Submitted:
16 May 2023
Posted:
17 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
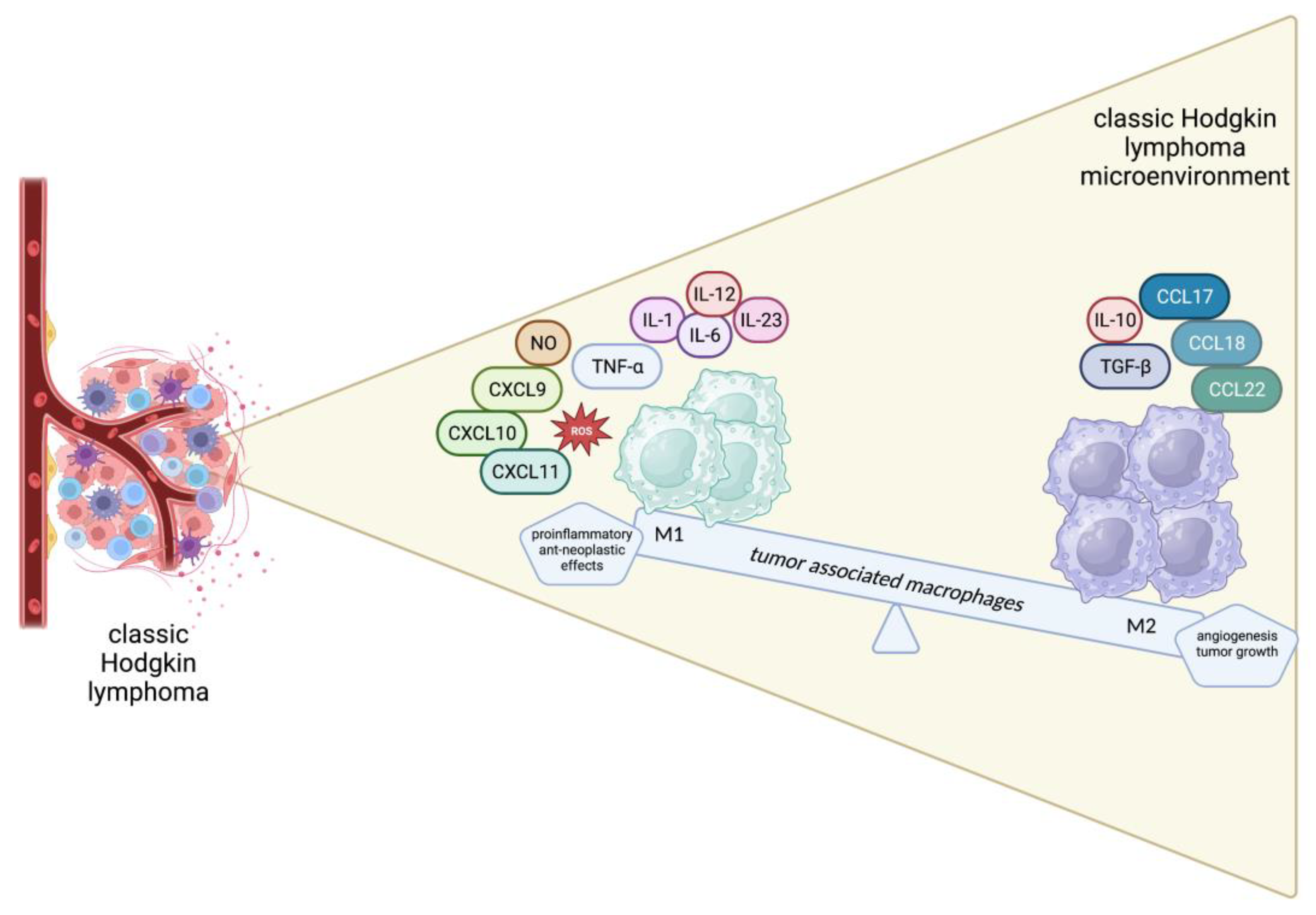
2. Overview of cellular components of TME in cHL
3. Immune Evasion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Eichenauer, D.A.; Aleman, B.M.P.; Andre, M.; Federico, M.; Hutchings, M.; Illidge, T.; Engert, A.; Ladetto, M.; Committee, E.G. Hodgkin lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018, 29, iv19–iv29. [Google Scholar] [CrossRef]
- Ansell, S.M. Hodgkin lymphoma: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol 2022, 97, 1478–1488. [Google Scholar] [CrossRef]
- Rengstl, B.; Newrzela, S.; Heinrich, T.; Weiser, C.; Thalheimer, F.B.; Schmid, F.; Warner, K.; Hartmann, S.; Schroeder, T.; Kuppers, R.; et al. Incomplete cytokinesis and re-fusion of small mononucleated Hodgkin cells lead to giant multinucleated Reed-Sternberg cells. Proc Natl Acad Sci U S A 2013, 110, 20729–20734. [Google Scholar] [CrossRef]
- Weniger, M.A.; Kuppers, R. Molecular biology of Hodgkin lymphoma. Leukemia 2021, 35, 968–981. [Google Scholar] [CrossRef]
- Satou, A.; Takahara, T.; Nakamura, S. An Update on the Pathology and Molecular Features of Hodgkin Lymphoma. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Kuppers, R.; Schwering, I.; Brauninger, A.; Rajewsky, K.; Hansmann, M.L. Biology of Hodgkin's lymphoma. Ann Oncol 2002, 13 (Suppl. 1), 11–18. [Google Scholar] [CrossRef]
- Brauninger, A.; Schmitz, R.; Bechtel, D.; Renne, C.; Hansmann, M.L.; Kuppers, R. Molecular biology of Hodgkin's and Reed/Sternberg cells in Hodgkin's lymphoma. Int J Cancer 2006, 118, 1853–1861. [Google Scholar] [CrossRef]
- Roemer, M.G.; Advani, R.H.; Ligon, A.H.; Natkunam, Y.; Redd, R.A.; Homer, H.; Connelly, C.F.; Sun, H.H.; Daadi, S.E.; Freeman, G.J.; et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J Clin Oncol 2016, 34, 2690–2697. [Google Scholar] [CrossRef]
- Skinnider, B.F.; Mak, T.W. The role of cytokines in classical Hodgkin lymphoma. Blood 2002, 99, 4283–4297. [Google Scholar] [CrossRef]
- de Charette, M.; Houot, R. Hide or defend, the two strategies of lymphoma immune evasion: potential implications for immunotherapy. Haematologica 2018, 103, 1256–1268. [Google Scholar] [CrossRef]
- Takahara, T.; Satou, A.; Tsuzuki, T.; Nakamura, S. Hodgkin Lymphoma: Biology and Differential Diagnostic Problem. Diagnostics (Basel) 2022, 12. [Google Scholar] [CrossRef]
- Alonso-Alvarez, S.; Vidriales, M.B.; Caballero, M.D.; Blanco, O.; Puig, N.; Martin, A.; Penarrubia, M.J.; Zato, E.; Galende, J.; Barez, A.; et al. The number of tumor infiltrating T-cell subsets in lymph nodes from patients with Hodgkin lymphoma is associated with the outcome after first line ABVD therapy<sup/>. Leuk Lymphoma 2017, 58, 1144–1152. [Google Scholar] [CrossRef]
- Schreck, S.; Friebel, D.; Buettner, M.; Distel, L.; Grabenbauer, G.; Young, L.S.; Niedobitek, G. Prognostic impact of tumour-infiltrating Th2 and regulatory T cells in classical Hodgkin lymphoma. Hematol Oncol 2009, 27, 31–39. [Google Scholar] [CrossRef]
- Walker, J.A.; McKenzie, A.N.J. T(H)2 cell development and function. Nat Rev Immunol 2018, 18, 121–133. [Google Scholar] [CrossRef]
- Cader, F.Z.; Schackmann, R.C.J.; Hu, X.; Wienand, K.; Redd, R.; Chapuy, B.; Ouyang, J.; Paul, N.; Gjini, E.; Lipschitz, M.; et al. Mass cytometry of Hodgkin lymphoma reveals a CD4(+) regulatory T-cell-rich and exhausted T-effector microenvironment. Blood 2018, 132, 825–836. [Google Scholar] [CrossRef]
- Greaves, P.; Clear, A.; Owen, A.; Iqbal, S.; Lee, A.; Matthews, J.; Wilson, A.; Calaminici, M.; Gribben, J.G. Defining characteristics of classical Hodgkin lymphoma microenvironment T-helper cells. Blood 2013, 122, 2856–2863. [Google Scholar] [CrossRef]
- Romagnani, S. Th1/Th2 cells. Inflamm Bowel Dis 1999, 5, 285–294. [Google Scholar] [CrossRef]
- Nishimura, T.; Nakui, M.; Sato, M.; Iwakabe, K.; Kitamura, H.; Sekimoto, M.; Ohta, A.; Koda, T.; Nishimura, S. The critical role of Th1-dominant immunity in tumor immunology. Cancer Chemother Pharmacol 2000, 46 Suppl, S52–61. [Google Scholar] [CrossRef]
- Cretney, E.; Kallies, A.; Nutt, S.L. Differentiation and function of Foxp3(+) effector regulatory T cells. Trends Immunol 2013, 34, 74–80. [Google Scholar] [CrossRef]
- Alvaro, T.; Lejeune, M.; Salvado, M.T.; Bosch, R.; Garcia, J.F.; Jaen, J.; Banham, A.H.; Roncador, G.; Montalban, C.; Piris, M.A. Outcome in Hodgkin's lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells. Clin Cancer Res 2005, 11, 1467–1473. [Google Scholar] [CrossRef]
- Dehghani, M.; Kalani, M.; Golmoghaddam, H.; Ramzi, M.; Arandi, N. Aberrant peripheral blood CD4(+) CD25(+) FOXP3(+) regulatory T cells/T helper-17 number is associated with the outcome of patients with lymphoma. Cancer Immunol Immunother 2020, 69, 1917–1928. [Google Scholar] [CrossRef]
- Ferrarini, I.; Rigo, A.; Visco, C.; Krampera, M.; Vinante, F. The Evolving Knowledge on T and NK Cells in Classic Hodgkin Lymphoma: Insights into Novel Subsets Populating the Immune Microenvironment. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef]
- Koreishi, A.F.; Saenz, A.J.; Persky, D.O.; Cui, H.; Moskowitz, A.; Moskowitz, C.H.; Teruya-Feldstein, J. The role of cytotoxic and regulatory T cells in relapsed/refractory Hodgkin lymphoma. Appl Immunohistochem Mol Morphol 2010, 18, 206–211. [Google Scholar] [CrossRef]
- Karihtala, K.; Leivonen, S.K.; Karjalainen-Lindsberg, M.L.; Chan, F.C.; Steidl, C.; Pellinen, T.; Leppa, S. Checkpoint protein expression in the tumor microenvironment defines the outcome of classical Hodgkin lymphoma patients. Blood Adv 2022, 6, 1919–1931. [Google Scholar] [CrossRef]
- Le, K.S.; Ame-Thomas, P.; Tarte, K.; Gondois-Rey, F.; Granjeaud, S.; Orlanducci, F.; Foucher, E.D.; Broussais, F.; Bouabdallah, R.; Fest, T.; et al. CXCR5 and ICOS expression identifies a CD8 T-cell subset with T(FH) features in Hodgkin lymphomas. Blood Adv 2018, 2, 1889–1900. [Google Scholar] [CrossRef]
- Gandhi, M.K.; Moll, G.; Smith, C.; Dua, U.; Lambley, E.; Ramuz, O.; Gill, D.; Marlton, P.; Seymour, J.F.; Khanna, R. Galectin-1 mediated suppression of Epstein-Barr virus specific T-cell immunity in classic Hodgkin lymphoma. Blood 2007, 110, 1326–1329. [Google Scholar] [CrossRef]
- Jachimowicz, R.D.; Pieper, L.; Reinke, S.; Gontarewicz, A.; Plutschow, A.; Haverkamp, H.; Frauenfeld, L.; Fend, F.; Overkamp, M.; Jochims, F.; et al. Whole-slide image analysis of the tumor microenvironment identifies low B-cell content as a predictor of adverse outcome in patients with advanced-stage classical Hodgkin lymphoma treated with BEACOPP. Haematologica 2021, 106, 1684–1692. [Google Scholar] [CrossRef]
- Panico, L.; Tenneriello, V.; Ronconi, F.; Lepore, M.; Cantore, N.; Dell'Angelo, A.C.; Ferbo, L.; Ferrara, F. High CD20+ background cells predict a favorable outcome in classical Hodgkin lymphoma and antagonize CD68+ macrophages. Leuk Lymphoma 2015, 56, 1636–1642. [Google Scholar] [CrossRef]
- Calabretta, E.; d'Amore, F.; Carlo-Stella, C. Immune and Inflammatory Cells of the Tumor Microenvironment Represent Novel Therapeutic Targets in Classical Hodgkin Lymphoma. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- Tudor, C.S.; Distel, L.V.; Eckhardt, J.; Hartmann, A.; Niedobitek, G.; Buettner, M. B cells in classical Hodgkin lymphoma are important actors rather than bystanders in the local immune reaction. Hum Pathol 2013, 44, 2475–2486. [Google Scholar] [CrossRef]
- Gholiha, A.R.; Hollander, P.; Hedstrom, G.; Sundstrom, C.; Molin, D.; Smedby, K.E.; Hjalgrim, H.; Glimelius, I.; Amini, R.M.; Enblad, G. High tumour plasma cell infiltration reflects an important microenvironmental component in classic Hodgkin lymphoma linked to presence of B-symptoms. Br J Haematol 2019, 184, 192–201. [Google Scholar] [CrossRef]
- Thompson, C.A.; Maurer, M.J.; Cerhan, J.R.; Katzmann, J.A.; Ansell, S.M.; Habermann, T.M.; Macon, W.R.; Weiner, G.J.; Link, B.K.; Witzig, T.E. Elevated serum free light chains are associated with inferior event free and overall survival in Hodgkin lymphoma. Am J Hematol 2011, 86, 998–1000. [Google Scholar] [CrossRef]
- Chiu, J.; Ernst, D.M.; Keating, A. Acquired Natural Killer Cell Dysfunction in the Tumor Microenvironment of Classic Hodgkin Lymphoma. Front Immunol 2018, 9, 267. [Google Scholar] [CrossRef]
- Stannard, K.A.; Lemoine, S.; Waterhouse, N.J.; Vari, F.; Chatenoud, L.; Gandhi, M.K.; Martinet, L.; Smyth, M.J.; Guillerey, C. Human peripheral blood DNAM-1(neg) NK cells are a terminally differentiated subset with limited effector functions. Blood Adv 2019, 3, 1681–1694. [Google Scholar] [CrossRef]
- Raber, P.; Ochoa, A.C.; Rodriguez, P.C. Metabolism of L-arginine by myeloid-derived suppressor cells in cancer: mechanisms of T cell suppression and therapeutic perspectives. Immunol Invest 2012, 41, 614–634. [Google Scholar] [CrossRef]
- Elliott, L.A.; Doherty, G.A.; Sheahan, K.; Ryan, E.J. Human Tumor-Infiltrating Myeloid Cells: Phenotypic and Functional Diversity. Front Immunol 2017, 8, 86. [Google Scholar] [CrossRef]
- Bertuzzi, C.; Sabattini, E.; Agostinelli, C. Immune Microenvironment Features and Dynamics in Hodgkin Lymphoma. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Romano, A.; Parrinello, N.L.; Vetro, C.; Forte, S.; Chiarenza, A.; Figuera, A.; Motta, G.; Palumbo, G.A.; Ippolito, M.; Consoli, U.; et al. Circulating myeloid-derived suppressor cells correlate with clinical outcome in Hodgkin Lymphoma patients treated up-front with a risk-adapted strategy. Br J Haematol 2015, 168, 689–700. [Google Scholar] [CrossRef]
- Romano, A.; Parrinello, N.L.; Chiarenza, A.; Motta, G.; Tibullo, D.; Giallongo, C.; La Cava, P.; Camiolo, G.; Puglisi, F.; Palumbo, G.A.; et al. Immune off-target effects of Brentuximab Vedotin in relapsed/refractory Hodgkin Lymphoma. Br J Haematol 2019, 185, 468–479. [Google Scholar] [CrossRef]
- Axdorph, U.; Porwit-MacDonald, A.; Grimfors, G.; Bjorkholm, M. Tissue eosinophilia in relation to immunopathological and clinical characteristics in Hodgkin's disease. Leuk Lymphoma 2001, 42, 1055–1065. [Google Scholar] [CrossRef]
- von Wasielewski, R.; Seth, S.; Franklin, J.; Fischer, R.; Hubner, K.; Hansmann, M.L.; Diehl, V.; Georgii, A. Tissue eosinophilia correlates strongly with poor prognosis in nodular sclerosing Hodgkin's disease, allowing for known prognostic factors. Blood 2000, 95, 1207–1213. [Google Scholar] [CrossRef]
- Masucci, M.T.; Minopoli, M.; Carriero, M.V. Tumor Associated Neutrophils. Their Role in Tumorigenesis, Metastasis, Prognosis and Therapy. Front Oncol 2019, 9, 1146. [Google Scholar] [CrossRef]
- Koh, Y.W.; Kang, H.J.; Park, C.; Yoon, D.H.; Kim, S.; Suh, C.; Kim, J.E.; Kim, C.W.; Huh, J. Prognostic significance of the ratio of absolute neutrophil count to absolute lymphocyte count in classic Hodgkin lymphoma. Am J Clin Pathol 2012, 138, 846–854. [Google Scholar] [CrossRef]
- Molin, D.; Fischer, M.; Xiang, Z.; Larsson, U.; Harvima, I.; Venge, P.; Nilsson, K.; Sundstrom, C.; Enblad, G.; Nilsson, G. Mast cells express functional CD30 ligand and are the predominant CD30L-positive cells in Hodgkin's disease. Br J Haematol 2001, 114, 616–623. [Google Scholar] [CrossRef]
- Mizuno, H.; Nakayama, T.; Miyata, Y.; Saito, S.; Nishiwaki, S.; Nakao, N.; Takeshita, K.; Naoe, T. Mast cells promote the growth of Hodgkin's lymphoma cell tumor by modifying the tumor microenvironment that can be perturbed by bortezomib. Leukemia 2012, 26, 2269–2276. [Google Scholar] [CrossRef]
- Molin, D.; Edstrom, A.; Glimelius, I.; Glimelius, B.; Nilsson, G.; Sundstrom, C.; Enblad, G. Mast cell infiltration correlates with poor prognosis in Hodgkin's lymphoma. Br J Haematol 2002, 119, 122–124. [Google Scholar] [CrossRef]
- Keresztes, K.; Szollosi, Z.; Simon, Z.; Tarkanyi, I.; Nemes, Z.; Illes, A. Retrospective analysis of the prognostic role of tissue eosinophil and mast cells in Hodgkin's lymphoma. Pathol Oncol Res 2007, 13, 237–242. [Google Scholar] [CrossRef]
- Shodell, M.; Kempin, S.; Siegal, F.P. Plasmacytoid dendritic cell and CD4 + T cell deficiencies in untreated Hodgkin disease: implications for susceptibility to opportunistic infections. Leuk Lymphoma 2014, 55, 2656–2657. [Google Scholar] [CrossRef]
- Tudor, C.S.; Bruns, H.; Daniel, C.; Distel, L.V.; Hartmann, A.; Gerbitz, A.; Buettner, M.J. Macrophages and dendritic cells as actors in the immune reaction of classical Hodgkin lymphoma. PLoS One 2014, 9, e114345. [Google Scholar] [CrossRef]
- Alavaikko, M.J.; Blanco, G.; Aine, R.; Lehtinen, T.; Fellbaum, C.; Taskinen, P.J.; Sarpola, A.; Hansmann, M.L. Follicular dendritic cells have prognostic relevance in Hodgkin's disease. Am J Clin Pathol 1994, 101, 761–767. [Google Scholar] [CrossRef]
- Galati, D.; Zanotta, S.; Corazzelli, G.; Bruzzese, D.; Capobianco, G.; Morelli, E.; Arcamone, M.; De Filippi, R.; Pinto, A. Circulating dendritic cells deficiencies as a new biomarker in classical Hodgkin lymphoma. Br J Haematol 2019, 184, 594–604. [Google Scholar] [CrossRef]
- Hourani, T.; Holden, J.A.; Li, W.; Lenzo, J.C.; Hadjigol, S.; O'Brien-Simpson, N.M. Tumor Associated Macrophages: Origin, Recruitment, Phenotypic Diversity, and Targeting. Front Oncol 2021, 11, 788365. [Google Scholar] [CrossRef]
- Cencini, E.; Fabbri, A.; Sicuranza, A.; Gozzetti, A.; Bocchia, M. The Role of Tumor-Associated Macrophages in Hematologic Malignancies. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, H.; Yang, C.; He, L.; Zhang, X.; Peng, L.; Zhu, H.; Gao, L. Role and Mechanisms of Tumor-Associated Macrophages in Hematological Malignancies. Front Oncol 2022, 12, 933666. [Google Scholar] [CrossRef]
- Carey, C.D.; Gusenleitner, D.; Lipschitz, M.; Roemer, M.G.M.; Stack, E.C.; Gjini, E.; Hu, X.; Redd, R.; Freeman, G.J.; Neuberg, D.; et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 2017, 130, 2420–2430. [Google Scholar] [CrossRef]
- Patel, S.S.; Weirather, J.L.; Lipschitz, M.; Lako, A.; Chen, P.H.; Griffin, G.K.; Armand, P.; Shipp, M.A.; Rodig, S.J. The microenvironmental niche in classic Hodgkin lymphoma is enriched for CTLA-4-positive T cells that are PD-1-negative. Blood 2019, 134, 2059–2069. [Google Scholar] [CrossRef]
- Hancic, S.; Grskovic, P.; Gasparov, S.; Ostojic Kolonic, S.; Dominis, M.; Korac, P. Macrophage Infiltration Correlates with Genomic Instability in Classic Hodgkin Lymphoma. Biomedicines 2022, 10. [Google Scholar] [CrossRef]
- Werner, L.; Dreyer, J.H.; Hartmann, D.; Barros, M.H.M.; Buttner-Herold, M.; Grittner, U.; Niedobitek, G. Tumor-associated macrophages in classical Hodgkin lymphoma: hormetic relationship to outcome. Sci Rep 2020, 10, 9410. [Google Scholar] [CrossRef]
- Steidl, C.; Lee, T.; Shah, S.P.; Farinha, P.; Han, G.; Nayar, T.; Delaney, A.; Jones, S.J.; Iqbal, J.; Weisenburger, D.D.; et al. Tumor-associated macrophages and survival in classic Hodgkin's lymphoma. N Engl J Med 2010, 362, 875–885. [Google Scholar] [CrossRef]
- Touati, M.; Delage-Corre, M.; Monteil, J.; Abraham, J.; Moreau, S.; Remenieras, L.; Gourin, M.P.; Dmytruk, N.; Olivrie, A.; Turlure, P.; et al. CD68-positive tumor-associated macrophages predict unfavorable treatment outcomes in classical Hodgkin lymphoma in correlation with interim fluorodeoxyglucose-positron emission tomography assessment. Leuk Lymphoma 2015, 56, 332–341. [Google Scholar] [CrossRef]
- Cuccaro, A.; Annunziata, S.; Cupelli, E.; Martini, M.; Calcagni, M.L.; Rufini, V.; Giachelia, M.; Bartolomei, F.; Galli, E.; D'Alo, F.; et al. CD68+ cell count, early evaluation with PET and plasma TARC levels predict response in Hodgkin lymphoma. Cancer Med 2016, 5, 398–406. [Google Scholar] [CrossRef]
- Greaves, P.; Clear, A.; Coutinho, R.; Wilson, A.; Matthews, J.; Owen, A.; Shanyinde, M.; Lister, T.A.; Calaminici, M.; Gribben, J.G. Expression of FOXP3, CD68, and CD20 at diagnosis in the microenvironment of classical Hodgkin lymphoma is predictive of outcome. J Clin Oncol 2013, 31, 256–262. [Google Scholar] [CrossRef]
- Jakovic, L.R.; Mihaljevic, B.S.; Perunicic Jovanovic, M.D.; Bogdanovic, A.D.; Andjelic, B.M.; Bumbasirevic, V.Z. The prognostic relevance of tumor associated macrophages in advanced stage classical Hodgkin lymphoma. Leuk Lymphoma 2011, 52, 1913–1919. [Google Scholar] [CrossRef]
- Mohamed, O.; El Bastawisy, A.; Allahlobi, N.; Abdellateif, M.S.; Zekri, A.R.N.; Shaarawy, S.; Korany, Z.; Mohanad, M.; Bahnassy, A.A. The role of CD68+ macrophage in classical Hodgkin lymphoma patients from Egypt. Diagn Pathol 2020, 15, 10. [Google Scholar] [CrossRef]
- Yoon, D.H.; Koh, Y.W.; Kang, H.J.; Kim, S.; Park, C.S.; Lee, S.W.; Suh, C.; Huh, J. CD68 and CD163 as prognostic factors for Korean patients with Hodgkin lymphoma. Eur J Haematol 2012, 88, 292–305. [Google Scholar] [CrossRef]
- Tan, K.L.; Scott, D.W.; Hong, F.; Kahl, B.S.; Fisher, R.I.; Bartlett, N.L.; Advani, R.H.; Buckstein, R.; Rimsza, L.M.; Connors, J.M.; et al. Tumor-associated macrophages predict inferior outcomes in classic Hodgkin lymphoma: a correlative study from the E2496 Intergroup trial. Blood 2012, 120, 3280–3287. [Google Scholar] [CrossRef]
- Kamper, P.; Bendix, K.; Hamilton-Dutoit, S.; Honore, B.; Nyengaard, J.R.; d'Amore, F. Tumor-infiltrating macrophages correlate with adverse prognosis and Epstein-Barr virus status in classical Hodgkin's lymphoma. Haematologica 2011, 96, 269–276. [Google Scholar] [CrossRef]
- Agur, A.; Amir, G.; Paltiel, O.; Klein, M.; Dann, E.J.; Goldschmidt, H.; Goldschmidt, N. CD68 staining correlates with the size of residual mass but not with survival in classical Hodgkin lymphoma. Leuk Lymphoma 2015, 56, 1315–1319. [Google Scholar] [CrossRef]
- Kayal, S.; Mathur, S.; Karak, A.K.; Kumar, L.; Sharma, A.; Bakhshi, S.; Raina, V. CD68 tumor-associated macrophage marker is not prognostic of clinical outcome in classical Hodgkin lymphoma. Leuk Lymphoma 2014, 55, 1031–1037. [Google Scholar] [CrossRef]
- Klein, J.L.; Nguyen, T.T.; Bien-Willner, G.A.; Chen, L.; Foyil, K.V.; Bartlett, N.L.; Duncavage, E.J.; Hassan, A.; Frater, J.L.; Kreisel, F. CD163 immunohistochemistry is superior to CD68 in predicting outcome in classical Hodgkin lymphoma. Am J Clin Pathol 2014, 141, 381–387. [Google Scholar] [CrossRef]
- Azambuja, D.; Natkunam, Y.; Biasoli, I.; Lossos, I.S.; Anderson, M.W.; Morais, J.C.; Spector, N. Lack of association of tumor-associated macrophages with clinical outcome in patients with classical Hodgkin's lymphoma. Ann Oncol 2012, 23, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.A.; Wada, N.; Ikeda, J.; Shibayama, H.; Hashimoto, K.; Yamagami, T.; Tatsumi, Y.; Tsukaguchi, M.; Take, H.; Tsudo, M.; et al. Prognostic implication of types of tumor-associated macrophages in Hodgkin lymphoma. Virchows Arch 2011, 459, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Gusak, A.; Fedorova, L.; Lepik, K.; Volkov, N.; Popova, M.; Moiseev, I.; Mikhailova, N.; Baykov, V.; Kulagin, A. Immunosuppressive Microenvironment and Efficacy of PD-1 Inhibitors in Relapsed/Refractory Classic Hodgkin Lymphoma: Checkpoint Molecules Landscape and Macrophage Populations. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Cen, H.; Tan, X.; Ke, Q. Meta-analysis of the prognostic and clinical value of tumor-associated macrophages in adult classical Hodgkin lymphoma. BMC Med 2016, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Karihtala, K.; Leivonen, S.K.; Bruck, O.; Karjalainen-Lindsberg, M.L.; Mustjoki, S.; Pellinen, T.; Leppa, S. Prognostic Impact of Tumor-Associated Macrophages on Survival Is Checkpoint Dependent in Classical Hodgkin Lymphoma. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef]
- Locatelli, S.L.; Careddu, G.; Serio, S.; Consonni, F.M.; Maeda, A.; Viswanadha, S.; Vakkalanka, S.; Castagna, L.; Santoro, A.; Allavena, P.; et al. Targeting Cancer Cells and Tumor Microenvironment in Preclinical and Clinical Models of Hodgkin Lymphoma Using the Dual PI3Kdelta/gamma Inhibitor RP6530. Clin Cancer Res 2019, 25, 1098–1112. [Google Scholar] [CrossRef]
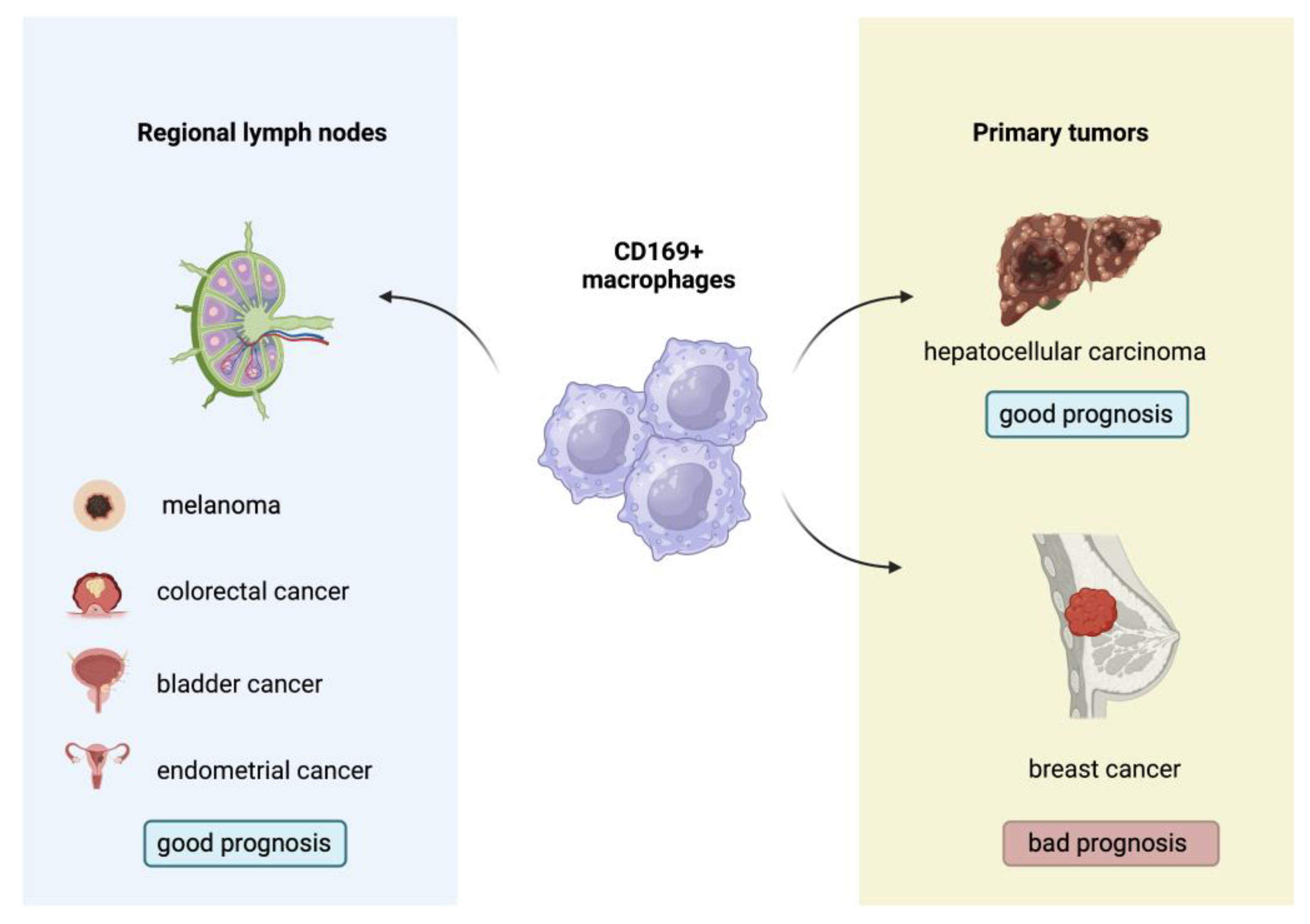
- Liu, Y.; Xia, Y.; Qiu, C.H. Functions of CD169 positive macrophages in human diseases (Review). Biomed Rep 2021, 14, 26. [Google Scholar] [CrossRef]
- Grabowska, J.; Lopez-Venegas, M.A.; Affandi, A.J.; den Haan, J.M.M. CD169(+) Macrophages Capture and Dendritic Cells Instruct: The Interplay of the Gatekeeper and the General of the Immune System. Front Immunol 2018, 9, 2472. [Google Scholar] [CrossRef]
- Affandi, A.J.; Olesek, K.; Grabowska, J.; Nijen Twilhaar, M.K.; Rodriguez, E.; Saris, A.; Zwart, E.S.; Nossent, E.J.; Kalay, H.; de Kok, M.; et al. CD169 Defines Activated CD14(+) Monocytes With Enhanced CD8(+) T Cell Activation Capacity. Front Immunol 2021, 12, 697840. [Google Scholar] [CrossRef]
- Asano, K.; Nabeyama, A.; Miyake, Y.; Qiu, C.H.; Kurita, A.; Tomura, M.; Kanagawa, O.; Fujii, S.; Tanaka, M. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity 2011, 34, 85–95. [Google Scholar] [CrossRef]
- Komohara, Y.; Ohnishi, K.; Takeya, M. Possible functions of CD169-positive sinus macrophages in lymph nodes in anti-tumor immune responses. Cancer Sci 2017, 108, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Ohnishi, K.; Shiota, T.; Motoshima, T.; Sugiyama, Y.; Yatsuda, J.; Kamba, T.; Ishizaka, K.; Komohara, Y. CD169-positive sinus macrophages in the lymph nodes determine bladder cancer prognosis. Cancer Sci 2018, 109, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.Q.; Jiang, Z.Z.; Li, L.; Wu, Y.; Zheng, L. CD169 identifies an anti-tumour macrophage subpopulation in human hepatocellular carcinoma. J Pathol 2016, 239, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Briem, O.; Kallberg, E.; Kimbung, S.; Veerla, S.; Stenstrom, J.; Hatschek, T.; Hagerling, C.; Hedenfalk, I.; Leandersson, K. CD169(+) Macrophages in Primary Breast Tumors Associate with Tertiary Lymphoid Structures, T(regs) and a Worse Prognosis for Patients with Advanced Breast Cancer. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, K.; Komohara, Y.; Saito, Y.; Miyamoto, Y.; Watanabe, M.; Baba, H.; Takeya, M. CD169-positive macrophages in regional lymph nodes are associated with a favorable prognosis in patients with colorectal carcinoma. Cancer Sci 2013, 104, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Ohnishi, K.; Miyashita, A.; Nakahara, S.; Fujiwara, Y.; Horlad, H.; Motoshima, T.; Fukushima, S.; Jinnin, M.; Ihn, H.; et al. Prognostic Significance of CD169+ Lymph Node Sinus Macrophages in Patients with Malignant Melanoma. Cancer Immunol Res 2015, 3, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, K.; Yamaguchi, M.; Erdenebaatar, C.; Saito, F.; Tashiro, H.; Katabuchi, H.; Takeya, M.; Komohara, Y. Prognostic significance of CD169-positive lymph node sinus macrophages in patients with endometrial carcinoma. Cancer Sci 2016, 107, 846–852. [Google Scholar] [CrossRef]
- Marmey, B.; Boix, C.; Barbaroux, J.B.; Dieu-Nosjean, M.C.; Diebold, J.; Audouin, J.; Fridman, W.H.; Mueller, C.G.; Molina, T.J. CD14 and CD169 expression in human lymph nodes and spleen: specific expansion of CD14+CD169- monocyte-derived cells in diffuse large B-cell lymphomas. Hum Pathol 2006, 37, 68–77. [Google Scholar] [CrossRef]
- Liu, W.R.; Shipp, M.A. Signaling pathways and immune evasion mechanisms in classical Hodgkin lymphoma. Blood 2017, 130, 2265–2270. [Google Scholar] [CrossRef]
- Kawashima, M.; Higuchi, H.; Kotani, A. Significance of trogocytosis and exosome-mediated transport in establishing and maintaining the tumor microenvironment in lymphoid malignancies. J Clin Exp Hematop 2021, 61, 192–201. [Google Scholar] [CrossRef]
- Roemer, M.G.M.; Redd, R.A.; Cader, F.Z.; Pak, C.J.; Abdelrahman, S.; Ouyang, J.; Sasse, S.; Younes, A.; Fanale, M.; Santoro, A.; et al. Major Histocompatibility Complex Class II and Programmed Death Ligand 1 Expression Predict Outcome After Programmed Death 1 Blockade in Classic Hodgkin Lymphoma. J Clin Oncol 2018, 36, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Menendez, V.; Solorzano, J.L.; Fernandez, S.; Montalban, C.; Garcia, J.F. The Hodgkin Lymphoma Immune Microenvironment: Turning Bad News into Good. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Hatic, H.; Sampat, D.; Goyal, G. Immune checkpoint inhibitors in lymphoma: challenges and opportunities. Ann Transl Med 2021, 9, 1037. [Google Scholar] [CrossRef] [PubMed]
- Vardhana, S.; Younes, A. The immune microenvironment in Hodgkin lymphoma: T cells, B cells, and immune checkpoints. Haematologica 2016, 101, 794–802. [Google Scholar] [CrossRef] [PubMed]
- El Halabi, L.; Adam, J.; Gravelle, P.; Marty, V.; Danu, A.; Lazarovici, J.; Ribrag, V.; Bosq, J.; Camara-Clayette, V.; Laurent, C.; et al. Expression of the Immune Checkpoint Regulators LAG-3 and TIM-3 in Classical Hodgkin Lymphoma. Clin Lymphoma Myeloma Leuk 2021, 21, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Abdul Razak, F.R.; Terpstra, M.; Chan, F.C.; Saber, A.; Nijland, M.; van Imhoff, G.; Visser, L.; Gascoyne, R.; Steidl, C.; et al. The mutational landscape of Hodgkin lymphoma cell lines determined by whole-exome sequencing. Leukemia 2014, 28, 2248–2251. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Schneider, S.; Zuhlke-Jenisch, R.; Klapper, W.; Sundstrom, C.; Hartmann, S.; Hansmann, M.L.; Siebert, R.; Kuppers, R.; Giefing, M. Alterations of the CD58 gene in classical Hodgkin lymphoma. Genes Chromosomes Cancer 2015, 54, 638–645. [Google Scholar] [CrossRef]
- Casagrande, N.; Borghese, C.; Aldinucci, D. Current and Emerging Approaches to Study Microenvironmental Interactions and Drug Activity in Classical Hodgkin Lymphoma. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Choe, J.Y.; Yun, J.Y.; Jeon, Y.K.; Kim, S.H.; Park, G.; Huh, J.R.; Oh, S.; Kim, J.E. Indoleamine 2,3-dioxygenase (IDO) is frequently expressed in stromal cells of Hodgkin lymphoma and is associated with adverse clinical features: a retrospective cohort study. BMC Cancer 2014, 14, 335. [Google Scholar] [CrossRef]
- Kim, M.S.; Park, T.I.; Son, S.A.; Lee, H.W. Immunohistochemical Features of Indoleamine 2,3-Dioxygenase (IDO) in Various Types of Lymphoma: A Single Center Experience. Diagnostics (Basel) 2020, 10. [Google Scholar] [CrossRef]
- Masaki, A.; Ishida, T.; Maeda, Y.; Ito, A.; Suzuki, S.; Narita, T.; Kinoshita, S.; Takino, H.; Yoshida, T.; Ri, M.; et al. Clinical significance of tryptophan catabolism in Hodgkin lymphoma. Cancer Sci 2018, 109, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Marshall, N.A.; Christie, L.E.; Munro, L.R.; Culligan, D.J.; Johnston, P.W.; Barker, R.N.; Vickers, M.A. Immunosuppressive regulatory T cells are abundant in the reactive lymphocytes of Hodgkin lymphoma. Blood 2004, 103, 1755–1762. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




