1. Introduction
According to (ICTV, 2023) classification, ZIKV belongs to Flavivirus genus and Flaviviridae Family. Its genome is presented as a positive sense single strand RNA (ssRNA+) that has around 11 kb. The genomic RNA is constituted by one strand and presents an ORF that codes for a single polyprotein. When it is cleaved, originates structural and non-structural proteins.
Zika virus was discovered in 1947 during a research expedition about Yellow Fever in the Ziika Forest located in Uganda, Africa [
1,
2,
3]. Only in 1964, Simpson et al. reported the first human case of ZIKV infection in Uganda [
4]. In 1966, the presence of the virus in the
Aedes aegypti mosquito circulating in the urban area was registered, for the first time, in Malaysia [
5]. Furthermore, the first recorded outbreak of Zika Fever occurred between 1977-1978 in Indonesia [
6,
7]. Until then, the infection was only identified by serological test in humans [
7]. Between 1947 and 2007, Posen et al. (2016) reported the presence of the ZIKV in 29 African, eight Asian and one European countries [
8,
9,
10]. Between 2013 and 2016, ZIKV spread to a larger global circulation being the causal agent for several outbreaks of Zika Fever in 2016 [
8,
9,
10]. Brazil was one of the most affected countries and registered one of the highest numbers of infected people, Guillain Barre Syndrome (GBS) in adults and congenital ZIKV Syndrome (CZS) in newborns [
9,
10,
11,
12].
The major ZIKV transmission is vectorial, with focus of infection on the nervous system [
9,
13]. Considering the viral detection in the amniotic fluid of pregnant women and in the female and male sexual organs, it is possible that ZIKV can be transmitted by means of sexual relation and maternal-fetal attachment [
13,
14,
16,
17]. Among flaviviruses, there are cases of vertical transmission (maternal-fetal) for CHIKV, DENV and WNV [
8,
9], but its relevance for the spread and disease of such viruses is still not conclusive. ZIKV cell tropism includes central and peripheral nervous system, epithelial cells, immune system, and reproductive system [
8,
9,
13]. In placental tissues, ZIKV has been identified in syncytiotrophoblasts, cytotrophoblasts and endothelial cells of the maternal immune system (dendritic cells and macrophages) [
14]. Receptor for ZIKV is present in female reproductive tissues an in the placenta, the receptors are Gas6-AXL receptor tyrosine kinase complex and AXL tyrosine kinase protein [
15].
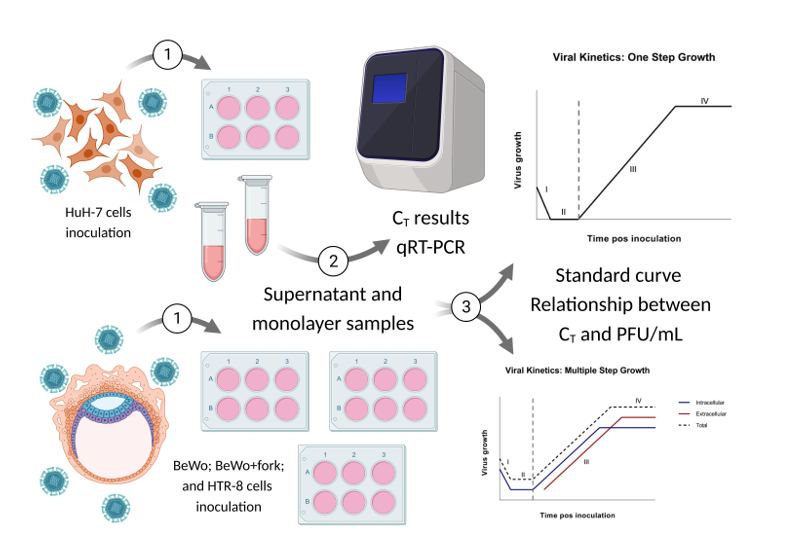
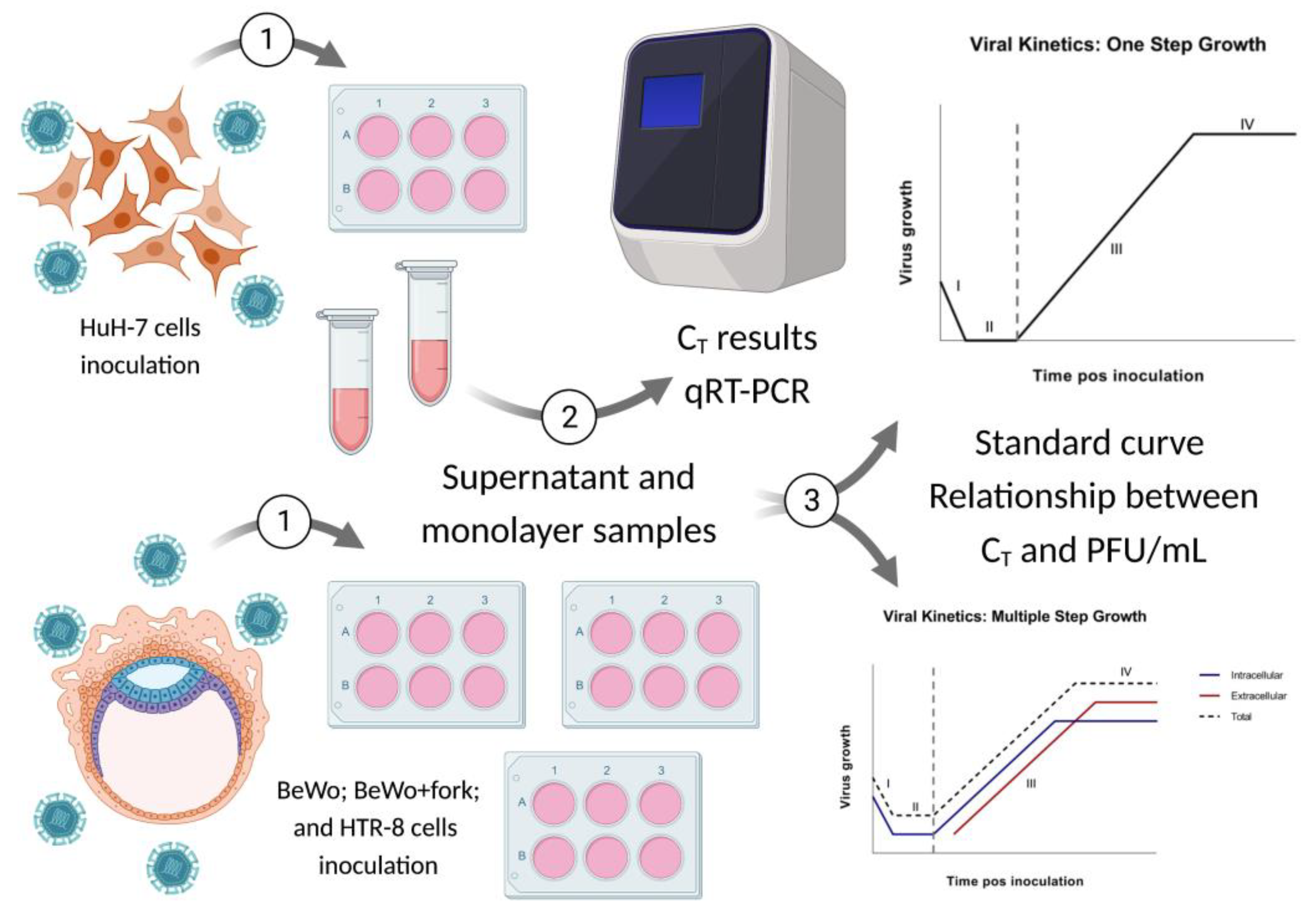
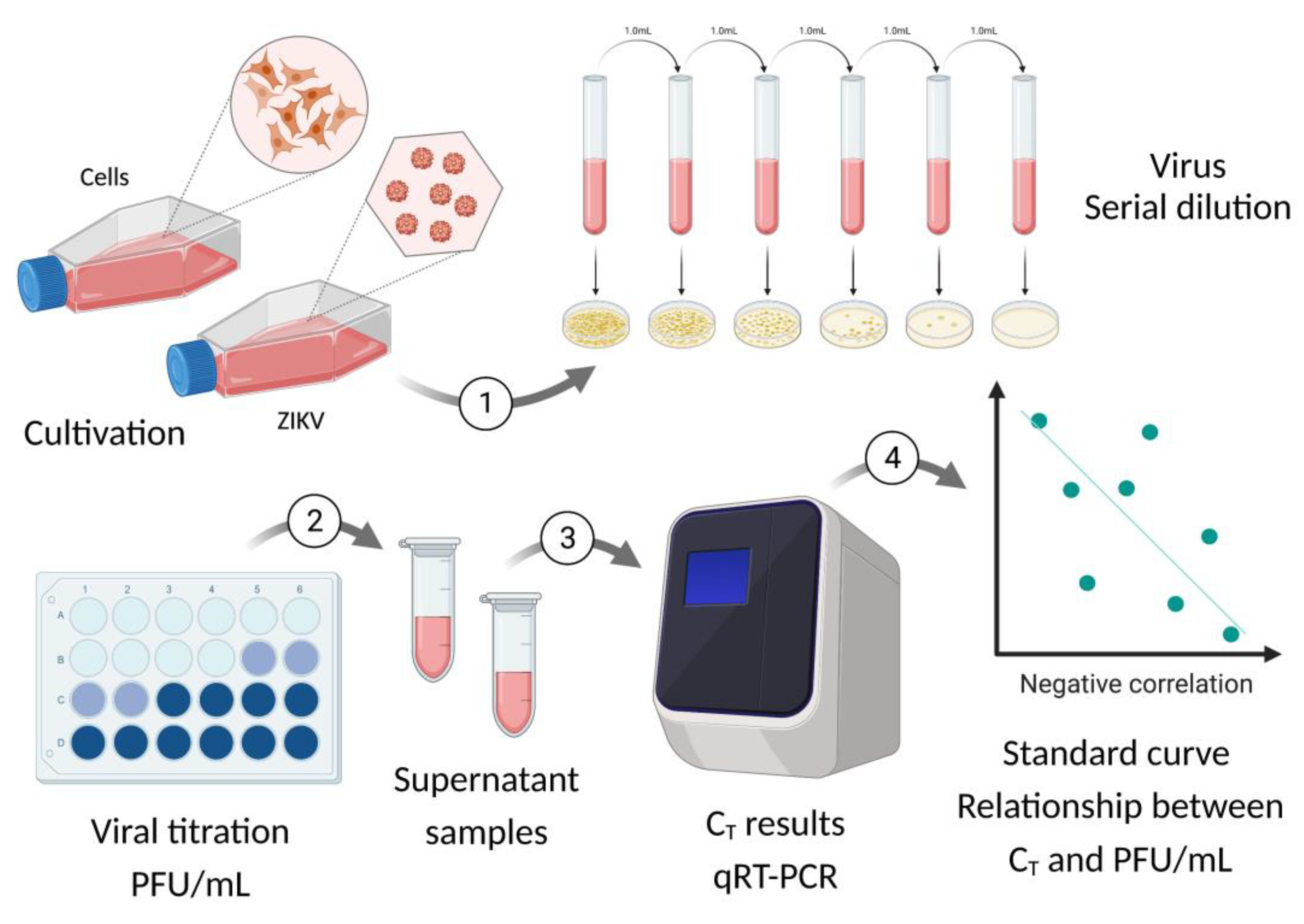
The Step Growth Curve (SGC) methodology was defined as standard to observe virus behavior over time, since such can be used in silico, in vitro, in/ex situ, in/ex vivo [
18,
19,
20,
21,
29]. Moreover, the data obtained through SGC allows application in different studies, such as: vaccines, antivirals, epidemiology, viral evolution, comparison between strains/variants/serological of the same species or different species, double or more infections, intra and inter- viral species in the same host, among others. Since sexual and maternal-fetal transmission is uncommon for Flaviviruses this study aimed to evaluate selected BeWo and HTR-8 placental cells to verify whether Asian-Brazilian strain of ZIKV has the same behavioral dynamics as the original African strain in sustained replication (viral kinetics).
2. Materials and Methods
2.1. Viral Isolation and Formation of Viral Stocks
ZIKV-MR766lp. Low passage (lp) strain isolated from
Macaca mulatta in 1947 in Ziika Forest, Uganda, Africa (GenBank: AY632535.2) [
22,
23,
24]. It presents the term low passage because has a smaller number of passages in mice, according to information provided by Dr. Amadou Alpha Sall, from the Pasteur Institute of Dakar, Senegal, Africa, who kindly provided samples for this study.
ZIKV-IEC-Paraíba. Strain isolated from a patient in Paraíba, Brazil, clinically diagnosed with ZIKV in the 2015 outbreak (GenBank: KX2800260) [
25]. Sample kindly provided by professor Dr. Pedro Vasconcelos from the Evandro Chagas Institute in Belém, Pará.
Both strains were cultivated in Vero CCL-81 cells (ATCC
®) to form viral stocks and to get the necessary amount for complete experimentation. Such stock was produced in three steps to obtain the maximum yield of infective virions. In the first step, there was cultivation in five cylindrical tubes of glass with volume of 1 mL of cells. The second occur in five culture flasks with 25 cm
2 Corning
® and volumes between 5-7 mL; and the third in five culture flasks with 75 cm
2 Corning
® filter and volumes between 15-20 mL. For each step, supernatant, debris and cell monolayer were collected together; gathered in a single container; and homogenized to be stored in 1.8 mL Corning
® cryotube, frozen in freezer at -80°C. We separated a small aliquot from the three steps for titration by PFU/mL and tested for the presence of mycoplasma according Timenetsky et al. [
26].
2.2. Cells Cultures
For Vero CCL-81, HTR-8/SVneo and BeWo, we followed the cell maintenance as described by the ATCC
® cell bank; while for HuH-7, JCRB Cell Bank recommendations were used. Cellular, viral stocks and samples from viral kinetics experiments were tested for mycoplasma [
26] and RNA contaminations by ZIKV [
27]. All results were negative.
BeWo is a fusiogenic choriocarcinoma cell type—they form syncytium with human villous trophoblastic cells properties. It shows features common to normal trophoblasts; in addition to expressing IL-6, IL-10, IFN-α, IFN-β, hCG, steroids, estrogens and progesterone. For this reason, it is a good model to study the dynamics of viral infection (ATCC
®). Considering that BeWo in vitro has low rate of spontaneous fusion, we used forskolin (chemical compound that accelerates the fusiogenic capacity of such cell lineage) [
28].
HTR8/SVneo is a cell type of human extravillous trophoblast immortalized by SV40 virus, and originary from chorionic villi, present between 6 and 12 weeks of gestation (first gestational trimester). Furthermore, it is characterized as a type of epithelial cell, with hCG production and invasion of maternal uterine tissue (ATCC®).
Both cell lineages were kindly provided by Prof. Dr. Estela M. A. F. Bevilacqua, from the Maternal-Fetal and Placenta Interaction Studies Laboratory, ICB-USP, São Paulo, SP—Brazil. (
Appendix A).
Vero CCL-81 is a type of epithelial adherent cell from the kidney of a normal adult monkey which belongs to the African species Chlorocebus aethiops (ATCC®).
HuH-7 (JCRB0403) is an adherent cell lineage that is immortalized and originary from tumorigenic epithelium of Homo sapiens—a hepatocellular carcinoma commonly used for HCV studies (JCRB).
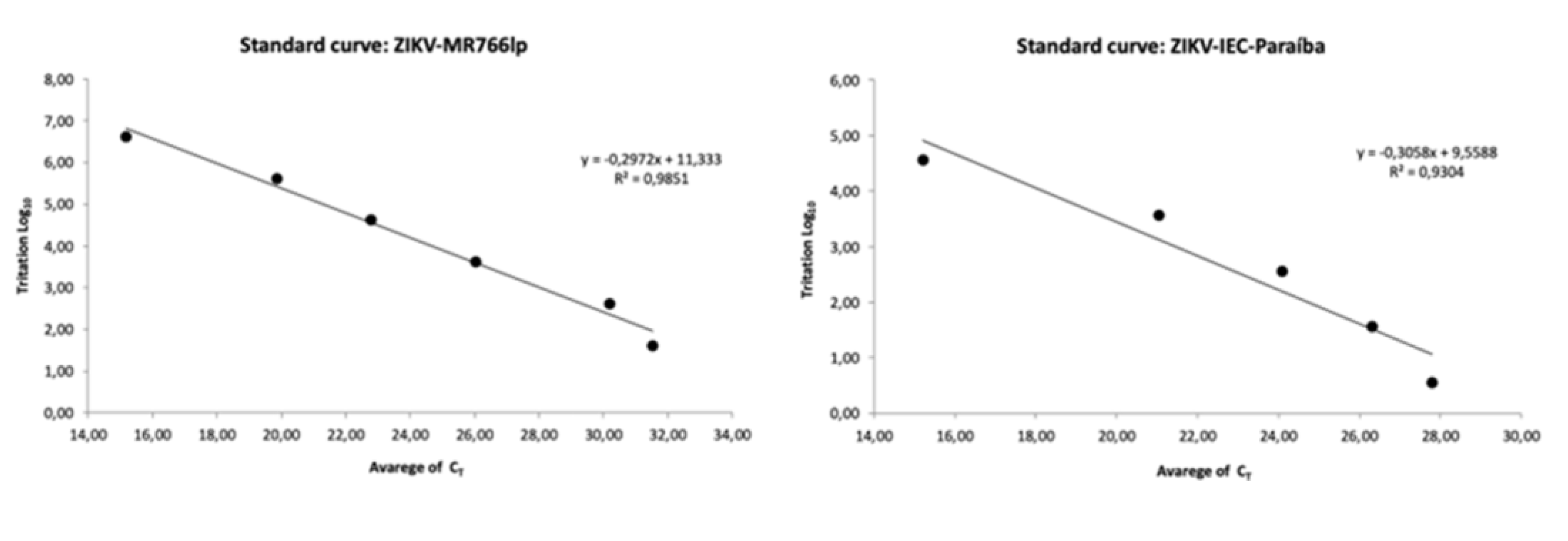
2.3. Standard Curve for PFU/mL Determination
The correlation between C
T and PFU/mL or standard curve was established through titration by PFU/mL performed in triplicate for ZIKV-MR766 and ZIKV-IEC, in plates of 24 wells with a cell concentration of 1x10
5 cells/mL per well of Vero cells. Moreover, the stock was titrated using 0.2 mL of the viral sample in a serial dilution of 10-fold (10
−1 to 10
−11). Then, we incubated the plates for five days in an oven with temperature at 37°C and 5% of CO
2. Thereafter, the supernatant was collected and stored for quantification by qRT-PCR (
Appendix B) and the cells monolayers were fixed to determine the viral titer. Based on a conversion table, C
T values were calculated and obtained from viral titers in PFU/mL (
Appendix H).
2.4. Viral Kinetics (Step Growth Curve)
In this experiment, we verified the capacity of infectivity and viral multiplication that both viral strains of ZIKV (African and Asian-Brazilian) have, considering the following cell lineages: BeWo, BeWo treated with forskolin, HTR-8 and HuH-7.
The step growth curve presented seven time-points, determined as hour post infection (hpi), starting from two hpi in the incubator and a 24 interval until 144 hours. Therefore, we obtained the following hpi: 2h, 24h, 48h, 72h, 96h, 120h, 144h. Sample collection for each time-point was performed in duplicate, with separation of samples between supernatant and cell monolayer, which was stored in a freezer at -80°C for later analysis (
Appendix C). The multiplicity of infection (MOI) It is the ratio of the number of viral particles to the number of host cells. The MOI = 1 implies that for each cell unit there is a single viral particle. The encounter of a particle by the host cell is a chance encounter, therefore statistically it can be represented by the Poisson distribution [
18,
19,
20,
29]. The variation was determined between 0.1 to 1 due to the different replicative capacity of both strains. According to the Poisson distribution, there are few differences within this variation when applied in the assay.
The data obtained from the first complete growth curve was used as standard for the analysis of the two remaining replicates at 24, 48 and 72 hpi. Thereafter, we calculated PFU/mL for each time-point from qRT-PCR data based on the standard curve, constructed before the beginning of kinetics experiment (
Appendix D and H). Then, it was performed a description of cytopathic effect noticed for all time-points of three replicates. Since it was not possible to fulfill the replications at the same time and in the same passage of cells, we chose that the cells had two passages of difference for each replication of the kinetic assay. Images for all kinetics were seen on the Thermo Fisher Scientific EVOS™ FL inverted light microscope at 20x (200 μM). The viral growth curve is based on the theoretical curves of Delbruck and Ellis Ellis [
19] and Burleson [
18] (
Figure 1, step 3).
3. Results
For the standard curve we used the average of the titers to construct the standard curve for each strain—conversion from C
T to PFU/mL (
Appendix D to F and
Table S1).
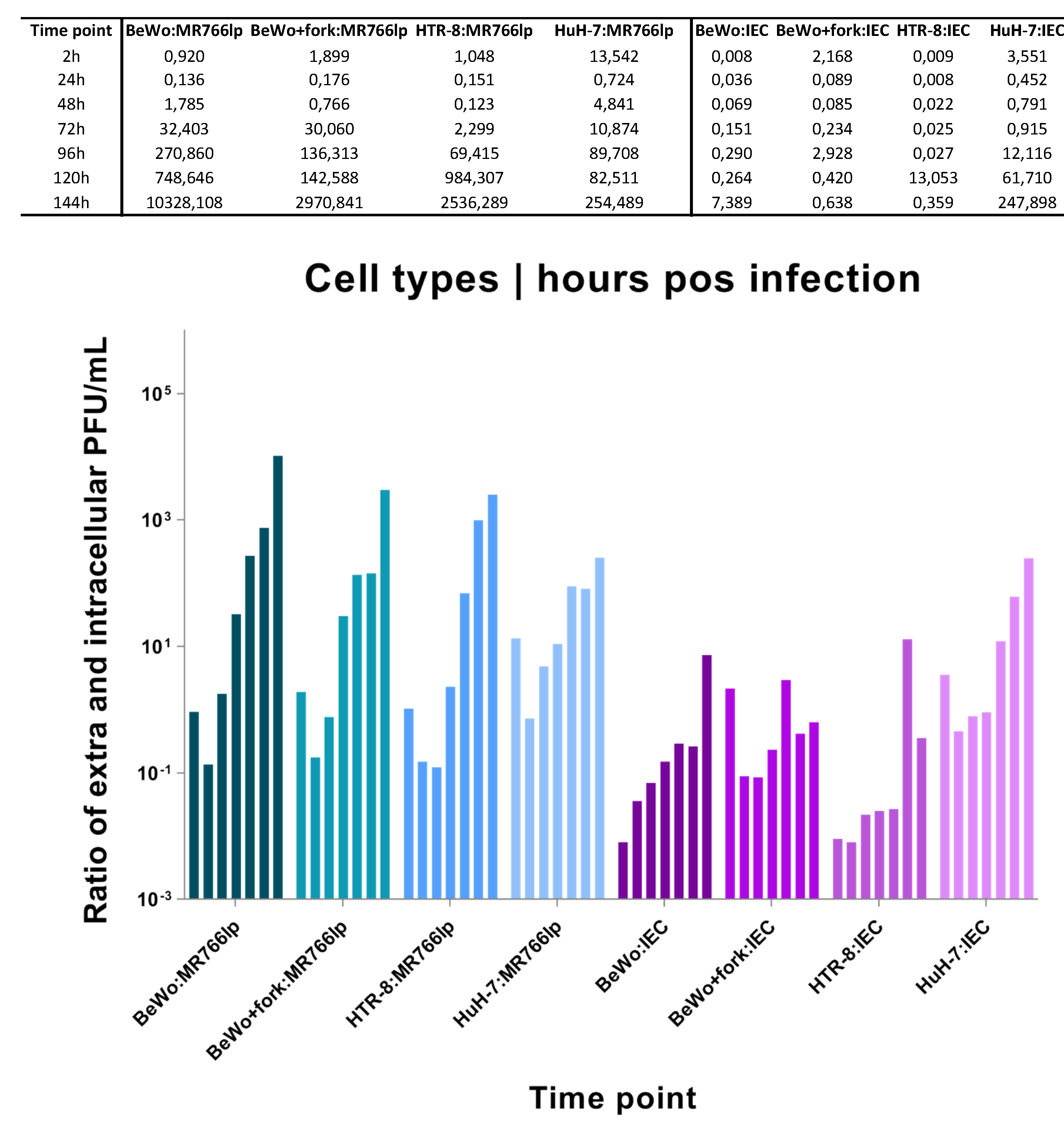
Viral Kinetics ZIKV-MR766(lp). The titer of the viral stocks was 1.7x108 PFU/mL. We utilized MOI = 1 and it were found the following averages of cell concentration: BeWo = 3.14x106 cell/mL, BeWo+fork = 1.7x106 cell/mL, HTR-8 = 1.7x106 cell/mL, and HuH-7 = 0.85x106 cell/mL.
Viral Kinetics ZIKV-IEC-Paraíba. The titer of the viral stocks was 1.5x106 PFU/mL. We used MOI = 0.5 for ZIKV-IEC, with the following averages of cell concentration: BeWo = 2.3x106 cell/mL, BeWo+forsk = 1.7x106 cell/mL, HTR-8 = 1.6x106 cell/mL, and HuH-7 = 0.32x106 cell/mL.
The data obtained in our study should be analyzed independently for each ZIKV strain as we used different MOIs due to the lack of ZIKV-IEC titers to reach MOI = 1. Regarding BeWo lineage, due to addition of forskolin, we performed cultivation of treated and untreated cells two days before, and such compound was added one day before inoculation. Eight assays of viral kinetics were fulfilled: four for each ZIKV strain, in triplicates, with seven hpi. Considering triplicates, there were a total of 208 samples, including negative controls for each hpi from intra and extracellular media (
Appendix C).
For standardization, we quantified the C
T of viral RNA for all hpi of triplicate one. Based on these results, we also quantified triplicates two and three but only for three time-points: 24h, 48h and 72h. Therefore, in the end, the quantification of 96 samples was obtained by the C
T result converted into PFU/mL. Since the results obtained after by the analysis of triplicates two and three validated those of triplicate one, we proceed the study (
Figure 1, step 1 and 2). The complete conversion table is found in (
Table S1).
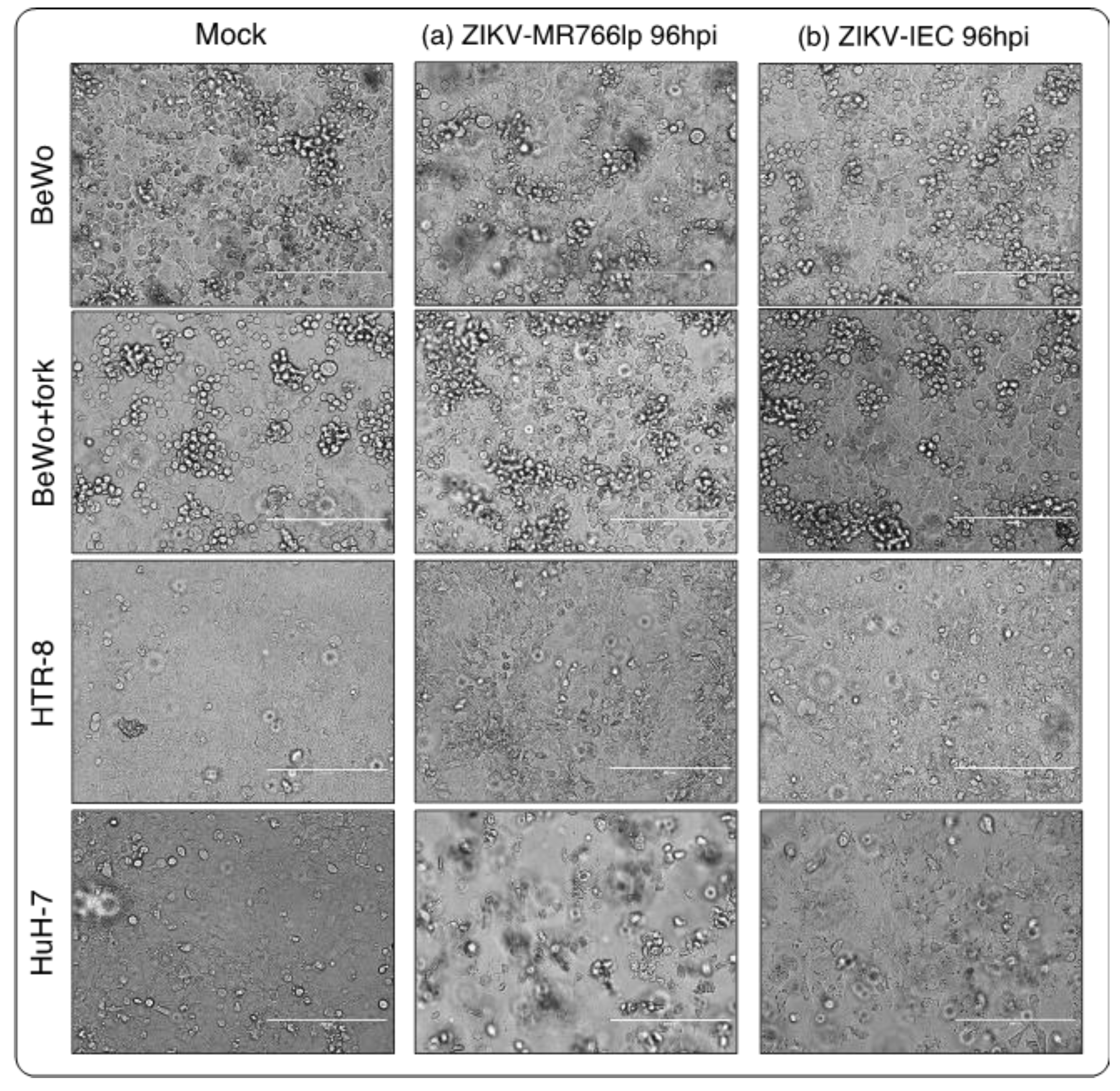
In general, all BeWo, HTR-8 and HuH-7 controls presented similar features in all hpi. At 2 and 24 hpi, the monolayer was intact, the cells had visible boundaries and absence of deformities, cell division, observable nucleus and nucleolus. At 48 hpi, the monolayer was more closed, with the appearance of cells in the supernatant (refringent cells), and also the presence of characteristics found in the previous hpi. Furthermore, at hpi between 72 and 144, there were saturation of the space occupied by monolayer, augmentation of the cells in supernatant, and same characteristics of 48 hpi (
Appendix I).
On the other hand, BeWo/HTR-8/HuH-7-infected cells displayed cytopathic effects (CPEs) that increased during the time points analyzed. Monolayer detachment, focal degeneration with rounded and refractory cells, partial and total destruction of the monolayer inoculated, as well as formation of cellular debris, morphological alterations, swelling and cluster of cells could be observed (
Figure 2a,b). Thus, we considered the three cell lineages susceptible to the ZIKV-MR766lp and ZIKV-IEC viruses.
3.1. Inoculation of ZIKV-MR766lp (a)
Regarding kinetics curve after infection with ZIKV-MR766lp (
Figure 3 and
Appendix I), BeWo and BeWo+fork had the following similarities: peak of intracellular viral production between hpi 24 and 48, in addition to decay soon after this period. Considering the presence of virus in the extracellular medium, there was growth in the first three time points. After these periods, a constant stability remained until the end.
BeWo showed a fast proliferation, although the onset of monolayer stayed with low density. After 3-4 days, 100% confluence was observed, with detachment of older cells (refringent cells). Concurrently with CPE, the cell growth was maintained at the beginning of kinetics. HTR-8 showed similar dynamics to other cell types. At 24 and 48 hpi, there was a peak of intracellular viral production, followed by a rapid decline. In contrast, extracellular medium remained constant throughout the kinetics. Furthermore, after 72 hpi, a plenty of cells still in the monolayer. HuH-7 presented a peak of intracellular viral production after 24h hpi and continued to decline until 144 hpi. In the extracellular medium showed a lot of cellular debris after the pick of infection. (
Figure 2a and
Figure S1).
3.2. Inoculation of ZIKV-IEC-Paraíba (b)
After ZIKV-IEC-Paraíba infection, BeWo and BeWo+fork showed different growth curves (
Figure 3 and
Appendix I). The amount of PFU/mL of BeWo cells remained higher intracellularly until 120 hpi, and the peak production occurred at 48 hpi. Regarding the extracellular medium, the PFU/mL was maintained lower the intracellularly until 144 hpi, when it became higher. The CPE was observed at 72 hpi and augmented at 144 hpi, but part of the monolayer retained the characteristics of non-infected (
Figure S1). BeWo+fork showed intense viral production between 24 and 72 hpi, with a drop at 96 hpi and an increase between 120 and 144 hpi. During all the kinetics, the amount of virus in the extracellular medium remained constant, with few fluctuations over the time. CPE was observed at 48 hpi and progressively increased in posterior hpi. Furthermore, there was monolayer detachment and cellular debris (
Figure 3b and
Figure S1).
HTR-8 maintained a high PFU/mL in the intracellular environment until 96 hpi. In contrast, at 120 and 144 hpi, the number of virions augmented in the extracellular medium and decreased in the intracellular medium, probably indicating cell bursts. CPE started was observed at 96 hpi, but debris formation only happened at next hpi. Most cells remained adhered to monolayers and showed signs of infection (
Figure 3b and
Figure S1).
HuH-7 viral production in intracellular and extracellular medium were high between 24 and 72 hpi. A fast decline of growth occurred in the intracellular environment after this period, whereas the extracellular environment remained constant until 144 hpi. CPE became apparent at 48 hpi and increased up to 144 hpi, when all cells formed cellular debris and there was no longer a monolayer (
Figure 2b and
Figure S1).
We considered 7 time points to describe the viral infection, its capacity to generate new infectious progenies and to observe the CPE. After six days, at 144 hpi for ZIKV-MR766lp, the majority of the available cells were infected, which reflected in the considerable decay of alive cells. Regarding ZIKV-IEC, cell death by infection was only high for HuH-7; while in BeWo, BeWo+fork and HTR-8 there were persistence of alive cells—possible abortive infection? Moreover, BeWo+fork and HTR-8 cells sustained the infection and a possible new round of viral replication, even after the infectious peak. Another fact noticed in the kinetics, for all assays with the exception of HuH-7, is that the cell monolayer remained the same in the last three hpi, when compared to its non-inoculated controls. The most prominent CPEs in infected cells were morphological changes and death. Dead cells are also present in non-inoculated control, due to the fact that the control trophoblastic cells reach maximum confluence and lose space during monolayer expansion. These results may be an indication that during the replicative cycle of the virus there was no inhibition of cell multiplication. Moreover, in the kinetics end, the supernatant had both dead cells by the virus and cells, possibly alive, due to the continuity of monolayer.
4. Discussion
Considering the results obtained by us, all cell lineages are permissive and susceptible to a both lineages of ZIKV with commons characteristics CPEs observed in Flavivirus.
Regarding virus transmission by placental barrier, the first 12 gestational weeks are the most critical, because placenta is still developing and its protection depends on maternal antibodies [
13,
14,
16,
17,
31]. Among proposed routes, virus transmission can occur via trophoblasts [
32,
33], which are pluripotent cells that arise in blastula stage and beginning of the first cell differentiation and will originate the embryo and embryonic annexes [
34]. Studies were published about the development of ZIKV infection in embryonic stem cells, from the first gestational semester, and in human placental explant [
33,
39,
40,
41,
42]. These papers comparatively showed how the two strains infect and cross the placental protective barrier, and shows that both lineages are permissive and susceptible. But does not show why the Asian lineage can cross placental barrier and causes CZS infection in the embryo/fetus/newborn, and African lineage does not.
The percentage of maternal-fetal transmission is estimated by 20-30% of pregnant [
9]. The hypotheses about maternal-fetal route of transmission, excluding the primary route of transmission by vector or sex, occur through the placental tissue. Villous trophoblast cells (VT), present in the beginning of embryogenesis, differentiate into cytotrophoblasts (CTB) and then into syncytiotrophoblasts (STB), that correspond respectively to BeWo and BeWo+fork. Forskolin induces BeWo to form syncytial cells. Based on the VT, there is a subset of extravillous cells (EVT) which matches with the HTR-8 lineage. Such cells arise between the sixth and twelfth week, in the first trimester of pregnancy, and constitute an efficient barrier against microorganisms like viruses. Thus, since the onset of gestation, the interaction between the maternal uterine and embryonic placental tissues promotes a barrier against the entry of microorganisms [
39,
42]. The first trimester is also considered the period of greatest risk for the embryo/fetus to contract CZS [
9,
12,
37,
38,
46]. Another factor of gestational vulnerability in the first trimester is that maternal IgG crosses the placental barrier only from the second trimester of gestation [
38,
42,
44].
The comparison of African and Asian lineages in vitro (in different cell types),
in vivo, non-human primates and in the most common species of
Aedes spp is presented and discussed in the scientific literature [
18]. The majority of such experiments showed that the African strain is more infectious than the Asian [
32,
33,
36]. However, only the Asian strain have the potential to cause CZS and GBS [
8,
11,
33].
Considering the hypotheses emerged for ZIKV to have become an infectious agent capable of infecting neural progenitor cells (NPC), one is due to point mutations in structural genes D67N [
35] and S139N [
36], and non-structural T2634V [
37]. When considering ZIKV-MR766 (African), ZIKV-Asian (2010) and ZIKV-Asian (2013-15) genomes, through in vivo and in vitro experiments that exist, at least such mutations that are considered hot- spot [
35,
36,
37]. Such mutations are related to increased infectivity, cellular tropism for NPC and NS cells, and less virulence, but with greater persistence in tissues [
43]. Considering that in our study the BeWo and HTR-8 cells that showed similar dynamics, it is possible that these same genes also favor viral tropism in placental cells and infect the embryo/fetus. But need more studies.
Another hypothesis is related of the target cells can be modified by ZIKV and induced an apoptosis, necrosis and paraptosis [
9]. Considering the results of Asian-Brazilian lineage one supposes the lineage cells BeWo and HTR-8 stay alive for more time, maybe because the pathway apoptosis is not activated or block for some reason not investigate yet.
5. Conclusions
In summary, our study shows that first trimester placenta cells are permissive and susceptible to African and Asian-Brazilian ZIKV infections in vitro. While ZIKV-MR766lp effectively infects placental cells leading to fast death, which can be an indication that infection by such strain is not persistent and is less transmissible to fetus, ZIKV-IEC-Paraíba presented sustained and longer replicative cycle, without inducing cell death.
Although it is emphasized here that this type of test does not represent the conditions of uterine environment, our method allows a better observation of the dynamics of ZIKV infection and, as such should be encouraged for further in-dept studies.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: Image result of cytopathic effect of viral kinetics (in .pdf extension); Table S1: Complete table of viral kinetics data (in .xlsx extension).
Author Contributions
M.D.B and A.C. they also contributed to the research design, execution and data analysis; M.D.B. preparation of writing the original draft; R.A.S.; C.M.P.; C.T.B. data review, writing review and editing; P.M.A.Z A. and L.M.J equal supervision and Principal Investigators. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
We encourage all authors of articles published in MDPI journals to share their research data. In this section, please provide details regarding where data supporting reported results can be found, including links to publicly archived datasets analyzed or generated during the study. Where no new data were created, or where data is unavailable due to privacy or ethical restrictions, a statement is still required. Suggested Data Availability Statements are available in section “MDPI Research Data Policies” at
https://www.mdpi.com/ethics.
Acknowledgments
We say thank you for Carla Torres Braconi, Marielton dos Passos Cunha, Telma Alves Monezi, Cila Ankier that they helped with this research in search way.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. BeWo treated with forskolin.
The treatment of BeWo cells with forskolin follow the protocol suggested by Profa.Dra. Estela M. A. F. Bevilacqua. Formation of 10 mM concentrated solution: 1 mL liquid DMSO (Merck™ Cas#67-68-5) plus 4.105 mg of forskolin in powder form (Sigma Aldrich® CAS# 66575-29-9). After, usage of DMSO 10 μM (0.2%). Later 24h incubation of BeWo cells grown in monolayer, add 1 μL of the diluted solution for every 1 ml of a new culture medium supplemented with 10% FBS. Return to the greenhouse for 24h at 37°C and 5% CO2, and then proceed with the inoculation.
Appendix B. Adaptation of qRT-PCR Performed by Lanciotti et al. 2008
Mix reagent of qRT-PCR for final volume of 15 μL do kit AgPath-ID™ One-Step RT-PCR—Applied Biosystems™ (Cat#AM1005): DEPC water (Sigma Aldrich® CAS#7732-18-5) 3,5 μL; 3,5 μL tampon; 0,5 μL sense and anti-sense primers for each; 0,5 μL enzyme; and 5 μL extracted RNA. StepOne™ Real-Time PCR System (Applied Biosystems™) reaction cycle: (i) one 45°C for 10 minutes; (ii) one 95°C for 10 min; and 40 cycles of 95°C for 15 seconds plus 60°C for 45s.
Appendix C. Viral Kinetics
1. Cell inoculation was performed according to the MOI determined from the viral stock of ZIKV-MR766 and ZIKV-IEC-Paraiba. 2. The MOI as established for ZIKV-MR766 = 1 and ZIKV-IEC = 0.5. 3. The cells were cultured in six-well plates for 24h before starting the assays, with exception of BeWo lineage treated with forskolin, started 48h before. 4. After the period of cultivation in the plate, we removed the medium and performed a new cell count. The monolayer was washed with PBS and the inoculation started per well. 5. We dripped the MOI value in μL of the inoculum throughout the monolayer cells. Then, we kept the cells in the greenhouse for two hours (adsorption phase, under ideal conditions for them); and minimum volume of medium was maintained on the monolayer together with the inoculum so that such cells did not dry out in the incubator. 6. After adsorption (2h in the incubator), it was washed again with PBS in order to remove cellular debris and non-adsorbed virions. 7. At zero time = 2 hpi, we collected from two wells and separately the supernatant (inoculum volume) and the monolayer cells. Before collecting the cell monolayer, it was washed with PBS to remove debris and virions. 8. Store in 2 mL microtubes the samples, identify and kept in the -80°C freezer until molecular analysis. 9. Repeat collection separately for each hpi (item 7 and 8). 10. Store in the freezer at -80°C for future analysis.
Appendix D. Standard Curve | Direct Relation between CT and PFU/mL
1. The standard curve stablished a direct relation between CT and PFU/mL, obtained from the viral stock titration of ZIKV-MR766 and ZIKV-IEC-Paraiba. Such titration was performed in triplicate and its mean values were gotten. The serial dilution for the titration started from 10−1 up to 10−11 fraction. 2. In a 24-well plate and in duplicate for each fraction. However, we collected the total volume for qRT-PCR because all cells generally die in the first fractions. 3. The extraction followed the TRIzol®Reagent (Invitrogen™ Cat#15596026) protocol. Extracted samples were quantified in the NanoDrop™ spectrophotometer and normalized to an average value of 200 ng/μL RNA contraction. The reaction and primers for qRT-PCR followed the protocol published by Lanciotti et al., 2008. 4. After obtaining the CT values for each fraction, we calculated the corresponding values of PFU/mL with them.
Appendix E. Standard Curve | Calculation of Direct Relation between CT and PFU/mL
1. Average CT obtained for each dilution fraction. 2. Mean titer of PFU/mL triplicate. 3. Conversion table. 3.1. First column. Numerical representation of dilution. 3.2. Second column. Values of mean dilution. The 10−1 fraction corresponded to the average value of the titer got in PFU/mL. The next fraction, 10−2, was the value from 10−1 divided for 10. Such division pattern was followed up to the 10−11 fraction. 3.3. Third column. Second column values multiplied by volume in mL used for RNA extraction with TRIzol per corresponding sample. 3.4. Fourth column. Values from third column multiplied by volume of RNA used in qRT-PCR in mL and its result divided by the resuspended volume of RNA after extraction with TRIzol. 3.5. Fifth column. Transformation of fourth column values into Log10. 3.6. Sixth column. Average of CT corresponding to each dilution fraction. 3.7. Chart of standard curve. The negative values of fifth column were excluded, and the equation of line and determination Coefficient was obtained. Follow the Excel table with the step by step described above. File in attachment.
Appendix F. Conversion of CT Values into PFU/mL of the Viral Kinetics Graph
The conversion of CT values obtained from the kinetics assay uses the equation of standard curve straight. 1. First column. Presentation of all hpi. 2. Second column. Values of all CT got from the supernatant and cell monolayer in their corresponding hpi. 3. Third column. Equation of the line referring to the ZIKV lineage, in which the x value corresponds to the CT of the second column. 4. Fourth column. Transformation of values to antiLog10, and multiplication of the previous column numbers by 10. 5. Fifth column. Conversion equation of PFU/mL value in volume of the sample collected for extraction with TRIzol in mL, x = PFU/0,25 mL. Equation: (volume in mL qRT-PCR) (x) = (volume in mL of extracted RNA) (PFU/antiLog10). 6. Sixth column. Conversion equation of PFU/mL value in volume in sampling one mL, x = PFU/mL. Equation: (volume in mL of sample for extraction) (x) = (sample volume of one mL) (PFU/0,25 mL). Chart: hpi abcissa and PFU/mL ordinate). Attached file.
Values table of triplicate tritation: the titers obtained for ZIKV-IEC were 1.30x106 PFU/mL, 1.40x106 PFU/mL, and 1.68x106 PFU/mL; while for ZIKV-MR766 were 1.57x108 PFU/mL, 1.57x108 PFU/mL, and 1.43x108 PFU/mL.
| Strain/Replicate (PFU/mL) |
First |
Second |
Third |
Average |
ZIKV-MR766
low passage
|
1.57x108
|
2.05x108
|
1.43x108
|
1.68x108
|
| ZIKV-IEC-Paraíba |
1.30x106
|
1.40x106
|
1.68x106
|
1.46x108
|
Appendix G. Calculation of the Extra and Intracellular Ratio
1. Average of free virions produced at time point—A value. Calculations of intracellular at the 2 hpi, and extracellular values at the 24 hpi. 2. Mean free virions at the time-point after burst, peak of highest extracellular production—B value. 3. The B minus A subtraction indicates full burst or release of new virions—C value. 4. Division of C by the number of virions average of the viral titer obtained in the construction of the standard curve.
Appendix H. Graph Results Standard Curve by qRT-PCR
Appendix I. Graphic Result of Complete Viral Kinetics

References
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus (II). Pathogenicity and physical properties. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Dick, G.W. Epidemiological notes on some viruses isolated in Uganda (Yellow fever, Rift Valley fever, Bwamba fever, West Nile, Mengo, Semliki Forest, Bunyamwera, Ntaya, Uganda S and Zika viruses). Trans. R. Soc. Trop. Med. Hyg. 1953, 47, 13–48. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.I. Zika virus infection in man. Trans. R. Soc. Trop. Med. Hyg. 1964, 58, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Marchette, N.J.; Garcia, R.; Rudnick, A. Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am. J. Trop. Med. Hyg. 1969, 18, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.G.; Ksiazek, T.G. Zika virus, a cause of fever in Central Java, Indonesia. Trans. R. Soc. Trop. Med. Hyg. 1981, 75, 389–393. [Google Scholar] [CrossRef]
- Posen, H.J.; Keystone, J.S.; Gubbay, J.B.; Morris, S.K. Epidemiology of Zika virus, 1947–2007. BMJ Glob. Health 2016, 1, e000087. [Google Scholar] [CrossRef]
- Musso, D.; Gubler, D.J. Zika virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef]
- Masmejan, S.; Musso, D.; Vouga, M.; Pomar, L.; Dashraath, P; Stojanov, M. ; Panchaud, A.; Baud, D. Zika Virus. Pathogens 2020, 9, 898. [Google Scholar] [CrossRef]
- Musso, D.; Ko, A.I.; Baud, D. Zika virus infection—After the pandemic. New Engl. J. Med. 2019, 381, 1444–1457. [Google Scholar] [CrossRef]
- Pardy, R.D.; Richer, M.J. Zika Virus Pathogenesis: From Early Case Reports to Epidemics. Viruses 2019, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Martines, R.B.; Bhatnagar, J.; de Oliveira Ramos, A.M.; Davi, H.P.; Iglezias, S.D.; Kanamura, C.T.; Keating, M.K.; Hale, G.; Silva-Flannery, L.; Muehlenbachs, A.; Ritter, J. Pathology of congenital Zika syndrome in Brazil: A case series. Lancet 2016, 388, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Miner, J.J.; Diamond, M.S. Zika virus pathogenesis and tissue tropism. Cell Host Microbe 2017, 21, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Rabelo, K.; De Souza, L.J.; Salomão, N.G.; Machado, L.N.; Pereira, P.G.; Portari, E.A.; Basílio-de-Oliveira, R.; Dos Santos, F.B.; Neves, L.D.; Morgade, L.F.; Provance, Jr. DW. Zika induces human placental damage and inflammation. Front. Immunol. 2020, 1, 2146. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.S.; Sevvana, M.; Kuhn, R.J.; Rossmann, M.G. Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 2018, 25, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Diamond, M.S.; Mysorekar, I.U. Maternal-fetal transmission of Zika virus: Routes and signals for infection. J. Interferon Cytokine Res. 2017, 37, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Mysorekar, I.U.; Diamond, M.S. Modeling Zika virus infection in pregnancy. New Engl. J. Med. 2016, 375, 481–484. [Google Scholar] [CrossRef]
- Burleson, F.G.; Chamber, T.M.; Widebrauk, D. Virology: A laboratory manual; Academic Press, Inc.: San Diego, California, 1992; pp. 100–106. [Google Scholar]
- Ellis, E.L.; Delbruck, M. The growth of bacteriophage. J. Gen. Physiol. 1939, 22, 365–384. [Google Scholar] [CrossRef]
- Falke, D. Virologia; E.P.U. Springer EDUSP: São Paulo, Brasil, 1979; pp. 13–34. [Google Scholar]
- Mautner, L.; Hoyos, M.; Dangel, A.; Berger, C.; Ehrhardt, A.; Baiker, A. Replication kinetics and infectivity of SARS-CoV-2 variants of concern in common cell culture models. Virol. J. 2022, 19, 1–11. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.J. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch. Virol. 2007, 152, 687–696. [Google Scholar] [CrossRef]
- Faye, O.; Freire, C.C.; Iamarino, A.; Faye, O.; de Oliveira, J.V.; Diallo, M.; Zanotto, P.M.; Sall, A.A. Molecular evolution of Zika virus during its emergence in the 20th century. PLoS Neglected Trop. Dis. 2014, 8, e2636. [Google Scholar] [CrossRef] [PubMed]
- Aubry, F.; Jacobs, S.; Darmuzey, M.; Lequime, S.; Delang, L.; Fontaine, A.; Jupatanakul, N.; Miot, E.F.; Dabo, S.; Manet, C.; Montagutelli, X. Recent African strains of Zika virus display higher transmissibility and fetal pathogenicity than Asian strains. Nat. Commun. 2021, 12, 916. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Azevedo, R.D.; Kraemer, M.U.; Souza, R.; Cunha, M.S.; Hill, S.C.; Thézé, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I.; Rocco, I.M. Zika virus in the Americas: Early epidemiological and genetic findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Timenetsky, J.; Santos, L.M.; Buzinhani, M.; Mettifogo, E. Detection of multiple mycoplasma infection in cell cultures by PCR. Braz. J. Med. Biol. Res. 2006, 39, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Orendi, K.; Gauster, M.; Moser, G.; Meiri. H.; Huppertz, B. The choriocarcinoma cell line BeWo: Syncytial fusion and expression of syncytium-specific proteins. Reproduction 2010, 140, 759–766. [Google Scholar] [CrossRef]
- Keogh, B.P. Adsorption, latent period and burst size of phages of some strains of lactic streptococci. J. Dairy Res. 1973, 40, 303–309. [Google Scholar] [CrossRef]
- Haddow, A.D.; Schuh, A.J.; Yasuda, C.Y.; Kasper, M.R.; Heang, V.; Huy, R.; Guzman, H.; Tesh, R.B.; Weaver, S.C. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLoS Neglected Trop. Dis. 2012, 6, e1477. [Google Scholar] [CrossRef]
- Sheridan, M.A.; Yunusov, D.; Balaraman, V.; Alexenko, A.P.; Yabe, S.; Verjovski-Almeida, S.; Schust, D.J.; Franz, A.W.; Sadovsky, Y.; Ezashi, T.; Roberts, R.M. Vulnerability of primitive human placental trophoblast to Zika virus. Proc. Natl. Acad. Sci. USA 2017, 114, 1587–1596. [Google Scholar] [CrossRef]
- Tabata, T.; Petitt, M.; Puerta-Guardo, H.; Michlmayr, D.; Wang, C.; Fang-Hoover, J.; Harris, E.; Pereira, L. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe 2016, 20, 155–166. [Google Scholar] [CrossRef]
- Tabata, T.; Petitt, M.; Puerta-Guardo, H.; Michlmayr, D.; Harris, E.; Pereira, L. Zika virus replicates in proliferating cells in explants from first-trimester human placentas, potential sites for dissemination of infection. J. Infect. Dis. 2018, 217, 1202–1213. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S. F.; Barresi, M.J.F. Developmental Biology, 11th ed.; Sinauer Associates, Inc.: Sunderland, Massachusetts, USA, 2016; pp. 143–180. [Google Scholar]
- Liu Liu. Z.; Zhang, Y.; Cheng, M.; Ge, N.; Shu, J.; Xu, Z.; Su, X.; Kou, Z.; Tong, Y.; Qin, C.; Jin, X. A single nonsynonymous mutation on ZIKV E protein-coding sequences leads to markedly increased neurovirulence in vivo. Virol. Sin. 2022, 37, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Huang, X.Y.; Liu, Z.Y.; Zhang, F.; Zhu, X.L.; Yu, J.Y.; Ji, X.; Xu, Y.P.; Li, G.; Li, C.; Wang, H.J. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science 2017, 358, 933–936. [Google Scholar] [CrossRef] [PubMed]
- King, E.L.; Irigoyen, N. Zika Virus and Neuropathogenesis: The Unanswered Question of Which Strain Is More Prone to Causing Microcephaly and Other Neurological Defects. Front. Cell. Neurosci. 2021, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.; Sadovsky, Y.; Dermody, T.S.; Coyne, C.B. Microbial vertical transmission during human pregnancy. Cell Host Microbe 2017, 21, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, M.A.; Yunusov, D.; Balaraman, V.; Alexenko, A.P.; Yabe, S.; Verjovski-Almeida, S.; Schust, D.J.; Franz, A.W.; Sadovsky, Y.; Ezashi, T.; Roberts, R.M. Vulnerability of primitive human placental trophoblast to Zika virus. Proc. Natl. Acad. Sci. USA 2017, 114, 1587–1596. [Google Scholar] [CrossRef]
- Sheridan, M.A.; Balaraman, V.; Schust. D.J.; Ezashi, T.; Roberts, R.M.; Franz, A.W. African and Asian strains of Zika virus differ in their ability to infect and lyse primitive human placental trophoblast. PLoS ONE 2018, 13, e0200086. [Google Scholar] [CrossRef]
- León-Juárez, M.; Martínez–Castillo, M.; González-García, L.D.; Helguera-Repetto, A.C.; Zaga-Clavellina, V.; García-Cordero, J.; Flores-Pliego, A.; Herrera-Salazar, A.; Vázquez-Martínez, E.R.; Reyes-Muñoz, E. Cellular and molecular mechanisms of viral infection in the human placenta. Pathog. Dis. 2017, 75, 1–15. [Google Scholar] [CrossRef]
- Quicke, K.M.; Bowen, J.R.; Johnson, E.L.; McDonald, C.E.; Ma, H.; O’Neal, J.T.; Rajakumar, A.; Wrammert, J.; Rimawi, B.H.; Pulendran, B.; Schinazi, R.F. Zika virus infects human placental macrophages. Cell Host Microbe 2016, 20, 83–90. [Google Scholar] [CrossRef]
- Simister, N.E. Placental transport of immunoglobulin G. Vaccine 2003, 21, 3365–3369. [Google Scholar] [CrossRef] [PubMed]
- Adibi, J.J.; Marques, Jr. E.T.; Cartus, A.; Beigi, R.H. Teratogenic effects of the Zika virus and the role of the placenta. Lancet 2016, 387, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- El Costa, H.; Gouilly, J.; Mansuy, J.M.; Chen, Q.; Levy, C.; Cartron, G.; Veas, F.; Al-Daccak, R.; Izopet, J.; Jabrane-Ferrat, N. ZIKA virus reveals broad tissue and cell tropism during the first trimester of pregnancy. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Meaney-Delman, D.; Oduyebo, T.; Polen, K.N.; White, J.L.; Bingham, A.M.; Slavinski, S.A.; Heberlein-Larson, L.; St George, K.; Rakeman, J.L.; Hills, S.; Olson, C.K. Prolonged detection of Zika virus RNA in pregnant women. Obstet. Gynecol. 2016, 128, 724–730. [Google Scholar] [CrossRef]
- Suy, A.; Sulleiro, E.; Rodó, C.; Vázquez, É.; Bocanegra, C.; Molina, I.; Esperalba, J.; Sánchez-Seco, M.P.; Boix, H.; Pumarola, T.; Carreras, E. Prolonged Zika virus viremia during pregnancy. New Engl. J. Med. 2016, 375, 2611–2613. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).