med. 2022, 12, e933. [Google Scholar] [CrossRef]
- Yokoyama, A. Transcriptional activation by MLL fusion proteins in leukemogenesis. Exp. Hematol. 2017, 46, 21–30. [Google Scholar] [CrossRef]
- Daigle, S.R.; Olhava, E.J.; Therkelsen, C.A.; Basavapathruni, A.; Jin, L.; Boriack-Sjodin, P.A.; Allain, C.J.; Klaus, C.R.; Raimondi, A.; Scott, M.P.; Waters, N.J.; Chesworth, R.; Moyer, M.P.; Copeland, R.A.; Richon, V.M.; Pollock, R.M. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 2013, 122, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.M.; So, C.W.E. Novel therapeutic strategies for MLL-rearranged leukemias. Biochim. Biophys. Acta, Gene Regul. Mech. 2020, 1863, 194584. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Li, D.D.; Chen, X.; Wang, Y.Z.; Xu, J.J.; Jiang, Z.Y.; You, Q.D.; Guo, X.K. Proton pump inhibitors selectively suppress MLL rearranged leukemia cells via disrupting MLL1-WDR5 protein-protein interaction. Eur. J. Med. Chem. 2020, 188, 112027. [Google Scholar] [CrossRef] [PubMed]
- Kessler, L.; Wu, T.; Grembecka, J.; Cierpicki, T.; Purohit, T.; Miao, H.; Kempinska, K.; Ely, T.; Hensen, R.; Li, S.; Patricelli, M.; Li, S.; Kucharski, J.; Zhang, J.; Yao, Y.; Yu, K.; Wang, Y.; Li, L.; Ren, P.; Liu, Y. Discovery of novel menin-MLL small molecule inhibitors that display high potency and selectivity in vitro and in vivo. Eur. J. Cancer 2016, 69, S88. [Google Scholar] [CrossRef]
- Jenkins, T.W.; Downey-Kopyscinski, S.L.; Fields, J.L.; Rahme, G.J.; Colley, W.C.; Israel, M.A.; Maksimenko, A.V.; Fiering, S.N.; Kisselev, A.F. Activity of immunoproteasome inhibitor ONX-0914 in acute lymphoblastic leukemia expressing MLL-AF4 fusion protein. Sci. Rep. 2021, 11, 10883. [Google Scholar] [CrossRef]
- Sandra, C.; Nicholas, G.; Owen, W.; Jasper, d.B. Drug induced MLL fusion degradation via Hsp90 inhibition. Blood 2015, 126, 4850. [Google Scholar]
- Wachtel, M.; Schafer, B.W. PAX3-FOXO1: zooming in on an "undruggable" target. Semin. Cancer Biol. 2018, 50, 115–123. [Google Scholar] [CrossRef]
- Raze, T.; Lapouble, E.; Lacour, B.; Guissou, S.; Defachelles, A.S.; Gaspar, N.; Delattre, O.; Pierron, G.; Desandes, E. PAX-FOXO1 fusion status in children and adolescents with alveolar rhabdomyosarcoma: Impact on clinical, pathological, and survival features. Pediatr. Blood Cancer 2023, 70, e30228. [Google Scholar] [CrossRef]
- Walters, Z.S.; Villarejo-Balcells, B.; Olmos, D.; Buist, T.W.; Missiaglia, E.; Allen, R.; Al-Lazikani, B.; Garrett, M.D.; Blagg, J.; Shipley, J. JARID2 is a direct target of the PAX3-FOXO1 fusion protein and inhibits myogenic differentiation of rhabdomyosarcoma cells. Oncogene 2014, 33, 1148–1157. [Google Scholar] [CrossRef]
- Bohm, M.; Wachtel, M.; Marques, J.G.; Streiff, N.; Laubscher, D.; Nanni, P.; Mamchaoui, K.; Santoro, R.; Schafer, B.W. Helicase CHD4 is an epigenetic coregulator of PAX3-FOXO1 in alveolar rhabdomyosarcoma. J. Clin. Invest. 2016, 126, 4237–4249. [Google Scholar] [CrossRef]
- Anderson, W.J.; Fletcher, C.D.M.; Hornick, J.L. Loss of expression of YAP1 C-terminus as an ancillary marker for epithelioid hemangioendothelioma variant with YAP1-TFE3 fusion and other YAP1-related vascular neoplasms. Mod. Pathol. 2021, 34, 2036–2042. [Google Scholar] [CrossRef]
- Szulzewsky, F.; Holland, E.C.; Vasioukhin, V. YAP1 and its fusion proteins in cancer initiation, progression and therapeutic resistance. Dev. Biol. 2021, 475, 205–221. [Google Scholar] [CrossRef]
- Pajtler, K.W.; Okonechnikov, K.; Vouri, M.; Brabetz, S.; Jones, D.T.W.; Korshunov, A.; Capper, D.; Chavez, L.; Pfister, S.M.; Kool, M.; Kawauchi, D. EPEN-24. YAP1 fusion proteins mediate oncogenic activity in ependymoma via interaction with tead transcription factors. Neuro-Oncology 2018, 20, i78. [Google Scholar] [CrossRef]
- Sekine, S.; Kiyono, T.; Ryo, E.; Ogawa, R.; Wakai, S.; Ichikawa, H.; Suzuki, K.; Arai, S.; Tsuta, K.; Ishida, M.; Sasajima, Y.; Goshima, N.; Yamazaki, N.; Mori, T. Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. J. Clin. Invest. 2019, 129, 3827–3832. [Google Scholar] [CrossRef]
- Frank, S.; Pia, H.; Hua-Jun, W.; J, C.P.; Franziska, M.; Patrick, P.; Valeri, V.; Eric, H. GENE-04. The oncogenic functions of yap1-gene fusions can be inhibited by disruption of yap1-tead interaction. Neuro-Oncology 2019, 21, vi98. [Google Scholar]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; Assempour, N.; Iynkkaran, I.; Liu, Y.; Maciejewski, A.; Gale, N.; Wilson, A.; Chin, L.; Cummings, R.; Le, D.; Pon, A.; Knox, C.; Wilson, M. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic. Acids. Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Wu, W.; Haderk, F.; Bivona, T.G. Non-canonical thinking for targeting ALK-Fusion onco-proteins in lung cancer. Cancers (Basel) 2017, 9, 164. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Cai, M.; Shao, L.; Zhang, J. Targeting protein kinases degradation by PROTACs. Front. Chem. 2021, 9, 679120. [Google Scholar] [CrossRef]
- Zhou, X.; Dong, R.; Zhang, J.Y.; Zheng, X.; Sun, L.P. PROTAC: A promising technology for cancer treatment. Eur. J. Med. Chem. 2020, 203, 112539. [Google Scholar] [CrossRef]
- Liu, J.P. Novel strategies for molecular targeting to cancer. Clin. Exp. Pharmacol. Physiol. 2016, 43, 287–289. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Q.; Xu, J.; Tang, L.; Zhang, Y.; Du, F.; Zhao, Y.; Wu, X.; Li, M.; Shen, J.; Ding, R.; Cao, H.; Li, W.; Li, X.; Chen, M.; Wu, Z.; Cho, C.H.; Du, Y.; Wen, Q.; Xiao, Z. PROTACs in gastrointestinal cancers. Mol. Ther. Oncolytics 2022, 27, 204–223. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Du, Y.; Huang, L.; Cui, J.; Niu, J.; Xu, Y.; Zhu, Q. Discovery of novel potent covalent inhibitor-based EGFR degrader with excellent in vivo efficacy. Bioorg. Chem. 2022, 120, 105605. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, J.; Liu, Y.; Xia, J.; Li, Y.; Wang, Z.P.; Wei, W. PROTACs: A novel strategy for cancer therapy. Semin. Cancer Biol. 2020, 67, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pu, W.; Zheng, Q.; Ai, M.; Chen, S.; Peng, Y. Proteolysis-targeting chimeras (PROTACs) in cancer therapy. Mol. Cancer 2022, 21, 99. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Xu, R.; Yan, B.; Li, F.; Liu, H. Elevation of tumor mutation burden in ROS1-fusion lung adenocarcinoma resistant to crizotinib: A case report. Medicine 2018, 97, e13797. [Google Scholar] [CrossRef]
- Hattori, T.; Maso, L.; Araki, K.Y.; Koide, A.; Hayman, J.; Akkapeddi, P.; Bang, I.; Neel, B.G.; Koide, S. Creating MHC-restricted neoantigens with covalent inhibitors that can be targeted by immune therapy. Cancer Discov. 2023, 13, 132–145. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, E.; Bai, L.; Li, Y. Autophagy in cancer immunotherapy. Cells 2022, 11, 2996. [Google Scholar] [CrossRef]

| Fusion proteins | Cancers | Inhibitors |
|---|---|---|
| ABL fusion (Mainly BCR-ABL) |
CML, ALL | Imatinib***, Dasatinib***, Nilotinib***, Bosutinib***, Ponatinib**, Radotinib**, Asciminib**, Vamotinib*/** |
| ALK-fusion | NSCLC, IMT | Crizotinib***, Ceritinib***, Alectinib***, Lorlatinib***, Brigatinib** |
| ROS1 fusion | NSCLC and Other solid tumors | Crizotinib***, Entrectinib** |
| NTRK1/2/3 fusion | GIST, NSCLC, IFS | Entrectinib***, Larotrectinib**, Taletrectinib* |
| RET fusion | NSCLC, THCA, HCC, MTC | Lenvatinib**, Vandetanib**, Cabozantinib**, Sorafenib**, Ponatinib*, Regorafenib*, Selpe Vamotinib rcatinib**, Pralsetinib** |
| EGFR fusion | NSCLC, PAAD | Gefitinib**, Erlotinib**, Afatinib**, Osimertinib** |
| BRAF fusion | NSCLC, CRC, HCC, GIST | Sorafenib**, Vemurafenib**, Dabrafenib**, Encorafenib** |
| RARα fusion | APL | Arsenic trioxide*** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




