1. Introduction
The COVID-19 pandemic, caused by SARS-CoV-2 infection, caused around 660 million cases and 6.7 million deaths worldwide until the end of 2022 [
1]. The different isolates of SARS-CoV-2 have been classified into lineages, and, among these lineages, 5 Variants of Concern (VOCs) have been recognized by the WHO [
2,
3,
4].
The last known VOC until the end of 2022 was Omicron. This VOC caused immediate concern for two reasons: its emergence in November 2021 was associated with an explosive increase in cases in South Africa, and it harbors a large number of mutations, compared to the previous ones [
5]. This VOC displaced all the other lineages circulating in the world, mainly Delta VOC. At the end of June 2022, the WHO designed the Omicron VOC as the only variant circulating in the World [
6,
7].
All international travelers arriving at Venezuelan airports were subjected to molecular testing for SARS-CoV-2 until December 2022; this testing allowed us to describe previously the introduction of the Omicron VOC in the country [
8]. This study aims to describe the dissemination of the Omicron VOC lineages in Venezuela, by monitoring the diversity of viral variants in community cases and international travelers returning to Venezuela.
2. Materials and Methods
This study was approved by the Human Bioethical Committee of IVIC. Of the five international commercial airports available in Venezuela in 2022, Maiquetia (close to Caracas, the capital of the country) is the largest and was chosen for this study. Samples from this airport were evaluated by LAMP (Evotech-Mirai Genomics, Innopolis City, Verkhneuslonsky Region, Republic of Tatarstan Russia). For sequence analysis, positive samples from January to December 2022 were re-tested by qRT-PCR (Sansure, Changsha, Hunan Province, P. R. China). In addition, samples positive by qRT-PCR during the routine COVID-19 diagnosis in Venezuela, from the same period, were analyzed. The identity of the patients was maintained anonymous.
RNA from samples positive by qRT-PCR was amplified with primers 75L and 76.8R to generate an amplicon of 614 nt, with the PCR conditions previously described [
8,
9]. PCR-purified fragments were sent to the Macrogen Sequencing Service (Macrogen, Korea). This fragment allows us to analyze amino acids 345 to 533 of the Spike gene, which includes several mutations of the Omicron VOC, and other variants, allowing the differentiation of the major sub-lineages BA.1 to BA.5. The 5´non-coding region of the SARS-CoV-2 genome was also analyzed for differentiating the sub-lineages BA.4 and BA.5, with an amplicon of ≈ 640 nt generated with primers 1L and 3R [
10].
Complete genome sequencing was performed on selected samples, by Next Generation sequencing. Libraries were prepared with a DNA Prep library preparation kit using the Nextera DNA CD Indexes (Illumina, Inc. USA) as previously reported [
9], or Illumina COVIDSeq Assay (96 Samples) (RUO Version, Document # 1000000126053 v05), using IDT for Illumina-PCR Indexes set 1 or 3. The libraries were pooled, quantified (Qubit DNA HS, Thermo Scientific), and their quality checked (Bio-Fragment Analyzer, Qsep1-Lite, BiOptic) before sequencing, which was performed with 10% PhiX control v3, using an iSeq 100 or MiSeq platform and a 300 cycle V2 kit with paired-end sequencing. Viral genome assembly was performed using the Dragen COVID-19 program or Genome Detective Virus tool. The variant assignment was performed using the Dragen COVID-19 program, Nextclade Web 1.14.1, or Pangolin COVID-19 Lineage Assigner. Nucleotide sequences of complete genomes have been deposited into the GISAID database.
Statistical differences were evaluated by Chi-Square tests: p values less than 0.05 were considered significant.
3. Results
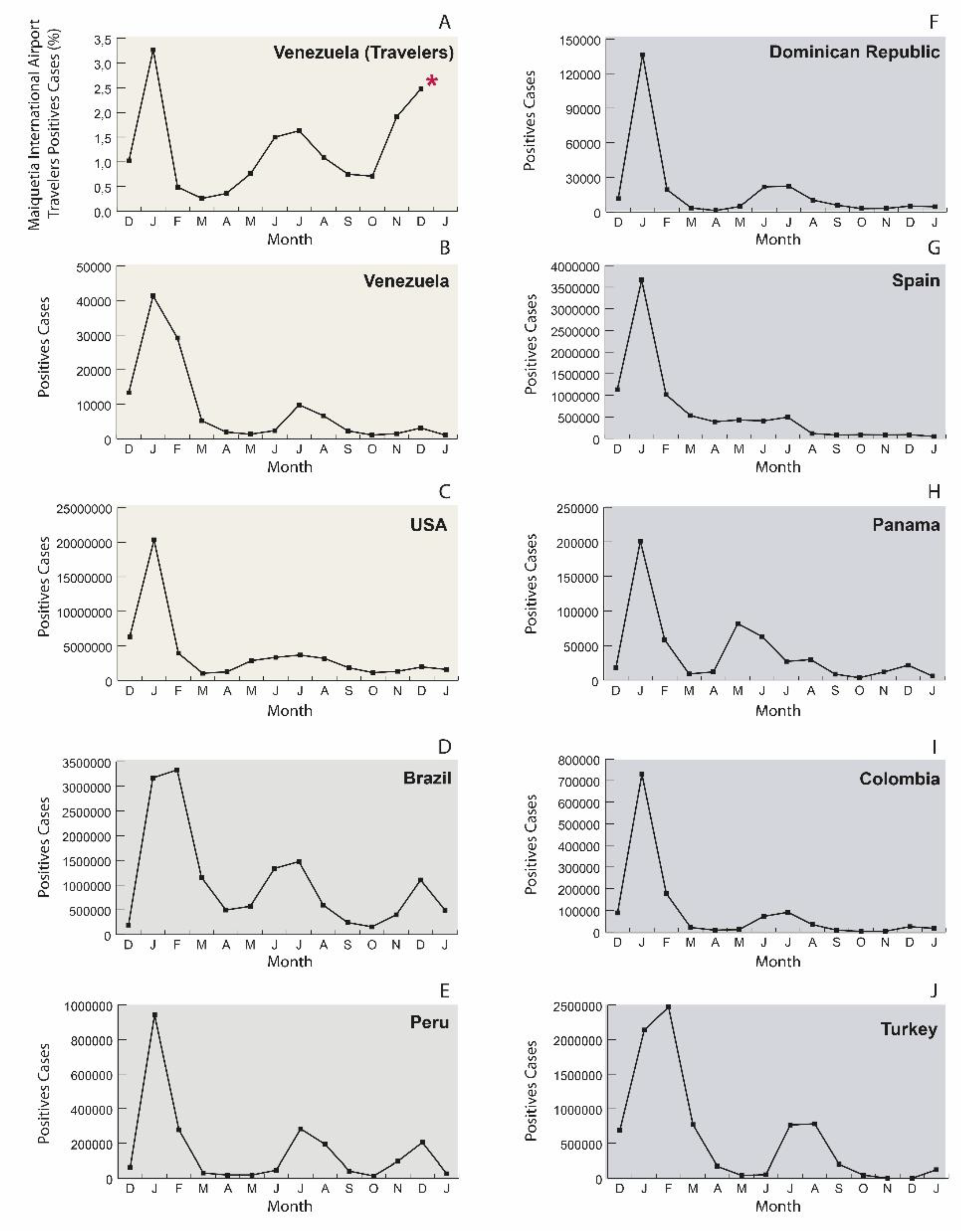
Figure 1 shows the evolution in the number of positive SARS-CoV-2 cases in the World and selected countries, and the percent positivity observed in travelers from Maiquetia airport from December 2021 to December 2022. Before December 2021, no peak of positive cases was observed among international travelers (IT), with a positivity rate for international travelers (PRIT) below or near 1% [
8]. In 2022, a different picture was observed in the frequency of PRIT (
Figure 1A). The first Omicron wave peaked around 7% at the beginning of January and then returned to the PRIT observed in 2021, below 1%, in March and April. An extended peak of PRIT was observed in June-August, and then PRIT peaked up to 3.6% at the end of 2022. The peaks of PRIT were not correlated with the ones observed in Venezuela (
Figure 1A vs. 1B): indeed, these peaks somehow correlate with the epidemic curve of the USA, and Brazil (
Figure 1 CD).
The frequency of samples with high viral content was also analyzed during the study period. No significant difference in the number of samples with Ct inferior to 25 (high viral load) by qRT-PCR, was observed during the whole year 2022 (data not shown).
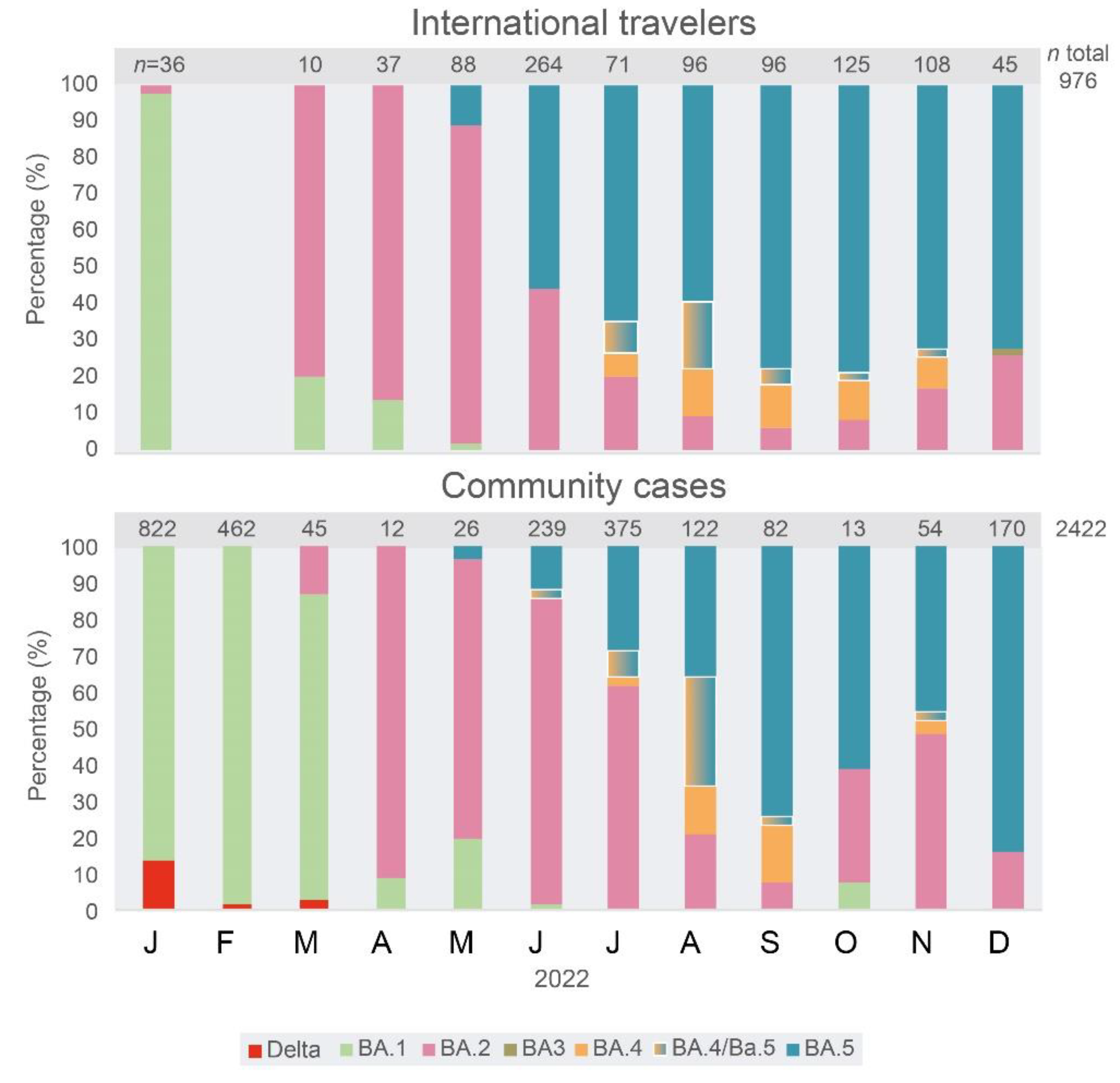
The frequency of Omicron VOC major sub-lineages was estimated by partial sequencing in samples from international travelers and community cases.
Table 1 shows the specific mutations analyzed to identify the different Omicron VOC sub-lineages.
Complete genome sequencing confirmed the assignment of the major sub-lineages by the rapid method in most cases. The concordance between partial and complete genome sequencing methods was 97.5% for the assignment of the major sub-lineages BA.1 to BA.5 (
Table 2). The most frequent discrepancy occurred in the discrimination between BA.4 and BA.5.
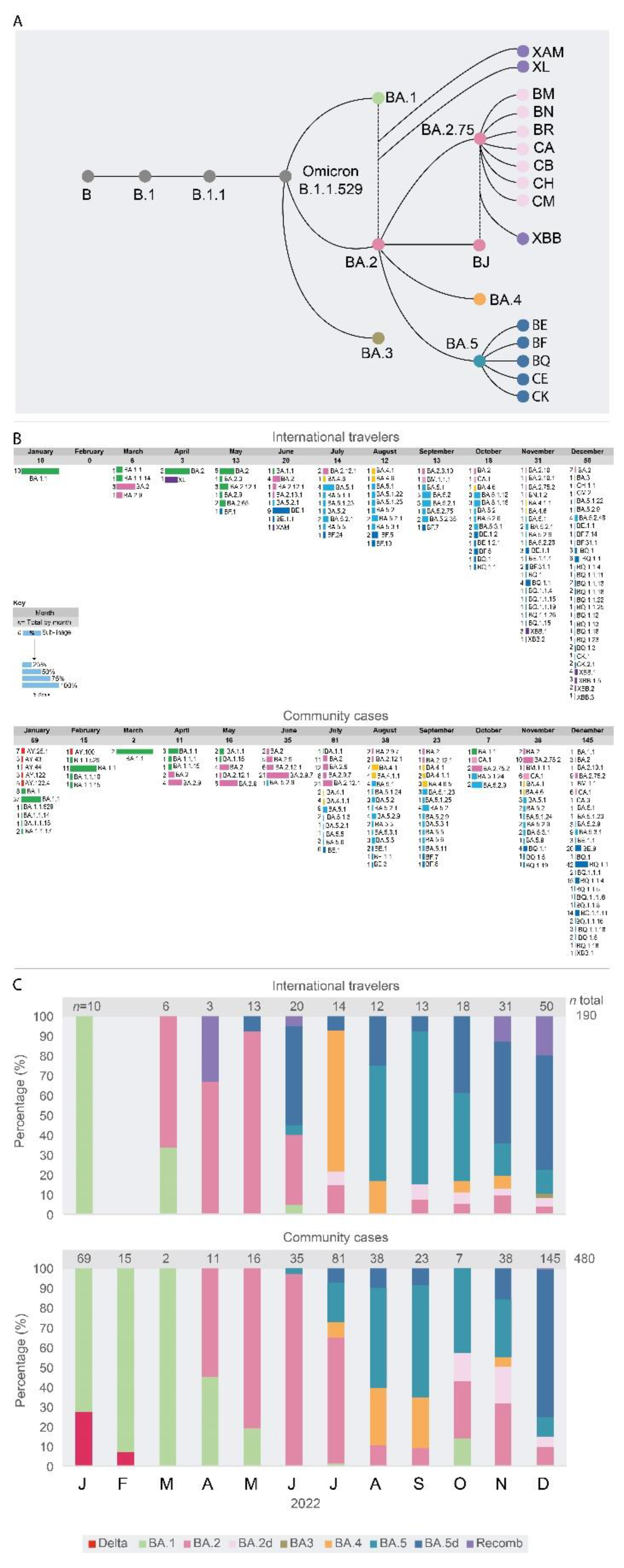
A total of 670 complete genome sequences were analyzed, 190 from IT and 480 from CC (
Figure 3). Since July 2022, a great diversity of sub-lineages was observed by complete genome sequencing of viral samples. In December, a large number of complete genome sequences could be obtained. Up to 29 different sub-lineages were found in 50 IT samples, a diversity significantly higher (p<0.001) than the one observed in CC (26 different sub-lineages in 145 samples,
Figure 3A), a situation expected, because of community transmission in CC. The wider diversity of sub-lineages and recombinant forms of the Omicron VOC was always first detected in samples from IT and then in CC Venezuela. However, the distribution of specific sub-lineages was different in CC, compared with that observed in IT. In CC, for example, the sub-lineage BA.2.75.2 was frequent in CC since October 2022, and BQ.1.1 in November and December (
Figure 3A), while in IT, this former lineage was never frequent (
Figure 3A). In the case of BQ.1 and sub-lineages, in December, the frequency of these sub-lineages was similar in CC (59%) and IT (46%, p>0.05). The BA.1.1 sub-lineage was still circulating in CC in December 2022, but it was not found after June in IT. Only one BA.3 sub-lineage isolate was identified, in IT, relatively late, in December 2022 (
Figure 3B). The sub-lineages of BA.5 were the most frequent lineages in IT since June 2022, while they prevailed in CC two months later, in August (
Figure 3B).
4. Discussion
Venezuela performed molecular testing of SARS-CoV-2 in international passengers arriving at the commercial airports of the country very soon after the declaration of the pandemic of COVID-19. An explosive increase of PRTI was observed since the second half of December 2021 in most of these airports, associated with the circulation of the Omicron VOC worldwide [
8].
The PRIT curve was correlated with the number of cases from the USA, and Brazil. This relation is in agreement with one of the most common destinations of Venezuelan international travel by airplane, i.e. Florida, USA [
12]. The PRIT and the RNA levels in the positive samples may be influenced by the changing regulations related to vaccination. On July 1, 2021, for example, the EU Digital COVID Certificate Regulation entered into the application [
13]. The absence of COVID-19 testing requirements in vaccinated individuals may have influenced the increase in PRIT in 2022. We were also expecting that the frequency of samples with high viral RNA levels could also increase with the reduction of the testing requirements. Contrary to our expectations, the frequency of samples with high viral RNA levels remained similarly high during the entire evaluation period.
The strategy adopted in this study (sequencing of a small genomic fragment of the RBD of SARS-CoV2, complemented with partial sequencing of the ORF1 region to differentiate BA.4 and BA.5), allowed us to analyze many samples with a good correlation (more than 97% in more than 350 complete genomes) for major sub-lineage assignment. However, since July 2022, a great diversity of sub-lineages and recombinant forms was detected, reducing the utility of the rapid sequencing method for major sub-lineage monitoring. This worldwide phenomenon has been called a swarm of variants or variant soup [
14], defined by the co-circulation of multiple sub-lineages of the Omicron VOC, with the appearance of common mutations, particularly associated with immune evasion, through convergent evolution [
15].
The results show the influence of air travelers on the introduction of the different sub-lineages of Omicron to the country. This situation contrasts with that observed for Gamma and Mu, for which, although the introduction by air route is not ruled out, the land route seems to have played a greater role, since the first states with the highest frequency of these variants were the border states, Eastern border for Gamma VOC and Western one for Mu VOC, according to the countries of origin of these variants, Brazil and Colombia respectively [
9,
16]. However, the introduction by land of the Omicron sub-lineages, although not evaluated in this study, certainly occurred and possibly influenced the distribution of the viral variants in the country. A limitation of this study is the number of positive samples that could be tested in 2022, compared to 2021. This is due to the fact that after the unprecedented spike in cases due to the first wave of the Omicron variant in December 2021 to January 2022, a significant reduction of cases occurred in the country. Afterward, despite a probable consistent number of cases restarting in July 2022, fewer people sought a diagnosis. This might be due in part to the combination in the eventual reduction in morbidity associated with the Omicron variant, compared to the Delta variant for example, and with the important herd immunity that was probably prevalent throughout the country [
7,
17,
18,
19,
20]. This implied that the waves of the major sub-lineages could not be evaluated by geographic regions in the country, so the study was limited to evaluating the country as a single region.
Even if the number of complete genomes is relatively reduced, we could appreciate a high diversity of sub-lineages circulating in Venezuela, with one or two major clades each month in CC. Two sub-lineages were found frequently since the second half of the year 2022: BA.2.75.2 and BQ.1.1 and descendants of BQ.1, this last in turn derived from the BA.5 major sub-lineage [
15,
21]. BA.2.75.2 and BQ.1.1 harbor several mutations associated with reduced neutralization both by sera from immunized individuals, and by several monoclonal antibodies [
22]. They were reported among the ones more resistant to the bivalent BNT162b2 vaccine-induced neutralizing antibodies [
23]. The circulation of these sub-lineages is in consonance with a presumed high level of herd hybrid immunity in the Venezuelan population in the second semester of the year 2022, after two waves of the Omicron variants in the country (
Figure 1).
The analysis of the frequency of sub-lineages in the main neighboring countries is not similar to the frequency of sub-lineages found in Venezuela. In Colombia, the sub-lineage BA.2.75.2 did not surpass 8% prevalence during 2022, although the sub-lineage BQ.1 did predominate (up to 85% in the middle of December 2022) [
24]. A pattern similar to the one observed in Colombia was observed in Brazil, where the frequency of BA.2.75.2 did not surpass 2% and up to 78% BQ.1.1 [
24].
Another important sub-lineage, XBB.1.5, emerged in the USA at the end of 2022 and predominated in February-March 2023. The XBB.1.5 sub-lineage is a recombinant form of two BA.2 sub-lineages. It exhibits high immune evasion as other previous sub-lineages but also was considered in March 2023 as the most transmissible of all the sub-lineages known to this date, surpassing the BQ.1.1 [
25]. It began to be found in South America in January 2023 and predominated in the region in March 2023. This sub-lineage was not found in community cases in Venezuela until February 2023 [
26], although found in a few international travelers since December 2022. This sub-lineage largely predominated in May 2023 in the World [
24]. However, molecular and phylodynamic studies do not predict a particularly high risk of XBB.1.5 expansion to become a new global public health threat, although the acquisition of additional mutations cannot be ruled out [
27].
Then, once the ¨soup of sub-lineages¨ was introduced in the country, community transmission was responsible for generating a characteristic distribution of them, with greater frequency of certain sub-lineages, not necessarily similar to those that predominated in travelers or neighboring countries.
5. Conclusions
International travelers returning to Venezuela were probably a significant source of the introduction of the Omicron sub-lineages in the country. A great diversity of sub-lineages was found both in IT and CC, particularly after July 2022. Once the different lineages were introduced in the country, some of them disseminate preferentially in the country at the end of 2022, such as BA.2.75.2 and BQ.1.1.
Author Contributions
Conceptualization, supervision, and project administration, F.H.P. and R.C.J.; investigation, R.C.J., F.H.P., Z.C.M., C.L.L. and Y.S.; resources, H.R.R., E.M., F.B., P.D.A. and L.R., writing—original draft preparation, F.H.P.; writing—review and editing, F.H.P., R.C.J. and F.L..; visualization, J.L.Z.; validation, F.H.P., funding acquisition, F.H.P. and H.R.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministerio del Poder Popular de Ciencia, Tecnología e Innovación of Venezuela.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Bioethical Committee of Instituto Venezolano de Investigaciones Cientificas (IVIC) (CBioEtIVIC-2022-001).
Informed Consent Statement
Patient consent was waived due to samples, from nasopharyngeal or nasal swabs and positive by qRT-PCR during the routine COVID-19 diagnosis in Venezuela. The identity of the patients was maintained anonymous.
Data Availability Statement
The complete genome sequences have been deposited in the GISAID database.
Acknowledgments
This study was supported by Ministerio del Poder Popular de Ciencia, Tecnología e Innovación of Venezuela. We are also indebted to the Pan American Health Organization (PAHO) and the Project ¨Diálogo sobre la Pandemia ̈ directed by the Charité Institute and financed by the Ministry of Foreign Affairs of Germany, in cooperation with the Deutsche Gesellschaft für Internationale Zysammenarbeit (GIZ), for the generous gift of reagents for NGS sequencing.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- COVID - Coronavirus Statistics – Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 31 December 2022).
- Rambaut, A.; Holmes, E.C.; O'Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020, 5, 5,1403–1407. [Google Scholar] [CrossRef] [PubMed]
- O'Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C. , Abu-Dahab, K.; Taylor, B.; Yeats, C.; du Plessis, L.; Maloney, D.; Medd, N.; Attwood, S.W.; Aanensen, D.M.; Holmes, E.C.; Pybus, O.G.; Rambaut, A. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Cov-Lineages. Available online: https://cov-lineages.org/lineage_list.html (accessed on 08 May 2023).
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, Ch.L.; Anyaneji, U.J.; Bester, Ph.A.; Boni, M.F.; Chand, M.; Choga, W.T.; Colquhoun, R.; Davids, M.; Deforche, K.; Doolabh, D.; du Plessis, L.; Engelbrecht, S.; Everatt, J.; Giandhari, J.; Giovanetti, M.; Hardie, D.; Hill, V.; Hsiao, N.Y.; Iranzadeh, A.; Ismail, A.; Joseph, Ch.; Joseph, R.; Koopile, L.; Kosakovsky Pond, S.L.; Kraemer, M.U.G.; Kuate-Lere, L.; Laguda-Akingba, O.; Lesetedi-Mafoko, O.; Lessells, R.J.; Lockman, Sh.; Lucaci, A.G.; Maharaj, A.; Mahlangu, B.; Maponga, T.; Mahlakwane, K.; Makatini, Z.; Marais, G.; Maruapula, D.; Masupu, K.; Matshaba, M.; Mayaphi, S.; Mbhele, N.; Mbulawa, M.B.; Mendes, A.; Mlisana, K.; Mnguni, A.; Mohale, Th.; Moir, M.; Moruisi, K.; Mosepele, M.; Motsatsi, G.; Motswaledi, M.S.; Mphoyakgosi, Th.; Msomi, N.; Mwangi, P.N.; Naidoo, Y.; Ntuli, N.; Nyaga, M.; Olubayo, L.; Pillay, S.; Radibe, B.; Ramphal, Y.; Ramphal, U.; San, J.E.; Scott, L.; Shapiro, R.; Singh, L.; Smith-Lawrence, P.; Stevens, W.; Strydom, A.; Subramoney, K.; Tebeila, N.; Tshiabuila, D.; Tsui, J.; van Wyk, S.; Weaver, S.; Wibmer, C.K.; Wilkinson, E.; Wolter, N.; Zarebski, A.E.; Zuze, B.; Goedhals, D.; Preiser, W.; Treurnicht, F.; Venter, M.; Williamson, C.; Pybus, O.G.; Bhiman, J.; Glass, A.; Martin, D.P.; Rambaut, A.; Gaseitsiwe, S.; von Gottberg, A.; de Oliveira, T. Rapid epidemic expansion of the SARSCoV-2 Omicron variant in Southern Africa. Nature 2022. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Tracking SARS-CoV-2 variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 08 May 2023).
- Zambrano, J.L.; Jaspe, R.C.; Hidalgo, M.; Loureiro, C.L.; Sulbarán, Y.; Moros, Z.C.; Garzaro, D.J.; Vizzi, E.; Rangel, H.R.; Liprandi, F.; Pujol, F.H. Sub-lineages of the Omicron variant of SARS-CoV-2: characteristic mutations and their relation to epidemiological behavior. Inv Clin (Venezuela) 2022, 63, 262–274. [Google Scholar] [CrossRef]
- Jaspe, R.C.; Sulbaran, Y.; Loureiro, C.L.; Moros, Z.C.; Marulanda, E.; Bracho, F.; Ramírez, N.A.; Canonico, Y.; D'Angelo, P.; Rodríguez, L.; Castro, J.; Liprandi, F.; Rangel, HR.; Pujol, F.H. Detection of the Omicron variant of SARS-CoV-2 in international travelers returning to Venezuela. Travel Med Infect Dis 2022, 48, 102326. [Google Scholar] [CrossRef] [PubMed]
- Jaspe, R.C.; Loureiro, C.L.; Sulbaran, Y.; Moros, Z.C.; D'Angelo, P.; Hidalgo, M.; Rodríguez, L.; Alarcón, V.; Aguilar, M.; Sánchez, D.; Ramírez, J.; Garzaro, D.J.; Zambrano, J.L.; Liprandi, F.; Rangel, H.R.; Pujol, F.H. Description of a One-Year Succession of Variants of Interest and Concern of SARS-CoV-2 in Venezuela. Viruses 2022, 14, 1378. [Google Scholar] [CrossRef] [PubMed]
- SARS-CoV-2 V4.1 Update for Omicron Variant - Laboratory - ARTIC Real-Time Genomic Surveillance Available online:. Available online: https://community.artic.network/t/sars-cov-2-v4-1-update-for-omicron-variant/342 (accessed on 16 May 2023).
- SARS-CoV-2 (hCoV-19) Mutation Reports, Lineage Comparison. Available online: https://outbreak.info/compare-lineages?%20pango=BA.1&pango=BA.2&pango=%20BA.3&pango=BA.4&pango=BA.5&gene=S&gene=ORF1a&threshold=75&nthresh=1&sub=true&dark=false (accessed on 08 May 2023).
- Marulanda, E. (CasaLab, Caracas, Miranda, Venezuela). Personal communication, 2023.
- EU Digital COVID Certificate. Available online: https://ec.europa.eu/info/live-work-travel-eu/coronavirus-response/safe-covid-19-vaccines-europeans/eu-digital-covid-certificate_en (accessed on 08 May 2023).
- Callaway, E. COVID 'variant soup' is making winter surges hard to predict. Nature 2022, 611, 213–214. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Quiroga, R.; McConnell, S.; Johnson, M.C.; Casadevall, A. Convergent Evolution in SARS-CoV-2 Spike Creates a Variant Soup from Which New COVID-19 Waves Emerge. Int J Mol Sci 2023, 24, 2264. [Google Scholar] [CrossRef] [PubMed]
- Jaspe, R.C.; Loureiro, C.L.; Sulbaran, Y.; Moros, Z.C.; D'Angelo, P.; Rodríguez, L.; Zambrano, J.L.; Hidalgo, M.; Vizzi, E.; Alarcon, V.; Aguilar, M.; Garzaro, D.J.; CoViMol Group. ; Rangel, H.R.; Pujol, F.H. Introduction and rapid dissemination of SARS-CoV-2 Gamma Variant of Concern in Venezuela. Infect Genet Evol 2021, 96, 105147. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Omicron Delta study group. Clinical progression, disease severity, and mortality among adults hospitalized with COVID-19 caused by the Omicron and Delta SARS-CoV-2 variants: A population-based, matched cohort study. PLoS One 2023, 18, e0282806. [Google Scholar] [CrossRef]
- Robinson, M.L.; Morris, C.P.; Betz, J.F.; Zhang, Y.; Bollinger, R.; Wang, N.; Thiemann, D.R.; Fall, A.; Eldesouki, R.E.; Norton, J.M.; Gaston, D.C.; Forman, M.; Luo, C.H.; Zeger, S.L.; Gupta, A.; Garibaldi, B.T.; Mostafa, H.H. Impact of SARS-CoV-2 variants on inpatient clinical outcome. Clin Infect Dis 2022, 19, ciac957. [Google Scholar] [CrossRef]
- Lorenzo-Redondo, R.; Ozer, E.A.; Hultquist, J.F. Covid-19: is omicron less lethal than delta? BMJ 2022, 378, o1806. [Google Scholar] [CrossRef] [PubMed]
- Adamoski, D.; Baura, V.A.; Rodrigues, A.C.; Royer, C.A.; Aoki, M.N.; Tschá, M.K.; Bonatto, A.C.; Wassem, R.; Nogueira, M.B.; Raboni, S.M.; Almeida, B.M.M.; Trindade, E.D.S.; Gradia, D.F.; Souza, E.M.; Carvalho de Oliveira, J. SARS-CoV-2 Delta and Omicron Variants Surge in Curitiba, Southern Brazil, and Its Impact on Overall COVID-19 Lethality. Viruses 2022, 14, 809. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Evans, J.P.; Faraone, J.N.; Zheng, Y.M.; Carlin, C.; Anghelina, M.; Stevens, P.; Fernandez, S.; Jones, D.; Lozanski, G.; Panchal, A.; Saif, L.J.; Oltz, E.M.; Xu, K.; Gumina, R.J.; Liu, S.L. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 2023, 31, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Bruel, T.; Staropoli, I.; Guivel-Benhassine, F.; Porrot, F.; Maes, P.; Grzelak, L.; Prot, M.; Mougari, S.; Planchais, C.; Puech, J.; Saliba, M.; Sahraoui, R.; Fémy, F.; Morel, N.; Dufloo, J.; Sanjuán, R.; Mouquet, H.; André, E.; Hocqueloux, L.; Simon-Loriere, E.; Veyer, D.; Prazuck, T.; Péré, H.; Schwartz, O. Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies. Nat Commun 2023, 14, 824. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Kurhade, C.; Patel, S.; Kitchin, N.; Tompkins, K.; Cutler, M.; Cooper, D.; Yang, Q.; Cai, H.; Muik, A.; Zhang, Y.; Lee, D.Y.; Şahin, U.; Anderson, A.S.; Gruber, W.C.; Xie, X.; Swanson, K.A.; Shi, P.Y. Neutralization of BA.4-BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with Bivalent Vaccine. N Engl J Med 2023, 388, 854–857. [Google Scholar] [CrossRef] [PubMed]
- Our World in Data. Available online: https://ourworldindata.org/ (accessed on 08 May 2023).
- Uriu, K.; Ito, J.; Zahradnik, J.; Fujita, S.; Kosugi, Y.; Schreiber, G. Genotype to Phenotype Japan (G2P-Japan) Consortium; Sato K. Enhanced transmissibility, infectivity, and immune resistance of the SARS-CoV-2 omicron XBB.1.5 variant. Lancet Infect Dis 2023, 23, 280–281. [Google Scholar] [CrossRef] [PubMed]
- GISAID. Available online: https://gisaid.org/ (accessed on 8 May 2023).
- Scarpa, F.; Azzena, I.; Locci, C.; Casu, M.; Fiori, P.L.; Ciccozzi, A.; Angeletti, S.; Imperia, E.; Giovanetti, M.; Maruotti, A.; Borsetti, A.; Cauda, R.; Cassone, A.; Via, A.; Pascarella, S.; Sanna, D.; Ciccozzi, M. Molecular In-Depth on the Epidemiological Expansion of SARS-CoV-2 XBB.1.5. Microorganisms 2023, 11, 912. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).