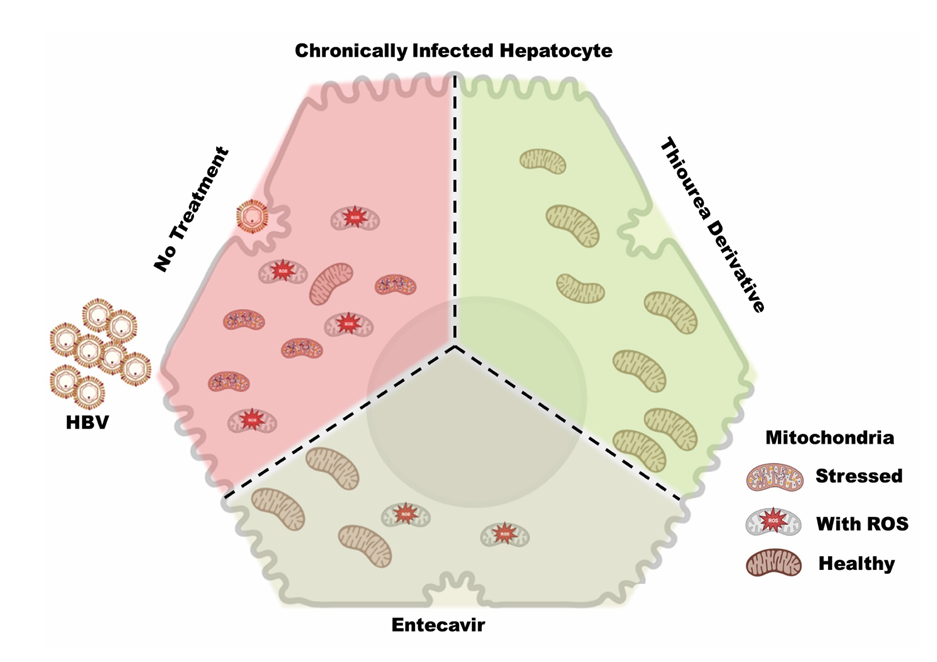
1. Introduction
Chronic hepatitis B (CHB) infection is caused by Hepatitis B Virus (HBV) that lasts for more than six months and leading to long-term liver disease. Currently, 257 million people around the world had a CHB infection [
1,
2]. CHB often leads to major problems with the liver, such as cirrhosis, liver failure, and liver cancer. As the illness persists longer, there is more liver inflammation and damage, with likelihood of aggravation of these issues [
3]. CHB can also alter mitochondrial and endoplasmic reticulum (ER) functions, which can contribute to liver damage and disease progression [
4,
5]. Patients with CHB show higher levels of mitochondrial DNA (mtDNA) in their serum than healthy, indicating increased mitochondrial dysfunction and damage [
6]. Alterations in mitochondrial morphology, and their membrane potential, metabolism, and biogenesis have been reported in the HBV-infected hepatocytes [
7,
8]. CHB has also been linked to ER stress and its response was found to be activated in liver biopsies from patients with persistent HBV infection, and this was linked to increased liver fibrosis and inflammation[
9,
10]. ER stress and dysfunction of mitochondria are associated to the progression to cirrhosis and end stage Hepatocellular carcinoma (HCC) [
11]. Several factors have been implicated in the development of HBV infection, including the HBx oncoproteins, immune-mediated death of HBV-infected hepatocytes, and liver regeneration [
12]. HBx regulates viral replication, viral and host genes transcription, various cell signaling activities, ubiquitin-proteasome machinery, cell cycle progression, and programmed cell deaths etc [
13].
The regulatory protein HBx, which plays an essential role in the viral life cycle, is encoded by the 0.7 kb RNA of HBV[
14]. HBV is a member of the Hepadnaviridae family and has a partially double-stranded DNA genome of 3.2 kb. Its genome contains four open reading frames (ORFs), which encode 3.5, 2.4, 2.1, and 0.7 kb RNAs and are translated into seven different proteins. The 3.5 kb RNA is responsible for encoding the core protein (HBcAg) and the pre-core protein (HBeAg), while the 2.4 and 2.1 kb RNAs encode the small (S), medium (M), and large (L) surface proteins. Previous research suggests that the regulatory protein HBx may be responsible for various effects attributed to HBV [
13]. HBx induced aberrant aggregation of mitochondrial components near the nucleus periphery triggered by HBx-driven p38 mitogen-activated protein kinase (MAPK), which may eventually lead to cell deaths [
15]. HBx damage mitochondria via interaction with voltage-dependent anion channel (HVDAC3), which is known as mitochondrial porins and forms pores in mitochondrial outer membranes[
16] and interaction alters mitochondrial transmembrane potential, resulting in the formation of reactive oxygen species (ROS) and cellular ATP/ADP ratio [
17]. Furthermore, HBx can suppress the expression of nucleus-encoded genes involved in mitochondrial beta-oxidation of fatty acids, resulting in a low level of cellular ATP due to energy source deficit. HBx-induced Raf-1 kinases pathway or the apoptosis regulator bcl-2-associated X protein (BAX) are translocated to mitochondria by HBx, resulting in hepatic cell proliferation or apoptosis, respectively [
6]. HBV-infected cells synthesize a lot of surface proteins in the endoplasmic reticulum (ER), disrupting ER homeostasis and causing ER stress [
10]. CHB patient’s biopsies reported with enlarge ER due to surface proteins (preS1 and preS2). ER stress or HBx can trigger autophagy [
18]. These findings imply that HBx-induced mitochondrial dysfunction and ER stress could be contributed to hepatocellular carcinoma pathogenesis, including proliferation, metastasis, and other features of carcinogenesis.
Biological networks have been used as a powerful approach for identifying novel dysregulated pathways driving molecular pathology. Weighted protein co-expression network analysis (WPCNA) is a multi-protein study that identifies groupings of proteins that are frequently altered and orchestrates them into modules without relying on a priori specified proteins sets or pathways. WPCNA is a useful technique for determining how networks of proteins and phenotypes are related [
19].
Thiourea and its derivatives have been used therapeutically as antioxidants[
20], anti-inflammatory drugs, anti-cancer [
21], fungicides [
22], as well as anti-viral [
23,
24]. Recently, a novel thiourea compound DSA-00 (IR-415) was identified by high-throughput screening of the Maybridge library that suppresses HBV replication like current antivirals [
23].
The present study is focused on the analysis of proteome alteration in cell (HepG2.2.15) under the influence of currently used anti-viral drugs in comparison to thiourea derivatives in order to identify key molecular changes linked to mitochondrial dysfunction and antiviral activity. We validated the proteomic finding of mitochondrial health using flow cytometry data with the ultimate goal of identifying whether repurposed thiourea derivatives could be improved mitochondrial and hepatocyte health.
2. Results
2.1. Alteration in protein profile following treatment with antiviral drug and thiourea derivatives
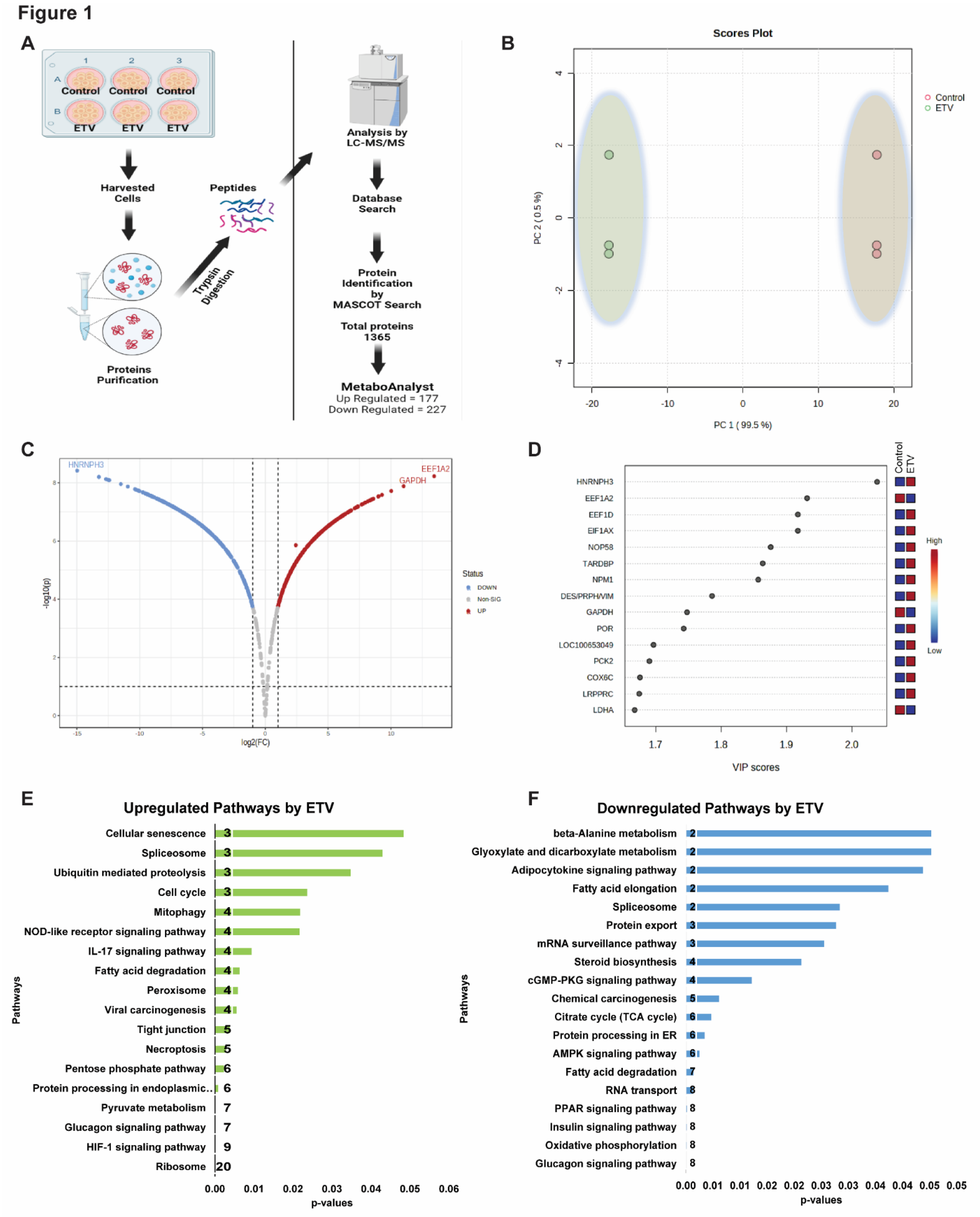
To investigate the alterations in protein profile during antiviral therapy in CHB infection, HepG2.2.15 cells were treated with standard antiviral drug Entecavir (ETV), which is a standard drug for treating CHB patients [
25]. A wide-scale comparison of the proteome ETV-treated and untreated cells showed complete separation of the samples based on their similarity (
Figure 1B). These was significant changes in the proteome of the ETV-treated group compared to the untreated group, with 607 proteins (274 UP and 246 Down-regulated) found to be significant (
Figure 1C, D). According to KEGG analysis [
26] the upregulated proteins were primarily linked to the ribosome, glycolysis, HIF-1 signaling, pyruvate metabolism, phagosome, pentose phosphate, necroptosis, tight junction, ferroptosis, peroxisome, fatty acid degradation, IL-17 signaling, mitophagy, and pathways related to the cell cycle (p<0.05) (
Figure 1E). On the other hand, the downregulated proteins were mostly linked to mRNA surveillance, TCA cycle, protein processing in the ER, oxidative phosphorylation, insulin signaling, glucagon signaling, and RNA transport (p<0.05) (
Figure 1F). Besides, a large proportion of the proteins that were significantly altered were host antiviral proteins, whereas the proteins involved in bioenergetics were downregulated. These findings suggest that novel approaches are needed to restore the bioenergetics function and modulate the host RNAi-mediated antiviral defense system.
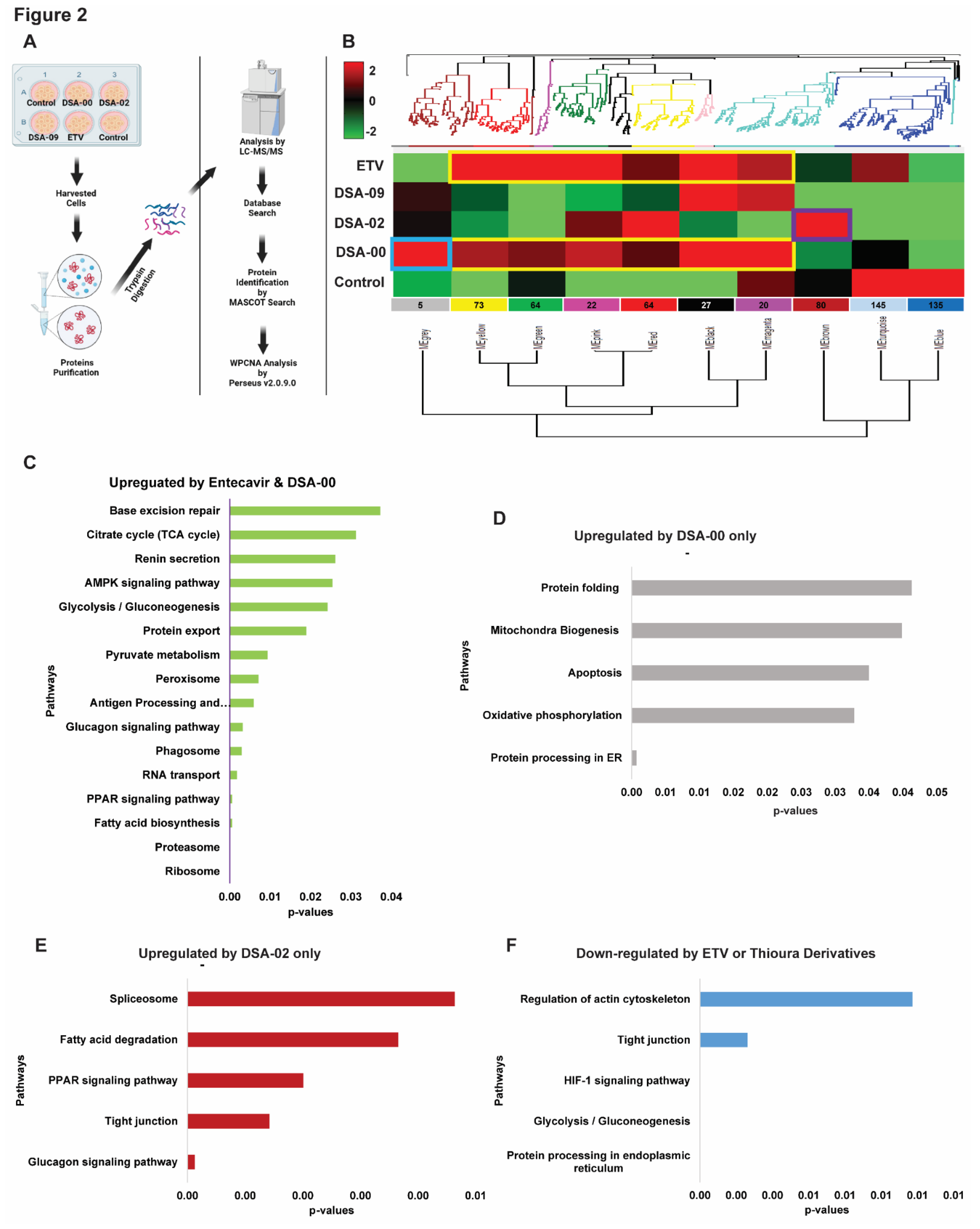
2.2. Thiourea derivatives induce specific protein modules to alter biological pathways
Though ETV and thiourea derivatives belonged to different classes of antivirals with distinct modes of action for suppressing HBV replication. ETV is known to have minimal impact on cellular energetics [
27] whereas, thiourea derivatives have shown potential in inhibiting HBV replication [
23] (Singh et al., 2023; Kumar et al., 2023 in communication). We used weighted protein co-expression network analysis (WPCNA) approach [
28] to identify proteins modules specific to DSA-00, DSA-02, DSA-09, ETV, and untreated cells for correlation patterns among proteins that may all work together in persistence of HBV infection (
Figure 2A). WPCNA revealed 10 clusters (modules), corresponding to a total of 636 strongly linked proteins in DSA-00, DSA-02, DSA-09, ETV, and untreated groups (
Figure 2B).
We got interested in modules whose expression pattern was similar to ETV and had some unique modules. DSA-00 has a similarity of approximately 70 percent with ETV (Black, Magenta, Red, Pink, Green, and Yellow) and also has unique module (Grey), but DSA-09 has two common modules with ETV and DSA-00 (Black and Magenta). The ETV, DSA-00, and DSA-09 shared modules with proteins involved in Ribosome, Proteasome, Fatty acid biosynthesis, PPAR signaling, and RNA transport pathways (
Figure 2C). DSA-02 showed unique Brown module proteins that were associated with biological pathways such as Glucagon signaling, tight junction, PPAR signaling, Fatty acid degradation, and Spliceosome (
Figure 2D). DSA-09 induced modules were common with DSA-00 and ETV modules. DSA-00 has a specific grey module other than common modules with ETV, DSA-02 and DSA-09. The grey module proteins were involved in protein processing in ER, Oxidative phosphorylation, Apoptosis, Mitochondria Biogenesis, and Protein folding pathways (
Figure 2E). Additionally, the untreated group has unique modules (Blue and Turquoise), and the proteins in these modules were linked with Regulation of actin cytoskeleton, Tight junction, HIF-1 signaling, Glycolysis/Gluconeogenesis, and protein processing in ER pathways (
Figure 2F). Based on these findings we conclude that, proteomic changes induced by DSA-00 are 70% similar to that seen under the treatment with ETV. In addition, our results highlight that thiourea derivatives modulate unique protein modules majorly linked to mitochondrial and ER activity suggesting that the thiourea derivatives not on provide anti-viral support but also rejuvenates the mitochondria and hepatocyte health in CHB.
2.3. Thiourea derivatives restore bioenergetics of exhausted hepatocytes
It is well established that HBx causes mitochondrial dysfunction and inflammation/injury in hepatocytes in CHB infection by disrupting mitochondrial respiration and membrane potential [
29]. Thiourea derivatives, on the other hands, have been shown antioxidant activity [
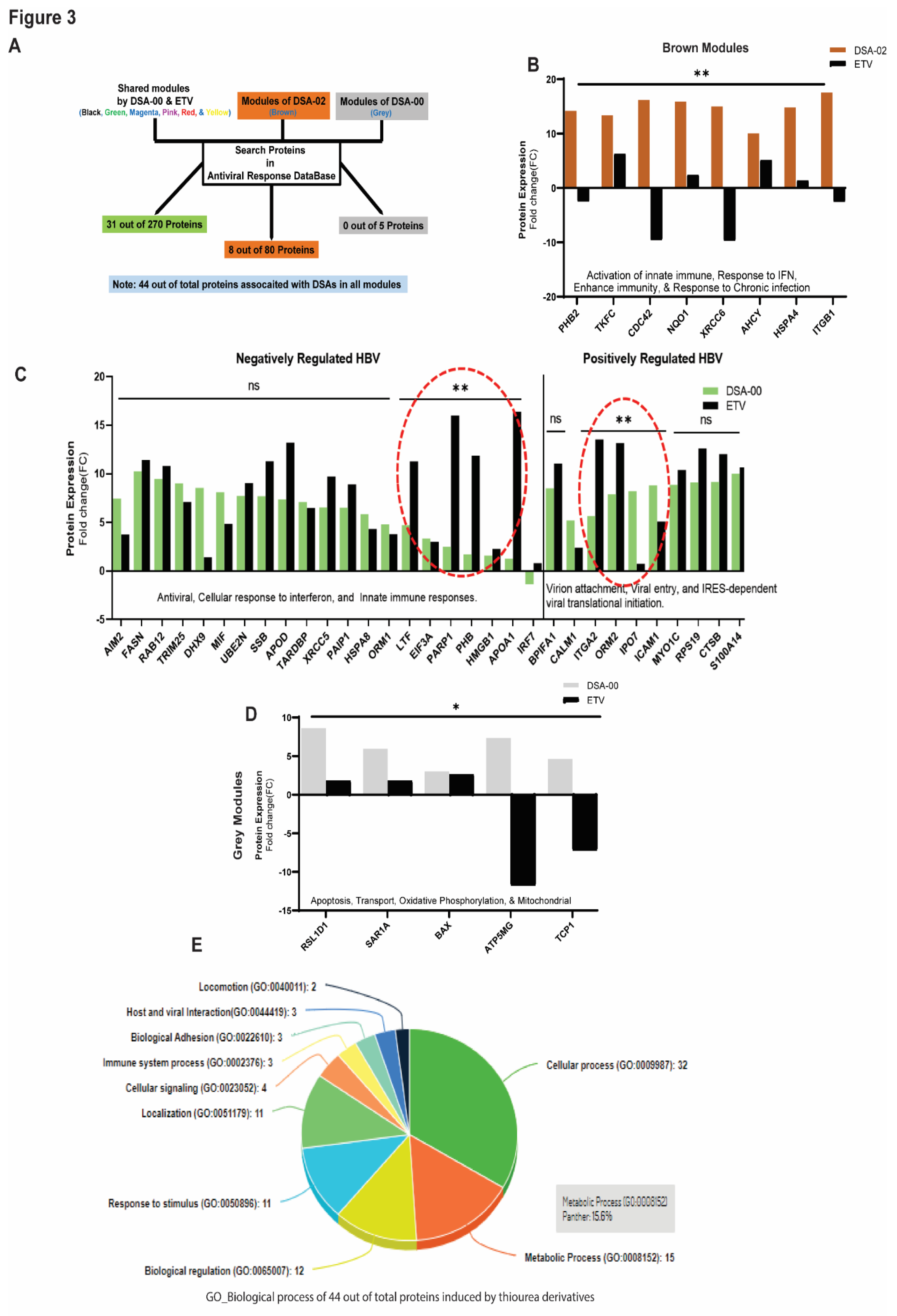
30] as well as bind to HBx and block the HBV-induced functions (Kumar et al., 2023 in communication). Based on these findings, we hypothesized that thiourea derivatives may have the potential to restore mitochondrial dysfunction in HBV-related liver diseases. To investigate this hypothesis, HepG2.2.15 cells were treated with DSA-00, DSA-02, DSA-09, and ETV. The proteome profile was subjected to WPCNA that led to the identification of several modules of proteins that were differentially expressed between the groups. Pathway analysis of these modules showed that 31 in DSA-00 and 8 proteins in DSA-02 were linked with the antiviral response (
Figure 3A). The brown module of DSA-02 contained eight proteins that were associated with enhanced host immunity and responsiveness to chronic infection (FIGURE 3B), whereas DSA-00 had twenty proteins providing negative regulation and ten associated with positive regulation to the HBV infection (
Figure 3C).
Furthermore, we observed that DSA-00 induced a unique grey module that contained only five proteins associated with mitochondrial functions like oxidative phosphorylation, membrane potential, and apoptosis (
Figure 3D). All antiviral response proteins that were up-regulated by DSA-00 (31 proteins), DSA-02 (8 proteins), and DSA-00 (5 proteins) were analyzed for their biological processes using the PANTHER classification system (v.14.0). This analysis revealed that nearly 50% of the proteins belonged to metabolic and cellular processes, suggesting that thiourea derivatives can restore bioenergetics in exhausted hepatocytes by targeting dysfunctional mitochondria (
Figure 3E).
2.4. Thiourea derivatives restore mitochondrial functions
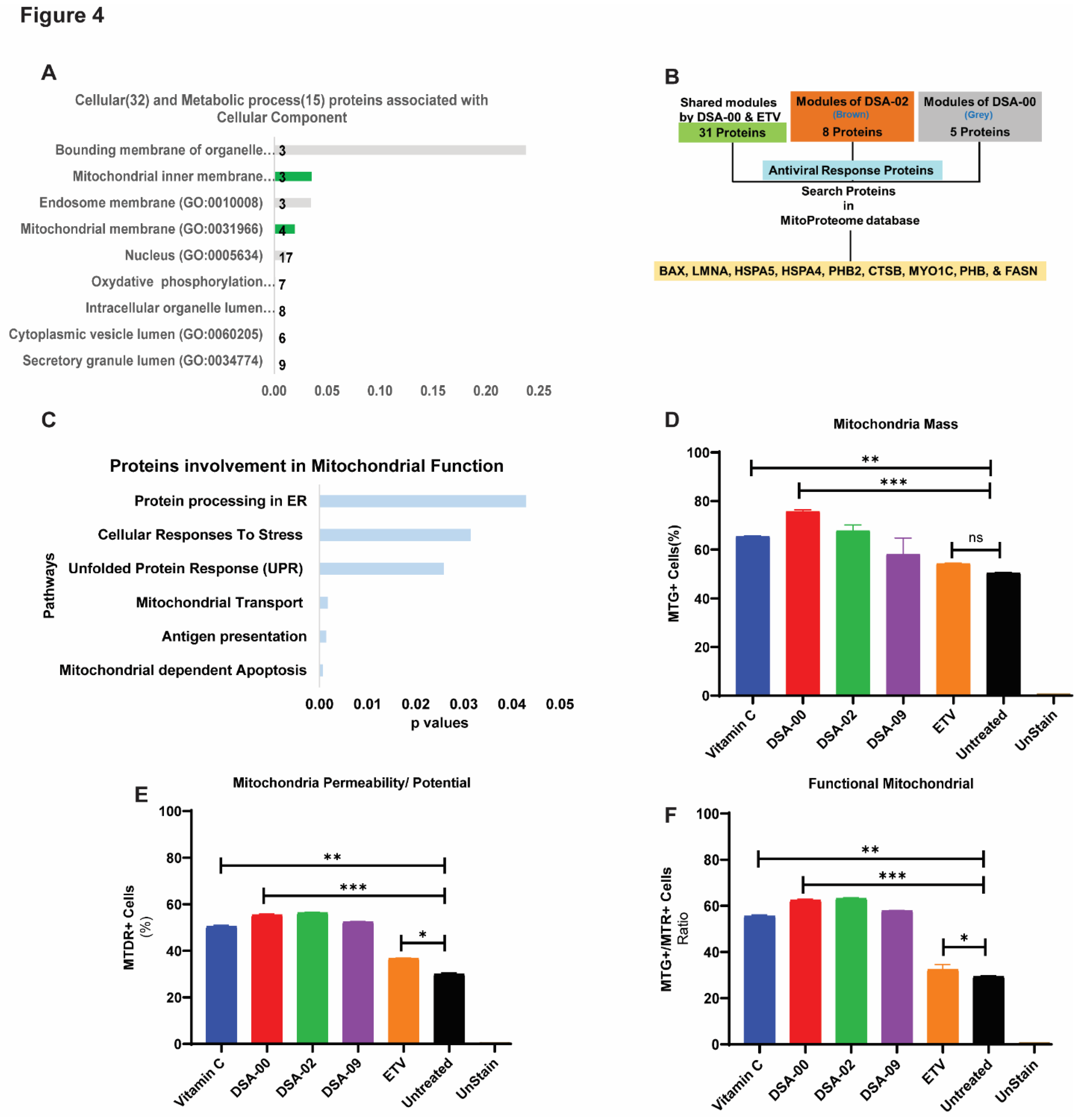
After analyzing the metabolic and cellular process associated proteins, we found that most of the proteins were linked to the mitochondria (
Figure 4A). Based on these results, we hypothesized that thiourea derivatives may have a role in mitochondrial-related functions. To validate these findings, we further blasted the proteins with the MitoProteome database [
31] and found nine proteins (BAX, LMNA, HSPA5, HSPA4, PHB2, CTSB, MYO1C, PHB, and FASN) were directly linked to mitochondria and its functions (
Figure 4B, C). These results suggested that thiourea derivatives may improving mitochondrial health and functionality. To validate our hypothesis, HepG2.2.15 cells were treated with thiourea derivatives. Vitamin C was used as a positive control whereas ETV was used as a control for HBV infection. FACS analysis revealed that thiourea derivatives had the potential to increase mitochondrial mass (
Figure 4D) and restore the membrane potential of mitochondria (
Figure 4E). Therefore, the ratio of mass and membrane potential of mitochondria represents the functionality of the mitochondrial pool. Moreover, these thiourea derivatives were able to restore and maintain the pool of functional mitochondria in chronic HBV infected hepatocytes (
Figure 4F). These observations clearly outline unique role of the thiourea derivative improving mitochondrial health not described earlier. Thus, the use of thiourea derivative in CHB care is could be complementary to the currently used antivirals.
3. Discussion
CHB is a major risk to the progression to HCC, the sustained presence of the virus in the liver causes liver inflammation and exhaustion of hepatocytes[
7]. Previous studies have shown that HBV replication and the expression of the non-structural protein HBx lead to mitochondrial dysfunction, including alterations in mitochondrial unfolded protein response, biogenesis, mitophagy, and increased production of reactive oxygen species (ROS) such as superoxide and peroxynitrite [
7,
32]. HBx has been shown to have diverse roles in regulatory HBV replication, cellular transcription and signal transduction pathways, proteasome activity, and cell cycle progression, and has also been linked to the progression of HBV-associated HCC [
33]. Furthermore, a portion of cytosolic HBx may localize to mitochondria of hepatocytes, where it can modulate mitochondrial membrane potential and alter cytosolic calcium levels via the mitochondrial permeability transition pore (mPTP) [
16,
34]. The current study has focused on HBx as a potential therapeutic target to block HBV replication, restore mitochondrial function, and reduce liver inflammation in CHB patients. One promising candidate for this approach is compound DSA-00 and its derivatives, that have been found to regulate HBx-induce activities, including HBV replication, HBx-induced kinase activity, and HBx-modulated host RNAi machinery [
23] (Kumar et al., 2023 communicated).
Now, using a proteomic approach, we have identified the proteomic perturbation associated with the decrease in HBV viral replication and improvement of mitochondrial functions and exhaustion of hepatocytes during CHB infection. To achieve this, proteome profile of cell culture-based model of chronically infected hepatocytes was compared to chronically infected hepatocytes treated with commonly used anti-viral ETV or different thiourea derivatives. Instead of performing conventional differential expression analysis based on predetermined protein sets or pathways, we conducted a modular analysis that used an unsupervised hierarchical clustering method to examine the link between groups of co-expressed proteins. We used the WPCNA method which enables the identification of protein co-expression and the determines of how well a particular protein is connected to other proteins pathways that are jointly involved in a biological process. The protein co-expression networks of intricate biological processes are segmented into modules by WPCNA, which identified 10 different modules (
Figure 2), with Black, Magenta, Red, Pink, Green, and Yellow modules most shared modules between DSA-00 and ETV treatment. According to enrichment analysis, these modules were mostly correlated with ribosome, proteasome, fatty acid biosynthesis, PPAR signaling, RNA transport pathways, Brown module was unique for the DSA-02 treatment, with proteins significantly associated with biological processes such as Glucagon signaling, tight junction, PPAR signaling, fatty acid degradation, spliceosome, etc. The Grey modules had only five proteins, which were upregulated by the treatment of DSA-00 (
Figure 2E). KEGG analysis of these proteins revealed that they were significantly involved in mitochondrial biogenesis, oxidative phosphorylation, and unfolded protein response.
Furthermore, we looked at proteins involved in the antiviral response by blasting with the antiviral response database, and found 44 proteins in Black, Magenta, Red, Pink, Green, Yellow, Brown, and Grey modules (
Figure 3D). Analysis of the correlation between the changed biological functions in DSA-00, DSA-02, and ETV groups and found that antiviral response proteins were associated with positively regulated and negatively regulated HBV in DSA-00+ETV groups. DSA-02 induced proteins that supported the antiviral response. Additionally, DSA-00 had a unique Grey module, which had five proteins associated with mitochondrial functions.
The aim of our study was to repurpose drugs that have the potential to reduce HBV infection and restore the exhausted hepatocytes that are notably exhausted during CHB infection. Restoration of dysfunctional mitochondria has been correlated with the improvement of cell health in the past, although not being previously linked to thiourea derivatives efficacy. Moreover, the blast hits with mitoproteome database [
31] revealed that thiourea derivatives are suitable candidates that have the efficacy to restore dysfunctional mitochondria as well as have the potential to reduce HBV infection. Our study provides valuable insights into the potential of thiourea derivatives for the treatment of CHB and restoration of exhausted hepatocytes.
Limitations of the investigation include the fact that it is a pilot study and that just one type of in-vitro CHB model (HepG2.2.15) was employed. Instead of measuring mitochondrial respiration, oxygen consumption rate (OCR), and ATP production in the validation research, just test the mitochondrial health using flow cytometry.
4. Material and Methods
4.1. Cell culture and reagents
HepG2.2.15 cells, which have integrated two copies of HBV genome, used in this study, were grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco of Invitrogen), GlutaMax, 10 mM HEPES, 100 unit/ml penicillin, 100 µg/ml streptomycin, and 10% FBS at 37°C in a humidified atmosphere containing 5% CO2.
DSA-00, DSA-02, DSA-09, Entecavir (ETV, Gilead Sciences), and Ascorbic Acid (Vitamin C, Sigma-Aldrich). All compounds were dissolved in dimethyl sulfoxide and stored at -20°C or -80°C until used.
4.2. Proteomics Analysis
Untargeted proteomic analysis of cell culture samples was performed using ultra-high performance liquid chromatography (UHPLC) coupled with high resolution tandem mass spectrometry (HRMS/MS)[
35]. HepG2.2.15 cells (10
5), were seeded in six well plates. Following day, cells were treated with 10μM of thiourea derivatives, ETV and Vitamin C. After 24 h, cells were trypsinized and collected by centrifugation at 5000 RPM. The cells pellet was then resuspended in RIPA lysis buffer to isolate total proteins. The scheme of proteomics analysis is shown in (
Figure 1A, and 2A). The detailed protocol for the untargeted proteomics analysis are provided in the supporting information (SI).
4.3. MTR and MTG HepG2.2.15 cell staining
MTR CM-H2XROS and MTG staining were done according to the manufacturer's instructions (Molecular Probes). Detailed methods are provided in the supporting information (SI).
4.4. Statistical and correlation analysis
The results are presented as mean ± S.D. Statistical analysis was performed using Graph Pad Prism, version 8 (GraphPad Software, La Jolla, CA, USA;
www.graphpad.com. A difference was considered to be statistically significant at *P< 0.05 and **P< 0.01, and not significant (NS).
5. Conclusions
Our research concentrated on CHB, which causes mitochondrial dysfunction, HBV reactivation, and HBV replication. Thiourea derivatives known for the antiviral activity and restoration of host RNA interference mediated defense machinery. Here, we repurposed thiourea derivatives, they have the potential to restoration of the dysfunctional mitochondria. Mitochondrion-targeted antioxidants considerably enhanced mitochondrial and antiviral hepatocytes functions, revealing a crucial role for reactive oxygen species (ROS) in cell exhaustion. our study provides valuable insights into the potential of thiourea derivatives for the treatment of CHB and restoration of exhausted hepatocytes. Further experimental validation of our findings is necessary to confirm their potential clinical application.
Author Contributions
Conceptualization, J.K., PT, and J.S.M.; methodology, J.K., and J.S.M.; software, J.K.; validation, J.K. and P.T.; formal analysis, J.K.; investigation, J.K..; resources, J.K., and AKS; data curation, J.K.; writing-original draft preparation, J.K.,; writing-review and editing, J.S.M., and V.K.; visualization, J.K.; supervision, V.K.; project administration, DS; funding acquisition, V.K. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
This work was supported by grants to VK from the Department of Biotechnology (DBT), Government of India (Grant No. BT/PR30082/Med/29/1341/2018) and J.C. Bose National Fellowship (Grant No. SR/S2/JCB-80)/2012) of the Department of Science and Technology, Government of India, New Delhi. JK, PT, and AKS received Senior Research Fellowships respectively from the Council of Scientific and Industrial Research, New Delhi and University Grants Commission, New Delhi for the period of this study.
References
- Jeng, W.J., G.V. Papatheodoridis, and A.S.F. Lok, Hepatitis B. Lancet 2023, 401, 1039–1052.
- WHO, World Health Statistics 2022. 2022. Retrieved from:https://www.who.int/news/item/20-05-2022-world-health-statistics-2022.
- Yeh, M.L., et al., Long-term outcome of liver complications in patients with chronic HBV/HCV co-infection after antiviral therapy: a real-world nationwide study on Taiwanese Chronic Hepatitis C Cohort (T-COACH). Hepatol Int 2021, 15, 1109–1121. [CrossRef]
- Li, D., et al., A potent human neutralizing antibody Fc-dependently reduces established HBV infections. Elife 2017, 6.
- Han, C.Y., et al., FXR Inhibits Endoplasmic Reticulum Stress-Induced NLRP3 Inflammasome in Hepatocytes and Ameliorates Liver Injury. Cell Rep 2018, 24, 2985–2999. [CrossRef]
- Li, L., et al., Cell-free circulating mitochondrial DNA content and risk of hepatocellular carcinoma in patients with chronic HBV infection. Sci Rep 2016, 6, 23992. [CrossRef]
- Loureiro, D., et al., Mitochondrial stress in advanced fibrosis and cirrhosis associated with chronic hepatitis B, chronic hepatitis C, or nonalcoholic steatohepatitis. Hepatology 2023, 77, 1348–1365. [CrossRef]
- Lin, C. and Q. Ou, Emerging role of mitochondria in response to HBV infection. J Clin Lab Anal 2022, 36, e24704. [CrossRef]
- Kim, J.Y., et al., ER Stress Drives Lipogenesis and Steatohepatitis via Caspase-2 Activation of S1P. Cell 2018, 175, 133–145. [CrossRef]
- Li, Y., et al., Hepatitis B Surface Antigen Activates Unfolded Protein Response in Forming Ground Glass Hepatocytes of Chronic Hepatitis B. Viruses 2019, 11.
- Ghany, M.G., et al., Serum alanine aminotransferase flares in chronic hepatitis B infection: the good and the bad. Lancet Gastroenterol Hepatol 2020, 5, 406–417. [CrossRef]
- Morikawa, K., et al., Hepatitis B: progress in understanding chronicity, the innate immune response, and cccDNA protection. Ann Transl Med 2016, 4, 337. [CrossRef]
- Slagle, B.L. and M.J. Bouchard, Role of HBx in hepatitis B virus persistence and its therapeutic implications. Curr Opin Virol 2018, 30, 32–38. [CrossRef] [PubMed]
- Yuen, M.F., et al., Hepatitis B virus infection. Nat Rev Dis Primers 2018, 4, 18035. [CrossRef]
- Kim, S., et al., Hepatitis B virus x protein induces perinuclear mitochondrial clustering in microtubule- and Dynein-dependent manners. J Virol 2007, 81, 1714–26. [CrossRef]
- Rahmani, Z., et al., Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J Virol 2000, 74, 2840–6. [CrossRef]
- Foo, J., et al., Mitochondria-mediated oxidative stress during viral infection. Trends Microbiol 2022, 30, 679–692. [CrossRef]
- Wang, D., et al., Identification of the risk for liver fibrosis on CHB patients using an artificial neural network based on routine and serum markers. BMC Infect Dis 2010, 10, 251.
- Stuart, J.M., et al., A gene-coexpression network for global discovery of conserved genetic modules. Science 2003, 302, 249–55. [CrossRef]
- Naz, S., et al., Synthesis, characterization, and pharmacological evaluation of thiourea derivatives. Open Chemistry 2020, 18, 764–777. [CrossRef]
- Ghorab, M.M., et al., Novel Thiourea Derivatives Bearing Sulfonamide Moiety as Anticancer Agents Through COX-2 Inhibition. Anticancer Agents Med Chem 2017, 17, 1411–1425.
- Antypenko, L., et al., Novel acyl thiourea derivatives: Synthesis, antifungal activity, gene toxicity, drug-like and molecular docking screening. Arch Pharm (Weinheim) 2019, 352, e1800275. [CrossRef] [PubMed]
- Ghosh, S., et al., An RNAi-based high-throughput screening assay to identify small molecule inhibitors of hepatitis B virus replication. J Biol Chem 2017, 292, 12577–12588. [CrossRef] [PubMed]
- Kucukguzel, I., et al., Synthesis of some novel thiourea derivatives obtained from 5-[(4-aminophenoxy)methyl]-4-alkyl/aryl-2,4-dihydro-3H-1,2,4-triazole-3-thiones and evaluation as antiviral/anti-HIV and anti-tuberculosis agents. Eur J Med Chem 2008, 43, 381–92. [CrossRef] [PubMed]
- Zhang, Z., et al., The effectiveness of TDF versus ETV on incidence of HCC in CHB patients: a meta analysis. BMC Cancer 2019, 19.
- Kanehisa, M., et al., KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 2023, 51, D587–D592. [CrossRef] [PubMed]
- Santantonio, T.A. and M. Fasano, Chronic hepatitis B: Advances in treatment. World J Hepatol 2014, 6, 284–92. [CrossRef]
- Seyfried, N.T., et al., A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer's Disease. Cell Syst 2017, 4, 60–72. [CrossRef]
- Ling, L.R., et al., Effect of HBx on inflammation and mitochondrial oxidative stress in mouse hepatocytes. Oncol Lett 2020, 19, 2861–2869.
- Arbuthnot, P. and M. Kew, Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol 2001, 82, 77–100. [CrossRef]
- Calvo, S.E., K.R. Clauser, and V.K. Mootha, MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 2016, 44, D1251–D1257. [CrossRef]
- Brenner, C., R. Ventura-Clapier, and E. Jacotot, Mitochondria and cytoprotection. Biochem Res Int 2012, 2012, 351264.
- Slagle, B.L. and M.J. Bouchard, Hepatitis B Virus X and Regulation of Viral Gene Expression. Cold Spring Harb Perspect Med 2016, 6, a021402. [CrossRef] [PubMed]
- Sivasudhan, E., et al., Hepatitis B Viral Protein HBx and the Molecular Mechanisms Modulating the Hallmarks of Hepatocellular Carcinoma: A Comprehensive Review. Cells 2022, 11.
- Tripathi, G., et al., Protocol for global proteome, virome, and metaproteome profiling of respiratory specimen (VTM) in COVID-19 patient by LC-MS/MS-based analysis. STAR Protoc 2022, 3, 101045. [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).