1. Introduction
One of the most damaging diseases to salmonid aquaculture is the infestation of the ectoparasitic copepods collectively called sea lice, which has been estimated to have cost the salmonid industry £700 million globally in 2015, and the impact is likely to keep increasing in the future [
1]. The deployment of lumpfish (Cyclopterus lumpus) as cleaner fish has gained popularity as a long-term biological control that effectively controls lice abundance in sea cages [
2]. This method reduces the need for other delousing methods such as chemotherapeutics, mechanical, and thermal treatments which can compromise the fish’s welfare [
3,
4,
5], the ecosystem [
6], and the public perception toward the aquaculture [
2]. Hence, salmonid farms are utilising lumpfish as a “green” alternative for sea lice management.
Despite the benefit, this alternative can potentially have an impact on the wild lumpfish populations since its production still mainly relies on the capture of wild stocks [
7]. Lumpfish life cycle has been closed and currently can spawn in captivity, opening the door for a sustainable alternative to the fisheries [
8]. However, lumpfish lack pronounced sexual dimorphism before maturation [
9]. This limitation could impede the management of the breeding stock's sex ratio, which is crucial for further developments of lumpfish aquaculture. Lumpfish exhibit a XX/XY genetic sex-determination system [
10], therefore, DNA markers located within the sex-determining region could potentially identify the sex of the organism regardless of the life stage [
11].
A recent genome-wide association study (GWAS) that described markers associated with phenotypical sex in lumpfish suggested that chromosome 13 is the sex chromosome which contains a male-specific region [
10]. Sequence comparison within this region with other teleost sex-determining (SD) genes has proposed the anti-Müllerian hormone (amh) as the putative SD gene in lumpfish [
10]. The available male lumpfish genome assemblies contain three paralogues of amh with ambiguous sex-specificity: amh1, amh2, and amh3, with amh1 and amh2 being inverted tandem repeats [
10]. Studies in other species have already utilised male-specific amh as a sex-specific molecular marker for sex identification [
12,
13,
14]. Thus, the region containing the male-specific amh in the lumpfish genome is expected to also allow their sex identification.
This study aims to describe molecular markers on the male-specific region in the lumpfish genome for the design of a polymerase chain reaction (PCR) test for sex identification. To achieve this goal, our study characterised the sex specificity of the three amh genes in the lumpfish genome, determined the size of the male-specific region in the lumpfish genome, and examined amh in related species within the Cyclopteridae family. This study provides male-specific markers to improve the sex identification process in lumpfish, which can assist in sustainable lumpfish production, and in addition, provides more insight into the mechanism and evolution of lumpfish’s sex-determination system.
2. Materials and methods
2.1 Samples
A total of 143 archived samples of lumpfish were utilised in this study, comprised of 67 males and 76 females. A total of 13 males and 34 females were sampled from the wild population of the British Isles. A total of 22 males and 17 females were sampled from the wild population of Iceland. A total of 32 males and 25 females were offspring maintained at Otter Ferry Seafish (OFS) Ltd, comprised of four families which were derived from wild Norwegian parents. The phenotypic sex of samples was determined by gonad inspection and the DNA of samples was collected by fin-clipping in 2017 for further molecular analysis. The detailed records and the identification code of the samples are available in
Table S1.
Tissue samples of two male and one female Aptocyclus ventricosus, all from the Sea of Japan, one male and female Eumicrotremus taranetzi, both from the Sea of Okhotsk, and one male and female E. asperrimus, from the Sea of Japan and the Sea of Okhotsk respectively, were also utilised for further marker and phylogenetic analyses.
2.2. DNA extraction
Genomic DNA was extracted from the samples using the SSTNE extraction protocol [
15]. The purity (260/280 and 260/230 ratios) and concentration of the extracted DNA were then measured using Nanodrop ND-1000 Spectrophotometer with ND-1000 version 3.8.1 software (Thermo Fisher Scientific, USA). DNA samples with a 260/280 ratio of approximately 1.8 and the 260/230 ratio between 2.0 and 2.2 were then diluted to 50 ng/μL for downstream applications. The DNA samples of A. ventricosus, E. taranetzi, and E. asperrimus were diluted to 20 ng/μL instead due to low concentration.
2.3. Genomic sequence retrieval
The putative genomic DNA sequences of the three amh genes, amh1 (Ensembl accession no. ENSCLMG00005014163), amh2 (Ensembl accession no. ENSCLMG00005014165), and amh3 (Ensembl accession no. ENSCLMG00005016820) were extracted from the Ensembl genome database project [
16].
The three sequences were entirely aligned with the reference genome (GenBank accession no. GCA_009769545.1), so two genomic regions containing the amh paralogues covering 30 kilobases (kb) upstream and downstream from the paralogues were extracted from the reference genome: the amh1 and amh2 region and the amh3 region. The sequences were then annotated for further bioinformatics analyses by Geneious Prime 2022.2 (
https://www.geneious.com), based on the annotations in the Ensembl database.
2.4. Primer design
Primers utilised in this study were designed with the online software, Primer3 [
17] and manufactured by Integrated DNA Technologies, Inc. (IDT, USA). The primers were based on the available genome sequences found in Ensembl and NCBI.
2.5. Determination of sex specificity of markers
Four primer sets were designed to bind to the three found amh paralogues (
Table 1). Primer set AMH1_E3I6 was designed to bind to exon 3 (forward) and intron 6 (reverse) of amh1 to amplify 1,410 bp amplicons. AMH2_I6E4 binds to intron 6 (forward) and exon 4 (reverse) of amh2 to amplify 1,804 bp amplicons. AMH3_E3I6 binds to exon 3 (forward) and intron 6 (reverse) of amh3 to amplify 1,136 bp amplicons. The AMH1+3_E6I6 binds to the shared regions of exon 6 (forward) and intron 6 (reverse) of amh1 and amh3, which contain amh1-specific 91 bp deletion, to amplify 481 bp and 566 bp amplicons, respectively.
PCR amplifications were performed with these primers on randomly selected 12 male and 12 female lumpfish to characterise the sex specificity of each amh paralogue. PCR reactions were performed at a volume of 12.5 µL per reaction, comprised of 6.25 µL of 2x MyTaq™ HS Mix (Meridian Life Science, USA), 0.25 µL of the forward primer (10 µM), 0.25 µL of the reverse primer (10 µM), 4.75 µL of nuclease-free water, and 1 µL template DNA. In addition, blanks were created with the same procedure, excluding the DNA, to serve as the negative control.
The PCR amplification condition for the primer set AMH1_E3I6, AMH2_I6E4, and AMH3_E3I6 was 1 min at 95 °C, followed by 28 cycles of 15 s at 95 °C, 15 s at 57 °C, and then 1 min at 72 °C, and a final extension of 5 min at 72 °C. The PCR amplification condition for the primer set AMH1+3_E6I6 was 1 min at 95 °C, followed by 28 cycles of 15 s at 95 °C, 15 s at 57 °C, and then 20 s at 72 °C, and a final extension of 5 min at 72 °C.
PCR products and GeneRuler 1 kb DNA Ladder (Thermo Fisher Scientific, USA) were loaded into a 2.0% agarose gel (Agarose, Molecular Grade [Meridian Life Science, USA]; 0.5× Tris-acetate-EDTA [TAE] buffer; 2.5 mg/mL ethidium bromide), submerged in 0.5× TAE buffer, and applied with 90 V for 40 min. The gel images were visualised by InGenius 3 Manual Gel Documentation System with the GeneSys software (Syngene, India).
To further validate the accuracy of the male-specific markers, primer sets that amplified the male-specific amh were chosen to test on the remaining lumpfish samples. Chi-square tests were performed to determine the significant association between the phenotypic sex and the genotype displayed by each primer set. Statistical analyses were performed in R version 4.1.3 [
18], using RStudio version 2022.02.1+461 [
19].
PCR amplifications were also performed for the A. ventricosus, E. taranetzi, and E. asperrimus samples using the primer set AMH1+3_E6I6. PCR reactions with a final volume of 12.50 µL per reaction, comprised of 6.250 µL of Q5® Hot Start High-Fidelity 2X Master Mix (New England Biolabs, the United Kingdom), 0.375 µL of Dimethyl sulfoxide, 0.625 µL of the forward primer (10 µM), 0.625 µL of the reverse primer (10 µM), 3.625 µL of nuclease-free water, and 1 µL template DNA (20 ng µL-1). The PCR amplification condition was 30 s at 98 °C, followed by 32 cycles of 10 s at 98 °C, 20 s at 55 °C, and then 15 s at 72 °C, and a final extension of 2 min at 72 °C. PCR products were run on agarose gels in similar conditions as described above but utilized Tris-borate-EDTA (TBE) buffer instead of TAE and applied with 120 V for 150 min instead.
2.6. DNA sequencing
Amplicons obtained from randomly chosen single male and female lumpfish amplified with the primer sets from
Table 1 were sequenced.
Amplicons of the primer set AMH1_E3I6, AMH2_I6E4, and AMH3_E3I6 were purified with GeneJET PCR Purification Kit (Thermo Fisher Scientific, USA). For the primer set AMH1+3_E6I6, gel band excision was performed to extract each amplicon for sequencing. The excised bands were purified with QIAquick Gel Extraction Kit (QIAGEN, Germany).
The samples were submitted to the LightRun Tube Sanger sequencing (Eurofins genomics, Germany) for Sanger sequencing. The generated consensus sequences were aligned with the lumpfish amh sequences from the database using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) alignment algorithm [
20] in the Geneious to calculate the percentage of identical sites to identify the most similar sequences.
Amplicons of the primer set AMH1+3_E6I6 from the A. ventricosus, E. taranetzi, and E. asperrimus samples were also purified and sequenced with similar protocols.
2.7. Estimation of the male-specific region coverage
Based on the sex-specificity of the amh, additional primer sets were designed to estimate the length of the male-specific region (
Table 2). Primer set 4K_Up, 6K_Up, and 10K_Up were designed to bind to approximately 4 kb, 6 kb, and 10 kb upstream of the male-specific amh region to amplify 683 bp, 540 bp, and 653 bp amplicons, respectively. Primer set 4K_Down, 14K_Down, and 16K_Down were designed to bind to approximately the 4 kb, 14 kb, and 16 kb downstream of the male-specific amh region to amplify 733 bp, 704 bp, and 670 bp amplicons, respectively.
PCRs were performed with these primer sets on six randomly selected male and female lumpfish. The PCR reactions were similar to the reaction utilizing 2x MyTaq™ HS Mix detailed in
Section 2.5. PCR amplification conditions for these primer sets were 1 min at 95 °C, followed by 28 cycles of 15 s at 95 °C, 15 s at 57 °C, and then 20 s at 72 °C, and a final extension of 5 min at 72 °C. Amplicons underwent the previous gel electrophoresis protocol (as detailed in
Section 2.5) but ran at 80 V for 30 min.
2.8. Phylogenetic analysis
The autosomal amplicon sequences from primer set AMH1+3_E6I6 from A. ventricosus, E. taranetzi, and E. asperrimus, the sequences of the three amh paralogues from lumpfish from this study, autosomal amh (GenBank accession no. KP686074.1) and male-specific amhy sequences (GenBank accession no. KP686073.1) from lingcod (Ophiodon elongatus), and autosomal amh sequence (Ensembl accession no. ENSDARG00000014357) from zebrafish (Danio rerio) as an outgroup, were utilized for phylogeny reconstruction. Only the sequences from males were utilized due to the similarity between the autosomal amh in both sexes. Sequences of the amplicon from primer set AMH1+3_E6I6 from male and female A. ventricosus, E. taranetzi, and E. asperrimus are available in Table B.1 B).
Sequence alignments using the MUSCLE algorithm and the best substitution model determination were performed with MEGA11: Molecular Evolutionary Genetics Analysis ver 11.0 [
21]. Phylogenetic trees were reconstructed using maximum likelihood (ML) and Bayesian inference (BI) methods. ML tree was reconstructed using Tamura 3-parameter model [
22] with 1000 bootstrap replicates by MEGA11. BI tree was reconstructed using the same model (lset nst=2 rates=equal) by MrBayes ver. 3.2.7a [
23]. Bayesian posterior probabilities were estimated using the Metropolis-coupled Markov Chain Monte Carlo method with four chains (three heated and one cold) with the temperature set to 0.2. Chains were run for 1,000,000 generations and sampled every 1000 trees, with 25% of the trees discarded as burn-in. The resulting phylogenies were processed by Figtree ver. 1.4.4 [
24].
3. Results
3.1. Male-specificity of the markers
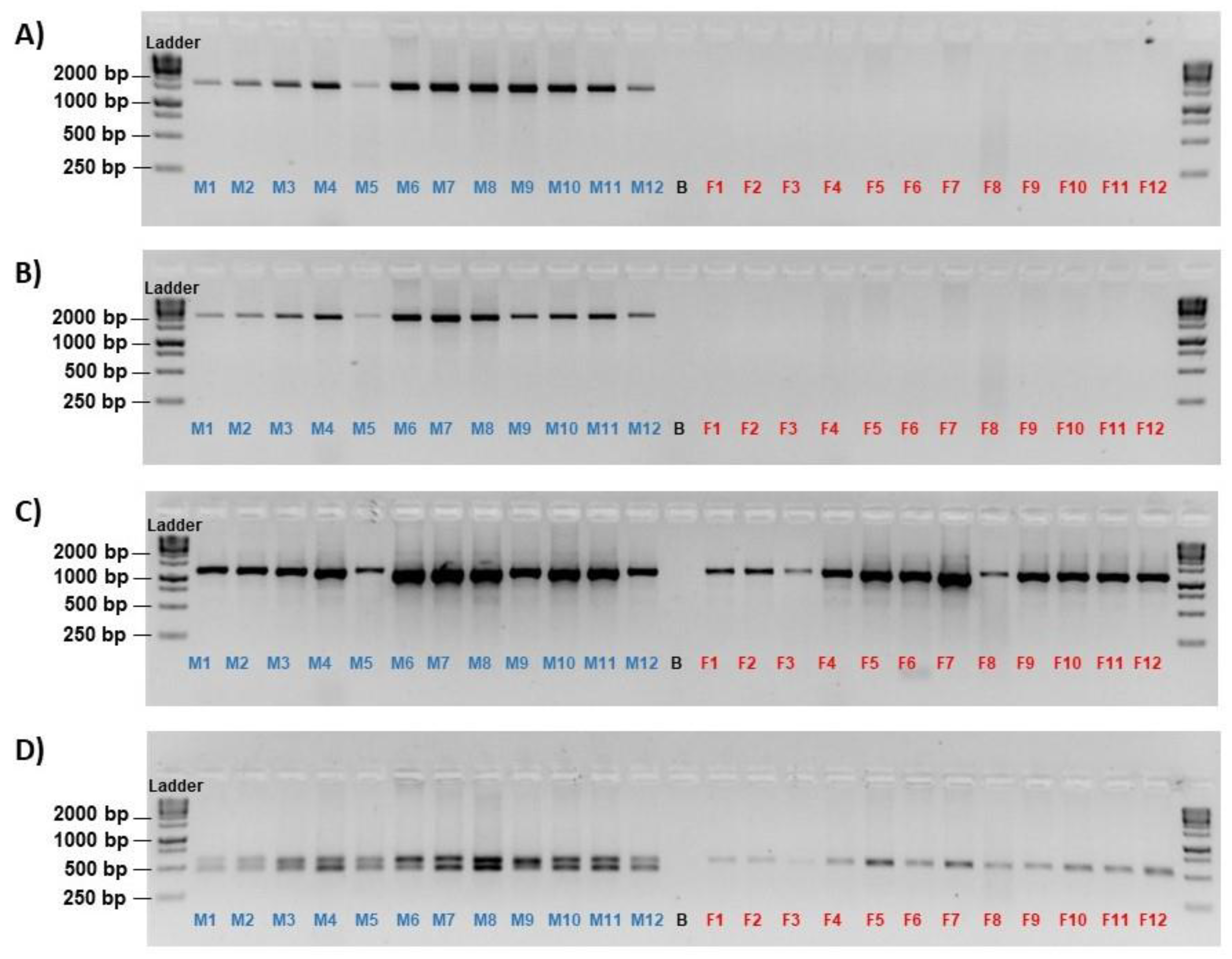
On the 12 males and 12 females initially tested, only primer set AMH1_E3I6 and AMH2_I6E4 amplified the anticipated 1,410 bp and 1,804 bp amplicons, respectively, in the males exclusively, while primer set AMH3_E3I6 amplified an 1,136 bp band in both sexes (
Figure 1A–C). The primer set AMH1+3_E6I6 amplified two bands of the anticipated sizes only in males, designated as the upper (566 bp) and the lower (481 bp) band, but amplified one band with a similar size to the upper band in females (
Figure 1D).
Every generated consensus sequence from the amplicons has the highest pairwise percentage identity with their expected paralogue, with the amplicon of AMH1_E3I6 matching 99.6% with amh1, AMH2_I6E4 matching 99.5% with amh2, AMH3_E3I6 matching 99.2% for male and 99.4% for female with amh3, AMH1+3_E6I6 matching 94.1% for the male upper band with amh3, 99.0% for the male lower band with amh1, and 85.8% for the female fragment with amh3. The consensus DNA sequences of these amplicons are available in
Figure S1.
3.2. Accuracy of the male-specific markers
Based on the preliminary tests, the primer sets that amplified male-specific amplicons (AMH1_E3I6, AMH2_I6E4, and AMH1+3_E6I6) were further assessed with the DNA samples of 67 male and 76 female lumpfish from different sources. The three chosen primer sets all have correctly identified 65 out of 67 males (97.0%) and 76 out of 76 females (100%). Note that the two samples which were identified as male but exhibit genotypes like the female are identical throughout the three tests. Summary of the results is available in
Table S1.
The genotype exhibited by the three primer sets and phenotypic sex are significantly associated, based on the chi-square test (P < 2.2 × 10-16).
3.3. Determination of the male-specific region length
Additional primers for estimating the size of male-specific region were based on the flanking regions upstream of amh1 and downstream of amh2 (
Table 2).
On the six males and the six females utilized for this purpose, only primer set 4K_Up exhibited male-specific amplification among the upstream primer sets, suggesting the extension of the male specific region upstream is at least 4 kb from amh1. Similarly, only the primer set 4K_Down, and 14K_Down exhibited male-specific amplifications among the downstream primer sets, suggesting the extension of the male specific region downstream is at least 14 kb from amh2. The gel images can be found in
Figures S2–S3.
Based on these results and the reference genome, the male-specific region of lumpfish was estimated to cover at least 26.4 kb but no more than 30.4 kb.
3.4. Conservation of the candidate sex determining gene in Cyclopteridae
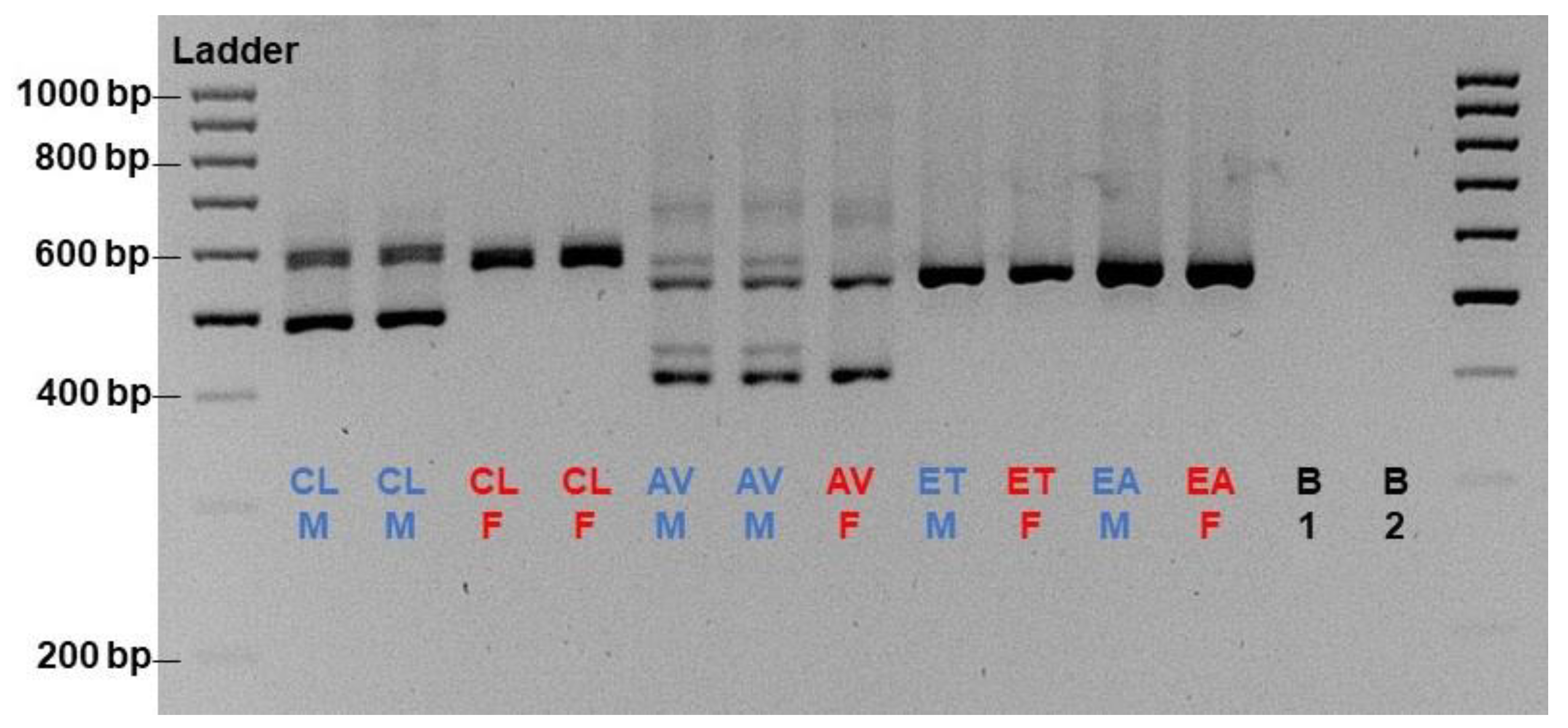
By utilizing the primer set AMH1+3_E6I6, only A. ventricosus showed a male-specific pattern on the gel, other than lumpfish, among the available Cyclopteridae samples (
Figure 2). Though, the size and the number of male-specific fragments are different between lumpfish and A. ventricosus. The DNA sequences of the amplicon from the non-lumpfish Cyclopteridae which were successfully sequenced are available in
Figure S1.
3.5. Phylogeny of the amh markers in Cyclopteridae
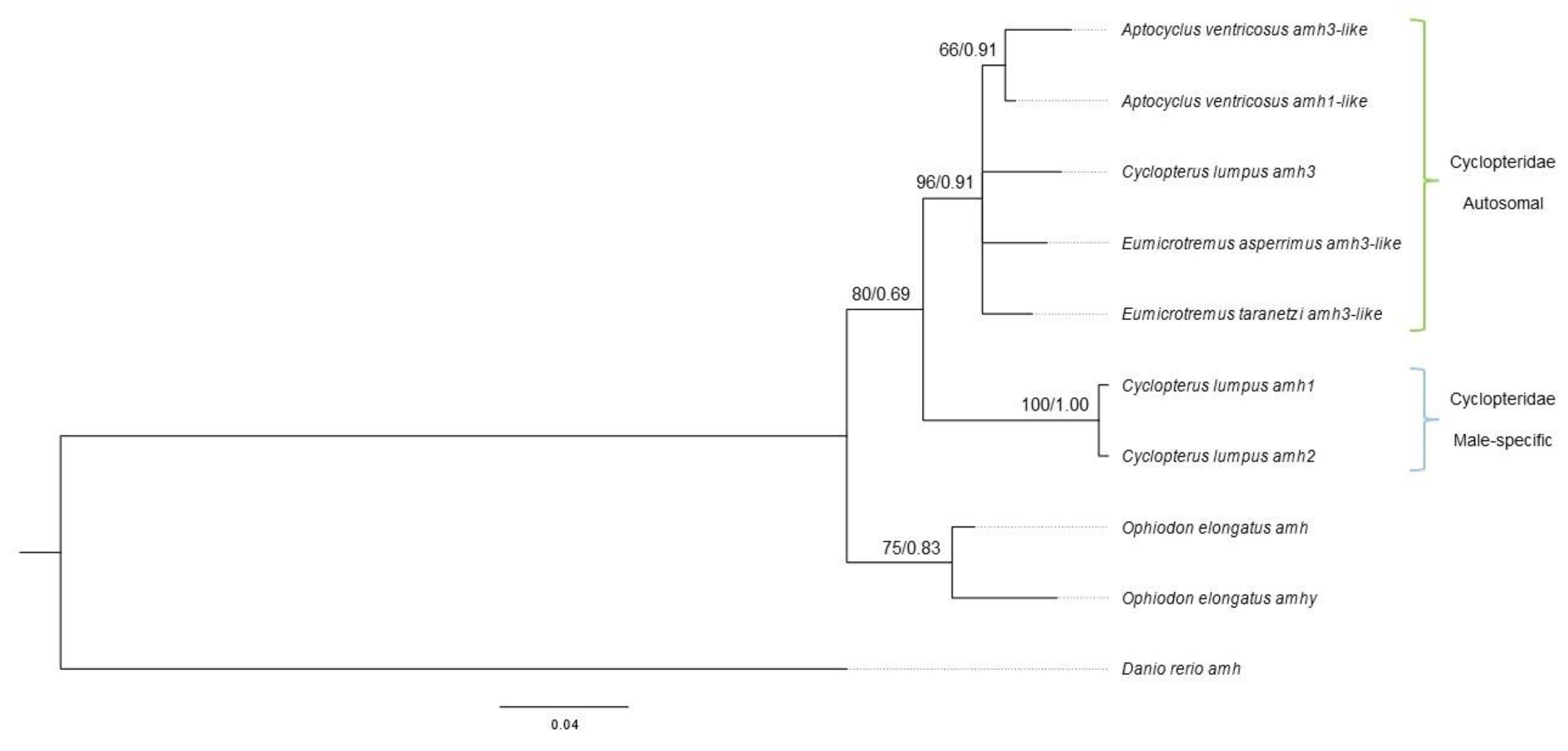
After gap deletion, a final alignment of 200 sites with 100 variable sites (50%) and 27 parsimony-informative sites (13.5%) was generated from ten amh sequences of Scorpaeniformes fishes and zebrafish. The ML tree and BI tree reconstructed from this alignment share a similar topology, with the Cyclopteridae autosomal amh and male-specific amh segregated into two clades (bootstrap support = 80%, posterior probability = 0.69), so the topology of the BI consensus tree with the bootstrap values from the ML analysis was chosen to represent both analyses (
Figure 3). These topologies suggest that the male-specific amh of lumpfish emerged after the split of Cyclopteroidea and Hexagrammoidea and before the split of Aptocyclus, Cyclopterus, and Eumicrotremus lineages, rather than independently arose in the lineage of lumpfish.
4. Discussion
Our findings suggest that amh1 and amh2 are found only in male lumpfish, while amh3 seems to be of an autosomal nature. The amh1- and amh2-based PCR tests exhibited consistent trends between genotypic and phenotypic sex, reaching an agreement in over 97% of the samples across the three assays. Furthermore, the DNA regions adjacent to amh1 and amh2 also exhibited male-specific amplifications. We have also observed male-specific amh in another closely related species, A. ventricosus, albeit seemingly different from lumpfish. In addition, the confirmation of a male-specific marker in lumpfish suggested by our results agrees with previous research on lumpfish sex GWAS, where a male-specific region in chromosome 13 was described [
10,
25]. Based on these results and previous studies, we can confirm that amh1 and amh2 can function as male-specific molecular markers for sex identification in lumpfish.
4.1. Application in aquaculture
Lumpfish lack pronounced sexual dimorphism before maturity. Once they mature, sex identification by visual inspection is possible, as the mature lumpfish exhibit sexual dichromatism during the spawning season [
9]. However, lumpfish may require five to eight years before they mature and display this dimorphism [
26]. This sex identification method for lumpfish is ill-suited for aquaculture production due to the time and resources required to hold the potential broodstock until it reaches maturity. Sex identification is essential for managing the sex ratio of broodstock to develop successful breeding schemes for offspring production, therefore, the ability to identify the sex of lumpfish at early stages will facilitate more efficient production. Genetic sex identification, like the PCR-based techniques applied in this study, is a powerful alternative for sex identification, and ultimately improve the management of the sex ratio within the breeding population [
27].
PCR-based sex identification has been developed in many important aquaculture species with identified SD genes, such as in various salmonids [
28], with an aim to improve selective breeding programs. Such molecular tools are also desirable in certain species with production-related sexual dimorphism, such as the large yellow croaker (Larimichthys crocea) for which the mono-sex female production is favoured [
29]. Accordingly, the developed molecular tools could also assist in the mono-sex production of lumpfish, although, a previous study did not find any significant difference in grazing efficacy between sexes in juvenile lumpfish [
30] but the production of mono-sex female lumpfish could potentially boost the profitability of lumpfish culture by the additional by-product as the roe of lumpfish is a delicacy in many countries, such as France, Germany, and the USA [
31].
Similar to the accuracy of our sex identification tools, PCR-based sex identification techniques using male-specific amh duplicates in other teleost species with similar homologues have been demonstrated to identify the sex with high accuracy (97.5-100%), given both wild-caught and lab-reared samples raised in controlled environments [
12,
13,
14]. The consistent presence of male-specific amh in the male of many species might be due to the role of male-specific amh as a male SD gene or its involvement in testis development. Therefore, our molecular tools which utilize male-specific amh paralogues of lumpfish should be suitable for genetic sex identification to improve the efficiency of lumpfish breeding program.
4.2. Implication in the sex-determination system of lumpfish
The agreement between genotypic and phenotypic sex in our PCR tests supports that lumpfish have a genetic sex-determination system that is unlikely to be majorly influenced by environmental factors.
A male-specific region in the lumpfish genome suggests a XX/XY sex-determination system, as supported by the previous GWAS analysis [
10,
25] and observation from a feminisation experiment [
32]. Our study has also identified amh1 and amh2 as the male-specific amh paralogues, which may function as the SD gene in lumpfish.
Presently, male-specific amh duplicates have been found in various teleost species, including the Odontesthes clade of the order Atheriniformes [
33], notably Patagonian silverside (O. hatcheri) [
13] and Argentinian silverside (O. bonariensis) [
34], Nile tilapia (Oreochromis niloticus) [
35,
36,
37], lingcod (Ophiodon elongatus) [
12], cobaltcap silverside (Hypoatherina tsurugae) [
38], northern pike (Esox lucius) [
14], Korean rockfish (Sebastes schlegelii) [
39], and the Gasterosteus clade of the family Gasterosteidae [
40,
41]; some of these homologs have also been recognized as the male SD gene. Among these species, Korean rockfish, belonging to the order Scorpaeniformes, is the closest species to lumpfish in evolutionary terms with amh as their SD gene. Even considering that, our phylogenetic analyses suggest that the male-specific amh in lumpfish is more likely to have emerged independently in the Cyclopteridae lineage rather than being a shared ancestral trait in the Scorpaeniformes lineage. In addition, our phylogeny shares a similar topology to another neighbour-joining phylogeny reconstructed with amh sequences from lumpfish and their related species [
10], further supporting this theory.
Phylogenetic analyses of amh sequences from other teleosts also indicated that most sex-determining amh genes emerged independently in clades of closely related species as a form of convergent evolution rather than due to shared ancestry of teleost [
14,
34,
39]. One of the theories for these independent recruitments is that amh encodes a protein belonging to the Tgf-β family, which is composed of versatile and flexible signalling networks that interact with the gonad development cascade in vertebrates [
42]. Due to this connectivity, the male-specific amh paralogues are prime candidates for the SD gene in lumpfish.
Most male-specific amh paralogues of teleost species were theorised to arise as an SD gene through neofunctionalization via the gene duplication (GD) model [
13,
14,
39]. This model is well-described in the SD gene of Japanese medaka, dmrt1bY, which was proposed to emerge via the events of gene duplication from its ancestral gene, translocation to a new chromosome, and neo-functionalisation by acquiring a pre-existing cis-regulatory element necessary for rewiring its expression for the sex determination through a transposable element [
43]. Following this model, the male-specific amh duplicates in lumpfish could be derived from the autosomal amh3, translocated to their current sex chromosome, and neo-functionalised into an SD gene by acquiring a novel expression pattern, mutation, or a combination of both that can induce gonad development.
Contrary to our deduction, the previous research [
10] proposed amh3 as the potential SD gene, as it is the only amh transcript detected in the testis tissue of mature male lumpfish. However, this observation could be explained by differences in expression patterns between the matures and juveniles, as observed in male Japanese medaka which exhibited a decline in their SD gene expression as they matured [
44]. Further expression and functional analyses are recommended to determine the level of expression of both amh1 and amh2 during the different development stages of both males and females. If one of the amh functions as an SD gene in lumpfish, it is expected to exhibit male-specific expression before the sexual differentiation of the bipotential gonadal primordium that is requisite and adequate for testis development [
13,
14,
39]. Further understanding of the sex-determination mechanism in lumpfish is expected to improve the management of the stock’s sex ratio and fill in the knowledge gap in the sex-determination system of teleost, which is still limited to a few species.
4.3. Implication in the sex chromosome evolution of lumpfish
We have estimated that the male-specific region of lumpfish covers approximately 26-30 kb, which is considerably minuscule compared to the length of the putative sex chromosome. Nevertheless, a study on Korean rockfish has estimated the male-specific region in this species to be approximately 5 kb [
39], providing evidence of another small genomic region controlling such an important developmental process. Assuming that the male-specific amh is the SD gene of lumpfish, the relatively small male-specific region and the apparent homomorphic state of their chromosomes [
45] suggest that the sex chromosome in lumpfish is nascent. Worthwhile to mention that we have designed and tested multiple primers upstream and downstream from the amh region apart from those presented in our methods, but PCR amplification of most primers was not possible for an unknown reason. We suspected potential assembly errors within this region in the currently available genome are the cause therefore, the real male-specific region could be larger or shorter than we have estimated according to the currently available coordinates. Future studies should attempt to characterise the male-specific region using other sequencing methods such as Long-read Oxford Nanopore sequencing [
46], or new assembly algorithms that could provide a more accurate sequence and will ultimately allow a more comprehensive analysis between male and female lumpfish genome.
The nascent sex chromosome could be explained by the “high turnover” theory, in which a new SD gene frequently emerges on an autosome, replacing the ancestral sex chromosome, and preventing it from further degradation [
47]. This phenomenon can be observed in the genus Oryzias which contains various SD genes within the clade [
48]. However, our phylogenetic analyses disagree with this theory, as the consensus phylogeny implies that the male-specific amh of lumpfish might have emerged early in Cyclopteridae lineage and were retained in the lumpfish lineage. We may require additional samples from other members of the family, informative sites, and genomic analysis to better elucidate the evolution of amh in Cyclopteridae. Nevertheless, the species utilized in our study have been identified as the closest ones to lumpfish within the family, therefore, our phylogenetic analyses should reflect the actual evolutionary pathway of amh in Cyclopteridae family.
Another possible theory is the “jumping sex locus” theory, in which the existing SD gene is translocated to a new autosome by transposable elements, allowing a replacement of the ancestral sex chromosome without overhauling the current sex-determination mechanism, as observed in sdY of many salmonids [
28,
49]. Alternatively, the SD gene may also remain in the same sex chromosome after the latest turnover event without chromosomal degradation until the present day. As observed in the Northern pike, which has conserved their male-specific amh as their SD gene in the same chromosome for at least 45 million years yet still retains homomorphic sex chromosomes, possibly through the preservation mechanism of direct repeats flanking the SD locus [
42].
It would be informative to analyse the male-specific region and its flanking region to identify possible transposable elements that may facilitate translocation or potential novel mechanisms which prevent chromosomal degradation. With such a relatively small male-specific region, further studies on the sex chromosome of lumpfish may provide more insight into the sex chromosome evolution mechanism in teleost.
5. Conclusions
Our PCR amplification results suggest that amh1 and amh2 paralogues are male-specific, while amh3 is autosomal. PCR amplification in 143 samples showed significant associations between the male phenotype to the presence of both amh1 and amh2, with more than 97% of the males amplifying fragments from both genes while every female did not. Further analysis of the flanking regions adjacent to amh1 and amh2 revealed that the male-specific region spans approximately 26 kb. These results support that amh1 and amh2 can function as male-specific molecular markers for genetic sex identification in lumpfish. The development of this accurate PCR test could benefit the development of lumpfish breeding programs, which would lead to less reliance on wild lumpfish. Additionally, these findings may also benefit future studies regarding the lumpfish’s sex-determination system and sex chromosome evolution.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: Detailed record of the lumpfish (Cyclopterus lumpus) samples in this study, Figure S1: DNA sequences of the amplicons sequenced in this study, Figure S2: PCR amplification results of the primer set utilised for estimating the upstream coverage of the male-specific region in lumpfish, Figure S3: PCR amplification results of the primer set utilised for estimating the downstream coverage of the male-specific region in lumpfish.
Author Contributions
Conceptualisation: A.P.G and K.C.; Data curation: K.C.; A.P.G, and A.D.; Formal analysis: K.C. and A.P.G.; Funding acquisition: A.P.G and A.D. ; Investigation: K.C.; P.M.; A.P.G and J.P.; Methodology: K.C.; P.P. and A.P.G; Writing - original draft: K.C. and A.P.G.; Writing - review & editing: A.P.G, K.C. and P.P.
Funding
This research was funded by the AquaLeap project of the Biotechnology and Biological Sciences Research Council, grant numbers BB/S004416/1.
Institutional Review Board Statement
The study was conducted following the UK Animals (Scientific Procedures) Act 1986 (revised 2013) and approved by the Animal Welfare and Ethical Review Body of the University of Stirling (AWERB 2022 7735 636).
Acknowledgments
We would like to express our gratitude to Dr Kai Yoshiaki of the University of Kyoto who graciously shared tissue samples of Aptocyclus ventricosus, Eumicrotremus taranetzi, and E. asperrimus for this research.
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Brooker, A.J.; Skern-Mauritzen, R.; Bron, J.E. (2018) Production, mortality, and infectivity of planktonic larval sea lice, Lepeophtheirus salmonis (Krøyer, 1837): current knowledge and implications for epidemiological modelling. ICES Journal of Marine Science, 75 (4), pp. 1214–1234. [CrossRef]
- Bolton-Warberg, M. (2018) An overview of cleaner fish use in Ireland. Journal of Fish Diseases, 41 (6), pp. 935-939. [CrossRef]
- Østevik, L.; Stormoen, M.; Evensen, Ø.; Xu, C.; Lie, K.; Nødtvedt, A.; Rodger, H.; Skagøy, A.; Manji, F.; Alarcón, M. (2022) Effects of thermal and mechanical delousing on gill health of farmed Atlantic salmon (Salmo salar L.). Aquaculture, 552, pp. 738019. [CrossRef]
- Overton, K.; Dempster, T.; Oppedal, F.; Kristiansen, T.S.; Gismervik, K.; Stien, L.H. (2019) Salmon lice treatments and salmon mortality in Norwegian aquaculture: a review. Reviews in Aquaculture, 11 (4), pp. 1398-1417. [CrossRef]
- Barisic, J.; Cannon, S.; Quinn, B. (2019) Cumulative impact of anti-sea lice treatment (azamethiphos) on health status of Rainbow trout (Oncorhynchus mykiss, Walbaum 1792) in aquaculture. Scientific Reports, 9 (1), pp. 16217. [CrossRef]
- Parsons, A.E.; Escobar-Lux, R.; Sævik, P.N.; Samuelsen, O.B.; Agnalt, A. (2020) The impact of anti-sea lice pesticides, azamethiphos and deltamethrin, on European lobster (Homarus gammarus) larvae in the Norwegian marine environment. Environmental Pollution, 264, pp. 114725. [CrossRef]
- Brooker, A.J.; Papadopoulou, A.; Gutierrez, C.; Rey, S.; Davie, A.; Migaud, H. (2018) Sustainable production and use of cleaner fish for the biological control of sea lice: recent advances and current challenges. Veterinary Record, 183 (12), pp. 383. [CrossRef]
- Powell, A.; Treasurer, J.W.; Pooley, C.L.; Keay, A.J.; Lloyd, R.; Imsland, A.K.; Garcia de Leaniz, C. (2018) Use of lumpfish for sea-lice control in salmon farming: challenges and opportunities. Reviews in Aquaculture, 10 (3), pp. 683-702. [CrossRef]
- Davenport, J.; Bradshaw, C. (1995) Observations on skin colour changes in juvenile lumpsuckers. Journal of Fish Biology, 47 (1), pp. 143-154. [CrossRef]
- Holborn, M.K.; Einfeldt, A.L.; Kess, T.; Duffy, S.J.; Messmer, A.M.; Langille, B.L.; Brachmann, M.K.; Gauthier, J.; Bentzen, P.; Knutsen, T.M.; et al. (2022) Reference genome of lumpfish Cyclopterus lumpus Linnaeus provides evidence of male heterogametic sex determination through the AMH pathway. Molecular Ecology Resources, 22 (4), pp. 1427-1439. [CrossRef]
- Patil, J.G.; Hinze, S.J. (2008) Simplex PCR assay for positive identification of genetic sex in the Japanese medaka, Oryzias latipes. Marine Biotechnology, 10 (6), pp. 641-644. [CrossRef]
- Rondeau, E.B.; Laurie, C.V.; Johnson, S.C.; Koop, B.F. (2016) A PCR assay detects a male-specific duplicated copy of Anti-Müllerian hormone (amh) in the lingcod (Ophiodon elongatus). BMC Research Notes, 9 (1), pp. 230. [CrossRef]
- Hattori, R.S.; Murai, Y.; Oura, M.; Masuda, S.; Majhi, S.K.; Sakamoto, T.; Fernandino, J.I.; Somoza, G.M.; Yokota, M.; Strüssmann, C.A. (2012) A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proceedings of the National Academy of Sciences, 109 (8), pp. 2955-2959. [CrossRef]
- Pan, Q.; Feron, R.; Yano, A.; Guyomard, R.; Jouanno, E.; Vigouroux, E.; Wen, M.; Busnel, J.; Bobe, J.; Concordet, J.; et al. (2019) Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLOS Genetics, 15 (8), pp. e1008013. [CrossRef]
- Blanquer, A. (1990) Phylogéographie intraspécifique d'un poisson marin, le flet Platichthys flesus L. (Heterosomata) : polymorphisme des marqueurs nucléaires et mitochondriaux. PhD, University of Montpellier.
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. (2022) Ensembl 2022. Nucleic Acids Research, 50 (D1), pp. D988-D995. [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. (2012) Primer3—new capabilities and interfaces. Nucleic Acids Research, 40 (15), pp. e115. [CrossRef]
- R Core Team. (2022) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 6 September 2022).
- RStudio Team. (2022) RStudio: integrated development for R. RStudio, Inc.; Boston, MA. Available online: http://www.rstudio.com/ (accessed on 6 September 2022).
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32 (5), pp. 1792-1797. [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution, 38 (7), pp. 3022-3027. [CrossRef]
- Tamura, K. (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Molecular Biology and Evolution, 9 (4), pp. 678-687. [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology, 61 (3), pp. 539-542. [CrossRef]
- Rambaut, A. (2018) FigTree v1.4.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 6 February 2023).
- Gutierrez, A.P.; Counter Selly, S.-L.; Taggart, J.B.; Kokkinias, P.; Cavrois-Rogacki, T.; Jimenez Fernandez, F.; Migaud, H.; Lein, I.; Davie, A.; Bekaert, M. (2023) Development of genomic markers associated to production traits in lumpfish (Cyclopterus lumpus). bioRxiv, pp. 2023.05.10.540148. [CrossRef]
- Davenport, J. (1985) Synopsis of biological data on the lumpsucker, Cyclopterus lumpus (Linnaeus, 1758). Rome, Italy: Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/3/ap950e/ap950e.pdf (accessed on 6 May 2022).
- Martínez, P.; Viñas, A.M.; Sánchez, L.; Díaz, N.; Ribas, L.; Piferrer, F. (2014) Genetic architecture of sex determination in fish: applications to sex ratio control in aquaculture. Frontiers in Genetics, 5, pp. 340. [CrossRef]
- Yano, A.; Nicol, B.; Jouanno, E.; Quillet, E.; Fostier, A.; Guyomard, R.; Guiguen, Y. (2013) The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evolutionary Applications, 6 (3), pp. 486-496. [CrossRef]
- Lin, A.; Xiao, S.; Xu, S.; Ye, K.; Lin, X.; Sun, S.; Wang, Z. (2017) Identification of a male-specific DNA marker in the large yellow croaker (Larimichthys crocea). Aquaculture, 480, pp. 116-122. [CrossRef]
- Imsland, A.K.D.; Reynolds, P.; Hangstad, T.A.; Kapari, L.; Maduna, S.N.; Hagen, S.B.; Jónsdóttir, Ó.D.B.; Spetland, F.; Lindberg, K.S. (2021) Quantification of grazing efficacy, growth and health score of different lumpfish (Cyclopterus lumpus L.) families: Possible size and gender effects. Aquaculture, 530, pp. 735925. [CrossRef]
- Johannesson, J.; (2006) Lumpfish caviar: from vessel to consumer. Rome, Italy: Food & Agriculture Organization of the United Nations. Available online: https://www.fao.org/3/a0685e/a0685e.pdf (accessed on 25 July 2022).
- Martin-Robichaud, D.; Peterson, R.H.; Benfey, T.J.; Crim, L.W. (1994) Direct feminization of lumpfish (Cyclopterus lumpus L.) using 17β-oestradiol-enriched Artemia as food. Aquaculture, 123 (1), pp. 137-151. [CrossRef]
- Hattori, R.S.; Somoza, G.M.; Fernandino, J.I.; Colautti, D.C.; Miyoshi, K.; Gong, Z.; Yamamoto, Y.; Strüssmann, C.A. (2019) The duplicated Y-specific amhy gene is conserved and linked to maleness in Silversides of the genus Odontesthes. Genes, 10 (9), pp. 679. [CrossRef]
- Yamamoto, Y.; Zhang, Y.; Sarida, M.; Hattori, R.S.; Strüssmann, C.A. (2014) Coexistence of genotypic and temperature-dependent sex determination in pejerrey Odontesthes bonariensis. Plos One, 9 (7), pp. e102574. [CrossRef]
- Eshel, O.; Shirak, A.; Dor, L.; Band, M.; Zak, T.; Markovich-Gordon, M.; Chalifa-Caspi, V.; Feldmesser, E.; Weller, J.I.; Seroussi, E.; et al. (2014) Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics, 15 (1), pp. 774. [CrossRef]
- Li, M.; Sun, Y.; Zhao, J.; Shi, H.; Zeng, S.; Ye, K.; Jiang, D.; Zhou, L.; Sun, L.; Tao, W.; et al. (2015) A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y Chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLOS Genetics, 11 (11), pp. e1005678. [CrossRef]
- Liu, X.; Dai, S.; Wu, J.; Wei, X.; Zhou, X.; Chen, M.; Tan, D.; Pu, D.; Li, M.; Wang, D. (2022) Roles of anti-Müllerian hormone and its duplicates in sex determination and germ cell proliferation of Nile tilapia. Genetics, 220 (3), pp. iyab237. [CrossRef]
- Bej, D.K.; Miyoshi, K.; Hattori, R.S.; Strüssmann, C.A.; Yamamoto, Y. (2017) A duplicated, truncated amh gene is involved in male sex determination in an Old World silverside. G3 Genes|Genomes|Genetics, 7 (8), pp. 2489-2495. [CrossRef]
- Song, W.; Xie, Y.; Sun, M.; Li, X.; Fitzpatrick, C.K.; Vaux, F.; O'Malley, K.G.; Zhang, Q.; Qi, J.; He, Y. (2021) A duplicated amh is the master sex-determining gene for Sebastes rockfish in the Northwest Pacific. Open Biology, 11 (7), pp. 210063. [CrossRef]
- Sardell, J.M.; Josephson, M.P.; Dalziel, A.C.; Peichel, C.L.; Kirkpatrick, M. (2021) Heterogeneous histories of recombination suppression on stickleback sex chromosomes. Molecular Biology and Evolution, 38 (10), pp. 4403-4418. [CrossRef]
- Peichel, C.L.; McCann, S.R.; Ross, J.A.; Naftaly, A.F.S.; Urton, J.R.; Cech, J.N.; Grimwood, J.; Schmutz, J.; Myers, R.M.; Kingsley, D.M. (2020) Assembly of the threespine stickleback Y chromosome reveals convergent signatures of sex chromosome evolution. Genome Biology, 21 (1), pp. 177. [CrossRef]
- Pan, Q.; Kay, T.; Depincé, A.; Adolfi, M.; Schartl, M.; Guiguen, Y.; Herpin, A. (2021) Evolution of master sex determiners: TGF-β signalling pathways at regulatory crossroads. Philosophical Transactions of the Royal Society B: Biological Sciences, 376 (1832), pp. 20200091. [CrossRef]
- Herpin, A.; Braasch, I.; Kraeussling, M.; Schmidt, C.; Thoma, E.C.; Nakamura, S.; Tanaka, M.; Schartl, M. (2010) Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLOS Genetics, 6 (2), pp. e1000844. [CrossRef]
- Hornung, U.; Herpin, A.; Schartl, M. (2007) Expression of the male determining gene dmrt1bY and its autosomal coorthologue dmrt1a in medaka. Sexual Development, 1 (3), pp. 197-206. [CrossRef]
- Li, M.F.; Clyburne, S. (1977) New cell line from the marine lumpfish, Cyclopterus lumpus. Journal of the Fisheries Research Board of Canada, 34 (1), pp. 134-139. [CrossRef]
- Feron, R.; Zahm, M.; Cabau, C.; Klopp, C.; Roques, C.; Bouchez, O.; Eché, C.; Valière, S.; Donnadieu, C.; Haffray, P.; et al. (2020) Characterization of a Y-specific duplication/insertion of the anti-Mullerian hormone type II receptor gene based on a chromosome-scale genome assembly of yellow perch, Perca flavescens. Molecular Ecology Resources, 20 (2), pp. 531-543. [CrossRef]
- Meisel, R.P. (2020) Evolution of sex determination and sex chromosomes: a novel alternative paradigm. BioEssays, 42 (9), pp. 1900212. [CrossRef]
- Myosho, T.; Takehana, Y.; Hamaguchi, S.; Sakaizumi, M. (2015) Turnover of sex chromosomes in Celebensis group medaka fishes. G3 Genes|Genomes|Genetics, 5 (12), pp. 2685-2691. [CrossRef]
- Woram, R.; Gharbi, K.; Sakamoto, T.; Høyheim, B.; Holm, L.; Naish, K.; Mcgowan, C.; Ferguson, M.; Phillips, R.; Stein, J.; et al. (2003) Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Research, 13 (2), pp. 272-80. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).