Submitted:
16 May 2023
Posted:
18 May 2023
You are already at the latest version
Abstract
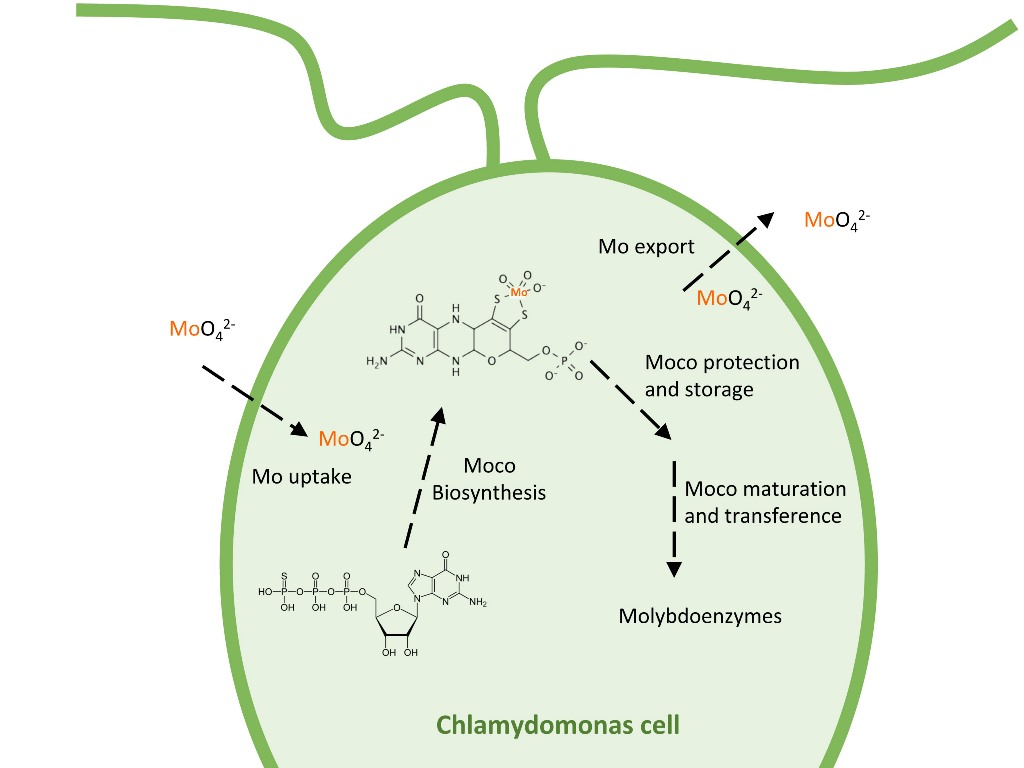
Keywords:
1. Chlamydomonas reinhardtii as a Model Microorganism
2. The Role of Molybdenum in Living Organisms
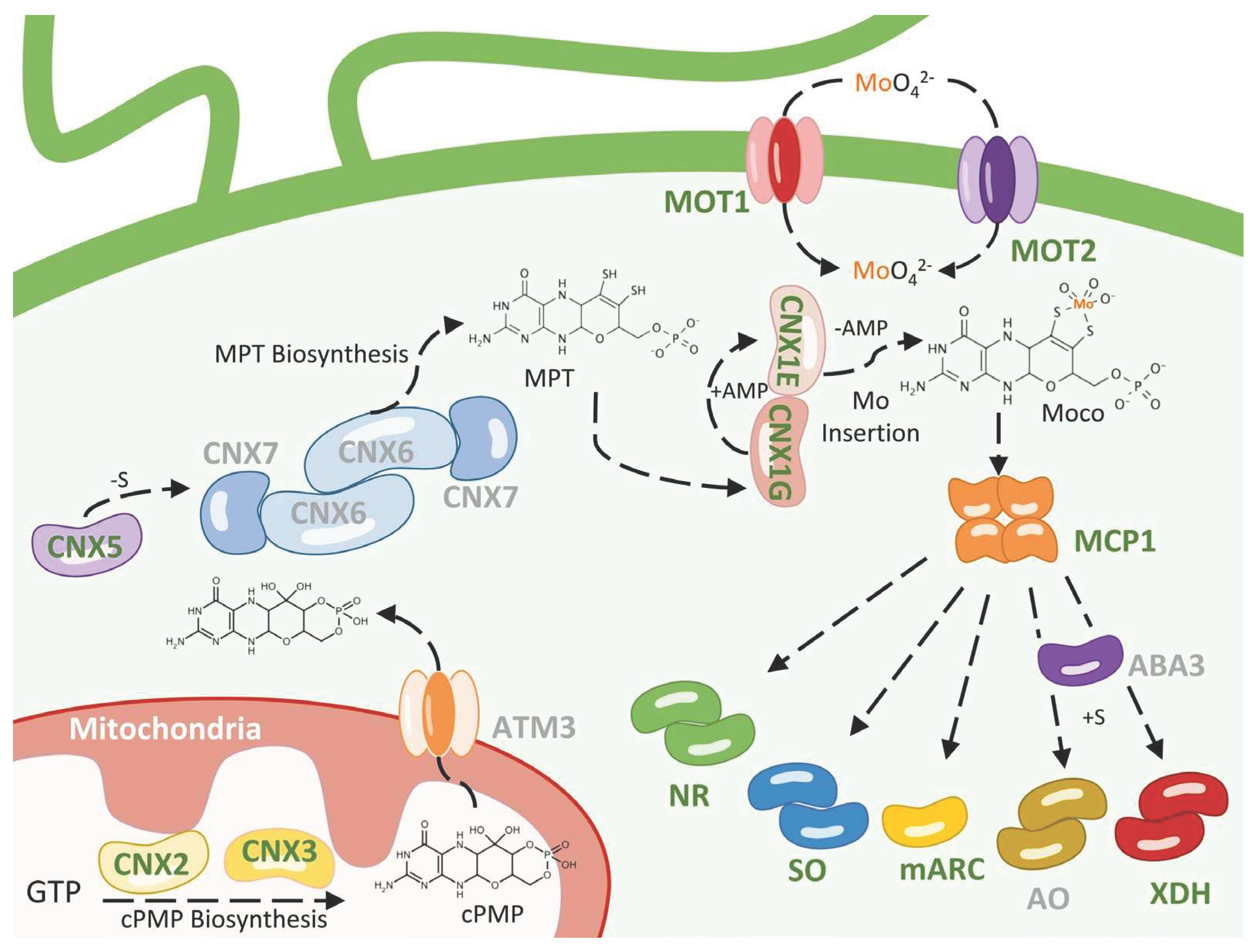
3. Molybdenum Uptake
4. Moco Biosynthesis
4.2. MPT Biosynthesis
4.3. Mo Insertion
5. Moco Storage
6. Chlamydomonas Moco Enzymes
6.1. Xanthine Dehydrogenase
6.2. Aldehyde Oxidase
6.3. Sulfite Oxidase
6.4. Nitrate Reductase
7. Concluding Remarks and Future Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Quintas-Nunes, F.; Brandão, P.R.; Barreto Crespo, M.T.; Glick, B.R.; Nascimento, F.X. Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium Rhizosphaerae Sp. Nov. Plants 2023, 12, 651. [CrossRef]
- Maire, J.; Buerger, P.; Chan, W.Y.; Deore, P.; Dungan, A.M.; Nitschke, M.R.; van Oppen, M.J.H. Effects of Ocean Warming on the Underexplored Members of the Coral Microbiome. Integr. Comp. Biol. 2022, 62, 1700–1709. [CrossRef]
- Grimm, M.; Grube, M.; Schiefelbein, U.; Zühlke, D.; Bernhardt, J.; Riedel, K. The Lichens’ Microbiota, Still a Mystery? Front. Microbiol. 2021, 12, 623839. [CrossRef]
- Cvetkovska, M.; Vakulenko, G.; Smith, D.R.; Zhang, X.; Hüner, N.P.A. Temperature Stress in Psychrophilic Green Microalgae: Minireview. Physiol. Plant. 2022, 174, e13811.. [CrossRef]
- Falkowski, P.G. The Role of Phytoplankton Photosynthesis in Global Biogeochemical Cycles. Photosynth. Res. 1994, 39, 235–258. [CrossRef]
- Tarafdar, A.; Sowmya, G.; Yogeshwari, K.; Rattu, G.; Negi, T.; Awasthi, M.K.; Hoang, A.T.; Sirohi, R. Environmental Pollution Mitigation through Utilization of Carbon Dioxide by Microalgae. Environ. Pollut. 2023, 328, 121623. [CrossRef]
- Kazmi, S.S.U.H.; Yapa, N.; Karunarathna, S.C.; Suwannarach, N. Perceived Intensification in Harmful Algal Blooms Is a Wave of Cumulative Threat to the Aquatic Ecosystems. Biology (Basel). 2022, 11, 852. [CrossRef]
- Gerotto, C.; Norici, A.; Giordano, M. Toward Enhanced Fixation of CO2 in Aquatic Biomass: Focus on Microalgae. Front. Energy Res. 2020, 8, 213. [CrossRef]
- Parmar, P.; Kumar, R.; Neha, Y.; Srivatsan, V. Microalgae as next Generation Plant Growth Additives: Functions, Applications, Challenges and Circular Bioeconomy Based Solutions. Front. Plant Sci. 2023, 14, 1073546. [CrossRef]
- Fabris, M.; Abbriano, R.M.; Pernice, M.; Sutherland, D.L.; Commault, A.S.; Hall, C.C.; Labeeuw, L.; McCauley, J.I.; Kuzhiuparambil, U.; Ray, P.; et al. Emerging Technologies in Algal Biotechnology: Toward the Establishment of a Sustainable, Algae-Based Bioeconomy. Front. Plant Sci. 2020, 11, 279. [CrossRef]
- Zhang, B.; Li, W.; Guo, Y.; Zhang, Z.; Shi, W.; Cui, F.; Lens, P.N.L.; Tay, J.H. Microalgal-Bacterial Consortia: From Interspecies Interactions to Biotechnological Applications. Renew. Sustain. Energy Rev. 2020, 118, 109563. [CrossRef]
- Harris, E.H. Introduction into Chlamydomonas and Its Laboratory Use. Chlamydomonas Sourcebook. Oxford Acad. Press 2009.
- Salomé, P.A.; Merchant, S.S. A Series of Fortunate Events: Introducing Chlamydomonas as a Reference Organism. Plant Cell 2019, 31, 1682–1707. [CrossRef]
- Llamas, A.; Tejada-Jiménez, M.; Fernández, E.; Galván, A. Molybdenum Metabolism in the Alga Chlamydomonas Stands at the Crossroad of Those in Arabidopsis and Humans. Metallomics 2011, 3, 578–590. [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science 2007, 318, 245–250. [CrossRef]
- Fauser, F.; Vilarrasa-Blasi, J.; Onishi, M.; Ramundo, S.; Patena, W.; Millican, M.; Osaki, J.; Philp, C.; Nemeth, M.; Salomamp, P.A.; et al. Systematic Characterization of Gene Function in the Photosynthetic Alga Chlamydomonas Reinhardtii. Nat. Genet. 2022, 54, 705–714. [CrossRef]
- Goodenough, U. The Chlamydomonas Sourcebook. Volume 1: Introduction to Chlamydomonas and Its Laboratory Use. Elsevier Acad. Press 2023. [CrossRef]
- Bellido-Pedraza, C.M.; Calatrava, V.; Sanz-Luque, E.; Tejada-Jiménez, M.; Llamas, A.; Plouviez, M.; Guieysse, B.; Fernández, E.; Galván, A. Chlamydomonas Reinhardtii, an Algal Model in the Nitrogen Cycle. Plants 2020, 9, 903. [CrossRef]
- Saroussi, S.; Sanz-Luque, E.; Kim, R.G.; Grossman, A.R. Nutrient Scavenging and Energy Management: Acclimation Responses in Nitrogen and Sulfur Deprived Chlamydomonas. Curr. Opin. Plant Biol. 2017, 39, 114–122. [CrossRef]
- Irihimovitch, V.; Yehudai-Resheff, S. Phosphate and Sulfur Limitation Responses in the Chloroplast of Chlamydomonas Reinhardtii. FEMS Microbiol. Lett. 2008, 283, 1–8. [CrossRef]
- Calatrava, V.; Hom, E.F.Y.; Llamas, Á.; Fernández, E.; Galván, A. Nitrogen Scavenging from Amino Acids and Peptides in the Model Alga Chlamydomonas Reinhardtii. The Role of Extracellular L-Amino Oxidase. Algal Res. 2019, 38, 101395. [CrossRef]
- Li-Beisson, Y.; Kong, F.; Wang, P.; Lee, Y.; Kang, B.-H. The Disassembly of Lipid Droplets in Chlamydomonas. New Phytol. 2021, 231, 1359–1364. [CrossRef]
- Rengel, R.; Giraldez, I.; Díaz, M.J.; García, T.; Vigara, J.; Leon, R. Simultaneous Production of Carotenoids and Chemical Building Blocks Precursors from Chlorophyta Microalgae. Bioresour. Technol. 2022, 351, 127035. [CrossRef]
- Sirohi, R.; Joun, J.; Choi, H.I.; Gaur, V.K.; Sim, S.J. Algal Glycobiotechnology: Omics Approaches for Strain Improvement. Microb. Cell Fact. 2021, 20, 163. [CrossRef]
- Calderon, R.H.; de Vitry, C.; Wollman, F.A.; Niyogi, K.K. Rubredoxin 1 Promotes the Proper Folding of D1 and Is Not Required for Heme B559 Assembly in Chlamydomonas Photosystem II. J. Biol. Chem. 2023, 299, 102968. [CrossRef]
- Przybyla-Toscano, J.; Couturier, J.; Remacle, C.; Rouhier, N. Occurrence, Evolution and Specificities of Iron-Sulfur Proteins and Maturation Factors in Chloroplasts from Algae. Int. J. Mol. Sci. 2021, 22, 3175. [CrossRef]
- Roach, T.; Baur, T.; Kranner, I. β-Cyclocitral Does Not Contribute to Singlet Oxygen-Signalling in Algae, but May Down-Regulate Chlorophyll Synthesis. Plants 2022, 11, 2155. [CrossRef]
- Yang, M.; Xie, X.; Kong, F.T.; Xie, K.P.; Yu, S.H.; Ma, J.Y.; Xue, S.; Gong, Z. Differences in Glycerolipid Response of Chlamydomonas Reinhardtii Starchless Mutant to High Light and Nitrogen Deprivation Stress Under Three Carbon Supply Regimes. Front. Plant Sci. 2022, 13, 860966. [CrossRef]
- Kreis, E.; Niemeyer, J.; Merz, M.; Scheuring, D.; Schroda, M. CLPB3 Is Required for the Removal of Chloroplast Protein Aggregates and Thermotolerance in Chlamydomonas. J. Exp. Bot. 2023, erad109. [CrossRef]
- Zou, Y.; Bozhkov, P. V. Chlamydomonas Proteases: Classification, Phylogeny, and Molecular Mechanisms. J. Exp. Bot. 2021, 72, 7680–7693. [CrossRef]
- Marchetti, G.M.; Füsser, F.; Singh, R.K.; Brummel, M.; Koch, O.; Kümmel, D.; Hippler, M. Structural Analysis Revealed a Novel Conformation of the NTRC Reductase Domain from Chlamydomonas Reinhardtii. J. Struct. Biol. 2022, 214, 107829. [CrossRef]
- Marshall, W.F. The Flagellar Length Control System: Exploring the Physical Biology of Organelle Size. Phys. Biol. 2023, 20, 021001. [CrossRef]
- Sasso, S.; Stibor, H.; Mittag, M.; Grossman, A.R. From Molecular Manipulation of Domesticated Chlamydomonas Reinhardtii to Survival in Nature. Elife 2018, 7, e39233. [CrossRef]
- Masi, A.; Leonelli, F.; Scognamiglio, V.; Gasperuzzo, G.; Antonacci, A.; Terzidis, chael A. Chlamydomonas Reinhardtii: A Factory of Nutraceutical and Food Supplements for Human Health. Molecules 2023, 28, 1185. [CrossRef]
- Dehghani, J.; Movafeghi, A.; Mathieu-Rivet, E.; Mati-Baouche, N.; Calbo, S.; Lerouge, P.; Bardor, M. Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants. Mar. Drugs 2022, 20, 657. [CrossRef]
- Rosales-Mendoza, S.; García-Silva, I.; González-Ortega, O.; Sandoval-Vargas, J.M.; Malla, A.; Vimolmangkang, S. The Potential of Algal Biotechnology to Produce Antiviral Compounds and Biopharmaceuticals. Molecules 2020, 25, 4049. [CrossRef]
- Fields, F.J.; Lejzerowicz, F.; Schroeder, D.; Ngoi, S.M.; Tran, M.; McDonald, D.; Jiang, L.; Chang, J.T.; Knight, R.; Mayfield, S. Effects of the Microalgae Chlamydomonas on Gastrointestinal Health. J. Funct. Foods 2020, 65, 103738. [CrossRef]
- Calatrava, V.; Tejada-Jimenez, M.; Sanz-Luque, E.; Fernandez, E.; Galvan, A.; Llamas, A. Chlamydomonas Reinhardtii, a Reference Organism to Study Algal–Microbial Interactions: Why Can’t They Be Friends? Plants 2023, 12, 788. [CrossRef]
- Fakhimi, N.; Gonzalez-Ballester, D.; Fernández, E.; Galván, A.; Dubini, A. Algae-Bacteria Consortia as a Strategy to Enhance H2 Production. Cells 2020, 9, 1353. [CrossRef]
- Torres, M.; Gonzalez-Ballester, D.; Gomez-Osuna, A.; Galván, A.; Fernandez, E.; Dubini, A. Chlamydomonas-Methylobacterium Oryzae Cooperation Leads to Increased Biomass, Nitrogen Removal, and Hydrogen Production. Bioresour. Technol. 2022, 352, 127088. [CrossRef]
- Kelterborn, S.; Boehning, F.; Sizova, I.; Baidukova, O.; Evers, H.; Hegemann, P. Gene Editing in Green Alga Chlamydomonas Reinhardtii via CRISPR-Cas9 Ribonucleoproteins. Plant Synth. Biol. Methods Mol. Biol. 2022, 2379, 45–65. [CrossRef]
- Tejada-Jiménez, M.; Llamas, A.; Sanz-Luque, E.; Galván, A.; Fernández, E. A High-Affinity Molybdate Transporter in Eukaryotes. Proc. Natl. Acad. Sci. USA 2007, 104, 20126–20130. [CrossRef]
- Tejada-Jiménez, M.; Galván, A.; Fernández, E. Algae and Humans Share a Molybdate Transporter. Proc. Natl. Acad. Sci. USA 2011, 108, 6420–6425. [CrossRef]
- Fischer, K.; Llamas, A.; Tejada-Jiménez, M.; Schrader, N.; Kuper, J.; Ataya, F.S.; Galván, A.; Mendel, R.R.; Fernández, E.; Schwarz, G. Function and Structure of the Molybdenum Cofactor Carrier Protein from Chlamydomonas Reinhardtii. J. Biol. Chem. 2006, 281, 30186–30194. [CrossRef]
- Ataya, F.S.; Witte, C.P.; Galván, A.; Igeño, M.I.; Fernández, E. Mcp1 Encodes the Molybdenum Cofactor Carrier Protein in Chlamydomonas Reinhardtii and Participates in Protection, Binding, and Storage Functions of the Cofactor. J. Biol. Chem. 2003, 278, 10885–10890. [CrossRef]
- Llamas, A.; Tejada-Jimenez, M.; González-Ballester, D.; Higuera, J.J.; Schwarz, G.; Galván, A.; Fernández, E. Chlamydomonas Reinhardtii CNX1E Reconstitutes Molybdenum Cofactor Biosynthesis in Escherichia Coli Mutants. Eukaryot. Cell 2007, 6, 1063–1067. [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galván, A.; Fernandez, E. Nitrate Reductase Regulates Plant Nitric Oxide Homeostasis. Trends Plant Sci. 2017, 22, 163–174. [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A Dual System Formed by the ARC and NR Molybdoenzymes Mediates Nitrite-Dependent NO Production in Chlamydomonas. Plant. Cell Environ. 2016, 39, 2097–2107. [CrossRef]
- Emsley, J. Nature’s Building Blocks. Oxford Univ. Press 2001.
- Lešková, A.; Javot, H.; Giehl, R.F.H. Metal Crossroads in Plants: Modulation of Nutrient Acquisition and Root Development by Essential Trace Metals. J. Exp. Bot. 2022, 73, 1751–1765. [CrossRef]
- Llamas, A.; Kalakoutskii, K.L.; Fernández, E. Molybdenum Cofactor Amounts in Chlamydomonas Reinhardtii Depend on the Nit5 Gene Function Related to Molybdate Transport. Plant, Cell Environ. 2000, 23, 1247–1255. [CrossRef]
- Mendel, R.R.; Haensh, R. Molybdoenzymes and Molybdenum Cofactor in Plants. J. Exp. Bot. 2002, 53, 1689–1698. [CrossRef]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. Characterization of Chlamydomonas 102 and 104 Mutants Reveals Intermolecular Complementation in the Molybdenum Cofactor Protein CNX1E. Protist 2013, 164, 116–128. [CrossRef]
- Schwarz, G.; Schulze, J.; Bittner, F.; Eilers, T.; Kuper, J.; Bollmann, G.; Nerlich, A.; Brinkmann, H.; Mendel, R.R. The Molybdenum Cofactor Biosynthetic Protein Cnx1 Complements Molybdate-Repairable Mutants, Transfers Molybdenum to the Metal Binding Pterin, and Is Associated with the Cytoskeleton. Plant Cell 2000, 12, 2455–2471. [CrossRef]
- Tomatsu, H.; Takano, J.; Takahashi, H.; Watanabe-Takahashi, A.; Shibagaki, N.; Fujiwara, T. An Arabidopsis Thaliana High-Affinity Molybdate Transporter Required for Efficient Uptake of Molybdate from Soil. Proc. Natl. Acad. Sci. 2007, 104, 18807–18812. [CrossRef]
- Ide, Y.; Kusano, M.; Oikawa, A.; Fukushima, A.; Tomatsu, H.; Saito, K.; Hirai, M.Y.; Fujiwara, T. Effects of Molybdenum Deficiency and Defects in Molybdate Transporter MOT1 on Transcript Accumulation and Nitrogen/Sulphur Metabolism in Arabidopsis Thaliana. J. Exp. Bot. 2011, 62, 1483–1497. [CrossRef]
- Gao, J.-S.; Wu, F.-F.; Shen, Z.-L.; Meng, Y.; Cai, Y.-P.; Lin, Y. A Putative Molybdate Transporter LjMOT1 Is Required for Molybdenum Transport in Lotus Japonicus. Physiol. Plant. 2016, 158, 331–340. [CrossRef]
- Tejada-Jiménez, M.; Gil-Díez, P.; León-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; González-Guerrero, M. Medicago Truncatula Molybdate Transporter Type 1 (MtMOT1.3) Is a Plasma Membrane Molybdenum Transporter Required for Nitrogenase Activity in Root Nodules under Molybdenum Deficiency. New Phytol. 2017, 216, 1223–1235. [CrossRef]
- Gil-Díez, P.; Tejada-Jiménez, M.; León-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; González-Guerrero, M. MtMOT1.2 Is Responsible for Molybdate Supply to Medicago Truncatula Nodules. Plant. Cell Environ. 2019, 42, 310–320. [CrossRef]
- Forsberg, S.K.G.; Andreatta, M.E.; Huang, X.Y.; Danku, J.; Salt, D.E.; Carlborg, Ö. The Multi-Allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance. PLoS Genet. 2015, 11, 1005648. [CrossRef]
- Li, W.; Fingrut, D.R.; Maxwell, D.P. Characterization of a Mutant of Chlamydomonas Reinhardtii Deficient in the Molybdenum Cofactor. Physiol. Plant. 2009, 136, 336–350. [CrossRef]
- Tejada-Jiménez, M.; Chamizo-Ampudia, A.; Calatrava, V.; Galván, A.; Fernández, E.; Llamas, A. From the Eukaryotic Molybdenum Cofactor Biosynthesis to the Moonlighting Enzyme MARC. Molecules 2018, 23, 3287. [CrossRef]
- Schwarz, G.; Mendel, R.R. Molybdenum Cofactor Biosynthesis and Molybdenum Enzymes. Annu. Rev. Plant Biol. 2006, 57, 623–647. [CrossRef]
- Kruse, I.; Maclean, A.E.; Hill, L.; Balk, J. Genetic Dissection of Cyclic Pyranopterin Monophosphate Biosynthesis in Plant Mitochondria. Biochem J 2018, 475, 495–509. [CrossRef]
- González-Ballester, D.; de Montaigu, A.; Higuera, J.J.; Galván, A.; Fernández, E. Functional Genomics of the Regulation of the Nitrate Assimilation Pathway in Chlamydomonas. Plant Physiol. 2005, 137, 522–533. [CrossRef]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. The Chlamydomonas Reinhardtii Molybdenum Cofactor Enzyme CrARC Has a Zn-Dependent Activity and Protein Partners Similar to Those of Its Human Homologue. Eukaryot. Cell 2011, 10, 1270–1282. [CrossRef]
- Teschner, J.; Lachmann, N.; Schulze, J.; Geisler, M.; Selbach, K.; Santamaria-Araujo, J.; Balk, J.; Mendel, R.R.; Bittner, F. A Novel Role for Arabidopsis Mitochondrial ABC Transporter ATM3 in Molybdenum Cofactor Biosynthesis. Plant Cell 2010, 22, 468–480. [CrossRef]
- Kaufholdt, D.; Baillie, C.K.; Meinen, R.; Mendel, R.R.; Hänsch, R. The Molybdenum Cofactor Biosynthesis Network: In Vivo Protein-Protein Interactions of an Actin Associated Multi-Protein Complex. Front. Plant Sci. 2017, 8, 1946. [CrossRef]
- Nakai, Y.; Harada, A.; Hashiguchi, Y.; Nakai, M.; Hayashi, H. Arabidopsis Molybdopterin Biosynthesis Protein Cnx5 Collaborates with the Ubiquitin-like Protein Urm11 in the Thio-Modification of TRNA. J Biol Chem 2012, 287, 30874–30884. [CrossRef]
- Veldman, A.; Santamaria-Araujo, J.A.; Sollazzo, S.; Pitt, J.; Gianello, R.; Yaplito-Lee, J.; Wong, F.; Ramsden, C.A.; Reiss, J.; Cook, I.; et al. Successful Treatment of Molybdenum Cofactor Deficiency Type A with CPMP. Pediatrics 2010, 125, e1249-54. [CrossRef]
- Farrell, S.; Karp, J.; Hager, R.; Wang, Y.; Adeniyi, O.; Wang, J.; Li, L.; Ma, L.; Peretz, J.; Summan, M.; et al. Regulatory News: Nulibry (Fosdenopterin) Approved to Reduce the Risk of Mortality in Patients with Molybdenum Cofactor Deficiency Type A: FDA Approval Summary. J. Inherit. Metab. Dis. 2021, 44, 1085–1087. [CrossRef]
- Kuper, J.; Llamas, A.; Hecht, H.-J.; Mendel, R.R.; Schwarz, G. Structure of the Molybdopterin-Bound Cnx1G Domain Links Molybdenum and Copper Metabolism. Nature 2004, 430, 803–806. [CrossRef]
- Kruse, T. Function of Molybdenum Insertases. Molecules 2022, 27, 5372. [CrossRef]
- Llamas, A.; Mendel, R.R.; Schwarz, G. Synthesis of Adenylated Molybdopterin: An Essential Step for Molybdenum Insertion. J. Biol. Chem. 2004, 279, 55241–55246. [CrossRef]
- Llamas, A.; Otte, T.; Multhaup, G.; Mendel, R.R.; Schwarz, G. The Mechanism of Nucleotide-Assisted Molybdenum Insertion into Molybdopterin. J. Biol. Chem. 2006, 281, 18343–18350. [CrossRef]
- Nichols, J.D.; Rajagopalan, K. V. In Vitro Molybdenum Ligation to Molybdopterin Using Purified Components. J. Biol. Chem. 2005, 280, 7817–7822. [CrossRef]
- Selles, B.; Moseler, A.; Caubrière, D.; Sun, S.K.; Ziesel, M.; Dhalleine, T.; Hériché, M.; Wirtz, M.; Rouhier, N.; Couturier, J. The Cytosolic Arabidopsis Thaliana Cysteine Desulfurase ABA3 Delivers Sulfur to the Sulfurtransferase STR18. J. Biol. Chem. 2022, 298, 101749. [CrossRef]
- Mendel, R.R. The History of the Molybdenum Cofactor-A Personal View. Molecules 2022, 27, 4934. [CrossRef]
- Aguilar, M.; Cárdenas, J.; Fernández, E. Quantitation of Molybdopterin Oxidation Product in Wild-Type and Molybdenum Cofactor Deficient Mutants of Chlamydomonas Reinhardtii. Biochim. Biophys. Acta 1992, 1160, 269–274. [CrossRef]
- Witte, C.-P.; Igeño, M.I.; Mendel, R.; Schwarz, G.; Fernández, E. The Chlamydomonas Reinhardtii MoCo Carrier Protein Is Multimeric and Stabilizes Molybdopterin Cofactor in a Molybdate Charged Form. FEBS Lett. 1998, 431, 205–209. [CrossRef]
- Aguilar, M.; Kalakoutskii, K.; Cárdenas, J.; Fernández, E. Direct Transfer of Molybdopterin Cofactor to Aponitrate Reductase from a Carrier Protein in Chlamydomonas Reinhardtii. FEBS Lett 1992, 307, 162–163. [CrossRef]
- Krausze, J.; Hercher, T.W.; Archna, A.; Kruse, T. The Structure of the Moco Carrier Protein from Rippkaea Orientalis. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2020, 76, 453–463. [CrossRef]
- Hercher, T.W.; Krausze, J.; Yang, J.; Kirk, M.L.; Kruse, T. Identification and Characterization of the Volvox Carteri Moco Carrier Protein. Biosci. Rep. 2020, 40, BSR20202351. [CrossRef]
- Kruse, T.; Gehl, C.; Geisler, M.; Lehrke, M.; Ringel, P.; Hallier, S.; Hänsch, R.; Mendel, R.R. Identification and Biochemical Characterization of Molybdenum Cofactor-Binding Proteins from Arabidopsis Thaliana. J. Biol. Chem. 2010, 285, 6623–6635. [CrossRef]
- Kruse, T. Moco Carrier and Binding Proteins. Molecules 2022, 27, 6571. [CrossRef]
- Lawson, D.M.; Williams, C.E.; White, D.J.; Choay, A.P.; Mitchenall, L.A.; Pau, R.N. Protein Ligands for Molybdate. Specificity and Charge Stabilisation at the Anion-Binding Sites of Periplasmic and Intracellular Molybdate-Binding Proteins of Azotobacter Vinelandii. J. Chem. Soc. Dalt. Trans. 1997, 3981–3984. [CrossRef]
- Fenske, D.; Gnida, M.; Schneider, K.; Meyer-Klaucke, W.; Schemberg, J.; Henschel, V.; Meyer, A.-K.; Knöchel, A.; Müller, A. A New Type of Metalloprotein: The Mo Storage Protein from Azotobacter Vinelandii Contains a Polynuclear Molybdenum-Oxide Cluster. ChemBioChem 2005, 6, 405–413. [CrossRef]
- Steinke, D.R.; Majak, W.; Sorensen, T.S.; Parvez, M. Chelation of Molybdenum in Medicago Sativa (Alfalfa) Grown on Reclaimed Mine Tailings. J. Agric. Food Chem. 2008, 56, 5437–5442. [CrossRef]
- Yesbergenova, Z.; Yang, G.; Oron, E.; Soffer, D.; Fluhr, R.; Sagi, M. The Plant Mo-Hydroxylases Aldehyde Oxidase and Xanthine Dehydrogenase Have Distinct Reactive Oxygen Species Signatures and Are Induced by Drought and Abscisic Acid. Plant J 2005, 42, 862–876. [CrossRef]
- Pineda, M.; Cardenas, J. Transport and Assimilation of Purines in Chlamydomonas Reinhardtii. Sci. Mar. 1996, 60, 195–201.
- Schmollinger, S.; Mühlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-Sparing Mechanisms in Chlamydomonas Affect the Transcriptome, the Proteome, and Photosynthetic Metabolism. Plant Cell 2014, 26, 1410–1435. [CrossRef]
- Perez-Vicente, R.; Pineda, M.; Cardenas, J. Isolation and Characterization of Xanthine Dehydrogenase from Chlamydomonas Reinhardtii. Physiol. Plant. 1988, 72, 101–107. [CrossRef]
- Seo, M.; Koiwai, H.; Akaba, S.; Komano, T.; Oritani, T.; Kamiya, Y.; Koshiba, T. Abscisic Aldehyde Oxidase in Leaves of Arabidopsis Thaliana. Plant J 2000, 23, 481–488. [CrossRef]
- Rodríguez-Trelles, F.; Tarrío, R.; Ayala, F.J. Convergent Neofunctionalization by Positive Darwinian Selection after Ancient Recurrent Duplications of the Xanthine Dehydrogenase Gene. Proc Natl Acad Sci USA 2003, 100, 13413–13417. [CrossRef]
- Abu-Ghosh, S.; Iluz, D.; Dubinsky, Z.; Miller, G. Exogenous Abscisic Acid Confers Salinity Tolerance in Chlamydomonas Reinhardtii During Its Life Cycle. J. Phycol 2021, 57, 1323–1334. [CrossRef]
- Al-Hijab, L.; Gregg, A.; Davies, R.; Macdonald, H.; Ladomery, M.; Wilson, I. Abscisic Acid Induced a Negative Geotropic Response in Dark-Incubated Chlamydomonas Reinhardtii. Sci. Rep. 2019, 9, 12063. [CrossRef]
- Feng, C.; Tollin, G.; Enemark, J.H. Sulfite Oxidizing Enzymes. Biochim. Biophys. Acta - Proteins Proteomics 2007, 1774, 527–539. [CrossRef]
- Randewig, D.; Hamisch, D.; Herschbach, C.; Eiblmeier, M.; Gehl, C.; Jurgeleit, J.; Skerra, J.; Mendel, R.R.; Rennenberg, H.; Hänsch, R. Sulfite Oxidase Controls Sulfur Metabolism under SO2 Exposure in Arabidopsis Thaliana. Plant. Cell Environ. 2012, 35, 100–115. [CrossRef]
- Mellis, A.T.; Roeper, J.; Misko, A.L.; Kohl, J.; Schwarz, G. Sulfite Alters the Mitochondrial Network in Molybdenum Cofactor Deficiency. Front. Genet. 2021, 11, 594828. [CrossRef]
- Nowak, K.; Luniak, N.; Witt, C.; Wüstefeld, Y.; Wachter, A.; Mendel, R.R.; Hänsch, R. Peroxisomal Localization of Sulfite Oxidase Separates It from Chloroplast-Based Sulfur Assimilation. Plant Cell Physiol 2004, 45, 1889–1894. [CrossRef]
- Eilers, T.; Schwarz, G.; Brinkmann, H.; Witt, C.; Richter, T.; Nieder, J.; Koch, B.; Hille, R.; Hänsch, R.; Mendel, R.R. Identification and Biochemical Characterization of Arabidopsis Thaliana Sulfite Oxidase: A New Player in Plant Sulfur Metabolism. J. Biol. Chem. 2001, 276, 46989–46994. [CrossRef]
- Gerin, S.; Mathy, G.; Blomme, A.; Franck, F.; Sluse, F.E. Plasticity of the Mitoproteome to Nitrogen Sources (Nitrate and Ammonium) in Chlamydomonas Reinhardtii: The Logic of Aox1 Gene Localization. Biochim. Biophys. Acta - Bioenerg. 2010, 1797, 994–1003. [CrossRef]
- Fernandez, E.; Schnell, R.; Ranum, L.P.; Hussey, S.C.; Silflow, C.D.; Lefebvre, P.A. Isolation and Characterization of the Nitrate Reductase Structural Gene of Chlamydomonas Reinhardtii. Proc. Natl. Acad. Sci. 1989, 86, 6449–6453. [CrossRef]
- Vidal, E.A.; Alvarez, J.M.; Araus, V.; Riveras, E.; Brooks, M.D.; Krouk, G.; Ruffel, S.; Lejay, L.; Crawford, N.M.; Coruzzi, G.M.; et al. Nitrate in 2020: Thirty Years from Transport to Signaling Networks. Plant Cell 2020, 32, 2094–2119. [CrossRef]
- Dean, J. V; Harper, J.E. The Conversion of Nitrite to Nitrogen Oxide(s) by the Constitutive NAD(P)H-Nitrate Reductase Enzyme from Soybean. Plant Physiol 1988, 88, 389–395. [CrossRef]
- Khan, M.; Ali, S.; Al Azzawi, T.N.I.; Yun, B.W. Nitric Oxide Acts as a Key Signaling Molecule in Plant Development under Stressful Conditions. Int. J. Mol. Sci. 2023, 24, 4782. [CrossRef]
- Gupta, K.J.; Kaladhar, V.C.; Fitzpatrick, T.B.; Fernie, A.R.; Møller, I.M.; Loake, G.J. Nitric Oxide Regulation of Plant Metabolism. Mol. Plant 2022, 15, 228–242. [CrossRef]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Caló, G.; Salerno, G.; Lamattina, L. Characterization of a Nitric Oxide Synthase from the Plant Kingdom: NO Generation from the Green Alga Ostreococcus Tauri Is Light Irradiance and Growth Phase Dependent. Plant Cell 2010, 22, 3816–3830. [CrossRef]
- Santolini, J.; André, F.; Jeandroz, S.; Wendehenne, D. Nitric Oxide Synthase in Plants: Where Do We Stand? Nitric Oxide 2017, 63, 30–38. [CrossRef]
- Havemeyer, A.; Bittner, F.; Wollers, S.; Mendel, R.; Kunze, T.; Clement, B. Identification of the Missing Component in the Mitochondrial Benzamidoxime Prodrug-Converting System as a Novel Molybdenum Enzyme. J Biol Chem 2006, 281, 34796–34802. [CrossRef]
- Hsieh, S.I.; Castruita, M.; Malasarn, D.; Urzica, E.; Erde, J.; Page, M.D.; Yamasaki, H.; Casero, D.; Pellegrini, M.; Merchant, S.S.; et al. The Proteome of Copper, Iron, Zinc, and Manganese Micronutrient Deficiency in Chlamydomonas Reinhardtii. Mol Cell Proteomics 2013, 12, 65–86. [CrossRef]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. Study of Different Variants of Mo Enzyme CrARC and the Interaction with Its Partners CrCytb5-R and CrCytb5-1. Int. J. Mol. Sci. 2017, 18, 670. [CrossRef]
- Ott, G.; Plitzko, B.; Krischkowski, C.; Reichmann, D.; Bittner, F.; Mendel, R.R.; Kunze, T.; Clement, B.; Havemeyer, A. Reduction of Sulfamethoxazole Hydroxylamine (SMX-HA) by the Mitochondrial Amidoxime Reducing Component (MARC). Chem Res Toxicol 2014, 27, 1687–1695. [CrossRef]
- Kotthaus, J.; Wahl, B.; Havemeyer, A.; Schade, D.; Garbe-Schönberg, D.; Mendel, R.; Bittner, F.; Clement, B. Reduction of N(ω)-Hydroxy-L-Arginine by the Mitochondrial Amidoxime Reducing Component (MARC). Biochem J 2011, 433, 383–391. [CrossRef]
- Sparacino-Watkins, C.E.; Tejero, J.; Sun, B.; Gauthier, M.C.; Thomas, J.; Ragireddy, V.; Merchant, B.A.; Wang, J.; Azarov, I.; Basu, P.; et al. Nitrite Reductase and Nitric-Oxide Synthase Activity of the Mitochondrial Molybdopterin Enzymes MARC1 and MARC2. J. Biol. Chem. 2014, 289, 10345–10358. [CrossRef]
- Bender, D.; Schwarz, G. Nitrite-Dependent Nitric Oxide Synthesis by Molybdenum Enzymes. FEBS Lett 2018, 592, 2126–2139. [CrossRef]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of Nitric Oxide (NO) Production by Plant Nitrate Reductase in Vivo and in Vitro. J. Exp. Bot. 2002, 53, 103–110. [CrossRef]
- Maiber, L.; Koprivova, A.; Bender, D.; Kopriva, S.; Fischer-Schrader, K. Characterization of the Amidoxime Reducing Components ARC1 and ARC2 from Arabidopsis Thaliana. FEBS J. 2022, 289, 5656–5669. [CrossRef]
- Bellido-Pedraza, C.M.; Calatrava, V.; Llamas, A.; Fernandez, E.; Sanz-Luque, E.; Galvan, A. Nitrous Oxide Emissions from Nitrite Are Highly Dependent on Nitrate Reductase in the Microalga Chlamydomonas Reinhardtii. Int. J. Mol. Sci. 2022, 23, 9412. [CrossRef]
- Llamas, A.; Chamizo-Ampudia, A.; Tejada-Jimenez, M.; Galvan, A.; Fernandez, E. The Molybdenum Cofactor Enzyme MARC: Moonlighting or Promiscuous Enzyme? BioFactors 2017, 43, 486–494. [CrossRef]
- Gupta, M.N.; Uversky, V.N. Moonlighting Enzymes: When Cellular Context Defines Specificity. Cell. Mol. Life Sci. 2023, 80, 130. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




