Submitted:
17 May 2023
Posted:
19 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
| Agriculture research domains | ||
|---|---|---|
| fertilizers | different natures of agricultural production communities such as the winter-rainfall desert community | restoration and management of plant systems |
| microalgae refineries | first reports on plant findings | investigation of biochemical activities in plant species |
| climatic factors such as fires affecting food production | appraisals of agricultural products | in vitro cultivation of plant species |
| land degradation and cultivation research | competitive growth advantages of paired cultivation | characterizing seeds or plant species |
| antibacterial and chemical byproducts from plants | creation of taxonomic lists of crops | importing new plant species across regions |
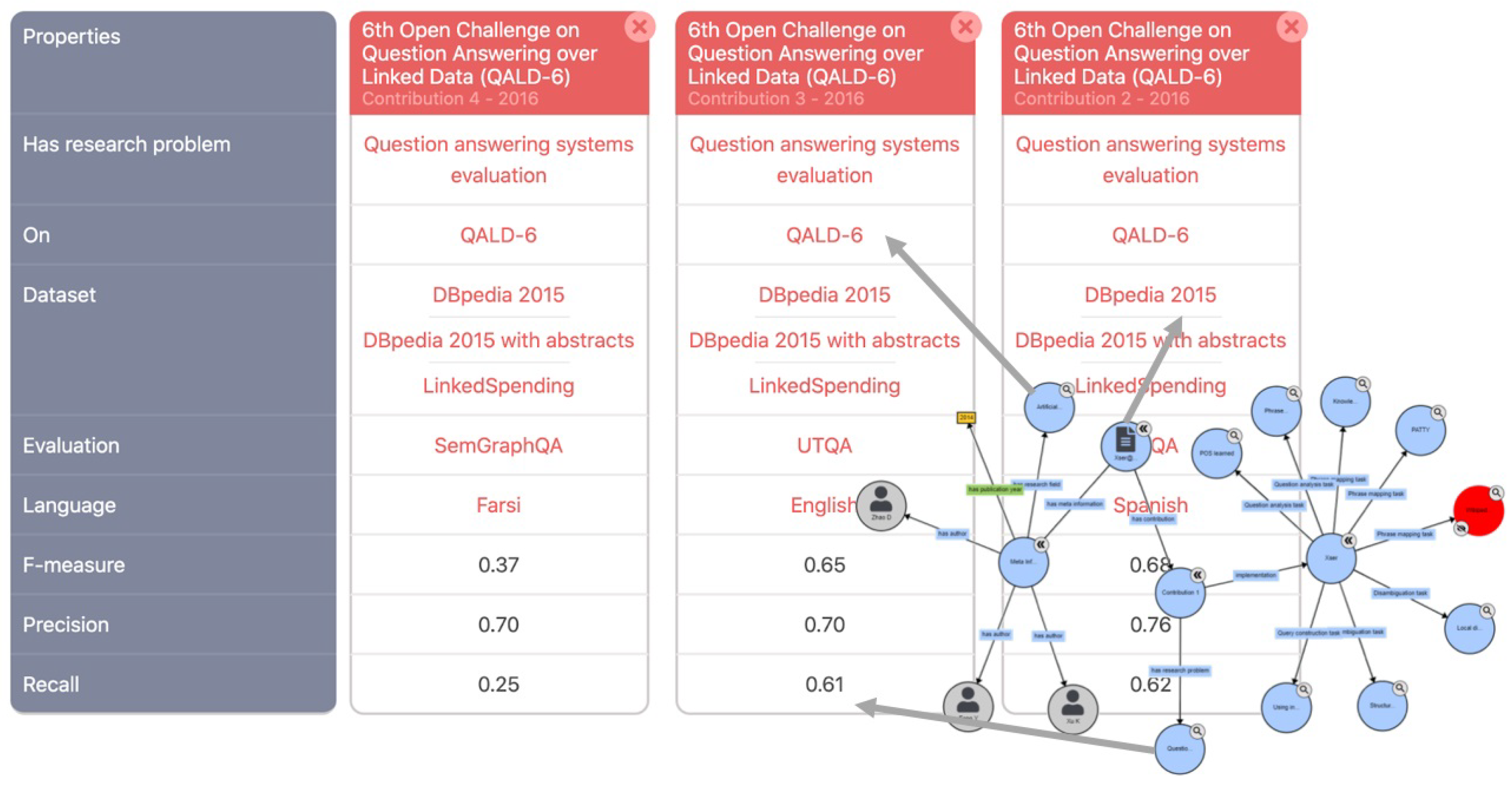
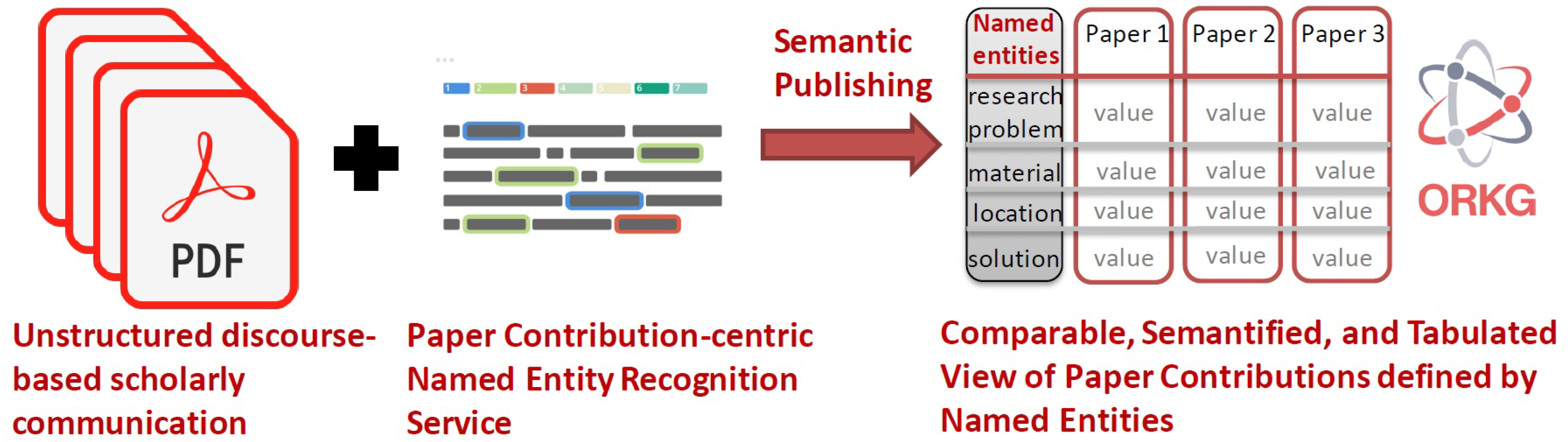
2. Background
2.1. Ontological structuring of scholarly publications
2.2. Entity-centric annotation models of scholarly publications
3. Materials and Methods
3.1. Context: The AGROVOC Ontology and the ORKG Agri-NER Model
3.2. The ORKG Agri-NER Specifications
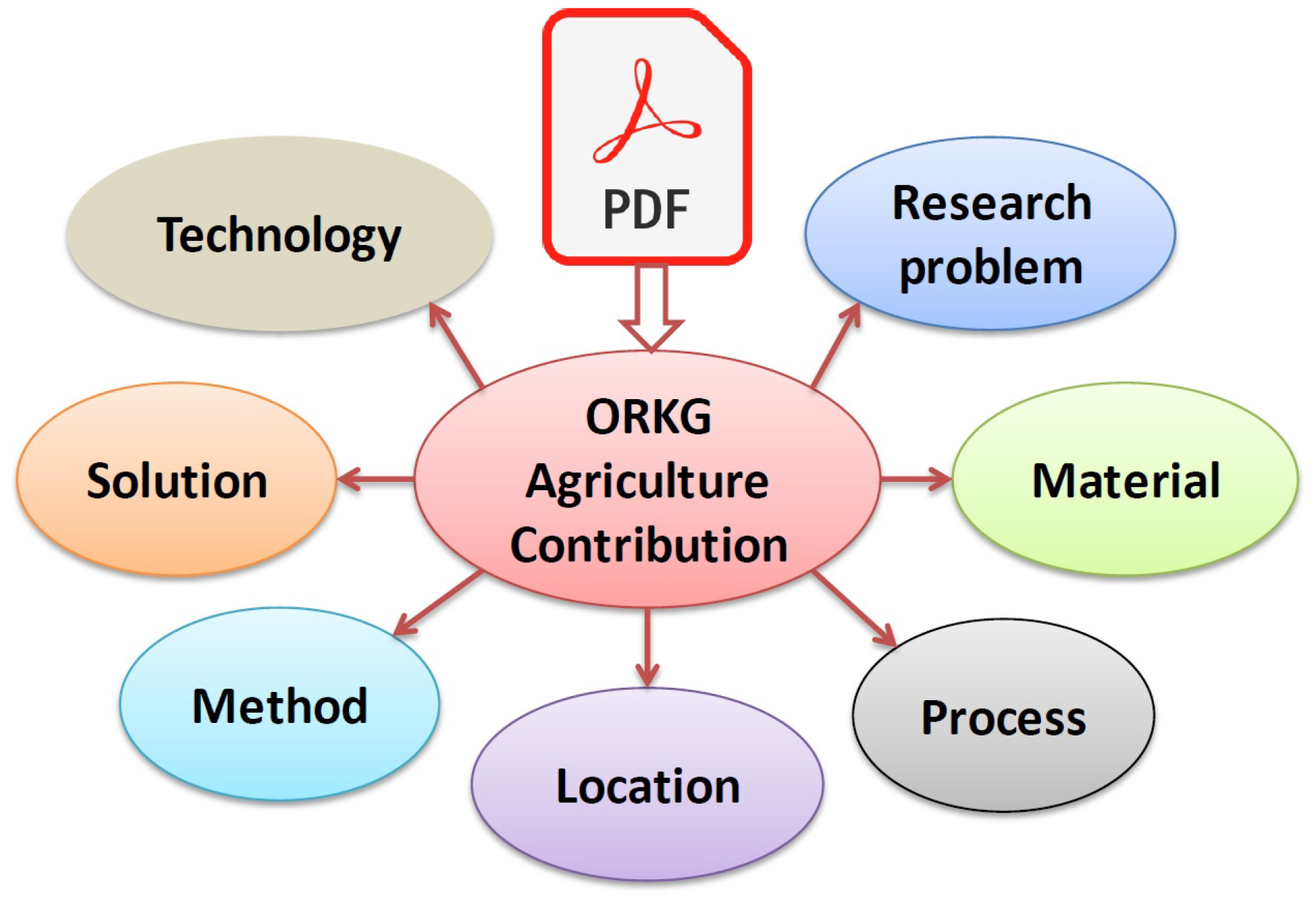
3.2.1. The seven ORKG Agri-NER entity type definitions
- research problem. It is a natural language mention phrase of the theme of the investigation in a scholarly article [41]. Alternatively, in the Computer Science domain it is referred to as task [17] or focus [7]. An article can address one or more research problems. E.g., seed germination, humoral immunity in cattle, sunbird pollination, seasonal and inter-annual soil CO2 efflux, etc. Generally, research problem mentions are often found in the article Title, Abstract, or Introduction in the context of discourse discussions on the theme of the study; otherwise in the Results section in the context of findings discussions on the theme of the study.
- resource. They are either man-made or naturally occurring tangible objects that are directly utilized in the process of a research investigation as material to facilitate a study’s research findings. “Resources are things that are used during a production process or that are required to cover human needs in everyday life.” [47] E.g., resource `pesticides’ used to study the research problem `survival of pines’; the process `repeated migrations’ studied over resource `southern African members of the genus Zygophyllum’; resource `Soil aggregate-associated heavy metals’ studied in location `subtropical China’. Resources are used to either address the research problem or to obtain the solution.
- process. It is defined as an event with a continuous time frame that is pertinent with a specific function or role to the theme of a particular investigation or research study. As defined in the AGROVOC ontology [47], a process can be a set of interrelated or interacting activities which transforms inputs into outputs, or simply a naturally occurring phenomenon that is studied. E.g., irradiance, environmental gradient, seasonal variation, quality control, salt and alkali stresses, etc.
- location. Includes all geographical locations in the world seen similar to the AGROVOC location concept [46] as a `point in space.’ Often location, in terms of relevance to the research theme, is the place where the study is conducted or a place studied for its resource or processes w.r.t. a research problem. location mentions can be as fine-grained as having regional boundaries or as broad as having continental boundaries. E.g., Cape Floristic Region of South Africa, winter rainfall area of South Africa, sahel zone of Niger, southern continents, etc.
- method. This concept imported from the Computer Science domain pertains to existing protocols used to support the solution [19]. The interpretation or definition of the concept similarly holds for the agriculture domain. It is a predetermined way of accomplishing an objective in terms of prespecified set of steps. E.g., On-farm comparison, semi-stochastic models, burrows pond rearing system, bradyrhizobium inoculation, electronic olfaction, systematic studies, etc.
- solution. It is a phrasal succinct mention of the novel contribution or discovery of a work that solves the research problem [19]. The solution entity type is characterized by a long-tailed distribution of mentions determined by the new research discoveries made. The solution of one work can be used as a method or technology or comparative baselines in subsequent research works. Of all the entity types introduced in this work, solution like research problem is specifically tailored to the ORKG contribution model. E.g., radiation-induced genome alterations, artificially assembled seed dispersal system, commercial craftwork, integrated ecological modeling system, the MiLA tool, next generation crop models etc.
- technology. Practical systems realized as tools, machinery or equipment based on the systematized application of reproducible scientific knowledge to reach a specifiable, repeatable goal. In the context of the agriculture domain, the goals would pertain to agricultural and food systems. E.g., stream and riverine ecosystem services, hyperspectral imaging, biotechnology, continuous vibrating conveyor, low exchange water recirculating aquaculture systems, etc.
3.2.2. The ORKG Agri-NER corpus annotation methodology
Raw dataset
Corpus annotation
- A list of entity types used in our prior work [19] on contribution-centric NER for the Computer Science (CS) domain was created as a reference list. This list included the following CS-domain-specific contribution-centric types, viz. solution, research problem, method, resource, tool, language, and dataset. We identified this as a suitable first step owing to the strong overlap of the annotation aim between our prior work on the CS domain and our present work on the agriculture domain, i.e. that of identifying contribution-centric entities from paper titles. We hypothesized that some entity types, e.g., research problem, that satisfy the functional role of reflecting the contribution of scholarly articles by nature of their genericity could be applicable across domains. As such the listed CS-domain contribution-centric entity types were tested for this hypothesis. Furthermore, based on the successful annotation outcomes of paper titles offering a rich store of contribution-centric entities, this work focusing on a new domain, i.e. agriculture, similarly based its entity annotation task on paper titles. Thus, with an initial set of entities in place, our task was then to identify the entities that were generic enough to be transferred from the CS domain to the domain of agriculture.
- Considering that some new agriculture domain-specific entity types would also need to be introduced, a list of the 24 top-level concepts in the AGROVOC ontology [47] as the reference standard was drawn up. This list included concepts such as features (https://tinyurl.com/agrovoc-features), location (https://tinyurl.com/agrovoc-location), measure (https://tinyurl.com/agrovoc-measure), properties (https://tinyurl.com/agrovoc-properties), strategies (https://tinyurl.com/agrovoc-strategies), etc. The focus was maintained only on the top-level concepts, since traversing lower levels in the ontology led to specific terminology defined as a concept space such as Maize https://agrovoc.fao.org/browse/agrovoc/en/page/c_12332. Since specific terminology do not serve the purpose of reflecting a functional role, hence by their inherent nature were ruled out as conceptual candidates for contribution-centric entity types.
- Given the two reference lists of generic CS domain entity types and domain-specific AGROVOC concepts from steps 1 and 2, respectively, the third step involved selecting and pruning the lists to arrive at a final set of contribution-centric entity types to annotate agricultural domain paper titles with. There were two prerequisites defined for arriving at the final set of entity types: a) it needed to include as many of the generic entities as were semantically applicable; and b) introduce new domain-specific types complementing the semantic interpretation of the generic types such that the final set could be used as a unit for contribution-centric entity recognition. Concretely, these requisites were realized as a pilot annotation task over a set of 50 paper titles performed by a postdoctoral researcher. Starting with the CS domain inspired list of generic entities, the pilot annotation task showed that the CS domain tool, language, and dataset types were not applicable to agricultural domain. This left a set of four types, viz. solution, research problem, method, and resource for the final annotation task. For the domain-specific entities, via the pilot annotation exercise, it was fairly straightforward to prune out most of the AGROVOC concepts on the basis of the following three criteria. a) Six concepts did not fit in the criteria of offering a functional role that reflected the contribution of a work. These were entities, factors, groups, properties, stages, state. b) Nine concepts indicated that they were more paper content-specific than title-specific. These were activities, events, features, measure, phenomena, products, site, systems, and time. And, c) since our objective was to capture the most generic entity satisfying the functional role of reflecting the paper contribution, some of the top-level AGROVOC concepts could be subsumed by others. Specifically, the four types viz. objects, organisms, subjects, substances were subsumed as AGROVOC resources. Also strategies was subsumed as AGROVOC methods. In the end, from an initial list of 25 types, pruning out 15 types and subsuming 5 types, we were left with a set of five types for the final annotation task, viz. location, methods, processes, resources, technology. Then the generic and domain-specific lists were resolved as follows: solution and research problem originating from the CS domain were retained as is for the agriculture domain; AGROVOC methods was resolved to the generic method type and AGROVOC resources was resolved to resource; the remaining AGROVOC entities were first lemmatized for plurals (e.g., processes → process) and otherwise retained as is for location and technology types.

The Agri-NER corpus statistics
| Statistic parameter | Counts |
|---|---|
| # Title Tokens overall | 71,632 |
| Max., Min., Avg. # Tokens/Title | 65, 2, 13.75 |
| # Entity Tokens overall | 47,608 |
| Max., Min., Avg. # Tokens/Entity | 15, 1, 3.12 |
| # Entities | 15,261 |
| # Unique Entities | 10,406 |
| Max., Min., Avg. # Entities/Title | 9, 0, 2.93 |
| Entity type | Counts |
|---|---|
| # resource | 5,490 (4,073) |
| # research problem | 4,707 (3,403) |
| # process | 1,789 (1,525) |
| # location | 1,525 (776) |
| # method | 1,364 (940) |
| # solution | 250 (221) |
| # technology | 136 (113) |
4. Results
4.1. Experimental Setup
4.1.1. Dataset
4.1.2. Models
4.1.3. Evaluation metrics
4.2. Experiments
| IOBES | IOB | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exact Match | Inexact Match | Exact Match | Inexact Match | |||||||||
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| no features | 56.38 | 62.27 | 59.18 | 59.1 | 65.27 | 62.03 | 54.62 | 59.47 | 56.94 | 58.52 | 63.71 | 61.0 |
| +GloVe | 57.74 | 62.79 | 60.16 | 60.86 | 66.19 | 63.41 | 57.9 | 63.49 | 60.57 | 61.64 | 67.59 | 64.48 |
| POS | 57.11 | 61.88 | 59.40 | 60.24 | 65.27 | 62.66 | 56.0 | 60.53 | 58.18 | 60.42 | 65.3 | 62.76 |
| +GloVe | 57.5 | 63.58 | 60.38 | 60.09 | 66.45 | 63.11 | 56.63 | 63.11 | 59.7 | 60.45 | 67.38 | 63.73 |
| NER | 56.12 | 61.1 | 58.5 | 58.39 | 63.58 | 60.88 | 56.43 | 61.59 | 58.9 | 60.44 | 65.96 | 63.08 |
| +GloVe | 57.5 | 63.58 | 60.38 | 60.09 | 66.45 | 63.11 | 58.32 | 63.49 | 60.8 | 62.09 | 67.59 | 64.72 |
| POS + NER | 56.66 | 61.75 | 59.09 | 59.52 | 64.88 | 62.09 | 55.93 | 61.77 | 58.71 | 59.88 | 66.14 | 62.85 |
| +GloVe | 58.23 | 63.71 | 60.85 | 60.98 | 66.71 | 63.72 | 55.93 | 61.77 | 58.71 | 59.88 | 66.14 | 62.85 |
| IOBES | IOB | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exact Match | Inexact Match | Exact Match | Inexact Match | |||||||||
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| BERT-Base-Cased | 58.74 | 66.71 | 62.47 | 62.64 | 71.15 | 66.63 | 60.58 | 68.25 | 64.19 | 64.09 | 72.2 | 67.91 |
| SciBERT-SciVocab-Cased | 58.78 | 66.06 | 62.2 | 62.49 | 70.23 | 66.13 | 59.15 | 66.09 | 62.43 | 63.63 | 71.11 | 67.17 |
5. Discussion
| Statistic parameter | Counts |
|---|---|
| % Entities resolved (% Entities resolved as subphrases) | 16.06% (53.75%) |
| Max., Min., Avg. phrase length resolved | 5, 1, 1.55 |
| Max., Min., Avg. subphrase length resolved | 5, 1, 1.23 |
| % location resolved (% location resolved as subphrases) | 31.82% (41.57%) |
| % technology resolved (% technology resolved as subphrases) | 17.99% (46.04%) |
| % process resolved (% process resolved as subphrases) | 16.57% (52.22%) |
| % method resolved (% method resolved as subphrases) | 15.11% (41.07%) |
| % research problem resolved (% research problem resolved as subphrases) | 13.77% (60.5%) |
| % resource resolved (% resource resolved as subphrases) | 13.68% (55.35%) |
| % solution resolved (% solution resolved as subphrases) | 3.19% (60.56%) |
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ORKG | Open Research Knowledge Graph |
| Agri | Agriculture |
| NER | Named Entity Recognition |
References
- Johnson, R.; Watkinson, A.; Mabe, M. The STM Report: An overview of scientific and scholarly publishing 2018.
- Renear, A.H.; Palmer, C.L. Strategic reading, ontologies, and the future of scientific publishing. Science 2009, 325, 828–832. [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; others. The FAIR Guiding Principles for scientific data management and stewardship. Scientific data 2016, 3, 1–9. [CrossRef]
- Ammar, W.; Groeneveld, D.; Bhagavatula, C.; Beltagy, I.; Crawford, M.; Downey, D.; Dunkelberger, J.; Elgohary, A.; Feldman, S.; Ha, V.; others. Construction of the Literature Graph in Semantic Scholar. Proceedings of NAACL-HLT, 2018, pp. 84–91.
- Auer, S.; Oelen, A.; Haris, M.; Stocker, M.; D’Souza, J.; Farfar, K.E.; Vogt, L.; Prinz, M.; Wiens, V.; Jaradeh, M.Y. Improving access to scientific literature with knowledge graphs. Bibliothek Forschung und Praxis 2020, 44, 516–529. [CrossRef]
- Kim, S.N.; Medelyan, O.; Kan, M.Y.; Baldwin, T. Semeval-2010 task 5: Automatic keyphrase extraction from scientific articles. Proceedings of the 5th International Workshop on Semantic Evaluation, 2010, pp. 21–26.
- Gupta, S.; Manning, C. Analyzing the Dynamics of Research by Extracting Key Aspects of Scientific Papers. Proceedings of 5th International Joint Conference on Natural Language Processing; Asian Federation of Natural Language Processing: Chiang Mai, Thailand, 2011; pp. 1–9.
- QasemiZadeh, B.; Schumann, A.K. The ACL RD-TEC 2.0: A Language Resource for Evaluating Term Extraction and Entity Recognition Methods. Proceedings of the Tenth International Conference on Language Resources and Evaluation (LREC’16); European Language Resources Association (ELRA): Portorož, Slovenia, 2016; pp. 1862–1868.
- Moro, A.; Navigli, R. Semeval-2015 task 13: Multilingual all-words sense disambiguation and entity linking. Proceedings of the 9th international workshop on semantic evaluation (SemEval 2015), 2015, pp. 288–297.
- Augenstein, I.; Das, M.; Riedel, S.; Vikraman, L.; McCallum, A. SemEval 2017 Task 10: ScienceIE - Extracting Keyphrases and Relations from Scientific Publications. Proceedings of the 11th International Workshop on Semantic Evaluation (SemEval-2017); Association for Computational Linguistics: Vancouver, Canada, 2017; pp. 546–555. [CrossRef]
- Gábor, K.; Buscaldi, D.; Schumann, A.K.; QasemiZadeh, B.; Zargayouna, H.; Charnois, T. Semeval-2018 Task 7: Semantic relation extraction and classification in scientific papers. Proceedings of The 12th International Workshop on Semantic Evaluation, 2018, pp. 679–688.
- Luan, Y.; He, L.; Ostendorf, M.; Hajishirzi, H. Multi-Task Identification of Entities, Relations, and Coreferencefor Scientific Knowledge Graph Construction. Proc. Conf. Empirical Methods Natural Language Process. (EMNLP), 2018.
- Hou, Y.; Jochim, C.; Gleize, M.; Bonin, F.; Ganguly, D. Identification of Tasks, Datasets, Evaluation Metrics, and Numeric Scores for Scientific Leaderboards Construction. Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics; Association for Computational Linguistics: Florence, Italy, 2019; pp. 5203–5213. [CrossRef]
- Dessì, D.; Osborne, F.; Reforgiato Recupero, D.; Buscaldi, D.; Motta, E.; Sack, H. Ai-kg: an automatically generated knowledge graph of artificial intelligence. International Semantic Web Conference. Springer, 2020, pp. 127–143.
- D’Souza, J.; Hoppe, A.; Brack, A.; Jaradeh, M.Y.; Auer, S.; Ewerth, R. The STEM-ECR Dataset: Grounding Scientific Entity References in STEM Scholarly Content to Authoritative Encyclopedic and Lexicographic Sources. Proceedings of the 12th Language Resources and Evaluation Conference; European Language Resources Association: Marseille, France, 2020; pp. 2192–2203.
- D’Souza, J.; Auer, S.; Pedersen, T. SemEval-2021 Task 11: NLPContributionGraph - Structuring Scholarly NLP Contributions for a Research Knowledge Graph. Proceedings of the 15th International Workshop on Semantic Evaluation (SemEval-2021); Association for Computational Linguistics: Online, 2021; pp. 364–376. [CrossRef]
- Kabongo, S.; D’Souza, J.; Auer, S. Automated Mining of Leaderboards for Empirical AI Research. International Conference on Asian Digital Libraries. Springer, 2021, pp. 453–470.
- D’Souza, J.; Auer, S. Pattern-based acquisition of scientific entities from scholarly article titles. International Conference on Asian Digital Libraries. Springer, 2021, pp. 401–410.
- D’Souza, J.; Auer, S. Computer Science Named Entity Recognition in the Open Research Knowledge Graph. arXiv preprint arXiv:2203.14579 2022.
- SUNDHEIM, B. Overview of results of the MUC-6 evaluation. Proc. Sixth Message Understanding Conference (MUC-6), 1995, 1995.
- Chinchor, N.; Robinson, P. MUC-7 named entity task definition. Proceedings of the 7th Conference on Message Understanding, 1997, Vol. 29, pp. 1–21.
- Sang, E.T.K.; De Meulder, F. Introduction to the CoNLL-2003 Shared Task: Language-Independent Named Entity Recognition. Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL 2003, 2003, pp. 142–147.
- Hovy, E.; Marcus, M.; Palmer, M.; Ramshaw, L.; Weischedel, R. OntoNotes: the 90% solution. Proceedings of the human language technology conference of the NAACL, Companion Volume: Short Papers, 2006, pp. 57–60.
- Batbayar, E.E.T.; Tsogt-Ochir, S.; Oyumaa, M.; Ham, W.C.; Chong, K.T. Development of ISO 11783 Compliant Agricultural Systems: Experience Report. In Automotive Systems and Software Engineering; Springer, 2019; pp. 197–223.
- Le Bourgeois, T.; Marnotte, P.; Schwartz, M. The use of EPPO Codes in tropical weed science. EPPO Codes Users Meeting 5th Webinar, 2021.
- Shotton, D. Semantic publishing: the coming revolution in scientific journal publishing. Learned Publishing 2009, 22, 85–94. [CrossRef]
- Lis-Balchin, M.T. A chemotaxonomic reappraisal of the Section Ciconium Pelargonium (Geraniaceae). South African Journal of Botany 1996, 62, 277–279. [CrossRef]
- Berners-Lee, T.; Hendler, J.; Lassila, O. The semantic web. Scientific american 2001, 284, 34–43. [CrossRef]
- Fathalla, S.; Vahdati, S.; Auer, S.; Lange, C. SemSur: a core ontology for the semantic representation of research findings. Procedia Computer Science 2018, 137, 151–162. [CrossRef]
- Vogt, L.; D’Souza, J.; Stocker, M.; Auer, S. Toward Representing Research Contributions in Scholarly Knowledge Graphs Using Knowledge Graph Cells. JCDL ’20, August 1–5, 2020, Virtual Event, China, 2020.
- Initiative, D.C.M.; others. Dublin core metadata initiative dublin core metadata element set, version 1.1, 2008.
- Baker, T. Libraries, languages of description, and linked data: a Dublin Core perspective. Library Hi Tech 2012. [CrossRef]
- Constantin, A.; Peroni, S.; Pettifer, S.; Shotton, D.; Vitali, F. The document components ontology (DoCO). Semantic web 2016, 7, 167–181. [CrossRef]
- Groza, T.; Handschuh, S.; Möller, K.; Decker, S. SALT-Semantically Annotated LATEX for Scientific Publications. European Semantic Web Conference. Springer, 2007, pp. 518–532.
- Ciccarese, P.; Groza, T. Ontology of rhetorical blocks (orb). editor’s draft, 5 june 2011. World Wide Web Consortium. http://www. w3. org/2001/sw/hcls/notes/orb/(last visited March 12, 2012) 2011.
- Sollaci, L.B.; Pereira, M.G. The introduction, methods, results, and discussion (IMRAD) structure: a fifty-year survey. Journal of the medical library association 2004, 92, 364.
- Soldatova, L.N.; King, R.D. An ontology of scientific experiments. Journal of the Royal Society Interface 2006, 3, 795–803. [CrossRef]
- Simperl, E. Reusing ontologies on the Semantic Web: A feasibility study. Data & Knowledge Engineering 2009, 68, 905–925.
- Peroni, S.; Shotton, D. FaBiO and CiTO: ontologies for describing bibliographic resources and citations. Journal of Web Semantics 2012, 17, 33–43. [CrossRef]
- Di Iorio, A.; Nuzzolese, A.G.; Peroni, S.; Shotton, D.M.; Vitali, F. Describing bibliographic references in RDF. SePublica, 2014.
- Fathalla, S.; Vahdati, S.; Auer, S.; Lange, C. Towards a knowledge graph representing research findings by semantifying survey articles. International Conference on Theory and Practice of Digital Libraries. Springer, 2017, pp. 315–327.
- Baglatzi, A.; Kauppinen, T.; Keßler, C. Linked science core vocabulary specification. Technical report, Tech. rep. available at, http://linkedscience. org/lsc/ns, 2011.
- Dessí, D.; Osborne, F.; Reforgiato Recupero, D.; Buscaldi, D.; Motta, E. CS-KG: A Large-Scale Knowledge Graph of Research Entities and Claims in Computer Science. International Semantic Web Conference. Springer, 2022, pp. 678–696.
- Jain, S.; van Zuylen, M.; Hajishirzi, H.; Beltagy, I. SciREX: A Challenge Dataset for Document-Level Information Extraction. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics; Association for Computational Linguistics: Online, 2020; pp. 7506–7516. [CrossRef]
- Mondal, I.; Hou, Y.; Jochim, C. End-to-End Construction of NLP Knowledge Graph. Findings of the Association for Computational Linguistics: ACL-IJCNLP 2021; Association for Computational Linguistics: Online, 2021; pp. 1885–1895. [CrossRef]
- Subirats-Coll, I.; Kolshus, K.; Turbati, A.; Stellato, A.; Mietzsch, E.; Martini, D.; Zeng, M. AGROVOC: The linked data concept hub for food and agriculture. Computers and Electronics in Agriculture 2022, 196, 105965. [CrossRef]
- AGROVOC Webpage. https://www.fao.org/agrovoc/home, 2022. [Online; accessed 12-October-2022].
- Soergel, D.; Lauser, B.; Liang, A.; Fisseha, F.; Keizer, J.; Katz, S. Reengineering thesauri for new applications: the AGROVOC example. Journal of digital information 2004, 4, 1–23.
- Lauser, B.; Sini, M.; Liang, A.; Keizer, J.; Katz, S. From AGROVOC to the Agricultural Ontology Service/Concept Server. An OWL model for creating ontologies in the agricultural domain. Dublin Core Conference Proceedings. Dublin Core DCMI, 2006.
- Mietzsch, E.; Martini, D.; Kolshus, K.; Turbati, A.; Subirats, I. How Agricultural Digital Innovation Can Benefit from Semantics: The Case of the AGROVOC Multilingual Thesaurus. Engineering Proceedings 2021, 9, 17.
- Auer, S. Towards an Open Research Knowledge Graph, 2018. [CrossRef]
- Sang, E.F.T.K.; De Meulder, F. Introduction to the CoNLL-2003 Shared Task: Language-Independent Named Entity Recognition. Development 1837, 922, 1341.
- Qi, P.; Zhang, Y.; Zhang, Y.; Bolton, J.; Manning, C.D. Stanza: A Python Natural Language Processing Toolkit for Many Human Languages. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics: System Demonstrations, 2020.
- Ramshaw, L.A.; Marcus, M.P. Text chunking using transformation-based learning. In Natural language processing using very large corpora; Springer, 1999; pp. 157–176.
- Krishnan, V.; Ganapathy, V. Named entity recognition. Stanford Lecture CS229 2005.
- Manning, C.D. Computational linguistics and deep learning. Computational Linguistics 2015, 41, 701–707. [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural computation 1997, 9, 1735–1780. [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation applied to handwritten zip code recognition. Neural computation 1989, 1, 541–551. [CrossRef]
- Huang, Z.; Xu, W.; Yu, K. Bidirectional LSTM-CRF models for sequence tagging. arXiv preprint arXiv:1508.01991 2015.
- Kim, Y.; Jernite, Y.; Sontag, D.; Rush, A.M. Character-aware neural language models. Thirtieth AAAI conference on artificial intelligence, 2016.
- Ma, X.; Hovy, E. End-to-end Sequence Labeling via Bi-directional LSTM-CNNs-CRF. Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), 2016, pp. 1064–1074.
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural Architectures for Named Entity Recognition. Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, 2016, pp. 260–270.
- Chiu, J.P.; Nichols, E. Named entity recognition with bidirectional LSTM-CNNs. Transactions of the Association for Computational Linguistics 2016, 4, 357–370. [CrossRef]
- Peters, M.; Ammar, W.; Bhagavatula, C.; Power, R. Semi-supervised sequence tagging with bidirectional language models. Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), 2017, pp. 1756–1765.
- Pennington, J.; Socher, R.; Manning, C. GloVe: Global Vectors for Word Representation. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); Association for Computational Linguistics: Doha, Qatar, 2014; pp. 1532–1543. [CrossRef]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural language processing (almost) from scratch. Journal of machine learning research 2011, 12, 2493–2537.
- Yang, J.; Zhang, Y. NCRF++: An Open-source Neural Sequence Labeling Toolkit. Proceedings of ACL 2018, System Demonstrations, 2018, pp. 74–79.
- Kenton, J.D.M.W.C.; Toutanova, L.K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of NAACL-HLT, 2019, pp. 4171–4186.
- Zhu, Y.; Kiros, R.; Zemel, R.; Salakhutdinov, R.; Urtasun, R.; Torralba, A.; Fidler, S. Aligning books and movies: Towards story-like visual explanations by watching movies and reading books. Proceedings of the IEEE international conference on computer vision, 2015, pp. 19–27.
- Beltagy, I.; Lo, K.; Cohan, A. SciBERT: Pretrained Language Model for Scientific Text. EMNLP, 2019, [arXiv:1903.10676].
- Bizer, C.; Heath, T.; Berners-Lee, T. Linked data: The story so far. In Semantic services, interoperability and web applications: emerging concepts; IGI global, 2011; pp. 205–227.
- Auer, S.; Bizer, C.; Kobilarov, G.; Lehmann, J.; Cyganiak, R.; Ives, Z. Dbpedia: A nucleus for a web of open data. In The semantic web; Springer, 2007; pp. 722–735.
| 1 | |
| 2 | |
| 3 | |
| 4 | |
| 5 | |
| 6 | |
| 7 | |
| 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





