1. Introduction, Background, and Mission Space
In December 2019, a novel coronavirus disease (COVID-19) emerged in Wuhan, Hubei province, China [
1,
2,
3]. A cluster of patients with severe respiratory illnesses, such as viral pneumonia and lung failure were observed around that time. The causal agent, unidentified at that time, has since been named the “Severe Acute Respiratory Syndrome Coronavirus” - (SARS-CoV-2) virus. While initially there were few reports that person-to-person transmission was possible, it soon became evident that transmission from asymptomatic individuals or individuals having mild infection to others was observed to be possible [
4,
5,
6]. This clearly was a factor in allowing SARS-CoV-2 to be disseminated across the borders of many nations in less than six months from discovery and resulted in the global spread of the COVID-19 pandemic in 2020 [
7,
8,
9,
10]. Due to its high transmissivity, COVID-19 was declared a global pandemic by the World Health Organization (WHO) on 11 March 2020 [
11]. The causative agent, SARS-CoV-2, was confirmed via genome analysis to be a close relative of zoonotic coronaviruses, and of a prior outbreak strain, SARS-CoV, which caused an epidemic in 2003. Clinical observations of SARS-CoV-2 infections normally manifest themselves as respiratory syndromes, although there is a degree of intestinal involvement, and the most severe symptoms are interstitial pneumonia and acute respiratory distress syndrome (ARDS) [
11,
12,
13]. ARDS is considered a major driver in mortality and morbidity. Early in the outbreak, the taxonomy and language used were not finalized, and hence “Novel Coronavirus”, “2019-nCOV”, and sometimes “COVID-19” were used to describe the virus that is, now, known as SARS-nCOV-2 [
14]. For the purposes of discussion here, we will collectively refer to any coronavirus disease as COVID and will define any number of coronavirus viral strains, that may cause it. Due to the immense spread of COVID-19 in the first year of its first observation in Wuhan, the pandemic resulted in enormous human casualties and caused serious social, economic, educational, and productivity losses. The progression of the disease was well documented by several organizations.
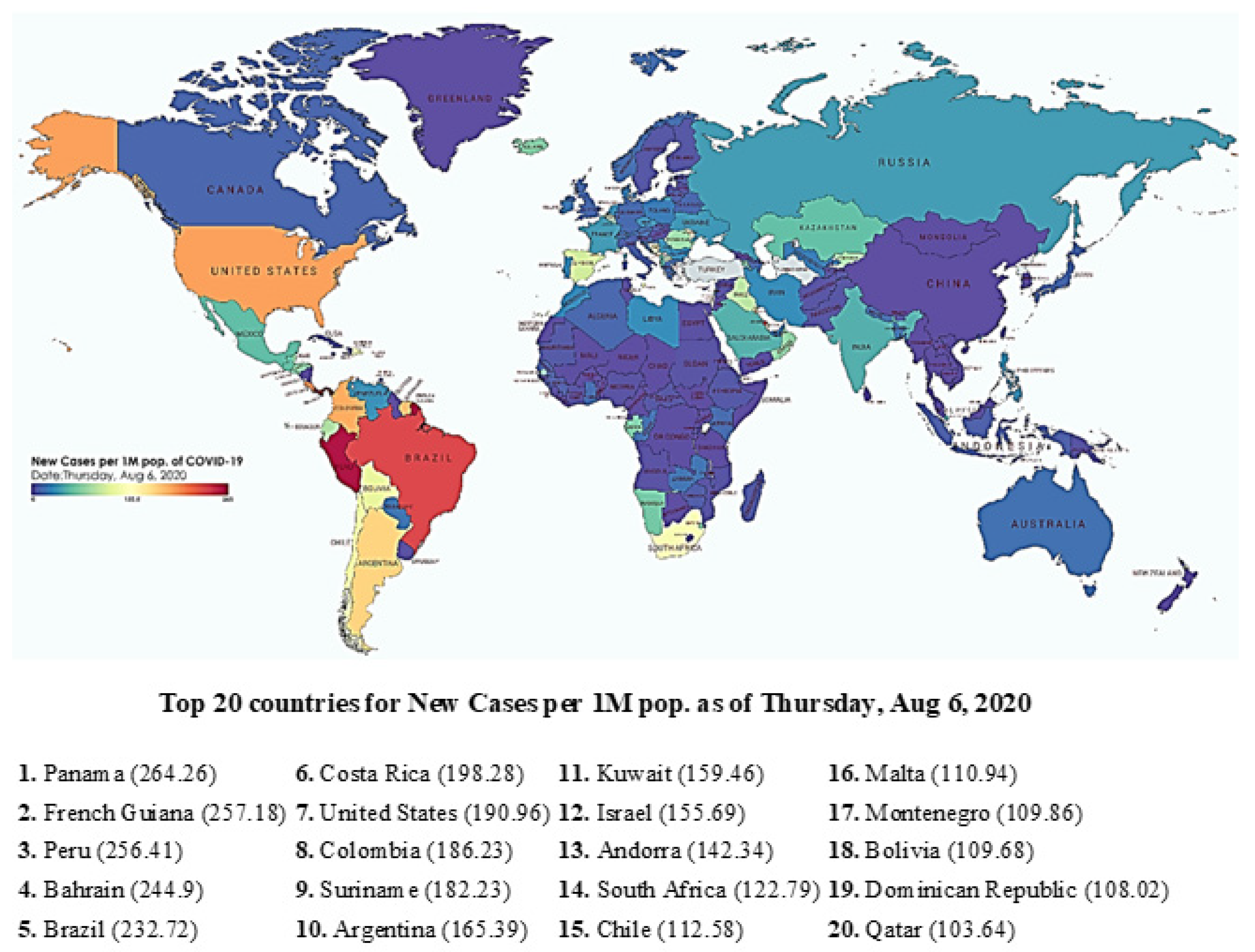
Figure 1 shows a global spread of SARS-nCOV2 recorded at the height of the peak, around mid-2020. With over 90 different variants currently reported, and second and third waves being reported in several countries, it is critical to have a holistic understanding of the ongoing situation and the development of strategies, not only to cope with the current but also the future spread of the virus. Such an approach is critical for us to be better prepared to deal with similar situations in the future.
Historical Context to Pandemics
Historically, communicable diseases have occurred during the existence of humankind, which was rendered possible by the shift to agrarian life about 10,000 years ago [
15]. There were several notable cases of pandemics, for example, Athens in 430 B.C., was gripped by a plague of unspecified etiology but possible typhoid [
16], which spread throughout Egypt, Libya, and Ethiopia during the Peloponnesian war, causing the loss of almost two-thirds of the population [
15,
17]. In 165 A.D., the Antonine plague of most probably smallpox started with the Huns. Huns then infected the Germans, who further passed the plague to the Romans and the Roman empire. This plague continued until about 180 A.D., claiming many lives including Emperor Marcus Aurelius [
15,
16,
17]. In 250 A.D., the diarrhea plague was named after the first known victim, the Christian bishop of Carthage. The Cyprian plague possibly started in Ethiopia and spread to Northern Africa, Rome, Egypt, and northward. The Cyprian plague lasted for some 20 years but a devastating impact on roman civilization [
18,
19]. In 541 A.D., the Justinian plague (Yersinia pestis) spread through Palestine, the Byzantine Empire, and then throughout the Mediterranean. It is believed to be the first significant appearance of the bubonic plague, recurring over the next two centuries and killing about 50 million people [
15].
During the 11th century, leprosy grew into a pandemic in Europe 20]. In 1347 (the black death), the second-largest outbreak of the bubonic plague possibly started in Asia and moved to the West. England and France were incapacitated by the plague, leading to a truce in their ongoing war, affecting as far as Greenland, and causing the death of about 60% Europe’s population [
15,
17].
Beginning in 1914 smallpox began to spread in Europe with a 30% mortality rate [
13,
21]. However, smallpox was lethal in the Americas. In 1492, following the arrival of the Spanish in the Caribbean, diseases such as smallpox, measles and typhus were passed along to the native populations by the Europeans [
17,
22]. Due a lack of earlier exposure, as many as 50-90% of the population died throughout the north and south continents with depopulation reaching a peak around 1780 and 1838 [
16,
21,
22,
23,
24,
25]. Through 1520, Taino and Aztec populations were also severely affected by smallpox [
17]. Smallpox would later make no less than 5 major outbreaks globally before its eradication in the 1970’s due to vaccination [
16,
21].
In 1623, the appearance of another bubonic plague led to the deaths of some 25% of selected European populations [
25]. Bubonic plague would reemerge in 1647 and again in China in 1855 and moved on to India and Hong Kong and has largely remained endemic at low levels ever since [
15,
16,
25]. It is likely that 200 million people have died of the plague since its emergence however infections have died down since the 1950’s [
26].
Cholera emerged several times from 1817 to 1886 in no less than 5 waves [
15]. Over the course of the next 150 years two more waves would occur with the 7
th still ongoing, the Cholera pandemic that started in 1817 in India, has spread globally resulting in the deaths of millions of people [
17]. In 1875 Fiji had a measles outbreak causing the loss of one-third of its population [
27]. In 1889, the Russian Flu started in Siberia and Kazakhstan and spread through Moscow, Finland, and then on to Poland, and North America, resulting in over 360,000 deaths through 1890 [
28].
In 1918, the avian-borne Spanish flu resulted in 50 million deaths worldwide. In 1957, the Asian flu started in Hong Kong and spread throughout China and then into the United States. The Asian flu became widespread in England for over six months and resulted in the death of approx. 14,000 people. The second wave of Asian flu in early 1958, caused an estimated loss of lives of about 1.1 million globally [
29].
In 1981, Human immunodeficiency virus (HIV) infection and acquired immunodeficiency syndrome (AIDS) were first identified and were believed to have developed from a chimpanzee virus, Simian (monkey-hosted) Immunodeficiency Virus (SIV) from West Africa. The disease, which spreads through certain body fluids, moved to Haiti in the 1960s [
30], and then to New York[13s] and San Francisco in the 1970s [
31]. The disease attacked and destroyed T-cells (CD4), thereby reducing the body’s immune response [
32].
In 2003, severe acute respiratory syndrome (SARS), a viral respiratory illness, was first identified, which was followed by Middle East Respiratory Syndrome (MERS), first reported in Saudi Arabia in 2012 and has since spread to several other countries, including the United States causing many casualties [
33,
34]. These two pandemics: SARS and MERS have guided health professionals through outbreak responses and lessons learned from pandemics to keep diseases such as H1N1, Ebola, and Zika under control [
35].
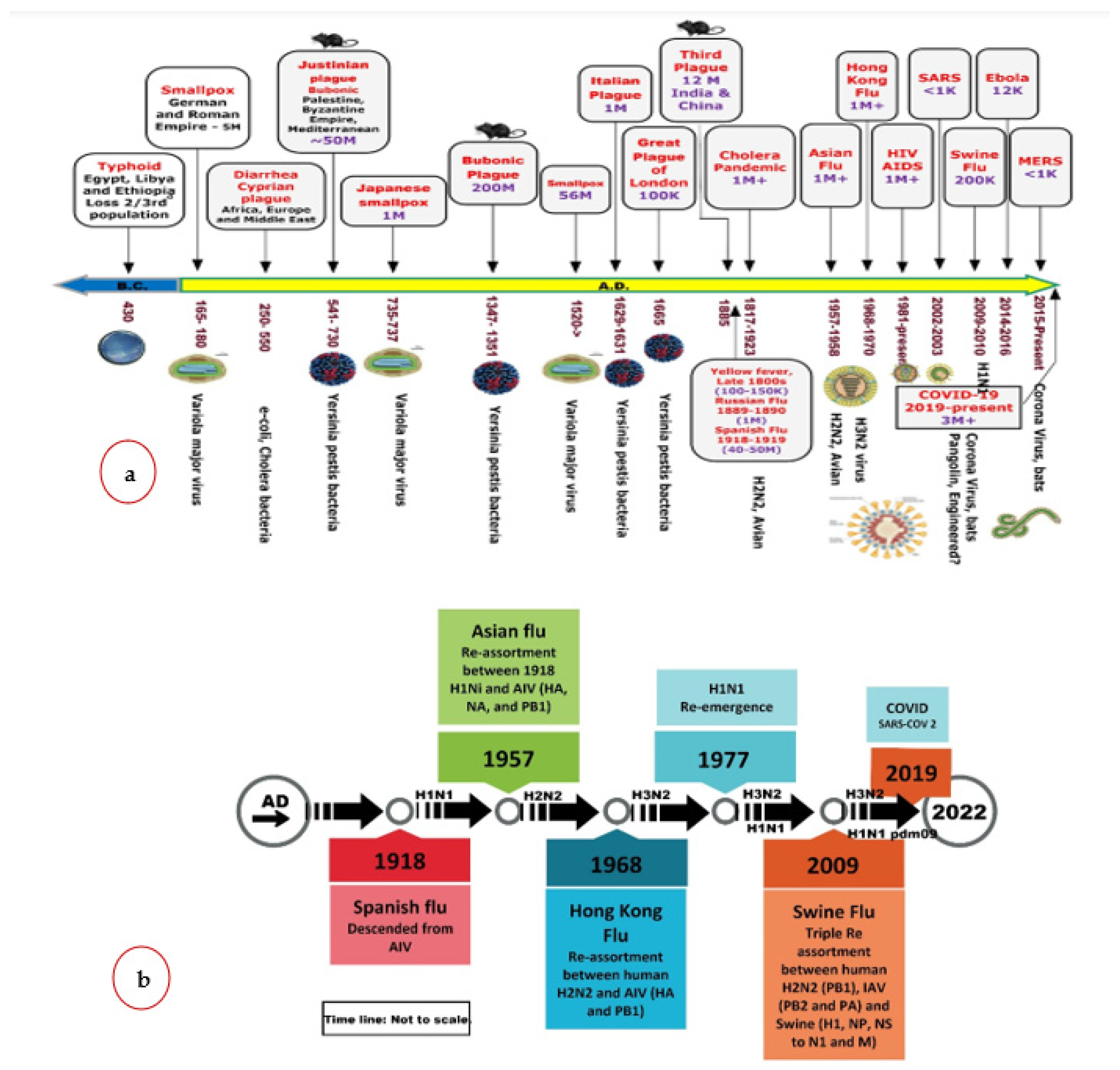
Figure 2a. shows a historical timeline of various pandemics since the records were kept until the present.
COVID-19 caused by the SARS-CoV-2 virus, which started in Wuhan, China in 2019 [
36,
37,
38], and remained very active throughout 2021 with multiple variations, remains a viable health challenge even now. In over three years duration, it has infected over 180 million people worldwide, causing a significant loss of life for over 3.5 million people. Although the impact of the spread of the virus has eased, the loss of economy, education, and productivity will take a long time to recover. Knowing what we now know, it is only prudent to take a proactive approach and learn from the lessons emerging from this pandemic, which will help guide us through future pandemics. In addition, with the use of artificial intelligence (AI) and bioinformatics tools [
39] for the detection of a wide spectrum of threat vectors, we should be able to not only detect but find remedies for unknown viruses, such as SARS-CoV-2 (or also known as (a.k.a) Coronavirus).
Figure 2b shows the evolution of the flu, while the evolution of the coronavirus is provided in the next section.
2. Overview of Coronavirus Family
Coronaviruses belong to the
Coronaviridae family, in the order
Nidovirales. This family includes two subfamilies:
Letovirinae and
Orthocoronavirinae - subfamily which includes the genera
Alphacoronvirus,
Betacoronavirus,
Gammacoronavirus, and
Deltacoronavirus.
Alphacoronaviruses and
betacoronaviruses, typically infect only mammals, whereas
gammacoronoviruses and
deltacoronaviruses, infect avian species and sometimes mammals.
Betacoronaviruses include SARS-CoV, MERS-CoV, bat coronavirus HKU4, mouse hepatitis coronavirus (MHV), bovine coronavirus (BCoV), and human coronavirus OC43 [
39,
40,
41].
2.1. Coronavirus Structure
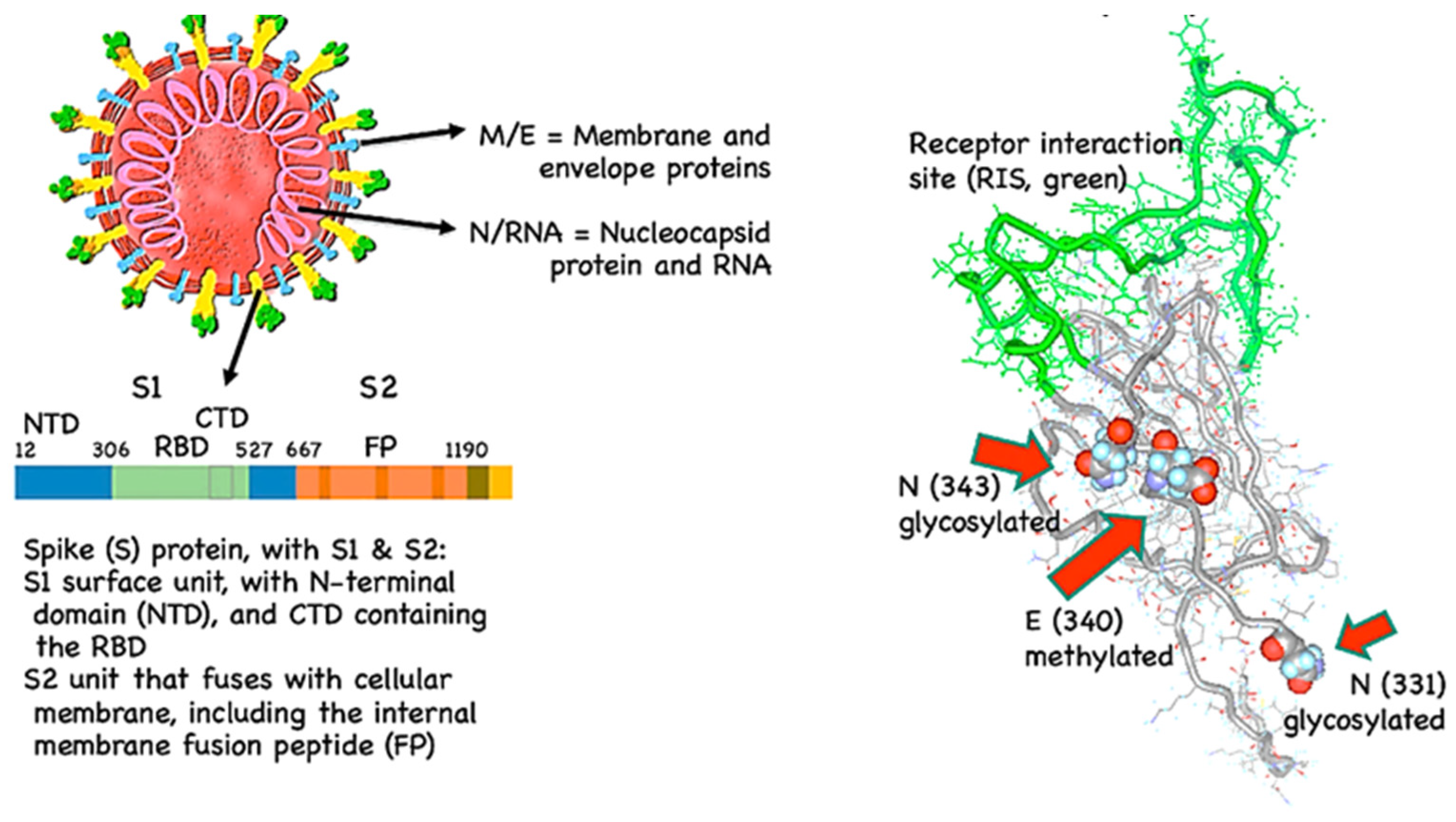
Coronaviruses are enveloped positive-sense single-stranded non-segmented RNA viruses. Their genome ranges from 27 to 32 kilobases [
42]. The genome is packed inside a helical capsid formed by the nucleocapsid protein (N) and is further surrounded by an envelope. Three structural proteins are associated with the viral envelope: Membrane protein (M), Envelope protein (E), and Spike protein (S). M and E proteins are involved in virus assembly, whereas the S protein mediates virus entry into the host cells. Some coronaviruses also encode an envelope-associated hemagglutinin-esterase protein (HE). Among these structural proteins, the spike present on the surface of the virus forms large protrusions, giving coronaviruses the appearance of crowns. Spike protein forms homotrimers and the virus surface mediates attachment of the virus to host cell surface receptors [
40,
41,
43,
44].
Figure 3a,b shows S protein and its receptor binding domain (RBD) of SARS-CoV-2.
2.2. Host Cell Binding and Entry
The virus S protein has a high-affinity RBD for the human angiotensin-converting enzyme 2 (ACE2) receptor [
44,
45,
46,
47]. It should also be noted that the S protein also has a region that has a galectin fold and may be an active galactose binding domain (GDB) [
23,
24]. Collectively the virus will either adhere to the cell via ACE2 and/or sulfated polysaccharides, called glycosaminoglycans (GAG) [
44,
47,
48]. After host cell binding, the SARS-COV-2 viral particles are primed for cellular entry via a spike cleavage event with readily available furin proteases, rather than cellular proteases, such as in SARS-CoV infections [
44,
47]. The receptor binding and membrane fusion is the initial step in the coronavirus infection and, thus serve as the primary target for human interventions.
2.3. SARS-COV-2 Replication and Life Cycle
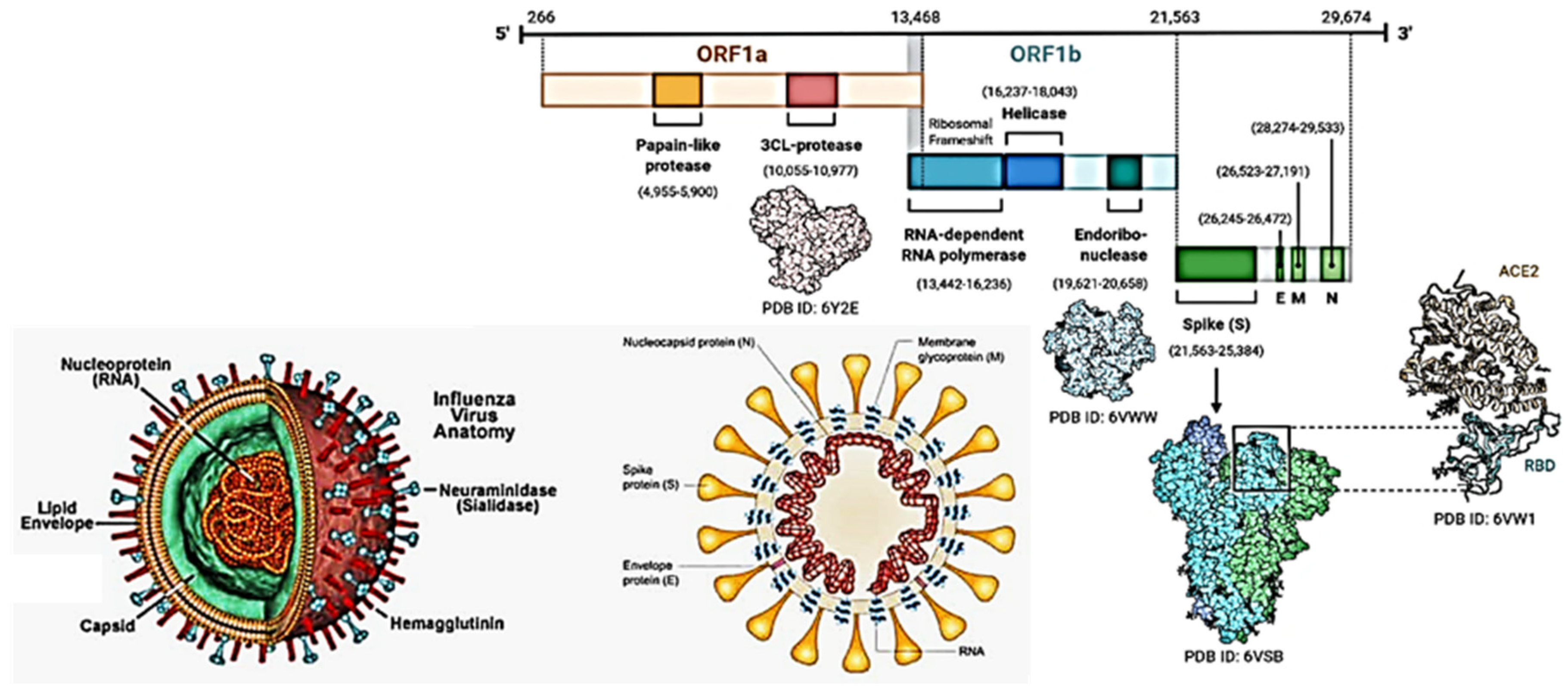
SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA
betacoronavirus of approximately 30 kb in length [
49,
50]. The RNA has a 5′-cap and a 3′-poly(A) tail and can act as an mRNA for immediate translation of the viral polyproteins. In addition, both 5′- and 3′-ends of the RNA present a highly structured untranslated region (UTR) that plays an important role in the regulation of RNA replication and transcription. Seven stem-loop structures are present at the 5′-UTR, while a stem-loop and a pseudoknot are present at the 3′-UTR. These two latter structures are mutually exclusive since their sequences overlap. It is hypothesized that the alternate forms of either the pseudoknot or the stem loop play some role in the transcriptional regulation [
51]. As shown in
Figure 4, the SARS-CoV-2 genome contains 14 open reading frames (ORFs), preceded by transcriptional regulatory sequences (TRSs). The two main transcriptional units, ORF1a and ORF1ab, encode replicase polyprotein 1a (PP1a) and polyprotein 1ab (PP1ab), respectively. The largest polyprotein PP1ab embeds non-structural proteins (Nsp1-16), which form complex replicase machinery. This includes enzyme activities that are rare or absent in other families of positive-stranded (+) RNA viruses [
52]. At the 3′ ends, the viral genome encodes four structural proteins (spike, envelope, membrane, nucleocapsid), which are components of the mature virus and play a crucial role in viral structure integrity, as in the case of the S protein, for viral entry into the host [
53,
54,
55]. Interspersed among the structural genes, the 3′ end of the genome also contains nine putative ORFs for accessory factors [
56]. The structural and accessory proteins are translated from a set of nested sub-genomic RNAs, all terminating with the 3′-end of the full-length RNA. The generation of these sub-genomic RNAs starting from negative-sense RNA intermediates is regulated by the TRSs. During minus-strand RNA synthesis, the viral RNA polymerase pauses at each TRS sequence. The pause can be resolved either by continuing the synthesis through the TRS into the adjacent gene, or it can lead to the termination of transcription with the generation of a sgRNA. The exact molecular mechanisms that determine either outcome are yet to be fully clarified but are likely involved in long-range RNA-RNA interactions between complementary sequences [
57,
58].
SARS-CoV-2 penetrates the cell through recognition by the S glycoprotein present on the surface of the virus envelope of the ACE2 receptors, as previously observed for SARS-CoV [
53,
54]. It is possible that other receptors mediate the entry of SARS-CoV-2 into host cells, such as CD147 [
59]. After attachment, the human transmembrane protease serine 2 (TMPRSS2) cleaves and activates the S protein [
60] in an event that allows SARS-CoV-2 to enter the cells by endocytosis or direct fusion of the viral envelope with the host membrane [
61,
62]. Once inside the cell, the infecting RNA acts as a messenger RNA (mRNA), which is then translated by host ribosomes to produce the viral replicative enzymes, which generate new RNA genomes and the mRNAs for the synthesis of the components necessary to assemble the new viral particles.
SARS-CoV-2 replication is a complex process that involves RNA synthesis, proofreading, and capping. Like other viruses, this process is likely to actively involve many host proteins, such as DEAD-box (DDX) helicases, which are exploited by the virus for more efficient replication [
63,
64,
65]. Understanding the molecular mechanisms that guide the replication of this coronavirus is essential to develop therapeutic tools to neutralize SARS-CoV-2. Here, we review structural information, mostly obtained through homology modeling based on the available structures for other coronaviruses, on the main protein actors of SARS-CoV-2 RNA replication and transcription.
2.4. Host Response to SARS-COV-2 Infection
A recent study [
66] identified a specific enzyme —sPLA2-IIA — that may be the key mechanism driving COVID-19 severity and mortality. Known for its elusive role in severe inflammation in the human body, sPLA2-IIA shares high sequence homology with the active enzyme in rattlesnake venom. The protein can bind to receptors at neuromuscular junctions and potentially disable the function of these muscles. In low concentrations in healthy individuals, sPLA2-IIA plays a critical role in destroying microbial cell membranes during bacterial infections. The enzyme sPLA2 is indicated in inflammatory disease, sepsis, and endotoxic shock and its suppression by indoxam can mitigate cellular damage and prolong survival, thus high circulation levels of sPLA2 may be any indicator and diver of collateral systemic damage [
67,
68]. While SARS-COV-2 was largely unknown to the medical community until late 2019, the family of viruses is not new, since historical outbreaks also share significant similarities to this current variant, and it is important to consider past lessons learned.
3. Historical Context to Coronavirus Diseases
3.1. SARS
In humans, coronaviruses generally cause mild respiratory infections, as observed in the common cold. But they can also cause gastrointestinal and central nervous system (CNS) diseases in humans and animals, threatening human health and lives, and the economy [
69,
70]. However, in 2002-2003 an emergent coronavirus that caused severe human disease was observed with the global outbreak of SARS (Severe Acute Respiratory Syndrome, SARS-CoV). SARS-CoV originated in Southern China and caused an epidemic in 2003 with a total of 8,098 SARS cases globally, including 774 associated deaths, and an estimated case-fatality rate of 14%–15% [
69,
71,
72,
73,
74,
75,
76].
Following on studies on seroprevalence (using serum collected no more than 4 months after the outbreak) showed that SARS asymptomatic prevalence in hospital workers was low, about 2.3-7.5% [
77,
78]. However low or asymptomatic seroprevalence rates in medical staff, a health-responsive group, may not closely compare to other groups due to differences in the pursuit rate for medical services for subclinical infections. Persons in the general populace may also actively avoid medical services in a pandemic and tolerate greater sub-clinical infection symptoms without seeking care, further decreasing detection rates. While there is little evidence of significant subclinical SARS [
79], one can presume there was "some" undercounting of the true infection burden during the SARS outbreak. While no cases of SARS-CoV infection have been reported since 2004, another newer outbreak, Middle East Respiratory Syndrome - MERS-CoV has caused sporadic outbreaks in different parts of the world.
3.2. MERS
In 2012 Middle East Respiratory Syndrome (MERS) emerged which led to the collection of some additional information. The first case of MERS-CoV was reported in Saudi Arabia in 2012. Since then, a total of 2,494 laboratory-confirmed cases of MERS-CoV have been reported until the end of November 2019, including 858 associated deaths, and an estimated high case-fatality rate of 34.4% globally [
70,
80,
81]. While this virus has a different spike protein than SARS and binds dipeptidyl peptidase-4 (DPP4) to infect cells, it shares many of SARS characteristics [
41,
82].
This outbreak included massive testing of asymptomatic persons and frequent screening of medical staff for infections. This virus demonstrated lower human-to-human transmissibility in a hospital setting than SARS, with less than 1% on average of medical workers seroconverted [
83]. This indicated that medical protective measures in use by health workers are reasonably effective. However, it also clearly showed that MERS has a potential for 12-25% lab-confirmed asymptomatic positive rate, although most studies in a review showed 1-4% [
84]. Again, one can presume there was "some" under-counting of the true infection burden of MERS.
MERS is often presumed to offer low human-to-human transmissivity and was mostly described as having limited nosocomial spread [
82,
85]. It is interesting to note that with such a low infection rate of the virus, it continues to cause sporadic infections to this day. Relatively higher than described human-to-human transmission in the community and more frequent limed symptoms might better explain how MERS continued to spread, presumptively aided by asymptomatic/low symptom carriers who chose not to seek care, much like SARS-COV-2 is known to spread today.
While MERS seems to act like SARS or SARS-COV-2, it does bind to a different receptor and has a markedly different genome [
85]. It is possible that at an immunological level, this virus should not be considered SARS-like at all. Having noted this possibility, for the purposes of our discussion, this virus will be grouped together with SARS, so we may later explore certain other hypotheses. However, it is interesting to note that several such studies used serology to determine if subclinical infections of MERS and SARS have occurred in the past. This was particularly true of the SARS outbreak [
86,
87]. While the SARS outbreak antibodies were presumed to be persistent for up to three years, it is obvious that this work was done in symptomatic cases [
88]. Upon reflection, after observing SARS-COV-2 patients and other common cold COVID infections, it has been shown that antibodies to this virus (SARS-COV-2) can wane and the strength of symptoms is related to the rate of immunoglobulin (Ig) response loss [
89,
90,
91]. At the very least this waning of Ig response was also observed in MERS asymptomatic cases [
89].
3.3. Common Cold Virus
There are also several well-known common cold coronavirus strains, including (but not limited to alpha (α) coronaviruses such as NL63 and 229E, β-coronaviruses such as OC43 and HKU1, however, these COVID strains are not known to cause severe illness most persons or pandemics [
41,
43]. These common cold coronaviruses frequently have different S proteins allowing diversity in cellular binding targets, NL63 uses the ACE2 receptor, OC43 uses glycans, 229E uses the APN receptor, HKU1 uses O-acetylated sialic acid [
92,
93].
3.4. SADS-CoV
In 2017 a novel swine acute diarrhea syndrome coronavirus (SADS-CoV, also known as swine enteric alphacoronavirus (SeACoV) or porcine enteric alphacoronavirus (PEAV) emerged, and more specifically, in Guangdong province, China [
86]. This virus ultimately caused the death of 24,693 piglets in 2017 [
94]. There has been a recent re-emergence of this virus in Southern China in February 2019 [
94,
95]. This virus is an alpha coronavirus that is likely zoonotic in origin and may have emerged from the same horseshoe bats (Rhinolophus sp.) that are thought to have been the original animal host for SARS-CoV and SARS-COV-2 [
96]. This alpha coronavirus has a relatively different S protein form, from other well know strains of coronavirus and does require glycans, ACE2, DPP4, or aminopeptidase N (APN) for cellular entry [
95]. There is also some concern that this virus may be able to infect human primary cells and thus has a potential risk of human contagion [
97].
3.5. PEDV and TGEV
While coronavirus is commonly thought to cause human disease, it also impacts the animal husbandry industry. The ailment, porcine epidemic diarrhea coronavirus (PEDV) and Transmissible Gastroenteritis Virus (TGEV) first isolated in 1946, has been killing pigs globally for many years [
98]. Since 2013, coronaviruses have swept throughout global industrial pig production facilities causing increasingly large losses [
98,
99,
100,
101]. While these strains do not infect humans, it causes an almost 100% fatality rate in piglets and wiped out more than 10% of America's pig population in less than a year [
98,
99,
100,
101]. Although mentioned here as an illustration, this review will focus on the virus that infects humans, but for the purpose of impact, it is intuitive as to what the spillover to humans of a strain would do.
3.6. Zoonotic Strains
Coronaviruses are widely dispersed in animal hosts that may encounter humans and presumptively produce spill-over infections. Past events with human casualties include civet cats, camels, and bats, all indicated in SARS, MERS, and now SARS-CoV-2, however, the risk is equally valid for animals [
8,
82]. Over the last two decades, several novel emergences of coronavirus have been widely associated with severe veterinary disease in animals. Besides being the costliest, PEDV, TGEV, and SADS-CoV, there are other notable veterinary concerns such as porcine respiratory coronavirus (PRCV), feline infectious peritonitis virus (FIPV), bovine coronavirus (BCoV), avian infectious bronchitis virus (IBV), porcine delta coronavirus (PdCoV) that cause significant economic loses. A detailed discussion is beyond the scope of this review; however, it suffices to state here, with history as our guide, that there are many unenumerated strains of coronavirus that can emerge, that are likely to threaten our population and economy. Furthermore, there is no clear delineation as to what an “animal” virus is, as many coronaviruses pose significant tropism for both human and animal cells depending on the receptor targets, they use to bind host cells [
64]. It is widely known that coronavirus can engage in recombination events and can thus develop new cellular or host tropisms with observable signs of such historical events seen by comparing the degree of conservation among various classes of spike proteins available in the coronavirus gene pool [
102].
3.7. Cryptic COVID Prevalence
There are several other well-known common cold coronavirus strains, however structurally many of these are significantly different from SARS-like viruses (β-coronavirus) [
41,
43]. Regardless of the reported transmission of the known prior "SARS-like" viruses, there is limited documented serological evidence of a much wider circulation of other unnamed variants of viruses such as these near animal vector sites [
103]. The symptomatology for these rare observations of the COVID variant is typically low or no symptoms have yet been observed in persons exposed to COVID, as backed up by serology [
103]. It is easy to envision that an unknown/unobserved SARS-like virus periodically would spread far wider than what is reported, provided the infection symptoms are low and detection tools such as strain-specific Polymerase Chain Reaction (PCR) tests are limited. Furthermore, if COVID symptoms are typically very mild on average, then this may well have been true of past known viruses such as SARS or MERS, if the contagion of these historical outbreaks was larger than described, abrogating the need for “other” strains to be circulating.
The serology evidence shows that pre-exposure would take prior exposure to "SARS-like" viruses that share similar homology in their protein-coding regions, particularly the spike protein to explant this serology evidence. Only a close relationship could promote cross-reactive antibody responses [
104,
105]. While there is only limited serology evidence of a population of cryptic SARS-like COVID infections, there is stronger adaptive immune evidence in T-cells. There is significant evidence that several patients may have cross-reactive T-cells response to SARS-CoV and SARS-CoV-2, long before they could have been exposed to the virus. Roughly 50% of T-cell "controls" were isolated before the SARS-CoV-2 outbreak and were already reactive to these antigens [
106,
107,
108,
109]. This cross-reaction also covers virus-specific T-cell receptors (TCR), so this response is somewhat less strain selective to SARS and presumably to COVID. However, it is now evident that while serology, and antibody levels, may wane rapidly over time, reactive T-cells long persist after prior SARS and MERS exposures [
110].
Interestingly, this pre-exposure reactive T-cell phenotype was also observed historically in persons without exposure history to SARS [
111], suggesting that reactive T-cells to spike protein epitopes were observed in unexposed persons. Reactive T-cells without known symptomatic infection were also observed in studies on MERS, since its outbreak in 2012 [
112,
113]. Thus, individuals possessing reactive T-cells to random coronavirus, with no symptomatic history of infection, is not a new observation. This implies that cryptic prior COVID infections have been commonly observed in the last decade. For the sake of discussion, rather than presuming massive SARS spread, the phenotype of reactive T-cells to SARS-CoV-2 and other COVID variants in persons with no known exposure is best explained by prior infection to other SARS-like strains of coronavirus (cryptic-COVID infections) not previously detected in our populations due to subclinical illness and uncomplicated recovery.
Cryptic-COVID infections could explain reactive T-cells in the population with no known history of any highly lethal and tracked strain of COVID. However, T cell reaction is based on short-presented peptides and not a full cognate antigen, thus it is hard to trace the source strain of any prior exposure using such reactive cells. Consequently, any source of the exposure that generated these reactive T cells will remain cryptic, and only a crude viral relationship can reliably be inferred. We can readily assume that some COVID strains have been common enough to explain this T-cell reactivity in these populations, and possibly by projection, to the world.
Getting the number of reactive T-cells in 20-60% unexposed individuals [
106,
107,
108,
109] in magnitude from least to greatest indicates cross-reactive T-cells recognition between circulating “common cold” coronavirus and SARS-CoV-2. Accepting 50% pre-reactive T cells as a conservative number (within range in 2 of the noted studies), this implies that there may be a minimum of 2 M cryptic COVID infections in the U.S./year (using approx. a population of 335 M people with 78-year life expectancy * 0.5 = 2.2 M), to get 150 M sensitized persons. There is likely quite a bit more than this limited calculation which would only give an average of 50% by the end of life, presumptively these reactive T-cells were observed in much younger patients and does not consider multiple reinfections and or reactive T-cells half-life. There is also more than one type of coronavirus (α and β) that would likely produce less overlapping reactive T-cell populations due to genetic differences and it is not clear to what extent such post-infection T-cells will cross-react across different strains of COVID. Thus, there are likely millions more cryptic COVID infections each year to provide this level of reactive T-cell phenotype observed in the naïve (with respect to (SARS-CoV-2) population.
During a regular COVID (not pandemic) season, we might have nearly as many multi-strain derived COVID infections of a sub-clinical nature with an uncomplicated recovery in mass in the US, as we had in the first part of the pandemic of SARS-CoV-2. This complicates the ability to use reactive T-cells as a surrogate for exposure history to a particular strain. It is likely that our global population is not so naive with respect to COVID strains that may cross-react to SARS-CoV-2 as we may have presumed at the start of the outbreak. Despite a large population with reactive T-cells to SARS-CoV-2, the scope of the current pandemic might suggest that 50% of persons with reactive T-cells do not translate into anything close to 50% herd immunity. So reactive T-cells may also not be a good indicator of protective immunity to any given strain of COVID, much less SARS-CoV-2.
4. Immunological Aspect of COVID-19 Disease
SARS-CoV-2 uses the same receptor, ACE2 as SARS-CoV, which primarily infects the respiratory tract. Bats are adapted to Coronaviruses (CoV) because of their high level of reactive oxygen species (ROS) and continual expression of interferon-stimulated genes, which have an advantage in suppressing CoV replication. They also have an attenuated nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome response, which leads to decreased viral virulence [
113]. Hence, infected bats generally have no symptoms or show moderate symptoms. The high levels of ROS in bats are mutagenic, affecting the proofreading capability of CoV polymerase, and this effect is compounded during the long-life span of bats (more than 25 years) [
113].
Toll-like receptors (TLR) 3, -7, -8, and -9 recognize viral RNA and DNA in the endosome which is present in the cytoplasm. The viral RNA receptor retinoic-acid inducible gene I (RIG-I), the cytosolic receptor melanoma differentiation-associated gene 5 (MDA5), and nucleotidyl transferase cyclic GMP-AMP synthase (cGAS) recognize both viral RNA and DNA in the cytoplasm [
114]. Activation of these receptor pathways activates the transcription factors NF-κB and Interferon regulatory factor 3 (IRF3), secretion of type I Interferons (IFN-α/β), and a gamut of pro-inflammatory cytokines [
114]. In a study conducted in ARDS, mouse models induced by multiple noxae, including SARS-CoV, and inhibition of the TLR-4 gene was observed, but no inhibition of TLR-3 or 9 genes was seen, resulting in alleviated acute lung injury. TLR-4 responds to bacteria, one hypothesis is that oxidized phospholipids due to SARS-CoV-2 can activate TLR-4 and result in the onset of ARDS. TLR-7 agonists may inhibit severe COVID-19 and reveal synergic activity with active anti-viral therapy [
115]. Interleukin-6 (IL-6) and Tissue Necrosis Factor-alpha (TNF-α) are important cytokines in SARS-CoV-2 infection and secretion after TLR-4 activation. A remarkable binding has been reported between the viral S protein and TLR-1, TLR-4, and TLR-6, and TLR-4 has the highest binding energy [
116].
It has been reported that after viral infection, host pattern recognition receptors (PRRs) including TLRs, RIG-I, and NOD-like receptors (NLR) detect the viral nucleic acid and induce the synthesis of type I interferons (IFNs) [
117]. The N-protein of SARS-CoV plays as an immune escape protein and even escapes interferon response [
117]. IFN-I levels associated with the severity of disease and COVID-19 can block the activation of IFN pathways, i.e., CoV proteins inhibit several steps of the signal transduction pathway that bridge the IFN receptor subunits (IFNAR1 and IFNAR2) to the STAT proteins that activate transcription [
118]. Subsequently, neutrophils and monocytes/macrophages are triggered, come to the site of infection, and induce hyperproduction of pro-inflammatory cytokines [
119]. Specific Th1 and Th17 cells are also initiated, and these further contribute to the exaggerated inflammatory response [
119].
In severe disease conditions, or when the viral load is high, the host immune system attempts to kill the virus. Which eventually results in the release of many inflammatory mediators and the production of cytokines [
120]. In return these cytokines induce organ damage and subsequently edema, ARDS, acute lung injury (ALI), acute cardiac injury, and secondary infection, leading to death [
121]. ACE2 receptors are abundantly expressed in the cardiovascular system, liver, digestive organs, and kidneys. In addition, all endothelial cells and smooth muscle cells across organs also express ACE2, enabling viral circulation and spread [
122]. In the early phase of CoV infection, dendritic cells and epithelial cells release pro-inflammatory cytokines and chemokines such as IL-1β, IL-2, IL-6, IL-8, both IFN-α/β, TNF, C-C motif chemokine 3 (CCL3), CCL5, CCL2 and IP-10 (CXCL 10), a chemokine of the CXC family [
123]. There is systemic lymphopenia, especially of Natural Killer (NK) cells, and atrophy of the spleen and lymph nodes. Furthermore, infiltration of activated monocytes, macrophages, and lymphocytes into the lung tissues and vascular system induces lesions in these organs [
9,
121,
123].
In COVID-19 infection, cytotoxic T-cells (CD8+), helper T-Cells (CD4+), and subsets of CD4+: CCR4+, CCR6+, and Th17 cells express a high level of Human Leukocyte Antigen-DR isotype (HLA-DR) [
124]. In some severe cases, the number of natural killer (NK) cells was very low or even undetectable [
96]. Memory helper T-cells, regulatory T-cells, and γδ T-cell numbers also decreased in severe cases [
118,
125]. In addition, the total number of lymphocytes, particularly CD4+ T-cells, CD8+ T-cells, and IFNγ-expressing CD4+ T-cells decreased considerably in severe conditions [
126]. Vγ9Vδ2 T cells are the dominant γδ T-cell subset in adults and with age, the numbers may vary. Elderly people with decreased numbers of Vγ9Vδ2 T-cells are vulnerable to SARS-CoV-2 infections [
127]. Overall, increased expression of cytokines (IL-6, IL-10, and TNF-α), systemic T-cell lymphopenia (CD4+ and CD8+ T-cells), and decreased IFN-γ expression in CD4+ T cells, play an important role in pulmonary damage, disease severity, and outcomes [
126].
Despite lower numbers of T-cells in severe COVID-19 patients, these T-cells are more activated and exhibit a tendency to exhaustion with the expression of PD-1 and TIM-3 markers. Whereas, recovering patients showed an increase in follicular helper CD4 T-cells (TFH) and decreased levels of inhibitory markers such as IFN with increased levels of granzyme and perforin [
118].
Most of the infiltrating cells in the lungs are monocytes and macrophages, with moderate numbers of multi-nucleated giant cells but few lymphocytes. Among the infiltrating lymphocytes, most of them are CD4+ T-cells [
121]. Peripheral blood of severe COVID-19 patients showed high numbers of CCR6+TH17 cells [
128]. TH17 cells produce IL-22 that upregulates antimicrobial peptides mucins and fibrinogen. Hence, the secretion of IL-22 may drive the formation of edema with abundant mucins and fibrin which is seen in SARS-CoV-2 and SARS-CoV patients [
128].
An increased number of monocyte subsets was seen in COVID-19 patients bearing CD14+ and CD16+ surface protein, and macrophages bearing CD68+, CD80+, CD163+, and CD206+ surface markers [
129]. Analysis of autopsies of COVID-19 patients showed that monocytes from infected patients that bear ACE2 receptors were associated with a delayed-type I INF response [
121].
There is also an increase in neutrophil-to-lymphocyte ratio (NLR) in severe COVID-19 patients when compared to mild cases [
125]. NLR is an indicator of systemic inflammation and infection and serves as a key indicator of bacterial infection. Increased NLR in infected patients associated with disease severity addresses a possible role in hyper-inflammatory responses to COVID-19 [
125]. The platelet-to-lymphocyte ratio (PLR) is another important response by the body to severe disease. Patients with increased platelets are hospitalized for a longer time [
130] and should be monitored because of the intensity of the cytokines produced [
130]. An increase in thymosin level could regulate immune responses to elevate lymphocytes and develop a situation that can prevent the development of a severe disease condition [
130]. Hence, understanding the host immune response against SARS-CoV 2 in COVID-19 patients can shed light on the immunopathogenesis of this disease, and help understand the molecular pathways for providing any medical intervention, which may provide long-term immunity by having circulated immune memory cells in the immune system, and enable designing of prophylactic and therapeutic measures to overcome future pandemics such as coronaviruses.
5. The Limitations of Serology as a Tool to Measure Historical Prevalence
Using seroprevalence to observe viral infection history in a population (viz. in the case of SARS) is dependent on reactive antibodies. However, it is now also known that like SARS, SARS-CoV-2 antibody detection rates seem to fall precipitously over a few months as in most MERS, SARS, and SARS-CoV-2 survivors [
89,
131]. This decay in Ig response is most marked in mild cases of SARS-CoV-2 [
132]. It may be safe to assume that the use of serology to detect the wider spread of various COVID viral strains may itself be prone to significant undercounts unless such serology is promptly undertaken after exposure.
It might be assumed that many modern COVID strains’ prevalence changed recently since we have no evidence in antiquity of its earlier spread. Such a change could have been the result of larger COVID antigenic/strain perturbations prevalent in the circulating population of human viruses such as those that followed the 1918 pandemic [
133]. Perhaps in the era of SARS, around 2003-2008, the silent spread was much wider than detected, perhaps even at the pandemic level, but paradoxically the rate of ARDS (SARS's showcase symptoms) was very low (like SARS-CoV-2 exhibits today). Alternatively, any number of coronavirus strains, yet to be enumerated, may collectively cause low levels of COVID for years to arrive at the same exposure phenotype. There does not seem to be much T-cell evidence before 2008, and such testing would be unlikely earlier than 2003 because COVID strain significance would be unknown prior to the SARS outbreak. Thus, it is likely we can never know if the true prevalence of COVID has changed in the last 20 years due to the lack of viable T-cell specimens.
6. SARS-COV Historical Transmission Context
Owing to the similarity between SARS-CoV and SARS-CoV-2, one may presume that they share very similar modes of transmission among human hosts [
134]. Let us examine SARS-CoV-2 by looking at the transmission behavior of SARS-COV. It is well known that SARS can spread between humans through routine contact with surfaces and fomites [
135]. The control of person-to-person contact and cleansing of potential fomites was a core activity in the early SARS outbreak [
136]. It soon emerged that SARS can be detected in many bodily fluids including urine and feces [
137]. SARS virus particles were shown to be stable in feces, thus fecal-oral transmission could never be ruled out [
138]. Later during the outbreak, it became known that SARS can be transmitted by either droplets or aerosols and can readily be detected in the air near the infected [
139,
140]. Later airborne transmission was observed to occur over relatively long distances such as in airplanes or apartment buildings in Hong Kong [
86,
139,
141]. Such long-distance transmission indicates that SARS can spread by aerosols [
86]. By the end of the SARS-CoV epidemic, we knew that the virus was easily transmitted between humans via casual contact, fomites, and inhalation of air carrying aerosol particles of the virus. The most protective containment strategy for healthcare workers was to use N95 masks in addition to standard gowns and gloves. In the case of SARS, the epidemic was contained and soon transmission was halted. Had it become more widespread, mask mandates would have been expected. All this information was known about SARS 15 years ago, yet even after the sequencing of SARS-CoV-2 and the observation that this was very closely related to SARS-CoV, the message was not clearly conveyed that this was, presumably directly, applicable to transmission behavior.
SARS-COV-2 Current Transmission Characteristics
Today SARS-CoV-2 looks very much like SARS, in its transmission profile, as it is stable on fomites [
142,
143]. Furthermore, it can be detected and is stable in feces and wastewater [
135,
144]. It is detectable in aerosols, as they are generated by casual breathing, coughing, and sneezing [
145]. It can be transmitted via both respiratory and fecal aerosols [
146,
147]. Furthermore, aerosol transmission appears to be the main route and is a frequent cause of super-spreading events [
148,
149,
150]. One new development is that transmission is possible before a patient is symptomatic [
151] and subclinical infections were known to also be common and infectious [
152] further compounding global control strategies. As we, presumably, approach the end of the COVID-19 pandemic, it took almost two years to re-discover the information that, by reflection, was learned already as wisdom from the SARS-CoV epidemic. Probably the most important difference between SARS-CoV-2 and SARS-CoV is that it is well known to cause cases with a less severity to be described by some asymptomatic [
102,
153,
154]. There is certainly some debate as to the prevalence and nature of these cases, whether the cases are truly asymptomatic or simply pre-symptomatic. Regardless of low symptoms carriers who are infectious could serve as a major factor driving the pandemic spread of SARS-COV-2 [
102].
As of October 2022, the novel SARS-CoV-2 has been reported to have infected about 98.5M persons with about 1.065M deaths in the US and over 626M people infected, and more than 6M deaths worldwide with the true toll likely to be much higher due to under-reporting and (continued pandemic spread), reported by the Institute for Health and Evaluation [
155]. It has caused an average incidence of ARDS of about 14% in the confirmed infected population [
156]. However, there are several reports about undercounts of both infected and dead presumptively due to pre/sub-clinical cases (sometimes described as asymptomatic or silent) and saturated testing situations in some jurisdictions. As an example, in the US this resulted in a significant excess for a reported period from a peak period of February 01, 2020, to October 09, 2020, in which there were 221,120 - 294,124 vs the reported 211K deaths, which implied a significant death undercount by at least 9-30% by the CDC 2020 [
157]; and Worldometers.info 2020[
158]. There is also mounting evidence of widespread asymptomatic/pre-symptomatic/sub-clinical cases, silent cases that are not being adequately enumerated but can harbor and spread the infection [
159,
160]. These silent cases could be 10 times larger than the lab-confirmed positive infections [
159,
161]. This 10-fold undercount in the US of the infected and potentially contagious is sported by our own internal modeling [data not shown]. Consequently, reported R0 values, percentage rates of ARDS, and other figures are likely to be inaccurate as they are related to the "true" infection rates that are widely under-counted.
Outside of the US, the B.1.617 (Delta) variant of SARS-CoV-2 was first identified in Maharashtra, India in late 2020, spread throughout India, and to other 60 countries, outcompeting pre-existing lineages including B.1.617.1 (Kappa) and B.1.1.7 (Alpha) [
162]. L452R and E484Q are in the critical RBD that interacts with ACE2 [
163]. All 3 lineages of B.1.617, showed a mutation in L452R, located in the RBD that interacts with ACE2. In vitro, findings demonstrated reduced sensitivity of the spike protein bearing RBD mutations in L452R and E484Q to BNT162b2 mRNA (Pfizer–BioNTech) vaccine-elicited antibodies [
162].
On November 26, 2021, WHO announced that a new SARS-CoV-2 Variant of Concern, Omicron (VOC) (initially named B.1.1.529), appeared in most of South Africa's provinces, particularly Gauteng. Because of the rapid spread of Omicron VOC, particularly among the younger age population, in Gauteng, South Africa, WHO showed concerns and announced an alert to global public health systems. The SARS-CoV-2 Omicron VOC was first reported to the WHO from South Africa on November 24, 2021. Cases of Omicron VOC were then identified in Botswana, Belgium, Hong Kong, and Israel. On November 29, 2021, three days after the announcement by WHO, cases of Omicron VOC were also detected in Austria, Australia, Belgium, Canada, the Czech Republic, Denmark, France, Germany, Italy, the Netherlands, and the United Kingdom [
164].
With the community infection rates so widely underreported, one must use estimated infections to calculate and develop a model to predict the spread of the virus. Presumptively using the death rates (or excess death) as a more accurate marker of infection rates as these reported figures are much more accurate and more relevant to the epidemiology. Thus, the accuracy of these estimates has a substantial bearing on any assumptions one may make about the "true" rates of any given occurrence. Thus, we attempt to describe the overall scenario in trends rather than absolute values and focus on differences in death rates. Simply because the accuracy and precise modeling of the COVID-19 pandemic are outside the scope of this discussion as the focus here is to synthesize trends using data across the spectrum of the sparse information available.
7. SARS-CoV-2, COVID-19 Diagnosis and Detection
The syndrome, COVID-19, caused by SARS-CoV-2 virus appeared initially as a flu-like illness [
2,
3,
160]. Symptoms include fever, chills, cough, shortness of breath or difficulty in breathing, fatigue, muscle or body aches, headache, loss of taste or smell, sore throat, congestion or runny nose, nausea or vomiting, and diarrhea [
160,
165]. During imaging, the lungs of infected patients exhibited a distinct ground-glass opacity in the lungs on chest computed tomography (CT) scans [
154]. It appears to have an incubation period of 2-14 days after exposure to the virus. It was the classic syndrome symptomology and image that was used as the initial case definition, early on in China. This was further compounded by the fact that the virus was not detectable by PCR at the time. This resulted in the sequencing of the virus, and on January 31, 2020, the Chinese laboratories emerged the genome of SARS-CoV-2, termed 2019-n-CoV. Soon accurate tests were developed to detect the virus. However, the initial low availability of tests mandated a focused testing program and hence lost many precious months globally while the capacity for mass testing grew.
Currently, there are many rapid tests available, according to WHO recommendations, samples should be collected from the respiratory tract of patients suspected of SARS-CoV-2, which include nasal and pharyngeal swabs, sputum, or bronchoalveolar lavage fluid. Samples are then subjected to nucleic acid testing, using real-time RT-PCR and next-generation sequencing as the main techniques used in the laboratory for the diagnosis of COVID-19. Other techniques include viral antigen and serological antibody testing [
59,
166,
167,
168]. The tests have different usage cases, however, with rapid antigen testing having a lower sensitivity, but a faster operation time. Lower sensitivity tests are best employed to confirm if a patient needed more sophisticated testing, which is not needed if the patient is infected.
7.1. Related Coronavirus Strains May Share Syndrome Criteria
SARS-like coronavirus (SARS-CoV-2, SARS-CoV, and any emergent zoonotic counterparts) typically appear immunologically similar, share infection and complication perimeters, and probably share more syndrome features than they diverge on, including the risk of ARDS, and hence should be strongly grouped. It may not be that the well-known COVID virus strains (SARS, MERS, SARS-CoV-2) are particularly exceptional, nor more lethal than the rest of the population of unenumerated SARS-like viruses. It is more likely that these viruses represent a relatively homogeneous group, like influenza strains, that collectively cause a common viral infection syndrome, the Flu. Influenza strains largely vary in transmission rates, presumptively driven by herd immunity features more than individual stain uniqueness. This is true for several other virus groups such as Dengue, rhinovirus, Respiratory Syncytial Virus (RSV), and many more, so why do we think of SARS-like coronaviruses independently?
It may be logical to articulate that most coronaviruses, much like the varied serological independent influenza strains (at a minimum SARS-like coronavirus and all other coronaviruses that use a common spike, viz. binding ACE2 receptor) should be epidemiologically grouped together in terms of expected gross impacts, responses, cellular tropisms, and predicted virus properties. This is likely true for even, yet unenumerated/novel versions - just as reflections, of SARS-CoV having very close behavior and capabilities of SARS-CoV-2. At least with SARS-CoV-2 infection, most illness is below the threshold that would cause the infected to seek medical care. Yet paradoxically there is a small, yet important, risk of ARDS with infection, this seems to be directly related to age [
169]. To a large extent, significant complications from ARDS syndrome that people experience due to this viral infection may be quite rare and atypical. It is reported that SARS-CoV-2 has about a 14% rate of ARDS [
156]. However, as asymptomatic cases are undetected or undiagnosed, perhaps as much as 10 times, the ARDS rate per infection may only be 1.4% [
159,
161]. Hence the question that arises is why COVID was so prevalent. Why did the frequency of ARDS remain so high during the recent pandemic? This could be possible since previously the ARDS frequency must have been very low, far less than 1%.
7.2. Phenotype of COVID-Triggered ARDS in Recent SARS-COV-2 Pandemic
During the recent pandemic, there were many ARDS-related complications with SARS-COV-2, either the virus was somehow more virulent, or there was some other interaction such as human genetics, predisposition, or immune system factors driving sporadic hypervirulence. It was also projected that many people may have reactive T-cells to this virus. Presumably, the lack of naiveté may be providing some response other than protection because these people were apparently getting sick (not exhibiting under-representation) assuming that the rate for T-cell reactive people is similar in Seattle [
170]. Perhaps the only real difference with SARS-COV-2 syndrome has been the most obvious one - exceptional prevalence, while the average COVID syndrome is mild. This leads to a concerning theory, “Could the severe cases of COVID-19 syndrome be directly related to prior virus exposure in the background of the exceptional prevalence of SARS-CoV-2?" Effectively, demonstrating the existence of secondary COVID syndrome. While the evidence remains certainly incomplete at this point, a summarization of why this concern emerged is essential and should be investigated before we experience another pandemic.
It has become abundantly clear that ARDS syndrome is driven by T-cell mediated cytotoxic responses, the exact details of which are outside the scope of this communication and is discussed elsewhere in the literature [
171,
172]. This has driven the prosed use of steroids and other immunomodulatory therapies in its treatment. If employing immunomodulatory therapies is an effective treatment, then perhaps having more reactive T-cells prior to infection is a factor in a poor prognosis. Furthermore, persons with an auto-immune disease and or acquired immunocompromised status due to cancer or transplant may never develop a "protective" humoral immunity, as defined by a population of neutralizing antibodies, and are routinely described at increased risk for viral infections like RSV and influenza [
173]. However, paradoxically immunocompromised persons can readily survive SARS-CoV-2 infection and have very similar (possibly slightly reduced) death rates when compared with cancer patients statistically developing the worst in infection and death rates [
174,
175].
It has also become clear that few studies in possession of neutralizing SARS-CoV-2 antibodies (example N122) were protective against infection in an outbreak on an isolated ship [
170]. Likewise, N17, N74, N122, and N149 are SARS-CoV-2 neutralizing antibodies, which target the N-terminal domain (NTD) of the viral spike [
176]. But not all persons make such effective antibodies, they may also wane over time, and the antibodies to one strain of COVID may be less effective or neutralizing the other strains. Thus, if someone was exposed to a SARS-like viral strain, some years later they may still possess their T-cell memory of the infection. Thus, a number of these survivors can be assumed if exposed to a new COVID strain (or the same one after antibody populations are lost) to not possess a neutralizing antibody phenotype, but very likely possess a trained adaptive T-cell population. Usually, immune memory including T-cell-only memory would be presumed to be a "good thing" and prompt faster clearance of the novel-related virus on exposure [
177]. However, there are well-known examples in literature, where this was not the case.
A classic example is secondary Dengue virus infections, in which an incomplete protective immunity shows a more severe disease phenotype that seems to be directly proportional to the T-cell activation based on past related strain exposure [
178,
179]. Some aspects of the immunopathy of secondary Dengue on synthesis look quite similar to ARD and COVID-19 pathologies [
171,
172,
180,
181]. While the tissues most manifesting cytotoxic damage differ between secondary Dengue and COVID-ARDS, the syndromes share surface pathologies such as entry into macrophages, activated T-cell populations, and a lack of neutralizing antibodies resulting in massively expanded T-cells leading to an overactive complement cytotoxic mediated response, the core common theme. Using a comparison of dengue, once the protective immunity of first COVID, as the primary case, wanes, it may set up a precedent for more severe secondary COVID cases.
7.3. Secondary COVID Syndrome -Theoretical Cause of ARDS in the Current SARS-CoV-2 Pandemic
It is interesting to note as it is well known that children (with less exposure history) around the age of 12 or less seem to have few symptoms when infected by SARS-COV-2. Whereas 50% of adults may have had prior SARS-like virus exposure at the T-cell level and suffered from more severe outcomes [
109,
182]. Perhaps ARDS, a secondary COVID syndrome caused by having a large reactive T-cell population to SARS-like virus but lacks any protective neutralizing antibodies. This state could set the stage to have an oversized hypersensitive T-cell response with a much larger than normal amplification of cytotoxic SARS reactive T-cell clonotypes upon reactivation by the presentation of SARS-CoV-2 antigen. The resulting over-amplification of certain T-cell types may then lead to subsequent cytokine storms during the resolution of the SARS-CoV-2 virus and significant tissue damage, not typically associated with a primary, cryptic COVID infection.
If secondary COVID syndrome is the main driver of ARDS in the era of the SARS-CoV-2 pandemic, it would explain why we see the impact today, at present we simply have a lot of "extra" infections. Cohorts that were protected from primary COVID through age or isolation or luck may be predisposed to get a mild primary COVID presentation with few symptoms, this may be observed in comparing Iceland's death rate on 20 August 2020 of about 0.5% (i.e., 2035 cases/10 deaths) vs the world’s average of 3.5% (22,732K cases/793K deaths) [
183]. It was observed that during the same time, the spread of the illness in Iceland in children under the age of 10 and females were low compared to adolescents or adults and males [
184]. Lower-than-expected Icelandic deaths do not conclude directly with the ARDS rate, but it does point to the overall severity trends of the illness. Iceland has had a larger than average COVID transmission rate as of August 2020 than the world (5,973 vs 2,916 per million), but paradoxically a much lower death rate (29 vs 101.6 deaths per million). Due to geographic isolation, one might presume (without other formal evidence) that they had a lower cryptic COVID history and were not exposed to some infection transmission opportunities in the past. This in theory would lead to lower rates of serial COVID infections of all types, and if secondary COVID was a syndrome, it would result in lower rates of severe illness, despite SARS-COV-2's widespread in that nation. While hardly confirmatory, other geographically isolated places show similar trends in August 2020, Madagascar 1.3%, Malta 0.6%, Trinidad and Tobago 1.6%, Bahrain 0.3%, New Zealand 1.3%, Cuba 2.4%, Taiwan 1.4%, yet collectively they have had more than 75K confirmed infections, with uncharacteristically low death rates adding credence to the theory.
Furthermore, Egorov and Romanova [
185] go a step further and associate the increased SARS-CoV-2 death rates observed in some countries with the prevalence of Rhinolophus bat populations of cryptic COVID strains. They theorize that presumptively prior exposure to strains like SARS-CoV-2 as spillover from these animal vectors may be causing antibody-dependent disease enhancement (ADE) and explain the enhanced death rates observed in certain countries. While Rhinolophus bats cannot fully explain the death rate spread as observed, clearly some regions of the world had a greater COVID exposure risk. This is posed by a combination of animal vectors, social factors, geographic isolation, and perhaps climate. However, collectively it seems safe to hypothesize that the severity of COVID-19 may have been more related to exposure history than to virus or host genetic factors or the standards of medicine in the country.
Theoretically, one acquires an infection with a strain of COVID virus that one has reactivity to but lacks neutralization immunity (such as exposure to a SARS-like strain in the past, then catch SARS-COV-2), this may lead to a high rate of secondary COVID and enhanced risk of death. However, it is not clear if this is a T-cell or antibody-dependent response as in the case of ADE. If secondary COVID infection is more severe, it may lead to progression to ARD and potentially for many other multiple organ syndromes associated with SARS-COV-2. Furthermore, if secondary COVID syndrome is the cause of severe disease, it may mean the severe illness is not tightly associated with the SARS-CoV-2 strain, merely triggered due to its exceptional prevalence. It could be a future concern that secondary COVID syndrome might just as easily be triggered by other strains now or in the future. It is also not clear if re-infection with SARS-CoV-2 will be common or have a different outlook as there have only been a few documented cases as per the European Centre for Disease Prevention and Control (ECDC) [
186]. While the authors lack finite resources, notably the cohorts, to perform most of these studies, it should be relatively straightforward for those already engaged in related research to non-invasively evaluate this theory.
7.4. Cohorts and Studies to Verify the Existence of Secondary COVID Syndrome
Identifying a set of experimental cohorts would require monitoring and isolation to ultimately determine the impact (if any) of secondary COVID syndrome. However, as the pandemic progresses there will be few isolated populations naturally. Thus, it will be necessary to watch groups that have varied susceptibilities. Collectively, several laboratories have confirmed COVID cases in SARS, ongoing MERS, and ongoing SARS-CoV-2 survivors in addition to any tested T-cell reactive persons. Regrettably, there are no recent infections with SARS, but some work was done on vaccine development, perhaps there may be a cohort for this study, or an animal model could be produced. These cases represent a spread of dates and a cross-section of the population, each with its own risk factors and dates of exposure. It is obvious that if secondary COVID existed, then as the pandemic progresses, we will get natural serial infections in these cohorts. All we will have to do is to ask if the patient is a member of any of these "risk factor" groups to ascertain if prior exposure is effective at protection, inducing complications, or has no impact.
Furthermore, the continued MERS periodic infections are also a relevant group, and the co-existence of MERS often with SARS, although they are highly genetically distinct, presents interesting challenges. They have different spikes and very different immunological signatures. It is possible that even if secondary COVID existed, it would not be induced/sensitized or agonized by MERS infections. Thus, this group is an interesting test bed for observations. However, the general prediction is that since some strains of Dengue are not associated with secondary dengue syndrome, this more distantly related virus will prove to be similar. An additional consequence of assuming that any cryptic SARS-like COVID virus may cause ARDS is that we should be able to see about a small rate of non-SARS-CoV-2 ARDS cases late in the pandemic. There should be no reason that a cryptic COVID infection after a lab-confirmed SARS-CoV-2 case should not risk ARDS. The observability of this should go up after we begin to achieve herd immunity to the monotype SARS-COV-2.
While it is less likely that the other common cold coronavirus that has different spikes such as α-coronaviruses (NL63 and 229E) would sensitize a secondary syndrome with β-coronaviruses, they may do so within their related groups. There are few laboratories that confirmed cases of ARDS that were traced to some of these other strains [
187,
188]. Thus, it might be presumed that if secondary COVID syndrome existed, there may be more set types such as APN binding strains of α-coronaviruses engaged in secondary α-COVID syndrome and SARS-like ACE2 viruses causing the secondary β-COVID syndrome. While again there is no firm evidence of this, if one phenotype is proven true then other related ones may exist. Further, advanced surveillance of cryptic COVID infections may demonstrate/disprove this. Ideal groups to evaluate for variation in ARDS rates are:
Successful post-2008 bone marrow transplants.
Recent or concurrent chemotherapy patents.
Persons taking immunomodulatory drugs.
Observations of persons with selected immune system mutations and variants.
The convalescent populations (of all 3 SARS-like outbreaks).
The convalescent populations (of other less related coronavirus infections and another virus).
Isolated populations that have had no history of bat vector or SARS-like exposure (Iceland).
Testing for T-cell cytokine responses in patients with SARS-like and other antigens in ARDS patients or serendipitous banked samples pre-ARDS.
Persons with a negative or cryptically positive SARS-CoV-2 test and presumptive ARDS.
7.5. Impacts of Theoretical Secondary COVID Syndrome
It cannot be overstated that the authors certainly do not recommend avoiding any of the SARS-CoV-2 vaccine products made available publicly, it is of paramount importance to vaccinate to stop the spread of this pandemic as soon as possible. We may need to be aware that if Ig responses do wane rapidly, the pandemic persists long enough to "re-infect", and such reinfection has a poor outcome, so we must be vaccinated. Failure to obtain good vaccine uptake would also result in a new wave(s) of COVID-19 (and any risk of subsequent secondary COVID) later. However, more unprotected COVID-19 infections now will result in more death in the short term, while not changing the long-term risk of exposure regardless. Thus, vaccine or no vaccine, we may be at risk of mass secondary COVID if that syndrome exists in the next coronavirus outbreak. If there is little risk of secondary COVID syndrome (theory rejected) then this also does not alter our immediate trajectory, viz. vaccination to save lives in the near term. We may, however, have to continue the vaccination program funding and make new strain-type boosters, as needed, like the influenza program, to compensate for the lives lost and save lives, in the future.
Presumptively if given enough time SARS-CoV-2 or another SARS-like virus will provide new emergent strains periodically through antigenic drift or recombination (or both). The infection rate and global distribution of the pandemic viral strain have now been such that "after-shocks" or waves of minor variants are virtually assured phenomena for the next several years if the behavior of past virus (H7N9 for example) outbreaks are projected on SARS-CoV-2 [
190]. Thus, we cannot know exactly what the SARS-CoV-2 pandemic will require of us in terms of fiscal resources in the near (1-3 years) or long-term (20 years) future. The impact of a larger population where secondary COVID syndrome becomes an established phenomenon (theory accepted) being potentially more susceptible to ARDS after any subsequent cryptic COVID infection is just one of these possible hurdles to overcome. Our medical social responsibility will not change, nor will the core tools at our disposal to deal with it.
8. COVID-19 Therapeutics, Treatment, and Vaccine Development
Intensive work was conducted in the US (under Operation warp speed) and internationally to develop novel treatments and vaccines to prevent COVID-19. Additionally, there have been several attempts to use existing medications to treat COVID-19. Most obviously other approved antivirals were nucleoside analogues and RNA-dependent RNA polymerase inhibitors. This important class of antiviral agents generally interferes with cellular nucleotide synthesis and terminates viral genome replication by targeting the RNA-dependent RNA polymerase, needed for RNA virus replication. There has been a growing understanding of SARS-CoV-2, and since then several drugs have been approved to mitigate SARS-COV2, albeit recently during a hearing on the European Union’s COVID-19 response, Pfizer’s president of internationally developed markets, stated [
191] that its vaccine had never been tested before its release to the general public on its ability to prevent the transmission of COVID. According to an article in August 2022 [
192], “The Pfizer BioNTech (BNT162b2) COVID-19 vaccine: What you need to know”, it states that there “is modest vaccine impact on transmission” to prevent COVID infection. We list below several vaccine development attempts by researchers.
8.1. Favipiravir (T-705)
A guanine nucleoside analogue, a viral RNA polymerase inhibitor approved for influenza virus and hemorrhagic fever virus infections treatment [
193], has been reported by a study to be a potential candidate for COVID-19 treatment in Vero E6 cells [
59]. For the treatment of COVID-19 patients, favipiravir has been used in combination with other antiviral agents like interferon-α (ChiCTR-2000029600) or baloxavir marboxil (ChiCTR-2000029544) [
194].
8.2. Ribavirin
It is also a guanine analogue, used for hepatitis C virus (HCV) and RSV infections treatment, and has been used to treat patients with SARS or MERS [
141,
195]. However, ribavirin has shown several side effects such as anemia when given in high doses for treatment [
141,
196]. Ribavirin has been administered for COVID-19 treatment in combination with pegylated interferon (ChiCTR2000029387) in lower doses to stimulate innate antiviral responses [
194].
8.3. Remdesivir (GS-5734)
It is an adenine analogue and an approved HIV reverse transcriptase inhibitor [
197]. This drug has also shown antiviral activity against MERS-CoV and SARS-CoV in human airway epithelial (HAE) cells and inhibited MERS-CoV replication in mice [
198]. Remdesivir was developed for the Ebola virus infection treatment [
199]. In the US, the first reported SARS-CoV-2-infected patient was admitted and administered Remdesivir [
200]. The antiviral activity of Remdesivir for SARS-CoV-2 was tested in Vero cells [
35]. Two phase III clinical trials were initiated to test Remdesivir efficiency and safety in patients with SARS-CoV-2 (NCT04252664 and NCT04257656) and have emerged as the most promising candidate for COVID-19 infection treatment [
201,
202].
8.4. Chloroquine
Chloroquine is a cheap unprotonated anti-malarial drug that can diffuse through the cell membrane, becomes protonated, accumulates in acidic organelles such as lysosomes, and increase the intracellular pH [
203,
204], and was used for the treatment of COVID-19 [
205]. Previous studies have shown that chloroquine also possesses broad-spectrum antiviral activity [
206,
207,
208]. An in vitro study revealed that chloroquine is a promising antiviral agent against SARS-CoV-2 infection in Vero E6 cells [
59]. To meet the urgent demand of health authorities, chloroquine was evaluated in clinical trials (ChiCTR2000029609). In China, COVID-19 patients were treated with chloroquine to test the efficacy and safety of this drug against SARS-CoV-2 infection. The results of these clinical trials demonstrated that chloroquine inhibits the severity of COVID-19 [
209]. Based on this finding, chloroquine is suggested to be effective against SARS-CoV-2 and has been recommended that this drug be included in the guidelines for the prevention, diagnosis, and treatment of pneumonia COVID-19 patients [
209]. Despite the advantages of chloroquine as an inhibitor of SARS, in practice, this treatment is no longer recommended for SARS-CoV-2 as it appears to offer no survival advantage, and its administration, like others, is not without a finite risk [
210,
211,
212].
8.5. Azithromycin
It is a macrolide-type antibiotic and is used to treat several bacterial infections. In a study, azithromycin showed promising results as an anti-influenza A (H1N1) virus drug [
181]. In vitro, evidence has also shown that azithromycin exhibits anti-viral activity against Zika virus [
213] and Ebola virus [
214]. Another clinical study has shown that azithromycin significantly reinforces the efficacy of hydroxychloroquine in patients with severe COVID-19 by eliminating the virus after the administration of the combination of this two-drug therapy [
215]. Despite the promise posed by the combination of hydroxychloroquine and azithromycin, in practice, this treatment is no longer recommended for SARS-COV-2, as it appears to offer no survival advantage (outside of lung infections that may warrant an antibiotic of this type) and its general usage is not without risk [
211].
8.6. Protease Inhibitors
Lopinavir and Ritonavir, are protease inhibitors used in the treatment of human immunodeficiency virus (HIV) patients. Lopinavir and Ritonavir regimen in combination with ribavirin was revealed to be effective in SARS-CoV patients and in vitro [
216,
217]. For the treatment of SARS-CoV-2, a clinical trial (ChiCTR2000029539) using Lopinavir and Ritonavir was initiated [
194].
8.7. Immunomodulatory
Dexamethasone and other steroids are becoming an interesting avenue of study for SARS-CoV-2 treatment. It has been reported in a press release for the RECOVERY trial, that 8-26% better outcomes are seen if 6 mg of Dexamethasone is provided in a study of over 4000 persons [
218]. Other studies, while not all showing the same level of effect, are describing this treatment as well-tolerated and likely to reduce the duration of hospital stays [
218,
219,
220]. This is one of the few success stories of the older already approved drugs suitable to treat SARS-CoV-2 infections.
8.8. REGN-COV2
Regeneron Pharmaceuticals, Inc., on September 29, 2020, announced the first data from a descriptive analysis of Phase 1/2/3 trials of its investigational antibody cocktail REGN-COV2 showing it reduced viral load and the time to alleviate symptoms in non-hospitalized patients with COVID-19. REGN-COV2 also showed positive trends in reducing medical visits. The ongoing, randomized, double-blind trial measured the effect of adding REGN-COV2 to the usual standard of care, compared to adding a placebo to standard-of-care. This trial was part of a larger program that also included studies of REGN-COV2 for the treatment of hospitalized patients and for the prevention of infection in people who have been exposed to COVID-19 patients. Hence, REGN-COV2 was the most advanced treatment in COVID-19 antiviral antibody drugs [
221].
8.9. LY-CoV555
AbCellera on August 3, 2020, announced that LY-CoV555 (a neutralizing antibody against SARS-CoV-2), a human antibody discovered by AbCellera in collaboration with the National Institute of Allergy and Infectious Diseases (NIAID) Vaccine Research Center (VRC) and co-developed with Eli Lilly and Company (Lilly) as a potential treatment and prophylaxis for COVID-19, progressed to Phase 3 clinical trials. A first-of-its-kind, the Lilly-sponsored trial used customized mobile research units to conduct the study at long-term care facilities across the United States. The study included up to 2,400 participants and assessed the ability of LY-CoV555 to prevent infection of long-term care residents and facility staff who were exposed to SARS-CoV-2 [
222].
8.10. LY-CoV555 and LY-CoV016
Eli Lilly and Company on October 7, 2020, announced that data from of the BLAZE-1 (NCT04427501) clinical trial showed that combination therapy with LY-CoV555 and LY-CoV016, Lilly's SARS-CoV-2 neutralizing antibodies reduced viral load, symptoms and COVID-related hospitalization and ER visits. The randomized, double-blind, placebo-controlled Phase 2 study evaluated LY-CoV555 and LY-CoV016, which bind complementary regions of the SARS-CoV-2 spike protein, for the treatment of symptomatic COVID-19 in the outpatient setting. The combination treatment was given to enrolled diagnosed patients with mild-to-moderate COVID-19. The combination treatment reduced viral levels at day 3 (p=0.016) and day 7 (p<0.001) [
223]. The improvement in symptoms was observed as early as three days after dosing and was similar in magnitude and timing to improvements previously seen with LY-CoV555 monotherapy. The rate of COVID-related hospitalization and ER visits was lower for patients treated with combination therapy (0.9%) versus placebo (5.8%), with a relative risk reduction of 84.5% (p=0.049). Combination therapy has been generally well tolerated with no drug-related serious adverse events. In LY-CoV555 monotherapy studies, there were isolated drug-related infusion reactions or hypersensitivity that were generally mild. Treatment-emergent adverse events were comparable to placebo for both LY-CoV555 monotherapy and combination therapy. For monotherapy, Lilly has been focused on the 700 mg dose of LY-CoV555 since similar clinical effects were seen across all dose levels tested in BLAZE-1. Lilly supplied as many as one million doses of 700 mg LY-CoV555 monotherapy in Q4 2020, and 50,000 doses of combination therapy in Q4 2020 with ramped-up production in Q1 2021 [
223].
8.11. AZD7442
AstraZeneca, which is also a frontrunner in the vaccine field, announced on October 9, 2020, that its long-acting antibody (LAAB) combination, AZD7442, will advance into two Phase III clinical trials in more than 6,000 participants at sites in and outside the US. LAAB was designed to increase the durability of the therapy for six to 12 months following a single administration. The combination of two LAABs was also designed to reduce the risk of resistance developed by the SARS-CoV-2 virus [
224].
AZD7442 is a combination of two LAABs derived from convalescent patients after SARS-CoV-2 infection. Discovered by Vanderbilt University Medical Center and licensed to AstraZeneca in June 2020, the LAABs were optimized by AstraZeneca with half-life extension and reduced Fc receptor binding. The half-life extended LAABs should afford six to 12 months of protection from COVID-19 [
225,
226,
227,
228]. The reduced Fc receptor binding aims to minimize the risk of antibody-dependent enhancement of disease - a phenomenon in which virus-specific antibodies promote, rather than inhibit, infection and/or disease [
229].
LAABs mimic natural antibodies and have the potential to treat and prevent disease progression in patients already infected with the virus, as well as to be given as a preventative intervention prior to exposure to the virus. A LAAB combination could be complementary to vaccines as a prophylactic agent, e.g., for people for whom a vaccine may not be appropriate or to provide added protection for high-risk populations. It could also be used to treat people who have been infected [
224].
The Company received the support of around
$486 million from the US government for the development and supply of AZD7442 under an agreement with the Biomedical Advanced Research and Development Authority (BARDA), part of the Office of the Assistant Secretary for Preparedness and Response (ASPER) at the US Department of Health and Human Services (DHHS), and the Department of Defense Joint Program Executive Office for Chemical, Biological, Radiological and Nuclear Defense (DoD-JPEO-CBRN) [
224]. One trial evaluated the safety and efficacy of AZD7442 to prevent infection for up to 12 months, in approximately 5,000 participants. The second trial evaluated post-exposure prophylaxis and pre-emptive treatment in approximately 1,100 participants. AstraZeneca planned additional trials to evaluate AZD7442 in approximately 4,000 patients for the treatment of COVID-19 [
224]. AstraZeneca supplied the US government with an additional one million doses in 2021 under a separate agreement [
224]. In a recent publication, the LAABs were shown in two pre-clinical murine models of SARS-CoV-2 infection to recognize unique sites, COV2-2196 and COV2-2130, that bound simultaneously to the receptor-binding domain of the S protein (SRBD) and synergistically neutralized the SARS-CoV-2 virus, blocking the binding of the SARS-CoV-2 virus to host cells and protected against infection in cell and animal models of disease [
230]. These results have identified protective epitopes on the SRBD and provide a structure-based framework for vaccine development and the selection of robust immunotherapeutic cocktails.
8.12. BRII-196 and BRII-198
China’s Brii Biosciences, on March 31, 2020, announced a collaboration with Tsinghua University, 3rd People’s Hospital of Shenzhen, to discover, develop, manufacture, and commercialize human-neutralizing monoclonal antibodies [
231]. Two monoclonal antibodies, BRII-196 (NCT04479631) and BRII-198 (NCT04479644) were tested in healthy subjects in Phase 1, with the potential for a combination to address the global pandemic of COVID-19. In both BRII-196 and BRII-198, monoclonal antibodies were isolated from a patient who had recovered from COVID-19. BRII-196 binds to a highly conserved epitope on the S protein and has the potential of becoming an effective therapy against the COVID-19 pandemic. BRII-198 binds to a different epitope on the S protein and has an additive to synergistic effect when combined with BRII-196. It has the potential of becoming an effective therapy against the COVID-19 pandemic [
232].
8.13. The Johnson & Johnson’s Janssen COVID-19 (J&J/Janssen) Vaccine
Johnson & Johnson, had temporarily paused further testing of all COVID-19 vaccine candidate clinical trials, including the Phase 3 ENSEMBLE trial, due to an unexplained illness in a study participant. Following their guidelines, the participant’s illness was reviewed and evaluated by the ENSEMBLE independent Data Safety Monitoring Board (DSMB) as well as their internal clinical and safety physicians [
233]. The temporary halt of the vaccine was lifted, and CDC and FDA recommended that the use of J&J/Janssen COVID-19 vaccine resume in the United States, effective April 23, 2021 [
234]. Johnson & Johnson on September 23, 2020, announced the launch of its large-scale, pivotal, multi-country Phase 3 trial (ENSEMBLE) for its COVID-19 vaccine candidate, JNJ-78436735, developed by its Janssen Pharmaceutical Companies. The initiation of the ENSEMBLE trial followed positive interim results from the Company’s Phase 1/2a clinical study, which demonstrated that the safety profile and immunogenicity after a single vaccination were supportive of further development. Based on these results and following discussions with the U.S. FDA, ENSEMBLE enrolled up to 60,000 volunteers 18 years old and older, across three continents (Argentina, Brazil, Chile, Colombia, Mexico, Peru, South Africa, and the United States) and studied the safety and efficacy of a single vaccine dose versus placebo in preventing COVID-19. In the US, COVID-19 Phase 3 trial program included a significant representation of Black, Hispanic/Latinx, American Indian, and Alaskan Native participants [
205]. Johnson & Johnson has continued the scaling up of its manufacturing capacity and remains on track to meet its goal of providing one billion doses of a vaccine each year. The company is committed to bringing an affordable vaccine to the public on a not-for-profit basis for emergency pandemic use and the first batches of a COVID-19 vaccine were available for emergency use authorization in early 2021 after proven to be safe and effective [
235].
ENSEMBLE is being initiated in collaboration with the BARDA, part of the Office of the Assistant Secretary for Preparedness and Response at the U.S. HHS under Other Transaction Agreement (OTA) HHSO100201700018C, and the National Institute of Allergy and Infectious Diseases (NIAID), part of the National Institutes of Health (NIH) at HHS [
205]. In parallel, the company has also agreed in principle to collaborate with the United Kingdom of Great Britain and Northern Ireland (the UK government) on a separate Phase 3 clinical trial in multiple countries to explore a two-dose regimen of Janssen’s vaccine candidate [
235].
8.14. Monoclonal and Polyclonal Antibodies
The rapid development of therapeutic drugs or vaccines to treat emerging diseases is challenged by time, to identify the drug compound, and to test its desired efficacy, safety, and pharmacokinetic profile. Antibodies have long been known to protect us against infections. A combination of antibodies targeting multiple epitopes on the virus has better viral neutralizing ability than the administration of single monoclonal antibodies (mAbs) [
236]. MAbs or polyclonal antibodies have been suggested as a prophy-lactic treatment tool for influenza, targeting hemagglutinin binding [
237]. An immune-based therapy study showed that human mAbs only provided protection against early-stage disease caused by MERS-CoV in mouse models [
238]. A human polyclonal antibody SAB-301 was generated in trans-chromosomic cattle for purified Al-Hasa strain MERS-CoV S protein. The results of this study showed that a single infusion of 50 mg/kg of SAB-301 was well tolerated and safe in healthy participants of the Phase 1 clinical trial [
239]. The use of human mAbs provides an attractive avenue to treat emerging diseases. However, for COVID-19, isolation of antibodies from survivors is limited due to the lack of plasma available, and combinational therapeutics with mAbs treatment are also limited because of the high cost of antibodies production and possible toxicity.
8.15. Convalescent Plasma
Convalescent plasma has been the last treatment resort to improve the survival rate of patients with various viral infections, such as SARS, avian influenza (H5N1), pandemic 2009 influenza A H1N1 (H1N1 pdm09), and severe Ebola virus infection [
240,
241]. One possible explanation for the efficacy of using convalescent plasma for treatment is that the antibodies in the patient’s plasma who have recovered from viral infection might suppress viremia. A study conducted in China [
241] reported case series of five critically ill patients with laboratory-confirmed COVID-19 and ARDS, who received transfusion with convalescent plasma obtained from five patients who recovered from COVID-19 with SARS-CoV-2–specific antibody (IgG). All five patients had received antiviral agents and methylprednisolone and at the time of treatment with convalescent plasma, they also received mechanical ventilation.
Following plasma transfusions, improvements were observed in their clinical condition, including the normalization of body temperature, a decrease in Sequential Organ Failure Assessment score, a rise in PaO2/FiO2, resolution of ARDS, a success of weaning from mechanical ventilation, the decline in viral loads, and increase in SARS-CoV-2–specific antibody and neutralizing antibody titers. Hence, the use of convalescent plasma transfusion is beneficial among patients infected with SARS-CoV-2, but further study needs to be conducted in large sample sizes to validate the efficacy of the usage of convalescent plasma. Despite the work proposed by Duan et al. [
242], in practice, this treatment appears to offer no survival advantage in a review of larger studies and its usage is not without risk [
243,
244]. Most notably there is a risk of antibody-dependent disease enhancement [
244].
8.16. Molnupiravir (Merck’s COVID-19 Pill)
Molnupiravir [Emory Institute for Drug Development [EIDD]-1931], is an oral ribonucleoside analog. It is a beta-D-N4-hydroxycytidine and an inhibitor of RNA-dependent RNA polymerase, which possesses significant activity against SARS-CoV-2. Its prophylactic efficacy has been evidenced in a ferret model by completely blocking SARS-CoV-2 transmission to untreated animals. Two Phase-I trials (NCT04392219 and NCT04746183) have demonstrated that oral molnupiravir is safe and well-tolerated at therapeutic doses [
245]. After taking four pills of molnupiravir orally twice a day for five days, the symptoms lessened, reducing the likelihood of being hospitalized, and reducing the death rate (
https://www.nationalgeographic.com/science/article/how-mercks-antiviral-pill-could-change-the-game-for-covid-19), by eliminating nasopharyngeal virus in patients with early and mild COVID-19 (NCT04405739 and NCT 04405570). Two Phase-II/III trials, NCT04575597 and NCT04575584, were completed, with estimated enrollments of 1850 and 304 cases, respectively. The NCT04575597 recently released that molnupiravir significantly reduced the risk of hospitalization or death in adults experiencing mild or moderate COVID-19 [
215].
The FDA in December 2021 approved emergency authorization [
246,
247] for Merck’s molnupiravir, a COVID-19 oral antiviral pill that reduces the number of hospitalization and death cases. It is an additional treatment option for some high-risk COVID-19 patients. The drug should not be administered to pregnant women, but physicians can prescribe it to pregnant women at their discretion. Females of childbearing age are required to use contraceptives during treatment for five days. Males of childbearing age are also advised to use contraception during treatment and for three months following their last dose. Molnupiravir is not authorized for patients younger than 18 years of age. Some data from animals indicate that the drug might affect bone and cartilage growth [
248].
9. SARS-CoV-2 Prevention
Vaccination is the most effective cornerstone strategy of public health for preventing infectious disease because vaccines stimulate an individual’s immune response to produce immunity to that specific disease. Vaccination is more cost-effective than other treatment strategies and reduces morbidity and mortality without long-lasting side effects [
249,
250]. Over the past two decades, three human coronaviruses, SARS-CoV, MERS-CoV, and SARS-CoV-2 have emerged globally, causing a considerable threat to public health [
251]. However, there are no approved vaccines for human coronaviruses. Scientists and pharmaceutical companies around the world accelerated the development of COVID-19 vaccines. Human ACE2 transgenic mouse model for SARS-CoV-2 has been well established for vaccine development, providing an understanding of the pathogenesis of COVID-19 [
252], and some SARS-CoV-2 vaccines were under clinical trial [
224,
253,
254].
Natural conformational proteins or even viruses can be inactivated and preserved as the basis for a vaccine. In the case of coronavirus, the S protein of coronaviruses is an important target for vaccine development because it mediates the disease by binding the receptor on the host cells [
255,
256,
257]. T- cell epitopes, mRNA, and S protein-RBD structure-based vaccines were proposed and development started [
157,
258]. Rapid reconstruction of SARS-CoV-2 has been reported using a yeast-based synthetic genomics platform, which can be a helpful technical tool in vaccine development [
259]. In the case of coronavirus, there are no known live attenuated strains suitable for vaccine development. However, recombinant vaccines can be developed by taking the same core epitopes (say the spike gene) that may be employed in inactivated vaccines and inserting them into the attenuated virus of other types (such as adnoivirus5) to make a live recombinant vaccine.
DNA vaccines have been used for coronavirus for over two decades when conventional protein/peptide-based vaccines were demonstrated to induce antigen-specific adaptive immune responses. Whereas plasmid DNA vaccines are more stable, cost-effective, easy to manufacture, safe in handling, have a long shelf life, and contribute autoimmunity to diseases, infections, cancer, and allergies [
260].
Recently, mRNA vaccines have generated significant interest as a rapidly developing technology to complement or even replace traditional vaccines because mRNA vaccines combine the advantages of subunit vaccines and live-attenuated vaccines without the risks associated with live-attenuated or DNA vaccines [
261]. Successful cytosolic delivery of mRNA, encoding for an antigen, results in vaccine epitope synthesis of the transfected host cells [
262,
263]. Hence, mRNA vaccines have advantages over conventional vaccines, by the absence of genome integration, improved immune responses due to the presence of specific antigens in the cytosol enabling the presentation of both endogenous and exogenous antigens, and providing T-cell activation while being safe, rapid development technology, and the production of multimeric antigens [
262,
263,
264].
9.1. Recombinant Vaccine AZD1222
A vaccine developed by Oxford University and AstraZeneca, known as AZD1222 (ChAdOx1 nCoV-19], an adenovirus-vectored vaccine, targets the S protein present on the surface of SARS-CoV-2. Phase 1/Phase 2 studies generated two immune responses: neutralizing antibodies and T-cell response in more than 1,000 participants from 18 to 55 years, who were not previously tested positive for COVID-19. AZD1222 is one of the 24 potential COVID-19 vaccines tested in clinical trials, according to WHO. AZD1222 was tested in phase 3 clinical trials by Oxford-AstraZeneca in 2020. In this study 20,000 participants received AZD1222 and 10,000 participants received saline placebo. The estimated date of the completion of the study was February 24, 2023.
9.2. Recombinant Vaccine Ad5
Recombinant adenovirus type-5 (Ad5) vectored COVID-19 vaccine targets the S protein present on the surface of SARS-CoV-2. Phase 1 of the study was conducted in Wuhan, China, in healthy adults aged 18 to 60 years. Ad5 vectored COVID-19 vaccine is reported to be tolerable and immunogenic at 28 days post-vaccination. Neutralizing antibodies increased significantly at day 14 and peaked 28 days post-vaccination. Specific T-cell response peaked at day 14 post-vaccination (NCT04313127). Ad5 vectored COVID-19 vaccine, was jointly developed by the Beijing Institute of Biotechnology, part of the Chinese government's Academy of Military Medical Sciences and CanSino Biologics [
265].
9.3. Recombinant Vaccine S-Trimer
Clover Biopharmaceutical was in pre-clinical testing of a recombinant COVID-19 S protein subunit trimer called S-Trimer [
266]. Clover Biopharmaceuticals confirmed that S-Trimer has no mutation to the amino acid sequences, generation of native-like trimer structure and antigenic epitopes of S protein and detected high titer of cross-reacting antibodies in the sera of previously infected COVID-19 patients in China. Clover Biopharmaceuticals and GSK announced a partnership to improve immune response by introducing GSK’s adjuvant system (AS03) to S-Trimer [
266].
9.4. Inactivated Vaccine CoronaVac
Sinovac’s Vaccine, CoronaVac is based on the whole inactivated virus, a vaccine technology that is also been used to produce influenza and polio vaccines, compared to the western COVID-19 vaccine producers focusing on DNA or RNA of the SARS-CoV-2. Two doses of CoronaVac, induced neutralizing antibodies 14 days after vaccination in more than 90% of 600 healthy volunteers in phase 2 clinical trials. Sinovac conducted its phase 3 trial in Brazil with Brazilian immunobiological producer, Instituto Butantan. Coro-naVac was also among the 24 COVID-19 vaccines being tested in phase 3 clinical trials [
267].
9.5. DNA Vaccine of S glycoprotein of SARS-CoV
A study was conducted in BALB/c mice using the plasmid DNA Vaccine encoding S glycoprotein of SARS-CoV. The results of the study showed that the expression vectors induced robust immune responses mediated by CD4 and CD8 T-cells, significant neutralizing antibody titers, and protection against the virus was mediated by a humoral immune mechanism and not by a T-cell-dependent immune mechanism. Hence, DNA-based vaccination for SARS-CoV elicited an effective immune response that generates protective immunity.
9.6. DNA Vaccine INO-4800
INOVIO Pharmaceuticals in partnership with Beijing Advaccine Biotechnology Company was on track for Phase II/III clinical trials for DNA vaccine (INO-4800) against COVID-19 S protein after generating positive Phase I result from (NCT04336410) [
268]. According to INOVIO, preliminary analyses of its Phase I results showed that 34 out of 36 (94%) trial participants demonstrated overall immune responses after receiving two doses of INO-4800 in the study. Participants were enrolled into 1.0 mg or 2.0 mg dose cohorts, with each participant receiving two doses of INO-4800 four weeks apart [
269]. INOVIO Pharmaceuticals with the support from the Coalition for Epidemic Preparedness Innovations (CEPI), the Bill and Melinda Gates Foundation, and the U.S. Department of Defense, contributed significant funding to the advancement and manufacturing of INO-4800 [
270]. INOVIO conducted the Phase II segment of the Phase II/III trial in the United States and obtained positive data. INO-4800 continues to be safe and well-tolerated and supports the body’s ability to generate neutralizing antibodies and T-cell responses, which can protect against current and emerging variants. INO-4800 does not require cold or ultra-cold chain transport [
271].
9.7. mRNA Vaccine mRNA-1273
The candidate vaccine mRNA-1273 by ModernaTX Inc., in partnership with the Biomedical Advanced Research and Development Authority (BARDA) and the National Institute of Allergy and Infectious Diseases (NIAID) Vaccine Research Center, encodes the stabilized prefusion SARS-CoV-2 S protein [
54,
272]. The mRNA-1273 vaccine induced anti–SARS-CoV-2 immune responses in all participants in the phase 1 clinical trial and no safety concerns were identified (NCT04283461) [
30,
241]. Moderna completed the phase 2 trial of mRNA-1273 in healthy adults, evaluating doses of 50 μg and 100 μg (NCT04405076) [
253,
273]. A large phase 3 mRNA-1273 efficacy trial expected to evaluate a 100 μg dose in collaboration with NIAID and BARDA started in the summer of 2020 [
253]. Moderna is one of the COVID-19 vaccines administered in the United States. Evidence from clinical trials shows that 94.1% of individuals aged 18 years and older, who received two doses of the Moderna vaccine were prevented from acquiring COVID-19 infection and had no evidence of being infected previously [
274].
9.7.1. BNT162b2
BNT162b2 (Pfizer-BioNTech. NCT04368728) is a lipid nanoparticle–formulated, nucleoside-modified RNA vaccine that encodes a prefusion stabilized, membrane-anchored SARS-CoV-2 full-length spike protein. A two-dose regimen of BNT162b2 conferred 95% protection against COVID-19 in individuals over 16 years of age [
275]. The two doses are given 21 days apart (Dose 1 Day 0 and dose 2 Day 21). BNT162b2 has been authorized by FDA to be given to children 5 years of age to teens 15 years of age.
9.7.2. Booster Doses
A booster dose is recommended to people who are fully vaccinated and whose immune response has weekend over time. Individuals 18 years and older, who have received two doses of BNT162b2 and Moderna, are recommended to receive a single booster dose at least six months after receiving the second dose. Whereas for Janssen/Johnson & Johnson COVID-19 Vaccine, people who are 18 years and older, who have received one dose of the vaccine, and has been two months after getting their first dose are recommended to get a single booster dose. Individuals can choose to get the booster dose of the same vaccine that they received previously or choose another vaccine.
10. Lessons Learned from COVID-19 – Rational for Preventing Transmission
The world had recently witnessed several outbreaks of rare and deadly diseases such as Ebola virus disease (EVD), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), Creutzfeldt-Jacob disease (CJD), also known as mad cow disease, etc., yet COVID-19 pandemic has left an indelible mark in our memory for a long time to come. Its impact on the world economy, health, and education has been massive and is likely to be long-lasting. Although the world is slowly recovering from such a massive shock, a few facts remain worth noting, viz. COVID-19 created a global recession that has been the deepest since the end of World War II, the global economy contracted significantly according to the World Economic Outlook Report published by the International Monetary Fund (IMF), and while virtually every country covered by the IMF posted negative growth, the downturn was more pronounced in the poorest parts of the world.
In addition to the overall economy, the impact on human health, social well-being, and global education have been pronounced. The health impact of COVID-19 is direct, as discussed earlier, and indirect. Coronaviruses spread among people through droplets from coughs, sneezes, or breathing. The droplets may land on another person on an item such as a door handle. If someone else touches the handle, the virus may pass on to them if they touch their mouth, nose, or eyes. Once inside the body, coronaviruses mostly affect the respiratory system, including the nose and lungs. However, some viruses and the immune reaction they trigger can have a wider impact. Indirect health impacts also include obesity, inactivity, high blood pressure (HBP), and computer vision syndrome (CVS) resulting in eye strain and discomfort. Research shows that between 50% and 90% of people who work at a computer screen have at least some symptoms.
A central early response to the spread of SARS-CoV-2 was the lockdown, an extreme restriction on personal movement to promote isolation and viral burnout due to non-transmission. Unfortunately, this approach largely failed to prevent the repeated global spread of multiple COVID-19 variants. However, the lockdowns due to COVID-19 have resulted in social and behavioral interaction impacts. As per Centers for Disease Control and Prevention, “Younger adults, racial/ethnic minorities, essential workers, and unpaid adult caregivers reported having experienced disproportionately worse mental health outcomes, increased use of controlled substances and elevated suicidal ideation.” Studies show that this pandemic is also associated with distress, anxiety, fear of contagion, depression, and insomnia in the general population and even among healthcare professionals. Social isolation, anxiety, fear of contagion, uncertainty, chronic stress, and economic difficulties may lead to the development or exacerbation of depression, anxiety, substance use, and other psychiatric disorders in vulnerable populations including individuals with pre-existing psychiatric disorders. Stress-related psychiatric conditions including mood and substance use disorders and are associated with suicidal behavior. Furthermore, the outbreak has affected all segments of the population and is particularly detrimental to members of those social groups in the most vulnerable situations continues to affect populations, including people living in poverty situations, older persons, persons with disabilities, youth, and indigenous peoples.
The impact of COVID-19 on education across the world has been unprecedented and devastating. Over 1.6 billion learners in 190 countries had been affected by school closures, which happened on a scale never seen before. This, however, is a short-term impact and the real impact of lockdowns will not be known for quite some time to come. While ad hoc solutions were implemented to address the sudden need for remote and online education, the pandemic highlighted how unprepared international education systems were to offer education in emergencies as disaster preparedness. However, with difficult decisions regarding public health, the economy, and other social impacts, widespread school closures are closely connected to the wider socio-economic effects of the COVID-19 pandemic.
As we transition out of the pandemic, it is vital to consider longer-term implications and actively seek support and collaboration in finding long-term solutions to address wider issues that society seemed so unprepared for. As established earlier, one can predict that there may be another pandemic very likely in the future. From the lessons learned from COVID-19, the world may be somewhat better prepared to counter another pandemic in the future. While it takes a while to effectively establish policies and guidelines for alternate means to offer education, conduct work remotely for non-required Jobsite employees, and ways to address social issues, one policy that does make the difference is how to prevent transmission. Do masks make any difference? What kind of masks are necessary? For social distancing, what is an optimum social spatial distance? What other precautions must be taken to prevent transmission – such as personal hygiene and what is the optimum? Hence it is critical to conduct aerosol modeling to reduce the probability of transmission. This may reduce, if not eliminate, the probability of transmission, which coupled with masks and other personal hygiene protocols will minimize the probability of inhaling any variant of the COVID-19 virus.
10.1. Aerosol Modeling based Social Distancing and Personal Protective Equipment
Upon observing COVID-19’s second wave and third waves, the logarithmic explosion of cases (in a matter of few weeks), one can only postulate mechanisms other than direct contact, viz. by aerosolized droplets – giving credence to “patient zero” theory, also discussed in Wuhan, either at the Wuhan Institute of Virology (WIV) or wet markets in Wuhan, as suggested in news media. Speculations aside, considering the respiratory nature of the virus, it is plausible to consider diffusion by aerosol to understand the social distancing protocols and if any personal protective equipment (PPE), such as masks can be used as a preventive measure.
In general, microbial aerosols are generated in indoor as well as outdoor environments because of a variety of natural and anthropogenic activities. Wind, rain, and wave splash spray irrigation, waste-water treatment activity, cooling towers and air handling water spray systems, and agricultural processes are examples of activities that generate bio-aerosols outdoors. A bio-aerosol is an airborne collection of biological- material comprised of bacterial cells and cellular fragments, fungal spores and fungal hyphae, viruses, and by-products of microbial metabolism. Pollen grains and other biological material can also be airborne as a bio-aerosol.
Table 1 below provides typical particle size and natural background concentration of bioaerosols.
Corona Viruses (CoV) are large enveloped non-segmented positive-sense RNA viruses. Viral- envelopes consist of proteins and lipid components and enveloped -viruses require the fusion of their lipid- envelope with the host cell membrane to enter the infected cells. Interaction of the SARS-CoV-2 envelope with particulate matter (PM) is possible in the aqueous surrounding. After drying, PM can serve as a carrier for transmission of SARS-CoV-2 immobilized at their surface. The lipid constituents of the viral envelope can be very important for the unspecific interaction of viral particles with different surfaces, including air-pollution PM. It should be emphasized that this interaction capability can be inherent mainly to enveloped viruses.
Several investigations have been conducted to study the diffusion of aerosolized particles with different, % RH (relative humidity), temperature, etc., have been conducted. In a controlled study conducted by Alford et al. [
276], volunteers were exposed to carefully titrated aerosolized influenza-virus suspensions by inhaling through a face- mask. The demonstration of infection in participants of the study was achieved by recovery of infectious viruses from throat swabs, taken daily, or by seroconversion, that is, the development of neutralizing- antibodies. The use of carefully titrated viral- stocks enabled the determination of the minimal -infectious dose by aerosol inoculation. The approximate 50% human-infectious dose (HID50-50% human infectious dose) of virus per volunteer was from 1 to 126 TCID50 (the tissue culture 50% infectious dose). The dose for half of the volunteers was 5 TCID50. The other half of the men, who had very low or non-detectable pre-inoculation antibody titers, were infected with 0.6 to 3 TCID50. The study reliably shows that the human-infectious dose of the influenza-A virus, when administered by aerosol to subjects free of serum-neutralizing antibodies, is approximately 3 TCID50. The approaches used in this study allow [
277] the precise number of infectious- particles in the total number of particles to be determined and show infection by aerosolized particulate matter.
A detailed lung-particle interaction model is necessary for a complete study, however, based on earlier experiments, it is conclusive to note that there is significant potential for inhalation exposure to viruses in microscopic- respiratory droplets (micro-droplets) at short to medium distances (up to several meters, or room scale). It is thus recommended to use preventive -measures to mitigate this route of airborne- transmission, especially in indoor and heavily occupied settings, realizing that viruses are released during exhalation, talking, and coughing in micro-droplets small enough to remain aloft in the air and pose a risk of exposure at distances beyond 1–2 m from an infected- individual. at typical indoor-air velocities, a 5-μm droplet will travel tens of meters, much greater than the scale of a typical room, while settling from a height of 1.5 m to the floor. While a mask and social distance of 1 meter may not be ideal, it is still better prevention than none. Increasing circulation air velocity and reduction in relative humidity may improve somewhat, as is evident from
Table 2 below shows the effect of relative humidity on the infectivity of the airborne virus.
10.2. Disease Surveillance: Bioinformatics, Contact Tracing
Disease surveillance is an information-based activity involving the collection, analysis, and interpretation of large volumes of data originating from a variety of sources. The information gathered is used in a variety of ways, such as to evaluate the effectiveness of control and preventative health measures, monitor the propagation of infectious agents, support health planning and the allocation of appropriate resources within the healthcare system, identify high-risk populations or areas to target interventions and provide a valuable progression of disease activity for archive and future reference.
Although the city of origin for COVID-19 is well documented, there is still a lot of discussion on the exact source and mode. The discussion has ranged from accidental release from WIV to wet markets around Wuhan and if the virus was zoonotic or non-zoonotic in nature. There have been several congressional debates over gain-of-function research, in which laboratories might engineer new properties into existing viruses that may theoretically render them more potent, as “superviruses”. In fact, the Intercept produced new evidence that the WIV and the nearby Wuhan University Center for Animal Experiment, along with their collaborator, the U.S.-based nonprofit EcoHealth Alliance, engaged in the “gain-of-function research of concern,”, as defined by the U.S. government in intentionally making viruses more pathogenic or transmissible to study them. This work was carried out despite stipulations from a U.S. funding agency that the money not be used for the purpose of “gain-of-function research”. Such activities may be categorized as dual-use research of concern (DURC), research that possesses both the properties of intellectual pursuit and those of a possible agent of concern or threat activity.
To fully consider the potential for SARS-CoV-2 to be a lab strain, viz. to have the functionality of an engineered strain, both source tracing, in the context of disease surveillance, and
in-silico bioinformatics analyses are needed to analyze and interpret whole-genome sequence data to develop an informed opinion. The
in-silico analyses can be implemented in a laboratory setting easily, affordably, and, in some cases, without the need for intensive computing resources and infrastructure. While SARS-CoV-2 certainly has novel features upon analysis and there is no clear consensus as to if it was an engineered viral strain or if it indeed escaped from a lab, however, Homes et al [
278] postulate that it is unlikely in both cases. The matter is still unsettled, at least in public debate, and future studies will likely continue to examine such possibilities.
Bioinformatics is an essential methodology in forecasting and responding to disease outbreaks and incidents of regional, national, and international significance. To be effective, the collection of surveillance data must be standardized on a national basis and made available at local, regional, and national levels. Baldwin et al. [
279] reported an automated sequencing analytics methodology suited for the detection/identification and classification of mixtures of Chemical, Biological, Radiological, and Nuclear (CBRN) agents of concern. The method provides simplified answers (or Go/No Go) responses), especially suited for a kinetic environment. The method operates regardless of the sequencer vendor and no special user skill or foreknowledge is required. Its operation in a standalone or cloud environment, coupled with artificial intelligence (AI) decision support, is likely to improve its readiness in disease surveillance and contact tracing – a close contact evaluation and monitoring hierarchy. Different protocols are in place but in general, contact tracing is the process used to identify those who encounter people who have tested positive for many contagious diseases – such as measles, HIV, and COVID-19. It is not the same as "exposure notification" or "digital alerting" tools. Consumer apps, such as those created with Google and Apple's API, are not contact tracing tools. These apps function as a way for the public to track if they have encountered a person who has tested positive and entered that information into their phone database.
10.3. Data Analytics for Informed Public Health Decision Making
Artificial intelligence (AI) has transformed the world around us and produced capabilities that have led to a wide range of innovations [280,281]. Complex algorithms and systems have evolved using logic rules and reasoning algorithms that mimic human thought processes. The introduction of decision-support tools provided the capability of complex and hierarchical reasoning but, unlike humans, they could not learn new rules to evolve and expand their decision-making capability. Biologically inspired software, also known as “neural networks,” provided the additional capability to identify complex patterns and complete complex tasks. These new capabilities are complemented by big data and data analytics – the process of examining raw datasets to find trends, draw conclusions and identify the potential for improvement. Healthcare analytics uses current and historical data to gain insights, macro, and micro, and support decision-making at both the patient and business levels. There are a variety of tools and systems used to gather and analyze health data gathered through various means, viz. Electronic Health Records (EHRs), Personal Health Records (PHRs), Electronic Prescription Services (E-prescribing), Patient Portals, Master Patient Indexes (MPI), Health-Related Smart Phone Apps, and more. With digital data collection, there is a vast amount of data to be analyzed in real-time. Big data and health data analytics have played an integral role in the fight against COVID-19 as analyzing data in real-time has allowed for a better understanding of trends, supply of PPEs, medical care availability, and hence decision-making in healthcare. The data analytics capability in conjunction with the wide-spectrum bio-threats identification and classification scheme, as reported by Baldwin et al. [
279], promises a tremendous capability to fight against such pandemics in the future.
11. Conclusions and Path Forward
Together with the understanding of COVID-19 as an emerging disease caused by SARS-CoV-2, diagnostics, therapeutics, and treatment platforms are available, and various pharmaceutical companies in partnership with research institutions worldwide have developed vaccines that are administered worldwide. There has been some progress in the treatment of the illness and globally death rates are now significantly reduced, and all remaining restrictions have been lifted globally. However, there is still much to do, there is no permanent cure for the COVID-19 infection currently. The care is largely supportive. Furthermore, as the pandemic evolves this infection will be a multiple-year event. Collectively, it is estimated that approximately 10% of the world had been exposed to COVID-19 during the peak period (around October 2020) and consequently, an estimated over 90% of the globe will remain susceptible to COVID-19. With the progression of time, a certain percentage of the population will see their antibody profile wane each month and potentially become susceptible again. If this occurs on the order of months and is frequent, herd immunity cannot be established naturally and will see a steady state of sporadic recurrences until a vaccine is widely available and more people get vaccinated. This is also confounded by the fact that there is significant evidence of continuous new novel COVID-19 variants emerging globally. Thus, either through the duration of the SARS-CoV-2 pandemic itself or through natural common cold COVID strains, we are likely to have a very good look at the theory of secondary COVID. It will be critical to accurately assess if prior coronavirus exposure will alter the impact (be protective, neutral, or harmful) with respect to the expected continued infections of SARS-COV-2.
The most important impact if secondary COVID syndrome is proven to exist is the immediate need to make a continuous coronavirus vaccine program based perhaps on the influenza vaccine program framework and release an annual or periodical COVID vaccine and or required boosters. This can be employed much like the Dengue or flu vaccines if the research supports it, with vaccines after the pandemic going to those known to be at risk of secondary COVID based on medical history or Ig titer testing. Descriptively, if one vaccinated a mass population versus dengue 1 in an outbreak to save many lives but becomes later aware of secondary dengue then one must immediately proceed to vaccinate, those previously treated for the new strains to prevent the syndrome. Thus, while theoretical secondary COVID syndrome is concerning, this should not change our vaccine trajectory in the short term.
If the outbreak lasts (this may be many years) news variants circulate, and the theoretical drop of coronavirus antibodies may also complicate things with required boosters. We may need to add a global ongoing program for the development of vaccines against COVID-19 and any future strains of this virus group to eradicate this disease. This may require a new annual or semiannual vaccination program to boost public immunity and provide protection, so we can once again reduce our social distance and regain our collective economic prosperity. However, for this pandemic to end, enormous funding and long-term clinical research studies will be required to compensate for the short and long-term impacts.
This also raises the question, that how we can, as humans, prepare for the unknown. While it is obvious that it is difficult to develop specific PCR tests for the unknown. But there are technologies like DNA/RNA sequencing which are more expensive but can detect novel agents. Considering the cost of this pandemic, would it not be prudent to spend more on a fraction of the testing and possibly prevent the next one? It is easy to presume that if China (or wherever the next outbreak starts) has used sequencing as the standard protocol of care for virus detection, then quarantine would have been much earlier and may have been vastly more effective. Regardless of the technology, we as humans need to be able to detect novel pathogens routinely in this highly mobile world to avert much of the risk of future pandemics. However, the bigger question remains, if the awareness of the historical fact that pandemics are a regular feature of human existence means that humans can somehow become sufficiently prescient to avoid virus surprises arising from novel circumstances.
Author Contributions
Conceptualization, A.V., S.N., and J.B.; methodology, A.V., S.N., and J.B.; software, N.A.; validation, A.V., S.N., and J.B.; formal analysis, A.V., S.N., and J.B.; investigation, A.V., S.N., and J.B.; resources, A.V., S.N., and J.B.; data curation, A.V., S.N., and J.B.; writing—original draft preparation, A.V., S.N., and J.B.; writing—review and editing, A.V., S.N., and J.B.; visualization, A.V., S.N., and J.B.; supervision, N.A.; project administration, A.V., S.N., and J.B.; funding acquisition, N.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Most of the data is available in the text. Additional data will be made available by the authors upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest - financial or otherwise.
References
- Zhou, F., Yu, T., Du, R., Fan, G., Liu, Y., Liu, Z., Xiang, J., Wang, Y., Song, B., Gu, X., et al., Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet, 2020, 395(10229): 1054-1062. PMID: 32171076. PMCID: PMC7270627. [CrossRef]
- Cheng, Z.J., Shan, J., 2019 Novel coronavirus: where we are and what we know. Infection, 2020, 48(2):155-163. PMID: 32072569. PMCID: PMC7095345. [CrossRef]
- Wu, Z., McGoogan, J.M., Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA, 2020, PMID: 32091533. [CrossRef]
- Pan, X., Chen, D., Xia, Y., Wu, X., Li, T., Ou, X., Zhou, L., Liu, J., Asymptomatic cases in a family cluster with SARS-CoV-2 infection. The Lancet Infectious Diseases, 2020, 20(4):410-411. PMID: 32087116. PMCID: PMC7158985. [CrossRef]
- Bai, Y., Yao, L., Wei, T., Tian, F., Jin, D.Y., Chen, L., Wang, M., Presumed asymptomatic carrier transmission of COVID-19. JAMA, 2020, 323(14):1406-1407. PMID: 32083643. PMCID: PMC7042844. [CrossRef]
- Rothe, C., Schunk, M., Sothmann, P., Bretzel, G., Froeschl, G., Wallrauch, C., Zimmer, T., Thiel, V., Janke, C., Guggemos, W., et al. Transmission of 2019-nCoV Infection from an asymptomatic contact in Germany. The New England Journal of Medicine, 2020, 382(10):970-971. PMID: 32003551. PMCID: PMC7120970. [CrossRef]
- Holshue, ML, DeBolt, C, Lindquist, S, Lofy, KH, Wiesman, J, Bruce, H, Spitters, C, Ericson, K, Wilkerson, S., Tural, A., et al. First Case of 2019 Novel Coronavirus in the United States. The New England Journal of Medicine, 2020, 382(10): 929-936. PMID: 32004427. PMCID: PMC7092802. [CrossRef]
- Lee, P.I., Hsueh, P.R., Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol Immunol Infect. 2020, 53(3):365-367. Epub 2020 Feb 4. PMID: 32035811; PMCID: PMC7102579. [CrossRef]
- Huang, W.H. Teng, LC, Yeh, TK, Chen, YJ, Lo, WJ, Wu, MJ, Cin, CD, Tsan, YT, Lin, TC, Chai, JW, et al. 2020. 2019 novel coronavirus disease (COVID-19) in Taiwan: reports of two cases from Wuhan, China. Journal of Microbiology, Immunology, and Infection, 2020, 53(3):481-484. PMID: 32111449. PMCID: PMC7102546. [CrossRef]
- Burke, RM, Midgley, CM, Dratch, A, Fenstersheib, M, Haupt, T, Holshue, M, Ghinai, I, Jarashow, MC, Lo, J, McPherson, TD, et al. 2020. Active monitoring of persons exposed to patients with confirmed COVID-19 - United States, January-February 2020. MMWR. Morbidity and Mortality Weekly Report, 2020, 69(9):245–246. PMID: 32134909. [CrossRef]
- WHO Director-General's opening remarks at the media briefing on COVID-19 -11 March 2020. 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020. Accessed on July 14, 2020.
- Hamming, I., Timens, W., Bulthuis, M.L.C., Lely, A.T., Navis, G.V., & van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland, 2004. 203(2), 631-637. [CrossRef]
- Lamers, M.M., Beumer, J., van der Vaart, J., Knoops, K., Puschhof, J., Breugem, T.I., Ravelli, R.B.G., van Schayck, J.P., Mykytyn, A.Z., Duimel, H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science, 2020, 369(6499):50-54. [CrossRef]
- Wu, Y., Ho, W., Huang, Y., Jin, D.Y., Li, S., Liu, S.L., Liu, X., Qiu, J., Sang, Y., Wang, Q., et al. SARS-CoV-2 is an appropriate name for the new coronavirus. The Lancet, 2020, 395(10228), 949-950. [CrossRef]
- Piret, J., Boivin, G. Pandemics throughout history. Frontiers in Microbiology, 2021, 11: 631736. [CrossRef]
- Samal, J. A historical exploration of pandemics of some selected diseases in the world. International Journal of Health Sciences and Research, 2014, 4(2):165-169.
- Sampath, S., Khedr, A., Qamar, S., Tekin, A., Singh, R., Green, R., Kashyap, R. Pandemics throughout history. Cureus, 2021, 13(9):e18136. [CrossRef]
- Huebner, S.R., The “Plague of Cyprian”: A revised view of the origin and spread of a 3rd-c. CE pandemic. Journal of Roman Archaeology, 2021, 34(1):151-174. [CrossRef]
- Harper, K., Pandemics and passages to late antiquity: rethinking the plague of c. 249–270 described by Cyprian. Journal of Roman Archaeology, 2015, 28:223-260. [CrossRef]
- Donoghue, H.D., Taylor, G.M., Marcsik, A., Molnár, E., Pálfi, G., Pap, I., Teschler-Nicola, M., Pinhasi, R., Erdal, Y.S., Velemίnsky, P., et al. A migration-driven model for the historical spread of leprosy in medieval Eastern and Central Europe. Infection, Genetics, and Evolution, 2015, 31: 250-256. 13m. [CrossRef]
- Geddes, A.M., The history of smallpox. Clinics in Dermatology, 2006, 24(3):152-157. [CrossRef]
- Patterson, K.B., Runge, T. Smallpox and the Native Americans. The American Journal of the Medical Sciences, 2002, 323(4):216-222. [CrossRef]
- Roberts, L., Disease and Death in the New World: Historians now agree that the European discovery of the Americas touched off waves of epidemics, but a debate is raging over the size of pre-Columbian populations. Science, 1989, 246(4935):1245-1247. [CrossRef]
- Carlos, A.M., Lewis, F.D., Smallpox and Native American mortality: The 1780s epidemic in the Hudson Bay region. Explorations in Economic History, 2012, 49(3):277-290. [CrossRef]
- Alfani, G., Murphy, T.E., Plague and lethal epidemics in the pre-industrial world. The Journal of Economic History, 2017, 77(1):314-343. [CrossRef]
- Glatter, K.A., Finkelman, P., History of the plague: An ancient pandemic for the age of COVID-19. The American Journal of Medicine, 2021, 134(2):176-181. [CrossRef]
- Morens, D.M., Measles in Fiji, 1875: thoughts on the history of emerging infectious diseases. Pacific Health Dialog, 1998, 5(1):119-128.
- Berche, P., The enigma of the 1889 Russian flu pandemic: A coronavirus? La Presse Médicale, 2022, 51(3):104111. [CrossRef]
- Lavigne, S.E., A history of pandemics: Lessons we can learn from the past. Canadian Journal of Dental Hygiene, 2020, 54(2): pp. 55-57.
- Junqueira, D.M., de Medeiros, R.M., Matte, M.C.C., Araújo, L.A.L., Chies, J.A.B., Ashton-Prolla, P., & Almeida, S.E.D.M., Reviewing the history of HIV-1: the spread of subtype B in the Americas. 2011, PloS one, 6(11):e27489. [CrossRef]
- Arno, P.S., Hughes, R.G., Local policy responses to the AIDS epidemic: New York and San Francisco. Acquired Immunodeficiency Syndrome: Current Issues and Scientific Studies, 1989, 11-19. [CrossRef]
- Lama, J., Planelles, V., Host factors influencing susceptibility to HIV infection and AIDS progression. Retrovirology, 2007, 4:52. [CrossRef]
- Zhong, N.S., Zheng, B.J., Li, Y.M., Poon, L.L.M., Xie, Z.H., Chan, K.H., Li, P.H., Tan, S.Y., Chang, Q., Xie, J.P., et al., Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February 2003. The Lancet, 2003, 362(9393):1353-1358. [CrossRef]
- de Wit, E., van Doremalen, N., Falzarano, D., Munster, V.J., SARS and MERS: recent insights into emerging coronaviruses. Nature Reviews Microbiology, 2016, 14(8):523-534. PMID: 27344959. PMCID: PMC7097822. [CrossRef]
- Reperant, L.A., Osterhaus, A.D.M.E., AIDS, Avian flu, SARS, MERS, Ebola, Zika… what next? Vaccine, 2017, 35(A):4470-4474. PMID: 28633891. PMCID: PMC7115365. [CrossRef]
- Zhu, H., Wei, L., Niu, P., The novel coronavirus outbreak in Wuhan, China. Global Health Research and Policy, 2020, 5:6. PMID: 32226823. PMCID: PMC7050114. [CrossRef]
- World Health Organization. (WHO), Naming the coronavirus disease (COVID-19) and the virus that causes it. Brazilian Journal of Implantology and Health Sciences, 2020, 2(3).
- Coronaviridae Study Group (of the International Committee on Taxonomy of Viruses), The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 2020, 5: 536–544. [CrossRef]
- Cui, J., Li, F., Shi, Z.L., Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology, 2019, 17(3):181-192. PMID: 30531947. PMCID: PMC7097006. [CrossRef]
- Hulswit, RJG, deHaan, CAM, Bosch, BJ., Coronavirus spike protein and tropism changes. Advances in Virus Research, 2016, 96:29-57. PMID: 27712627. PMCID: PMC7112277. [CrossRef]
- Fehr, A.R., & Perlman, S., Coronaviruses: an overview of their replication and pathogenesis. In Coronaviruses. 2015, pp. 1-23. Humana Press, New York, NY.
- Denison, MR, Graham, RL, Donaldson, EF, Eckerle, LD, Baric RS., An RNA proofreading machine regulates replication fidelity and diversity. RNA Biology, 2011, 8(2):270-279. PMID: 21593585. PMCID: PMC3127101. [CrossRef]
- Tortorici, M.A., & Veesler, D., Structural insights into coronavirus entry. In Advances in virus research, 2019, Vol. 105, pp. 93-116. Academic Press. [CrossRef]
- Walls, A.C., Park, Y.J., Tortorici, M.A., Wall, A., McGuire, A.T., & Veesler, D., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 Dec 10;183(6):1735. Erratum for: Cell. 2020 Apr 16;181(2):281-292.e6. PMID: 33306958; PMCID: PMC7833104. [CrossRef]
- Starr, T.N., Greaney, A.J., Hilton, S.K., Ellis, D., Crawford, K.H., Dingens, A.S., ... & King, N.P., Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell, 2020, 182(5), 1295-1310. [CrossRef]
- Kim, C.H., SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus–Host Interaction. International Journal of molecular sciences, 2020, 21(12), 4549. [CrossRef]
- Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F., Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences, 2020, 117(21), 11727-11734. [CrossRef]
- Kim, S.Y., Jin, W., Sood, A., Montgomery, D.W., Grant, O.C., Fuster, M.M., ... & Linhardt, R.J., Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Research, 2020, 181, 104873. [CrossRef]
- Pirone, L., Del Gatto, A., Di Gaetano, S., Saviano, M., Capasso, D., Zaccaro, L., Pedone, E., A multi-targeting approach to fight SARS-CoV-2 attachment. Frontiers in molecular biosciences, 2020, 7:186. PMID: 32850973; PMCID: PMC7416604. [CrossRef]
- Wu, A, Peng, Y, Huang, B, Ding, X, Wang, X, Niu, P, Meng, J, Zhu, Z, Zhang, Z, Wang, J., et al. Genome composition and divergence of the novel Coronavirus (2019-nCoV) originating in China. Cell Host Microbe, 2020, 27(3):325–328. PMID: 32035028. PMCID: PMC7154514. [CrossRef]
- Sola, I, Almazan, F, Zuniga, S., Enjuanes, L., Continuous and discontinuous RNA synthesis in Coronaviruses. Annual Review of Virology, 2015, 2(1):265–288. PMID: 26958916. PMCID: PMC6025776. [CrossRef]
- Gorbalenya, AE, Enjuanes, L, Ziebuhr, J, Snijder, EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Research 117(1):17–37. PMID: 16503362. PMCID: PMC7114179. [CrossRef]
- Luan, J, Lu, Y, Jin, X, Zhang, L., Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochemical and Biophysical Research Communications, 2020, 526(1):165-169. PMID: 32201080. PMCID: PMC7102515. [CrossRef]
- Wrapp, D, Wang, N, Corbett, KS, Goldsmith, JA, Hsieh, CL, Abiona, O, Graham, BS, McLellan, JS., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 2020, 367(6483):1260-1263. PMID: 32075877. PMCID: PMC7164637. [CrossRef]
- Wang, C, Horby, PW, Hayden, FG, Gao, GF., A novel coronavirus outbreak of global health concern. Lancet, 2020, 395(10223):470-473. PMID: 31986257. PMCID: PMC7135038. [CrossRef]
- Wu, F, Zhao, S, Yu, B, Chen, YM, Wang, W, Song, ZG, Hu, Y, Tao, ZW, Tian, JH, Pei, YY, et al., A new coronavirus associated with human respiratory disease in China. Nature, 2020, 579(7798):265–269. PMID: 32015508. PMCID: PMC7094943. [CrossRef]
- Dufour, D, Mateos-Gomez, PA, Enjuanes, L, Gallego, J, Sola, I., Structure and functional relevance of a transcription-regulating sequence involved in coronavirus discontinuous RNA synthesis. Journal of Virology, 2011, 85(10):4963–4973. PMID: 21389138. PMCID: PMC3126183. [CrossRef]
- Mateos-Gomez, PA, Morales, L, Zuniga, S, Enjuanes, L., Sola, I., Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. Journal of Virology, 2013, 87(1):177–186. PMID: 23055566. PMCID: PMC3536410. [CrossRef]
- Wang, K, Chen, W, Zhou, YS, Lian, JQ, Zhang, Z, Du, P, Gong, L, Zhang, Y, Cui, HY, Geng, JJ, et al., SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. 2020. [CrossRef]
- Hoffmann, M, Kleine-Weber, H, Schroeder, S, Krüger, N, Herrler, T, Erichsen, S, Schiergens, TS, Herrler, G, Wu, NH, Nitsche, A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and ss blocked by a clinically proven protease inhibitor. Cell, 2020, 181(2):271-280. PMID: 32142651. PMCID: PMC7102627. [CrossRef]
- Yang, N., Shen, HM. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int J. Biol Sci. 2020, 16(10):1724-1731. PMID: 32226290; PMCID: PMC7098027. [CrossRef]
- Xia, S, Liu, M, Wang, C, Xu, W, Lan, Q, Feng, S, Qi, F, Bao, L, Du, L, Liu, S, Qin, C, Sun, F, Shi, Z, Zhu, Y, Jiang, S, Lu, L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 4:343-355. Epub 2020 Mar 30. PMID: 32231345; PMCID: PMC7104723. [CrossRef]
- Gordon, DE, Jang, GM, Bouhaddou, M, Xu, J, Obernier, K, et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 2020, Jul;583(7816):459-468. Epub 2020 Apr 30. PMID: 32353859; PMCID: PMC7431030. [CrossRef]
- Brai, A, Martelli, F, Riva, V, Garbelli, A, Fazi, R, Zamperini, C, Pollutri, A, Falsitta, L, Ronzini, S., Maccari, L, et al., DDX3X helicase inhibitors as a new strategy to fight the West Nile Virus infection. Journal of Medicinal Chemistry, 2019, 62(5):2333–2347. PMID: 30721061. [CrossRef]
- Brai, A, Fazi, R, Tintori, C, Zamperini, C, Bugli, F, Sanguinetti, M, Stigliano, E, Este, J, Badia, R, Franco, S, et al., Human DDX3 protein is a valuable target to develop broad-spectrum antiviral agents. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(19):5388–5393. PMID: 27118832. PMCID: PMC4868442. [CrossRef]
- Snider, J.M., You, J.K., Wang, X., Snider, A.J., Hallmark, B., Zec, M.M., Seeds, M.C., Sergeant, S., Johnstone, L., Wang, Q., Sprissler, R., et al., Group IIA secreted phospholipase A2 is associated with the pathobiology leading to COVID-19 mortality. Journal of Clinical Investigation, 2021, 131(19):e149236. PMID: 34428181. PMCID: PMC8483752. [CrossRef]
- Nevalainen, T.J., Haapamäki, M.M., Grönroos, J.M., Roles of secretory phospholipases A2 in inflammatory diseases and trauma. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2000, 1488(1-2):83-90. [CrossRef]
- Yokota, Y., Hanasaki, K., Ono, T., Nakazato, H., Kobayashi, T., Arita, H., Suppression of murine endotoxic shock by sPLA2 inhibitor, Indoxam, through group IIA sPLA2-independent mechanisms. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 1999, 1438(2):213-222. [CrossRef]
- World Health Organization (WHO). 2003. Update 49 - SARS case fatality ratio, incubation period. https://www.who.int/csr/sars/archive/2003_05_07a/en/. Accessed on May 15, 2020.
- World Health Organization. 2019. Middle East respiratory syndrome coronavirus (MERS-CoV). https://www.who.int/emergencies/mers-cov/en/ Accessed on May 15, 2020.
- Ksiazek, TG, Erdman, D, Goldsmith, CS, Zaki, SR, Peret, T, Emery, S, Tong, S, Urbani, C, Comer, JA, Lim W, et al., A novel coronavirus associated with severe acute respiratory syndrome. The New England Journal of Medicine, 2003, 348(20):1953-1966. PMID: 12690092. [CrossRef]
- Peiris, JSM, Lai, ST, Poon, LLM, Guan, Y, Yam, LYC, Lim, W, Nicholls, J, Yee, WKS, Yan, WW, Cheung, MT et al., Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet, 2003, 361(9366):1319-1325. PMID: 12711465. PMCID: PMC7112372. [CrossRef]
- Marra, MA, Jones, SJM, Astell, CR, Holt RA, Brooks-Wilson A, Butterfield YSN, Khattra J, Asano JK, Barber SA, Chan SY.. The genome sequence of the SARS-associated coronavirus. Science, 2003, 300(5624):1399-1404. PMID: 12730501. [CrossRef]
- Centers for Disease Control (CDC) [2017] https://www.cdc.gov/dotw/sars/index.html Accessed on July 30, 2020.
- Centers for Disease Control (CDC) [2020a]. https://www.cdc.gov/nchs/nvss/vsrr/covid19/excess_deaths.htm Accessed on August 20, 2020.
- Centers for Disease Control (CDC) [2020b]. https://www.cdc.gov/nchs/nvss/vsrr/covid19/excess_deaths.htm Accessed on October 09, 2020.
- Ip, M., Chan, P.K.S., Lee, N., Wu, A., Ng, T.K.C., Chan, L., Ng, A., Kwan, H.M., Tsang, L., Chu, I., et al., Seroprevalence of antibody to severe acute respiratory syndrome (SARS)–associated coronavirus among health care workers in SARS and non-SARS medical wards. Clinical Infectious Diseases, 2004, 38(12):e116-e118. [CrossRef]
- Wilder-Smith, A., Teleman, M.D., Heng, B.H., Earnest, A., Ling, A.E., Leo, Y.S., Asymptomatic SARS coronavirus infection among healthcare workers, Singapore. Emerging Infectious Diseases, 2005, 11(7):1142-1145. PMID: 16022801. PMCID: PMC3371799.. [CrossRef]
- Zhou, Y.H., Prevalence of non-pneumonic infections with SARS-correlated virus. The Lancet, 2004, 363(9423):1826-1827. PMID: 15172787. PMCID: PMC7134706. [CrossRef]
- Zaki, AM, van Boheemen, S, Bestebroer, TM, Osterhaus, ADME, Fouchier, RAM., Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. The New England Journal of Medicine, 2012, 367(19):1814-1820. PMID: 23075143. [CrossRef]
- De Groot, RJ, Baker, SC, Baric, RS, Brown, CS, Drosten, C, Enjuanes, L, Fouchier, RAM, Galiano, M, Gorbalenya, AE, Memish, ZA, et al., Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. Journal of Virology, 2013, 87(14):7790-7792. PMID: 23678167. PMCID: PMC3700179. [CrossRef]
- De Wit, E., Van Doremalen, N., Falzarano, D., Munster, V.J., SARS and MERS: recent insights into emerging coronaviruses. Nature Reviews Microbiology, 2016, 14(8), 523. [CrossRef]
- Kim, C.J., Choi, W.S., Jung, Y., Kiem, S., Seol, H.Y., Woo, H.J., Choi, Y.H., Son, J.S., Kim, K.-H., Kim, Y.-S., et al., Surveillance of the Middle East respiratory syndrome (MERS) coronavirus (CoV) infection in healthcare workers after contact with confirmed MERS patients: incidence and risk factors of MERS-CoV seropositivity. Clinical Microbiology and Infection, 2016, 22(10):880-886. PMID: 27475739. PMCID: PMC128923. [CrossRef]
- Al-Tawfiq, J.A., Gautret, P., Asymptomatic Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infection: extent and implications for infection control: a systematic review. Travel Medicine and Infectious Disease, 2019, 27:27-32. PMID: 30550839. PMCID: PMC7110966. [CrossRef]
- Petrosillo, N., Viceconte, G., Ergonul, O., Ippolito, G., Petersen, E., COVID-19, SARS and MERS: are they closely related? Clinical Microbiology and Infection, 2020, 26(6):729-734. PMID: 32234451. PMCID: PMC7176926. [CrossRef]
- Yu, I.T., Li, Y., Wong, T.W., Tam, W., Chan, A.T., Lee, J.H., ... & Ho, T., Evidence of airborne transmission of the severe acute respiratory syndrome virus. New England Journal of Medicine, 2004, 350(17):1731-1739. [CrossRef]
- Leung, G.M., Chung, P.H., Tsang, T., Lim, W., Chan, S.K., Chau, P., ... & Ferguson, N.M., SARS-CoV antibody prevalence in all Hong Kong patient contacts. Emerging infectious diseases, 2004, 10(9):1653. [CrossRef]
- Mo, H., Zeng, G., Ren, X., Li, H., Ke, C., Tan, Y., ... & Zhong, N., Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology, 2006, 11(1):49-53. [CrossRef]
- Kellam, P., Barclay, W., The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. Journal of General Virology, 2020, 101(8):791-797. PMID: 32430094; PMCID: PMC7641391. [CrossRef]
- Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings, O., O'Byrne, A., et al., Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020 Dec;5(12):1598-1607. Epub: 2020 Oct 26. PMID: 33106674; PMCID: PMC7610833. [CrossRef]
- Edridge AWD, Kaczorowska J, Hoste ACR, Bakker M, Klein M, Loens K, Jebbink MF, Matser A, Kinsella CM, Rueda P, Ieven M, Goossens H, Prins M, Sastre P, Deijs M, van der Hoek, L. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020 Nov.;26(11):1691-1693. Epub 2020 Sep 14. PMID: 32929268. [CrossRef]
- Yu, J., Qiao, S., Guo, R., & Wang, X., Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution. Nature Communications, 2020, 11(1):3070. PMID: 32555182. PMCID: PMC7300015. [CrossRef]
- Huang, X., Dong, W., Milewska, A., Golda, A., Qi, Y., Zhu, Q.K., ... & Li, W., Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. Journal of Virology, 2015, 89(14), pp. 7202-7213. PMID: 25926653. PMCID: PMC4473545. [CrossRef]
- Zhou, P., Fan, H., Lan, T., Yang, X-L, Shi, W-F, Zhang, W., Zhu, Y., Zhang, Y.-W., Xie, Q.-M., Mani, S., et al., Fatal swine acute diarrhea syndrome caused by an HKU-2-related coronavirus of bat origin. Nature, 2018, 556:255-258. [CrossRef]
- Wang, Q, Vlasova, AN, Kenney, SP, Saif, LJ. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. 2019 Feb;34:39-49. Epub 2019 Jan 14. PMID: 30654269; PMCID: PMC7102852. [CrossRef]
- Zhou, L., Li, Q.N., Su, J.N., Chen, G.H., Wu, Z.X., Luo, Y., ... & Ma, J.Y., The re-emerging of SADS-CoV infection in pig herds in Southern China. Transboundary and emerging diseases, 2019, 66(5), 2180-2183. [CrossRef]
- Pontelli, MC, Castro, ÍA, Martins, RB, La Serra, L, Veras, FP, Nascimento, DC, Silva, CM, Cardoso, RS, Rosales, R, Gomes, R, Lima, TM, Souza, JP, Vitti, BC, Caetité, DB, de Lima, MHF, Stumpf, SD, Thompson, CE, Bloyet, LM, Toller-Kawahisa, JE, Giannini, MC, Bonjorno, LP, Lopes, MIF, Batah, SS, Siyuan, L, Luppino-Assad, R, Almeida, SCL, Oliveira, FR, Benatti, MN, Pontes, LLF, Santana, RC, Vilar, FC, Auxiliadora-Martins, M, Shi, PY, Cunha, TM, Calado, RT, Alves-Filho, JC, Zamboni, DS, Fabro, AT, Louzada-Junior, P, Oliveira, RDR, Whelan, SPJ, Cunha, FQ, Arruda, E. SARS-CoV-2 productively infects primary human immune system cells in vitro and in COVID-19 patients. J. Mol. Cell Biol. 2022 Aug 17;14(4):mjac021. PMID: 35451490; PMCID: PMC9384834. [CrossRef]
- Meng, F., Ren, Y., Suo, S., Sun, X., Li, X., Li, P., ... & Herrler, G., Evaluation on the efficacy and immunogenicity of recombinant DNA plasmids expressing spike genes from porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. PLoS One, 2013, 8(3), e57468. [CrossRef]
- Mole, B., Deadly pig virus slips through US borders. Nature, 2013, 499:388. [CrossRef]
- Stevenson, GW, Hoang, H, Schwartz, KJ, Burrough, ER, Sun, D, Madson, D, Cooper, VL, Pillatzki, A, Gauger, P, Schmitt, BJ, et al., Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. Journal of Veterinary Diagnostic Investigation, 2013, 25(5):649-654. PMID 23963154. [CrossRef]
- Chen, Q, Li G, Stasko, J, Thomas, JT, Stensland, WR, Pillatzki, AE, Gauger, PC, Schwartz, KJ, Madson, D., Yoon, KJ, et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. Journal of Clinical Microbiology, 2014, 52(1):234-243. PMID: 24197882. PMCID: PMC3911415. [CrossRef]
- Yu, X., Ran, D., Wang, J., Qin, Y., Liu, R., Shi, X., ... & Zhou, J., Unclear but present danger: An asymptomatic SARS-CoV-2 carrier. Genes and Diseases, 2020, 7(4):558-566. PMID: 32837986. PMCID: PMC7377777. [CrossRef]
- Wang, N., Li, S.Y., Yang, X.L., Huang, H.M., Zhang, Y.J., Guo, H., ... & Hagan, E., Serological evidence of bat SARS-related coronavirus infection in humans, China. Virologica Sinica, 2018, 33(1), 104-107. [CrossRef]
- Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., ... & Xiang, Z., Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications, 2020, 11(1), 1-12. [CrossRef]
- Tai, W, Zhang, X, He, Y, Jiang, S., Du, L. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res. 2020 Jul;179:104820. Epub 2020 May 13. PMID: 32405117; PMCID: PMC7219369. [CrossRef]
- Braun, J, Loyal, L, Frentsch, M, Wendisch, D, Georg, P, Kurth, F, Hippenstiel, S, Dingeldey, M, Kruse, B, Fauchere, F, Baysal, E, Mangold, M, Henze, L, Lauster, R, Mall, MA, Beyer, K, Röhmel, J, Voigt, S, Schmitz, J, Miltenyi, S, Demuth, I, Müller, MA, Hocke, A, Witzenrath, M, Suttorp, N, Kern, F, Reimer, U, Wenschuh, H, Drosten, C, Corman, VM, Giesecke-Thiel, C, Sander, LE, Thiel, A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020 Nov.;587(7833):270-274. Epub 2020 Jul 29. PMID: 32726801. [CrossRef]
- Weiskopf, D, Schmitz, KS, Raadsen, MP, Grifoni, A, Okba, NMA, Endeman, H, van den Akker, JPC, Molenkamp, R, Koopmans, MPG, van Gorp, ECM, Haagmans, BL, de Swart, RL, Sette, A., de Vries, RD., Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020 Jun 26;5(48):eabd2071. PMID: 32591408; PMCID: PMC7319493. [CrossRef]
- Sette, A., Crotty, S., Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol. 2020 Aug;20(8):457-458. Erratum in: Nat. Rev. Immunol. 2020 Oct;20(10):644. PMID: 32636479; PMCID: PMC7339790. [CrossRef]
- Grifoni, A, Weiskopf, D, Ramirez, SI, Mateus, J, Dan, JM, Moderbacher, CR, Rawlings, SA, Sutherland, A, Premkumar, L, Jadi, RS, Marrama, D, de Silva, AM, Frazier, A, Carlin, AF, Greenbaum, JA, Peters, B, Krammer, F, Smith, DM, Crotty, S., Sette, A., Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020 Jun 25;181(7):1489-1501.e15. Epub 2020 May 20. PMID: 32473127; PMCID: PMC7237901. [CrossRef]
- Zhao, M., Zhang, H., Liu, K., Gao, G.F., & Liu, W.J., Human T-cell immunity against emerging and re-emerging viruses. Science China Life Sciences, 2017, 60(12), 1307-1316. [CrossRef]
- Janice-Oh, H.L., Ken-En Gan, S., Bertoletti, A., & Tan, Y.J., Understanding the T cell immune response in SARS coronavirus infection. Emerging microbes & infections, 2012, 1(1), 1-6. PMID: 26038429. PMCID: PMC3636424. [CrossRef]
- Zumla, A., Chan, JFW, Azhar, EI, Hui, DSC, Yuen, KY., Coronaviruses - drug discovery and therapeutic options. Nature Reviews Drug Discovery, 2016, 15(5):327-347. PMID: 26868298. PMCID: PMC7097181. [CrossRef]
- Alshukairi, AN, Zheng, J, Zhao, J, Nehdi, A, Baharoon, SA, Layqah, L, Bokhari, A, Al Johani, SM, Samman, N, Boudjelal, M, Ten Eyck, P, Al-Mozaini, MA, Zhao, J, Perlman, S, Alagaili, AN. High Prevalence of MERS-CoV Infection in Camel Workers in Saudi Arabia. mBio. 2018 Oct 30;9(5):e01985-18. PMID: 30377284; PMCID: PMC6212820. [CrossRef]
- Guo, YR, Cao, QD, Hong, ZS, Tan, YY, Chen, SD, Jin, HJ, Tan, KS, Wang, DY, Yan, Y., The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res., 2020, 7(1):11. PMID: 32169119. PMCID: PMC7068984. [CrossRef]
- Onofrio, L, Caraglia, M, Facchini, G, Margherita, V, Placido, S., Buonerba, C., Toll-like receptors and COVID-19: a two-faced story with an exciting ending. Future Sci OA, 2020, 6(8):FSO605. PMID: 32974046. PMCID: PMC7434222. [CrossRef]
- Brandão, SCS, Ramos, JOX, Dompieri, LT, Godoi, ETAM, Figueiredo, JL, Sarinho, ESC, Chelvanambi, S., Aikawa, M., Is Toll-like receptor 4 involved in the severity of COVID-19 pathology in patients with cardiometabolic comorbidities? Cytokine Growth Factor Rev., 2021, 58:102-110. PMID: 32988728. PMCID: PMC7505161. [CrossRef]
- Li, G, Fan, Y, Lai, Y, Han, T, Li, Z, Zhou, P, Pan, P, Wang, W, Hu, D, Liu, X., Zhang, Q., Wu, J., Coronavirus infections and immune responses. J. Med. Virol, 2020, 92(4):424–432. PMID: 31981224. PMCID: PMC7166547. [CrossRef]
- Vabret, N, Britton, GJ, Gruber, C, Hegde, S, Kim, J, Kuksin, M, Levantovsky, R, Malle, L, Moreira, A, Park, MD. et al., Immunology of COVID-19: Current State of the Science. Immunity, 2020, 52(6):910–941. PMID: 32505227. PMCID: PMC7200337. [CrossRef]
- Prompetchara, E, Ketloy, C, Palaga, T., Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol, 2020, 38(1):1–9. PMID: 32105090. [CrossRef]
- Ye, Q., Wang, B., Mao, J., The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect., 2020, 80(6):607–613. PMID: 32283152. PMCID: PMC7194613. [CrossRef]
- Zhang, W, Zhao, Y, Zhang, F, Wang, Q, Li, T, Liu, Z, Wang, J, Qin, Y, Zhang, X, Yan, X., Zeng, X., Zhang, S., The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin. Immunol, 2020, 214:108393. PMID: 32222466. PMCID: PMC7102614. [CrossRef]
- Atluri, S, Manchikanti, L, Hirsch, JA., Expanded Umbilical Cord Mesenchymal Stem Cells (UC-MSCs) as a Therapeutic Strategy in Managing Critically Ill COVID-19 Patients: The Case for Compassionate Use. Pain Physician, 2020, 23(2): E71–E83. PMID: 32214286.
- Conti, P, Ronconi, G, Caraffa, A, Gallenga, C, Ross, R, Frydas, I, Kritas, S. 2020. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol Regul Homeost Agents, 2020, 34(2):327–331. PMID: 32171193. [CrossRef]
- Xu, Z, Shi, L, Wang, Y, Zhang, J, Huang, L, Zhang, C, Liu, S, Zhao, P, Liu, H, Zhu, L., et al., Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med., 2020, 8(4):420–422. PMID: 32085846. PMCID: PMC7164771. [CrossRef]
- Qin, C, Zhou, L, Hu, Z, Zhang, S, Yang, S, Tao, Y, Xie, C, Ma, K, Shnag, K, Wang, W., Tian, DS., Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis., 2020, 71(15):762–768. PMID: 32161940. PMCID: PMC7108125. [CrossRef]
- Pedersen, SF, Ho, YC., SARS-CoV-2: a storm is raging. J. Clin. Invest., 2020,130(5):2202–2205. PMID: 32217834. PMCID: PMC7190904. [CrossRef]
- Rijkers, G, Vervenne, T, van der Pol, P., More bricks in the wall against SARS-CoV-2 infection: involvement of γ9δ2 T cells. Cell Mol. Immunol, 2020, 17(7):771–772. PMID: 32467616. PMCID: PMC7331628. [CrossRef]
- Wu, D, Yang, XO., TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J. Microbiol Immunol Infect., 2020, 53(3):368–370. PMID: 32205092. PMCID: PMC7156211. [CrossRef]
- Zhang, D, Guo, R, Lei, L, Liu, H, Wang, Y, Wang, Y, Qian, H, Dai, T, Zhang, T, Lai, Y., et al., Frontline Science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J. Leukoc Biol, 2021, 109(1):13–22. PMID: 33040384. PMCID: PMC7675546. [CrossRef]
- Qu, R, Ling, Y, Zhang, YHZ, Wei, LY, Chen, X, Li, XM, Liu, XY, Liu, HM, Guo, Z., Ren, H., et al., Platelet-to-lymphocyte ratio is associated with prognosis in patients with coronavirus disease-19. J. Med. Virol, 2020, 92(9):1533–1541. PMID: 32181903. PMCID: PMC7228291. [CrossRef]
- Channappanavar, R., Zhao, J., & Perlman, S., T cell-mediated immune response to respiratory coronaviruses. Immunologic research, 2014, 59(1-3), 118-128. PMID: 24845462. PMCID: PMC4125530. [CrossRef]
- Ibarrondo, F.J., Fulcher, J.A., Goodman-Meza, D., Elliott, J., Hofmann, C., Hausner, M.A., ... & Yang, O.O., Rapid Decay of Anti–SARS-CoV-2 Antibodies in Persons with Mild Covid-19. New England Journal of Medicine, 2020,383(11):1085-1087. PMID: 32706954. PMCID: PMC7397184. [CrossRef]
- Morens, D.M., Fauci, A.S., The 1918 influenza pandemic: insights for the 21st century. The Journal of Infectious Diseases, 2007, 195(7):1018-1028. PMID: 17330739. [CrossRef]
- Rabaan, A.A., Al-Ahmed, S.H., Haque, S., Sah, R., Tiwari, R., Malik, Y.S., ... & Rodriguez-Morales, A.J., SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infezioni in Medicina, 2020, 28(2):174-184. PMID: 32275259.
- Xiao, F., Sun, J., Xu, Y., Li, F., Huang, X., Li, H., ... & Zhao, J., Infectious SARS-CoV-2 in feces of a patient with severe COVID-19. Emerging Infectious Diseases, 2020, 26(8):1920-1922. PMID: 32421494. PMCID: PMC7392466. [CrossRef]
- Lee, N., & Sung, J.J., Nosocomial transmission of SARS. Current infectious disease reports, 2003, 5(6), 473-476. [CrossRef]
- Lau, S.K., Che, X.Y., Woo, P.C., Wong, B.H., Cheng, V.C., Woo, G.K., ... & Yuen, K.Y., SARS coronavirus detection methods. Emerging Infectious Diseases, 2005, 11(7):1108-1111. www.cdc.gov/eid. [CrossRef]
- Conly, J.M., & Johnston, B.L., SARS: A tale of two epidemics. Canadian Journal of Infectious Diseases, 2003, 14(3):147-149. PMID: 18159447. PMCID: PMC2094926. [CrossRef]
- Booth, T.F., Kournikakis, B., Bastien, N., Ho, J., Kobasa, D., Stadnyk, L., ... & Mederski, B., Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. The Journal of Infectious Diseases, 2005, 191(9):1472-1477. PMID: 15809906. PMCID: PMC7202477. [CrossRef]
- Agranovski, I.E., Safatov, A.S., Pyankov, O.V., Sergeev, A.N., Agafonov, A.P., Ignatiev, G.M., ... & Rabenau, H.F., Monitoring of viable airborne SARS virus in ambient air. Atmospheric Environment, 2004, 38(23):3879-3884. PMID: 32288549. PMCID: PMC7129584. [CrossRef]
- McKinney, K.R., Gong, Y.Y., & Lewis, T.G., Environmental transmission of SARS at Amoy Gardens. Journal of Environmental Health, 2006, 68(9):26-30. PMID: 16696450.
- Pastorino, B., Touret, F., Gilles, M., de Lamballerie, X., & Charrel, R.N., Prolonged infectivity of SARS-CoV-2 in fomites. Emerging Infectious Diseases, 2020, 26(9):2256-2257. PMID: 32579874. PMCID: PMC7454106. [CrossRef]
- Ong, S.W.X., Tan, Y.K., Chia, P.Y., Lee, T.H., Ng, O.T., Wong, M.S.Y., & Marimuthu, K., Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA, 2020, 323(16):1610-1612. PMID: 32129805. PMCID: PMC7057172. [CrossRef]
- Orive, G., Lertxundi, U., & Barcelo, D., Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Science of The Total Environment, 2020, 732:139298. PMID: 32417555. PMCID: PMC7207139. [CrossRef]
- Liu, Z., Xiao, X., Wei, X., Li, J., Yang, J., Tan, H., ... & Liu, L., Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. Journal of Medical Virology, 2020, 92(6):595-601. PMID: 32100877. PMCID: PMC7228221. [CrossRef]
- Van Doremalen, N., Bushmaker, T., Morris, D.H., Holbrook, M.G., Gamble, A., Williamson, B.N., ... & Lloyd-Smith, J.O., Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. New England Journal of Medicine, 2020, 382(16):1564-1567. PMID: 32182409. PMCID: PMC7121658. [CrossRef]
- Kang, M., Wei, J., Yuan, J., Guo, J., Zhang, Y., Hang, J., ... Peng, X., Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Annals of Internal Medicine, 2020, 173(12):974-980. PMID: 32870707. PMCID: PMC7464151. [CrossRef]
- Miller, S.L., Nazaroff, W.W., Jimenez, J.L., Boerstra, A., Buonanno, G., Dancer, S.J., ...Noakes, C., Transmission of SARS-CoV-2 by inhalation of respiratory aerosol in the Skagit Valley Chorale superspreading event. Indoor Air, 2021, 31(2):314-323. PMID: 32979298. PMCID: PMC7537089. [CrossRef]
- Guenther, T., Czech-Sioli, M., Indenbirken, D., Robitailles, A., Tenhaken, P., Exner, M., Ottinger M, Fischer N, Grundhoff, A., Brinkmann, M. , Investigation of a superspreading event preceding the largest meat processing plant-related SARS-Coronavirus 2 outbreak in Germany, 2020, Available at SSRN https://ssrn.com/abstract=3654517.
- Kolinski, J.M., Schneider, T.M., Superspreading events suggest aerosol transmission of SARS-CoV-2 by accumulation in enclosed spaces. Physics Review, 2021, E 103(3-1):033109. PMID: 33862829. [CrossRef]
- Kenyon, C., The prominence of asymptomatic superspreaders in transmission means universal face masking should be part of COVID-19 de-escalation strategies. International Journal of Infectious Diseases, 2020, 97:21-22. PMID: 32497799. PMCID: PMC7263215. [CrossRef]
- Song, J.Y., Yun, J.G., Noh, J.Y., Cheong, H.J., & Kim, W.J., Covid-19 in South Korea—challenges of subclinical manifestations. New England Journal of Medicine, 2020, 382(19):1858-1859. PMID: 32251568. PMCID: PMC7154984. [CrossRef]
- Shental, N., Levy, S., Wuvshet, V., Skorniakov, S., Shalem, B., Ottolenghi, A., ...Goldhirsh, M., Efficient high-throughput SARS-CoV-2 testing to detect asymptomatic carriers. Science Advances, 2020, 6(37):eabc5961. PMID: 32917716. PMCID: PMC7485993. [CrossRef]
- Long, Q.X., Tang, X.J., Shi, Q.L., Li, Q., Deng, H.J., Yuan, J., ... & Su, K., Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nature Medicine, 2020, 26(8):1200-1204. PMID: 32555424. [CrossRef]
- Institute for Health Metrics and Evaluation. COVID-19 has caused 6.9 million deaths globally, more than double what official reports show. https://www.healthdata.org/news-release/covid-19-has-caused-69-million-deaths-globally-more-double-what-official-reports-show Accessed on May 2021.
- Sun, C, Chen, L, Yang, J, Luo, C, Zhang, Y, Li, J, Yang, J., Zhang, J, Xie, L., SARS-CoV-2 and SARS-CoV Spike-RBD Structure and Receptor Binding Comparison and Potential Implications on Neutralizing Antibody and Vaccine Development. 2020, bioRxiv. [CrossRef]
- Silverman, J.D., Hupert, N., & Washburne, A.D., Using influenza surveillance networks to estimate state-specific prevalence of SARS-CoV-2 in the United States. Science Translational Medicine, 2020, 12(554):eabc1126. PMID: 32571980. PMCID: PMC7319260. [CrossRef]
- Zhou, X., Li, Y., Li, T., & Zhang, W., Follow-up of asymptomatic patients with SARS-CoV-2 infection. Clinical Microbiology and Infection, 2020, 26(7):957-959. PMID: 32234453. PMCID: PMC7271011. [CrossRef]
- Al-Sadeq, D.W., & Nasrallah, G.K., The incidence of the novel coronavirus SARS-CoV-2 among asymptomatic patients: a systematic review. International Journal of Infectious Diseases, 2020, 98:372-380. PMID: 32623083. PMCID: PMC7330573. [CrossRef]
- Ferreira, IATM, Kemp, SA, Datir, R, Saito, A, Meng, B, Rakshit, P, Takaori-Kondo, A, Kosugi, Y, Uriu, K, Kimura, I, et al., SARS-COV-2 B.1.617 mutations L452R and E484Q are not synergistic for antibody evasion. The Journal of Infectious Diseases, 2021, 224(6):989-994. PMID: 34260717. PMCID: PMC8420622. [CrossRef]
- Greaney, AJ, Starr, TN, Gilchuk, P, Zost, SJ, Binshtein, E, Loes, AN, Hilton, SK, Huddleston, J, Eguia, R, Crawford, KHD, et al., Complete mapping of mutations to the SARS-CoV-2 spike receptor binding domain that escapes antibody recognition. Cell Host & Microbe, 2021, 29(1):44-57.e9. PMID: 33259788. PMCID: PMC7676316. [CrossRef]
- Petersen, E, Ntoumi, F, Hui, DS, Abubakar, A, Kramer, LD, Obiero, C, Tambyah, PA, Blumberg, L, Yapi, R, Al-Abri, S, et al., Emergence of new SRAS-COV-2 Variant of Concern Omicron (B.1.1.529) – highlights Africa’s research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. International Journal of Infectious Diseases, 2022, 114:268-272. PMID: 3463925. PMCID: PMC8634699. [CrossRef]
- Lin, L., Jiang, X., Zhang, Z., Huang, S., Zhang, Z., Fang, Z., ... & Liu, Y., Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut, 2020, 69(6):997-1001. PMID: 32241899. PMCID: PMC7316116. [CrossRef]
- Chen, Y, Chan, KH, Kang, Y, Chen, H, Luk, HKH, Poon, RWS, Chan, JFW, Yuen, KY, Xia, N., Lau, SKP, et al. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerging Microbes and Infections, 2015, 4(4):e26. PMID: 26421268. PMCID: PMC4575394. [CrossRef]
- Meyer, B, Drosten, C, Müller, M., Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Research, 2014, 194:175-183. PMID: 24670324. PMCID: PMC7114385. [CrossRef]
- Corman, VM, Landt, O, Kaiser, M, Molenkamp, R, Meijer, A, Chu, DK, Bleicker, T, Brünink, S, Schneider, J., Schidt, ML, et al., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveillance, 2020, 25(3):2000045. PMID: 31992387. PMCID: PMC6988269. [CrossRef]
- Zhang, J.J., Dong, X., Cao, Y.Y., Yuan, Y.D., Yang, Y.B., Yan, Y.Q., ... & Gao, Y.D., Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy, 2020, 75(7):1730-1741. PMID: 32077115. [CrossRef]
- Addetia, A., Crawford, K.H., Dingens, A., Zhu, H., Roychoudhury, P., Huang, M., ... Greninger, A., Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. Journal of Clinical Microbiology, 2020, 58(11):e02107=20. PMID: 32826322. PMCID: PMC7587101. [CrossRef]
- Merad, M., & Martin, J.C., Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nature Reviews Immunology, 2020, 20(6):355-362. PMID: 32376901. PMCID: PMC7201395. [CrossRef]
- Wong, J.J.M., Leong, J.Y., Lee, J.H., Albani, S., & Yeo, J.G., Insights into the immuno-pathogenesis of acute respiratory distress syndrome. Annals of translational medicine, 2019, 7(19):504. PMID: 31728357. PMCID: PMC6828790. [CrossRef]
- Hayden, F.G., Prevention and treatment of influenza in immunocompromised patients. The American Journal of Medicine,1997, 102(3A):55-60. PMID: 106868144. [CrossRef]
- Minotti, C., Tirelli, F., Barbieri, E., Giaquinto, C., & Donà, D., How is immunosuppressive status affecting children and adults in SARS-CoV-2 infection? A systematic review. Journal of Infection, 2020, 81(1):e-61-e66. PMID: 32335173. PMCID: PMC7179496. [CrossRef]
- D’Antiga, L., Coronaviruses and immunosuppressed patients: the facts during the third epidemic. Liver Transplantation, 2020, 26(6):832-834. PMID: 32196933. [CrossRef]
- Cerutti, G, Guo, Y, Zhou, T, Gorman, J, Lee, M, Rapp, M, Reddem, ER, Yu, U, Bahna, F, Bimela, J. et al., Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe, 2021, 29(5):819-833.e7. PMID: 33789084. PMCID: PMC7953435. [CrossRef]
- Jameson, S.C., & Masopust, D., Diversity in T cell memory: an embarrassment of riches. Immunity, 2009, 31(6):859-871. PMID: 20064446. PMCID: PMC2957815. [CrossRef]
- Simmons, C.P., Dong, T., Chau, N.V., Dung, N.T.P., Chau, T.N.B., Dung, N.T., ... & Farrar, J., Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. Journal of Virology, 2005, 79(9):5665-5675. PMID: 15827181. PMCID: PMC1082776. [CrossRef]
- Mangada, M.M., Endy, T.P., Nisalak, A., Chunsuttiwat, S., Vaughn, D.W., Libraty, D.H., ...Rothman, A.L., Dengue-specific T cell responses in peripheral blood mononuclear cells obtained prior to secondary dengue virus infections in Thai schoolchildren. The Journal of Infectious Diseases, 2002, 185(12):1697-1703. PMID: 12085313. [CrossRef]
- Green, S., & Rothman, A., Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Current opinion in infectious diseases, 2006, 19(5):429-436. PMID: 16940865. [CrossRef]
- Tian, Y., Grifoni, A., Sette, A., & Weiskopf, D., Human T cell response to dengue virus infection. Frontiers in Immunology, 2019, 10:2125. PMID: 31552052. PMCID: PMC6737489. [CrossRef]
- Lu, X., Zhang, L., Du, H., Zhang, J., Li, Y.Y., Qu, J., ... Wu, C., SARS-CoV-2 infection in children. New England Journal of Medicine, 2020, 382(17):1663-1665. PMID: 32187458. PMCID: PMC7121177. [CrossRef]
- Gudbjartsson, D.F., Helgason, A., Jonsson, H., Magnusson, O.T., Melsted, P., Norddahl, G.L., ... & Stefansson, K. , Spread of SARS-CoV-2 in the Icelandic population. New England Journal of Medicine, 2020, 382(24):2302-2315. PMID: 32289214. PMCID: PMC7175425. [CrossRef]
- Egorov, A., Romanova, J., The impact of the global distribution of bats on mortality in COVID-19 patients. [2020] Microbiology Independent Research Journal, 2020, 7(1):34-41.. [CrossRef]
- ECDC: https://www.ecdc.europa.eu/en/publications-data/operational-considerations-respiratory-virus-surveillance-europe, accessed: 18 October 2022.
- Parang, K., El-Sayed, N.S., Kazeminy, A.J., Tiwari, R.K., Comparative Antiviral Activity of Remdesivir and Anti-HIV Nucleoside Analogs against Human Coronavirus 229E (HCoV-229E). Molecules, 2020, 25(10):2343. PMID: 3249580. PMCID: PMC7287735. [CrossRef]
- Case, T., Khosravi, M., An Uncommon Cause of Adult Respiratory Distress Syndrome. Chest, 2014, 146(4):170A. PMCID: PMC7130358. [CrossRef]
- Canducci, F., Debiaggi, M., Sampaolo, M., Marinozzi, M.C., Berrè, S., Terulla, C., ... Clementi, M., Two-year prospective study of single infections and co-infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. Journal of Medical Virology, 2008, 80(4):716-723.PMID: 18297694. PMCID: PMC7167101. [CrossRef]
- Quan, C., Shi, W., Yang, Y., Yang, Y., Liu, X., Xu, W., ...Bi, Y., New threats from H7N9 influenza virus: spread and evolution of high-and low-pathogenicity variants with high genomic diversity in wave five. Journal of Virology, 2018, 92(11):E00301. PMID: 29563296. PMCID: PMC5952148. [CrossRef]
- COVID Transmissions: https://lynnwoodtimes.com/2022/10/11/covid-transmission-221011/.
- COVID Transmissions: https://www.who.int/news-room/feature-stories/detail/who-can-take-the-pfizer-biontech-covid-19--vaccine-what-you-need-to-know.
- De Clercq, E., New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chemistry An. Asian Journal, 2019, 14(22):3962-3968. PMID: 31389664. PMCID: PMC7159701. [CrossRef]
- Ahn, DG, Shin, HJ, Kim, MH, Lee, S, Kim, HS, Myoung, J, Kim, BT, Kim, SJ., Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). Journal of Microbiology and Biotechnology, 2020, 30(3):313-324. PMID: 32238757. [CrossRef]
- So, LK, Lau ACW, Yam LYC, Cheung TMT, Poon, E., Yung RWH, Yuen KY., Development of a standard treatment protocol for severe acute respiratory syndrome. Lancet, 2003, 361(9369):1615-1617. PMID: 12747883. PMCID: PMC7112408. [CrossRef]
- Chiou, HE, Liu, CL, Buttrey, MJ, Kuo, HP, Liu, HW, Kuo, HT, Lu, YT., Adverse effects of ribavirin and outcome in severe acute respiratory syndrome. Chest, 2005, 128(1):263-272. PMID: 16002945. PMCID: PMC7094379. [CrossRef]
- Choy, M., Pharmaceutical approval update. P T. 2016, 41(7):416-441. PMID: 27408516. PMCID: PMC4927014.
- Sheahan, TP, Sims, AC, Graham, RL, Menachery, VD, Gralinski, LE, Case, JB, Leist, SR, Pyrc, K, Feng, JY, Trantcheva, I., et al., Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Science Translational Medicine, 2017, 9(396):eaal3653. PMID: 28659436. PMCID: PMC5567817. [CrossRef]
- Mulangu, S, Dodd, LE, Davey, RT, Jr., Mbaya, OT, Proschan, M, Mukadi, D, Manzo, ML, Nzolo, D, Oloma, AT, Ibanda, A., et al., A randomized, controlled trial of Ebola virus disease therapeutics. The New England Journal of Medicine, 2019, 381(24):2293-2303. PMID: 31774950. [CrossRef]
- Holshue, ML, DeBolt, C, Lindquist, S, Lofy, KH, Wiesman, J, Bruce, H, Spitters, C, Ericson, K, Wilkerson, S., Tural, A., et al., First case of 2019 novel coronavirus in the United States. The New England Journal of Medicine, 2020, 382(10):929–936. PMID: 32004427. PMCID: PMC7092802. [CrossRef]
- 201. Capital Medical University, Bin Cao, China-Japan Friendship Hospital. (2020). A Trial of Remdesivir in Adults with Mild and Moderate COVID-19. (Clinicaltrials.gov Identifier NCT04252664). Retrieved from https://clinicaltrals.gov/ct2/show/NCT04252664 .
- 202. Capital Medical University, Bin Cao, China-Japan Friendship Hospital. (2020). A Trial of Remdesivir in Adults with Severe COVID-19. (Clinicaltrials.gov Identifier NCT04257656). Retrieved from https://clinicaltrials.gov/ct2/show/NCT04257656.
- Breckenridge, AM, Winstanley, PA., Clinical pharmacology and malaria. Annals of Tropical Medicine and Parasitology, 1997, 91(7):727–733. PMID: 9625927. [CrossRef]
- Solomon, VR, Lee, H., Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. European Journal of Pharmacology, 2009, 625(1-3):220–233. PMID: 19836374. [CrossRef]
- Devaux, CA, Rolain, JM, Colson, P., Raoult, D., New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? International Journal of Antimicrobial Agents, 2020, 55(5):105938. PMID: 32171740. PMCID: PMC7118659. [CrossRef]
- Keyaerts, E, Li, S, Vijgen, L, Rysman, E, Verbeeck, J, Ranst, MV, Maes, P., Antiviral activity of chloroquine against human coronavirus OC43 infection in newborn mice. Antimicrobial Agents and Chemotherapy, 2009, 53(8):3416–3421. PMID: 19506054. PMCID: PMC2715625. [CrossRef]
- Savarino, A, Boelaert, JR, Cassone, A, Majori, G, Cauda, R., Effects of chloroquine on viral infections: an old drug against today's diseases? The Lancet Infectious Diseases, 2003, 3(11):722–727. PMID: 14592603. PMCID: PMC7128816. [CrossRef]
- Savarino, A, Trani, DL, Donatelli, I, Cauda, R, Cassone, A. 2006. New insights into the antiviral effects of chloroquine. The Lancet Infectious Diseases, 2006, 6(2):67-69. PMID: 16439323. PMCID: PMC7129107. [CrossRef]
- Gao, J., Tian, Z., Yang, X., Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19-associated pneumonia in clinical studies. Bioscience Trends, 2020, 14(1):72-73. PMID: 32074550. [CrossRef]
- Borba, M.G.S., Val, F.F.A., Sampaio, V.S., Alexandre, M.A.A., Melo, G.C., Brito, M., ...Hajjar, L.A., Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA network open, 2020, 3(4), e208857-e208857. [CrossRef]
- Saleh, M., Gabriels, J., Chang, D., Kim, B.S., Mansoor, A., Mahmood, E., ... Beldner, S. et al., The effect of chloroquine, hydroxychloroquine, and azithromycin on the corrected QT interval in patients with SARS-CoV-2 infection. Circulation: Arrhythmia and Electrophysiology. 2020;13:e008662. [CrossRef]
- Schluenz, L.A., Ramos-Otero, G.P., & Nawarskas, J.J., Chloroquine or Hydroxychloroquine for Management of Coronavirus Disease 2019: Friend or Foe?. Cardiology in Review, 2020, 28(5), 266-271. [CrossRef]
- Bosseboeuf, E, Aubry, M, Nhan, T, de Pina, JJ, Rolain, JM, Raoult, D., Musso, D., Azithromycin inhibits the replication of zika virus. Journal of Antivirals and Antiretrovirals, 2018, 10(1):6-11. [CrossRef]
- Madrid, PB, Panchal, RG, Warren, TK, Shurtleff, AC, Endsley, AN, Green, CE, Kolokoltsov, A, Davey R, Manger ID, Gilfillan, L., et al., Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infectious Diseases, 2015, 1(7):317–326. PMID: 27622822. [CrossRef]
- 215. Gautret, P., Lagier, J.C., Parola, P., Meddeb, L., Mailhe, M., Doudier, B., ...Raoult, D., Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. International journal of antimicrobial agents, 2020, 56(1), 105949. [CrossRef]
- Chu, CM, Cheng, VCC, Hung, IFN, Wong, MML, Chan, KH, Chan, KS, Kao, RYT, Poon, LLM, Wong, CLP, Guan, Y., et al., Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax, 2004, 59(3):252–256. PMID: 14985565. PMCID: PMC1746980. [CrossRef]
- Chan, KS, Lai, ST, Chu, CM, Tsui, E, Tam, CY, Wong, MM, Tse, MW, Que, TL, Peiris, JSM, Sung, J., et al., Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicenter retrospective matched cohort study. Hong Kong Medical Journal, 2003, 9(6):399-406. PMID: 14660806.
- Johnson, R.M., Vinetz, J.M., Dexamethasone in the management of covid-19. BMJ; 2020, 370. [CrossRef]
- Selvaraj, V., Dapaah-Afriyie, K., Finn, A., & Flanigan, T.P., Short-term dexamethasone in sars-cov-2 patients. RI Med.
- . J., 2020, 103(6), 39-43.
- Cain, D.W., & Cidlowski, J.A., After 62 years of regulating immunity, dexamethasone meets COVID-19. Nature Reviews Immunology, 2020, 20(10), 587-588. [CrossRef]
- Regeneron. 2020. Regeneron’s REGN-COV2 antibody cocktail reduced viral levels and improved symptoms in non-hospitalized COVID-19 patients. https://investor.regeneron.com/news-releases/news-release-details/regenerons-regn-cov2-antibody-cocktail-reduced-viral-levels-and Accessed on October 17, 2020.
- AbCellera. (2020). As LY-CoV555 advances into Phase 3 COVID-19 prevention studies, AbCellera’s COVID-19 database reaches over 2,000 new antibodies against the virus, discovered from recovered patient samples. https://www.abcellera.com/news/2020-08-03-abcellera-provides-covid-19-program-update Accessed on October 17, 2020.
- Lilly. 2020. Lilly provides a comprehensive update on the progress of SARS-CoV-2 neutralizing antibody programs. https://investor.lilly.com/news-releases/news-release-details/lilly-provides-comprehensive-update-progress-sars-cov-2 Accessed on October 17, 2020.
- AstraZeneca. (2020). COVID-19 Long-Acting antibody (LAAB) combination AZD7442 rapidly advances into Phase III clinical trials. https://www.astrazeneca.com/media-centre/press-releases/2020/covid-19-long-acting-antibody-laab-combination-azd7442-rapidly-advances-into-phase-iii-clinical-trials.html Accessed on October 17, 2020.
- Robbie GJ, Criste R, Dall’acqua WF, Jensen K, Patel NK, Losonsky GA, Griffin MP. 2013. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrobial Agents and Chemotherapy 57(12):6147-6153. PMID: 24080653. PMCID: PMC3837853. [CrossRef]
- Griffin, MP, Khan, AA, Esser, MT, Jensen, K, Takas, T, Kankam, MK, Villafana, T. Dubovsky, F., Safety, tolerability, and pharmacokinetics of MEDI8897, the Respiratory Syncytial Virus prefusion F-targeting monoclonal antibody with an extended half-life, in healthy adults. Antimicrobial Agents and Chemotherapy, 2017, 61(3):e01714-16. PMID: 27956428. PMCID: PMC5328523. [CrossRef]
- Shi, Y., Wang, G., Cai, X.P., Deng, J.W., Zheng, L., Zhu, H, Zheng M, B Yang Chen, Z., An overview of COVID-19. Journal of Zhejiang University. Science. B, 2020, 21(5), 343. [CrossRef]
- Domachowske, JB, Khan, AA, Esser, MT, Jensen, K, Takas, T, Villafana, T, Dubovsky, F, Griffin, MP., Safety, tolerability and pharmacokinetics of MEDI8897, an extended half-life single-dose Respiratory Syncytial Virus prefusion F-targeting monoclonal antibody administered as a single dose to healthy preterm infants. The Pediatric Infectious Disease Journal, 2018, 37(9):886-892. PMID: 29373476. PMCID: PMC6133204. [CrossRef]
- Van Erp, EA , Luytjes, W. , Ferwerda, G., Van Kasteren, P., Fc-Mediated Antibody Effector Functions During Respiratory Syncytial Virus Infection and Disease, Front. Immunol. 2019 Mar. 22;10:548. PMID: 30967872. PMCID: PMC6438959. [CrossRef]
- Zost, SJ, Gilchuk, P, Case, JB, Binshtein, E, Chen, RE, Reidy, JX, Trivette, A, Nargi, RS, Sutton, RE, Suryadevara, N., et al., Potently neutralizing human antibodies that block SARS-CoV-2 receptor binding and protect animals. bioRxiv 2020. PMID: 32511409. PMCID: PMC7263556. [CrossRef]
- Hoy, S.M., Amubarvimab/romlusevimab: first approval. Drugs, 2022, 82(12), 1327-1331. [CrossRef]
- Brii Biosciences (2020). https://www.briibio.com/pipeline.php Accessed on October 18, 2020.
- Johnson & Johnson. 2020a. Johnson & Johnson temporarily pauses all dosing in our Janssen COVID-19 vaccine candidate clinical trials. https://www.jnj.com/our-company/johnson-johnson-temporarily-pauses-all-dosing-in-our-janssen-covid-19-vaccine-candidate-clinical-trials Accessed on October 18, 2020.
- Center for Disease Control and Prevention. 2021. CDC Recommends Use of Johnson & Johnson’s Janssen COVID-19 Vaccine Resume | Corporate Authors(s): National Center for Immunization and Respiratory Diseases (U.S.). Division of Viral Diseases. Published Date : Apr 30, 2021CDC. https://stacks.cdc.gov/view/cdc/105680. Accessed on Oct 8, 2021.
- Johnson & Johnson. 2020b. Johnson & Johnson initiates pivotal global phase 3 clinical trial of Janssen’s COVID-19 vaccine candidate. https://www.jnj.com/johnson-johnson-initiates-pivotal-global-phase-3-clinical-trial-of-janssens-covid-19-vaccine-candidate Accessed on October 18, 2020.
- Pascal, KE, Dudgeon, D, Trefry, JC, Anantpadma, M, Sakurai, Y, Murin, CD, Turner, HL, Fairhurst, J, Torres, M, Rafique, A., et al., Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in nonhuman primates. The Journal of Infectious Diseases, 2018, 218(suppl_5): S612–S626. PMID: 29860496. PMCID: PMC6249601. [CrossRef]
- Beigel, JH, Nam, HH, Adams, PL, Krafft, A, Ince, WL, El-Kamary, SS, Sims, AC., Advances in respiratory virus therapeutics - a meeting report from the 6th ISIRV Antiviral Group conference. Antiviral Research, 2019, 167:45–67. PMID: 30974127. PMCID: PMC7132446. [CrossRef]
- Cockrell, AS, Yount, BL, Scobey, T, Jensen, K, Douglas, M, Beall, A, Tang, XC, Marasco, WA, Heise, MT, Baric, RS., A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nature Microbiology, 2016, 2(2):16226. PMID: 27892925. PMCID: PMC5578707. [CrossRef]
- Beigel, JH, Voell, J, Kumar, P, Raviprakash, K, Wu, H, Jiao, JA, Sullivan, E, Luke, T, Davey, RT, Jr., Safety and tolerability of a novel, polyclonal human anti-MERS coronavirus antibody produced from transchromosomic cattle: a phase 1 randomized, double-blind, single-dose-escalation study. The Lancet Infectious Disease, 2018, 18(4):410–418. PMID: 29329957. PMCID: PMC5871563. [CrossRef]
- Chen, L, Xiong, J, Bao, L, Shi, Y., Convalescent plasma as a potential therapy for COVID-19. The Lancet Infectious Disease, 2020, 20(4):398-400. PMID: 32113510. PMCID: PMC7128218. [CrossRef]
- Shen, C, Wang, Z, Zhao, F, Yang, Y, Li, J, Yuan, J, Wang, F, Li, D, Yang, M, Xing, L., et al., Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA, 2020, PMID: 32219428. PMCID: PMC7101507. [CrossRef]
- Duan, K., Liu, B., Li, C., Zhang, H., Yu, T., Qu, J., ... Peng, C., Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proceedings of the National Academy of Sciences, 2020, 117(17), 9490-9496. [CrossRef]
- Valk, SJ, Piechotta, V, Chai, KL, Doree, C, Monsef, I, Wood, EM, Lamikanra, A, Kimber, C, McQuilten, Z, So-Osman, C, Estcourt, LJ, Skoetz, N. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: a rapid review. Cochrane Database Syst Rev. 2020 May 14;5(5):CD013600. Update in: Cochrane Database Syst Rev. 2020 Jul 10;7:CD013600. PMID: 32406927; PMCID: PMC7271896. [CrossRef]
- Fleming, A.B., Raabe, V., Current studies of convalescent plasma therapy for COVID-19 may underestimate risk of antibody-dependent enhancement. Journal of Clinical Virology.2020, Jun; 127: 104388. [CrossRef]
- Lee, CC, Hsieh, CC, Ko, WC., Molnupiravir- a novel oral anti-SARS-COV-2 agent. Antibiotics, 2021, 10(11):1294. PMID: 34827232. PMCID: PMC8614993. [CrossRef]
- 246. Landau, MD. National Geographic, How Merck's antiviral pill could change the game for COVID-19? October 1, 2021. https://www.nationalgeographic.com/science/article/how-mercks-antiviral-pill-could-change-the-game-for-covid-19 Accessed on December 9, 2021.
- Herpe,r M. (2021). FDA authorizes Merck’s COVID-19 pill but stresses its use should be limited. Dec. 23, 2021. https://www.statnews.com/2021/12/23/fda-authorizes-mercks-covid-19-pill-but-stresses-its-use-should-be-limited/ Accessed on June 27, 2022.
- U.S. Food and Drug Administration (2021). FDA News Release: Coronavirus (COVID-19) Update: FDA Authorizes Additional Oral Antiviral for Treatment of COVID-19 in Certain Adults. December 23, 2021. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-oral-antiviral-treatment-covid-19-certain accessed on April 18, 2023.
- Le, T.T., Andreadakis, Z., Kumar, A., Román, R.G., Tollefsen, S., Saville, M., & Mayhew, S., The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov, 2020, 19(5), 305-306. [CrossRef]
- André, FE, The future of vaccines, immunization concepts, and practice. Vaccine, 2001, 19(17-19):2206-2209. PMID: 11257334. [CrossRef]
- Guarner, J., Three emerging coronaviruses in two decades. American Journal of Clinical Pathology, 2020, 153(4):420-421. PMID: 32053148. PMCID: PMC7109697. [CrossRef]
- Bao, L, Deng, W, Huang, B, Gao, H, Liu, J, Ren, L, Wei, Q, Yu, P, Xu, Y, Qi, F, et al., The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. bioRxiv, 2020. [CrossRef]
- Moderna (2020). Moderna’s work on a COVID-19 vaccine candidate. 2020. https://www.modernatx.com/modernas-work-potential-vaccine-against-covid-19 Accessed on July 25, 2020.
- 254. AstraZeneca. (2021). Phase III Double-blind, Placebo-controlled Study of AZD1222 for the Prevention of COVID-19 in Adults. (Clinicaltrials.gov Identifier NCT04516746). Retrieved from https://clinicaltrials.gov/ct2/show/NCT04516746.
- Du, L, He, Y, Zhou, Y, Liu, S, Zheng, BJ, Jiang, S., The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nature Reviews Microbiology, 2009, 7(3):226-236. PMID: 19198616. PMCID: PMC2750777. [CrossRef]
- Coutard, B, Valle, C, de Lamballerie, X, Canard, B, Seidah, NG, Decroly, E., The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Research, 2020, 176:104742. PMID: 32057769. PMCID: PMC7114094. [CrossRef]
- Li, F., Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 2016, 3(1):237-261. PMID: 27578435. PMCID: PMC5457962. [CrossRef]
- Abdelmageed, MI, Abdelmoneim, AH, Mustafa, MI, Elfadol, NM, Murshed, NS, Shantier, SW, Makhawi, AM., Design of multi epitope-based peptide vaccine against E protein of human 2019-nCoV: an immunoinformatics approach. bioRxiv, 2020. [CrossRef]
- Nhu, TTT, Labroussaa F, Ebert N, V’kovski P, Stalder H, Portmann J, Kelly J, Steiner S, Holwerda M, Kratzel, A., et al., Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. bioRxiv, 2020. [CrossRef]
- Prazeres, DMF, Monteiro GA., Plasmid biopharmaceuticals. Microbiology Spectrum, 2014, 2(6). PMID: 26104457. [CrossRef]
- Crawford, NW, Bines, JE, Royle, J., Buttery JP., Optimizing immunization in pediatric special risk groups. Expert Review of Vaccines, 2011, 10(2):175–186. PMID: 21332267. [CrossRef]
- Liu, MA., Immunologic basis of vaccine vectors. Immunity, 2010, 33(4):504–515. PMID: 21029961. [CrossRef]
- Deering, RP, Kommareddy, S., Ulmer, J.B., Brito, L.A., Geall, A.J., Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opinion on Drug Delivery, 2014, 11(6):885–899. PMID: 24665982. [CrossRef]
- Pardi, N, Hogan, MJ, Porter, FW, Weissman, D., mRNA vaccines - a new era in vaccinology. Nature Review Drug Discovery, 2018, 17(4):261-279. PMID: 29326426. PMCID: PMC5906799. [CrossRef]
- Zhu, FC, Li, YH, Guan, XH, Hou, LH, Wang, WJ, Li, JX, Wu, SP, Wang, BS, Wang, Z., Wang, L., et al., Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomized, first-in-human trial. Lancet, 2020, 395(10240):1845-1854. PMID: 32450106. PMCID: PMC7255193. [CrossRef]
- Clover Biopharmaceuticals Vaccines Programs. 2020. https://www.cloverbiopharma.com/index.php?m=content&c=index&a=lists&catid=42 Accessed on July 27, 2020.
- China's Sinovac plots pivotal COVID-19 vaccine trial in Brazil after positive phase 2. 2020. https://www.fiercepharma.com/vaccines/china-s-sinovac-says-covid-19-vaccine-shows-early-positive-results-phase-2 Accessed on July 25, 2020.
- INOVIO Pharmaceuticals and Beijing Advaccine Biotechnology – INO-4800. 2020.y 18, 2020 at https://www.inovio.com accessed Oct 18, 2022.
- INVIO Pharmaceutical. 2020. DNA medicines pipelines. https://www.inovio.com/dna-medicines-pipeline/ Accessed on July 28, 2020.
- INVIO Pharmaceutical. 2020. INOVIO urgently focused on developing COVID-19 vaccine because the world can’t wait. https://www.inovio.com/our-focus-serving-patients/covid-19/ Accessed on July 28, 2020.
- Inovio Pharmaceuticals, Inc. - INOVIO Reports First Quarter 2021 Financial Results. Available at. https://www.inovio.com Accessed on August 11, 2021.
- Moderna's mRNA clinical trials: CMV, MMA, Zika, several types of cancer and other diseases. 2020. https://www.modernatx.com/pipeline/modernas-mrna-clinical-trials-cmv-mma-zika-several-types-cancer-and-other-diseases accessed 18 Oct, 2022.
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, et al., An mRNA vaccine against SARS-CoV-2-preliminary report. The New England Journal of Medicine, 2020. NEJMoa2022483. PMID: 32663912. PMCID: PMC7377258. [CrossRef]
- Centers for Disease Control (CDC) Moderna COVID-19 Vaccine Overview and Safety. https://www.cdc.gov/coronavirus/2019-ncov/vaccines/different-vaccines/Moderna.html Accessed August 11, 2021.
- Polack, F.P., Thomas, S.J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., Perez, J.L., Pérez Marc, G., Moreira, E.D., Zerbini, C., Bailey, R., Swanson, K.A., Roychoudhury, S., Koury, K., Li, P., Kalina, W.V., Cooper, D., Frenck, R.W. Jr, Hammitt, L.L., Türeci, Ö., Nell, H., Schaefer, A., Ünal, S., Tresnan, D.B., Mather, S., Dormitzer, P.R., Şahin, U., Jansen, K.U., Gruber, W.C.; C4591001 Clinical Trial Group. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J. Med. 2020 Dec 31;383(27):2603-2615. Epub 2020 Dec 10. PMID: 33301246; PMCID: PMC7745181. [CrossRef]
- Alford, R.H., Kasel, J.A., Gerone, P.J., Knight, V., Human Influenza Resulting from Aerosol Inhalation. Proceedings of the Society for Experimental Biology and Medicine. 1966; 122(3):800-804. [CrossRef]
- Nikitin, N, Petrova, E, Trifonova, E., Karpova, O., Influenza virus aerosols in the air and their infectiousness. Adv. Virol. 2014; 2014:859090. Epub 2014 Aug 13. PMID: 25197278; PMCID: PMC4147198. [CrossRef]
- Holmes, E.C., Goldstein, S.A., Rasmussen, A.L., Robertson, D.L., Crits-Christoph, A., Wertheim, J.O., ... Rambaut, A., The origins of SARS-CoV-2: A critical review. Cell, 2021, 184(19), 4848-4856. [CrossRef]
- Baldwin, J., Noorali, S., Vaseashta, A., Wide Spectrum Bio-threats Identification and Classification. Conference: Chemical, Biological, Radiological and Nuclear Defense - Modernizing the Future Fight: Accelerate & Adaptation, 2019, Delaware, USA.
- Vaseashta, A., Applying Resilience to Hybrid Threats in Infrastructure, Digital, and Social Domains Using Multisectoral, Multidisciplinary, and Whole-of-Government Approach. 2021, Pp: 42-59. NATO Science for Peace and Security Series - D: Information and Communication Security. Volume 61: Building Cyber Resilience against Hybrid Threats. [CrossRef]
- Vaseashta, A., Nexus of Advanced Technology Platforms for Strengthening Cyber-Defense Capabilities. pp: 14-31. NATO Science for Peace and Security Series - E: Human and Societal Dynamics. 2021, Vol. 155: Practical Applications of Advanced Technologies for Enhancing Security and Defense Capabilities: Perspectives and Challenges for the Western Balkans. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).