Submitted:
19 May 2023
Posted:
22 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampling and strains
2.2. Phenotypic analysis
2.3. DNA extraction, PCR amplification and sequencing
2.4. Phylogenetic analyses
| Species | Strain number | Substrate (country) | GenBank accesion numbers1 | Citation | |||
| ITS | LSU | tef1 | BenA | ||||
| A. cavernicola | CGMCC3.19571T | Bird faeces (China) | MK329056 | MK328961 | MK335997 | NA | [12] |
| LC 12560 | Animal faeces (China) | MK329061 | MK328966 | MK336002 | NA | [12] | |
| LC 12674 | Plant debris (China) | MK329065 | MK328970 | MK336006 | NA | [12] | |
| A. coprophila | CBS 247.82T (ex-type of O. coprophila) | Rabbit dung (England) | MH861494 | MH873238 | OQ954487 | pending | [41]; this study |
| CBS 424.88 (received as O. coprophila) | Chipmunk dung (Canada) | OQ942929 | OQ943166 | OQ954488 | pending | This study | |
| CBS 173.71 (received as B. felina) | Porcupine dung (Canada) | AY261368 | MH871833 | OQ954489 | pending | [41]; this study | |
| A. felina | CBS 250.34 (ex-type of I. cretacea) | Pressed yeast (England) | MH855498 | OQ943167 | OQ954490 | pending | [41]; this study |
| CBS 648.66 (received as B. felina) | Unknown, (Argentina) | OQ942930 | MH870575 | OQ954491 | pending | [41]; this study | |
| CBS 110.08 (received as B. felina) | Unknown | MH854578 | OQ943168 | OQ954492 | pending | [41]; this study | |
| A. guana | CGMCC3.17908T | Bat guano (China) | KU746665 | KU746711 | KX855211 | NA | [11] |
| CGMCC3.17909 | Bat guano (China) | KU746666 | KU746712 | KX855212 | NA | [11] | |
| CBS 312.50 (received as B. felina) | Rabbit dung (Unknown) | MH856641 | MH868150 | OQ954493 | pending | [41]; this study | |
| A. littoralis | FMR 17952 | Floating rubber tire (Spain) | OQ942925 | OQ943162 | OQ954483 | pending | This study |
| FMR 19404T | Marine sediment (Spain) | OQ942924 | OQ943161 | OQ954482 | pending | This study | |
| FMR 19611 | Marine sediment (Spain) | OQ942926 | OQ943163 | OQ954484 | pending | This study | |
| FMR 20067 | Marine sediment (Spain) | OQ942927 | OQ943164 | OQ954485 | pending | This study | |
| FMR 20149 | Marine sediment (Spain) | OQ942928 | OQ943165 | OQ954486 | pending | This study | |
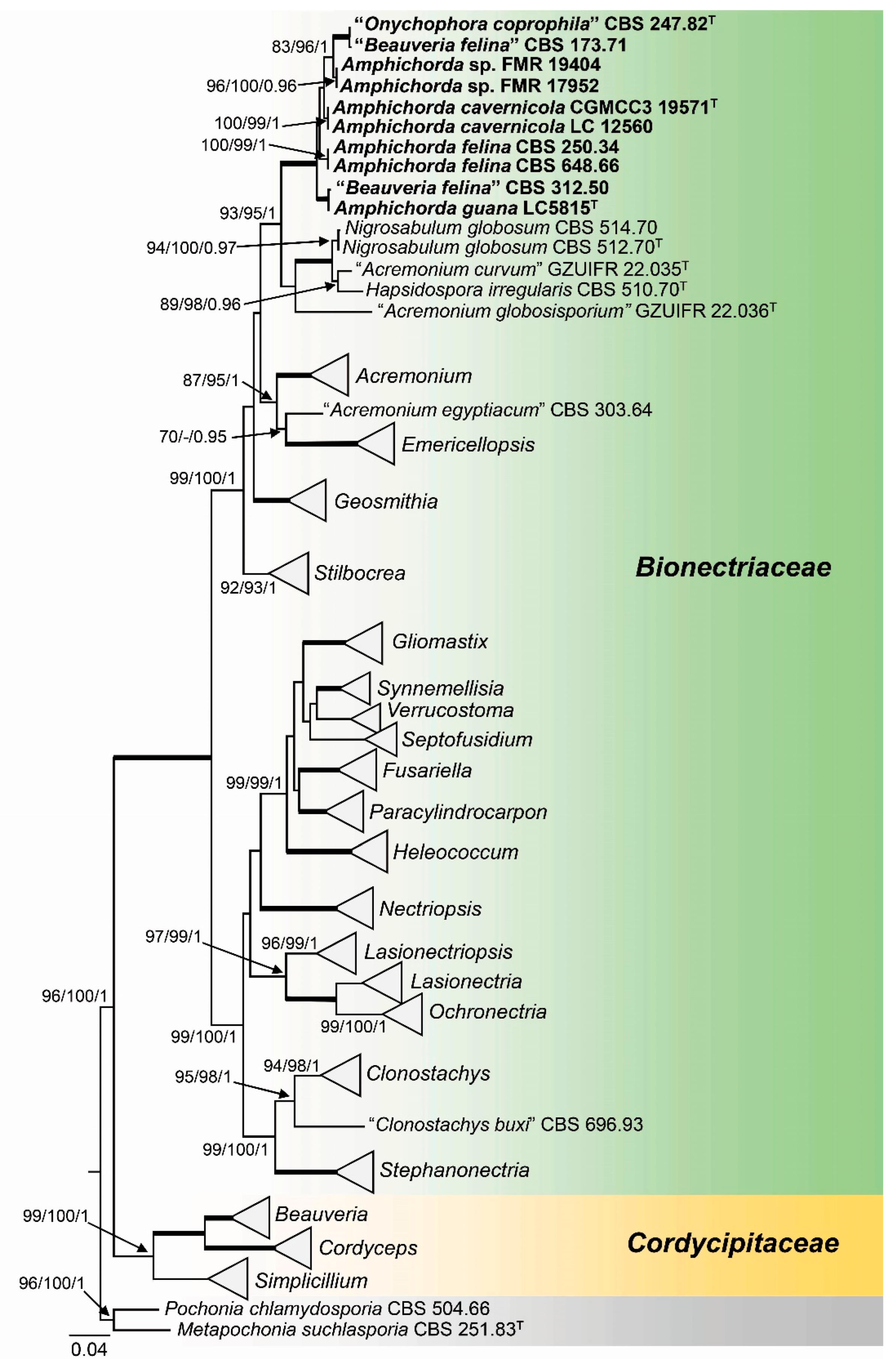
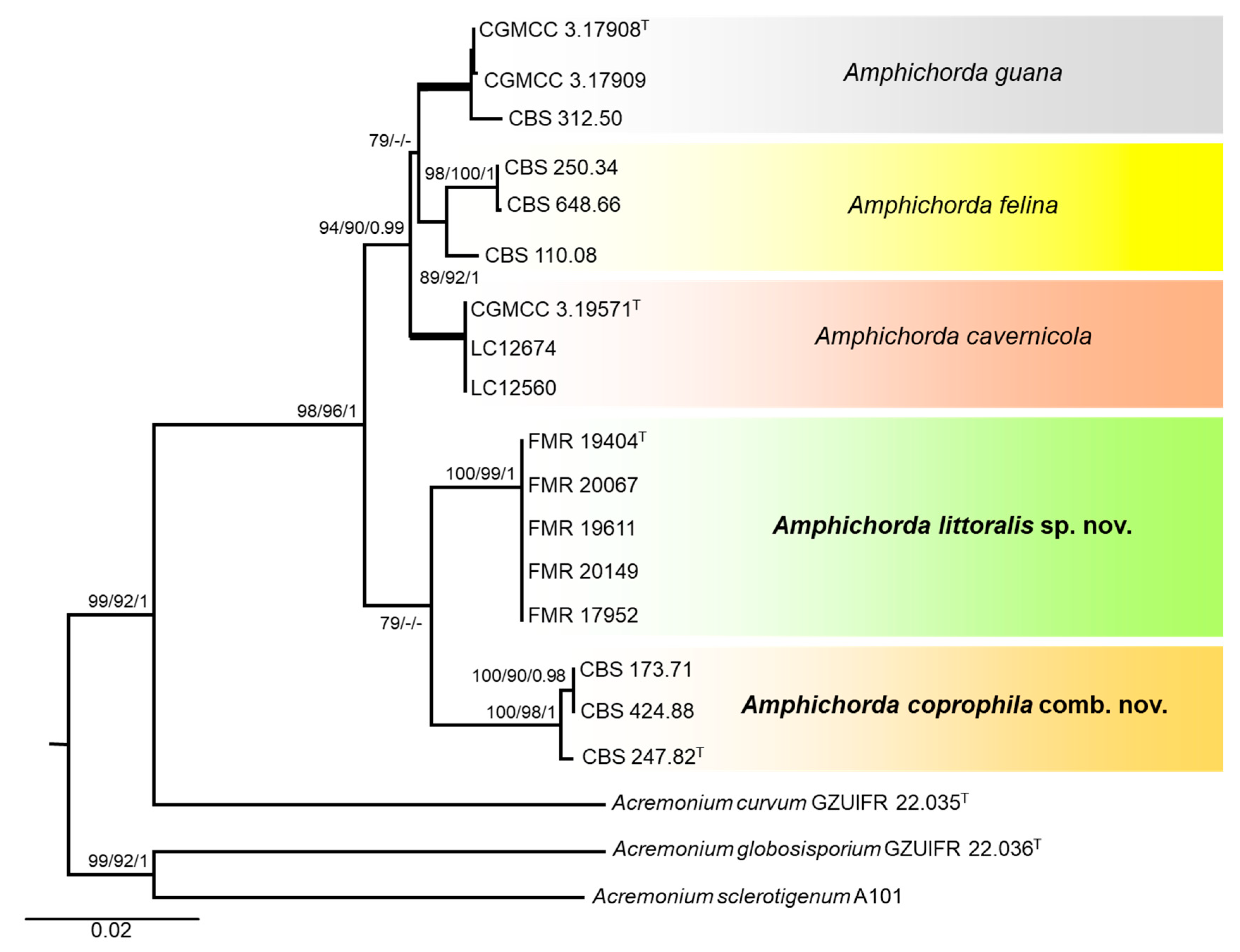
3. Results
3.1. Phylogeny
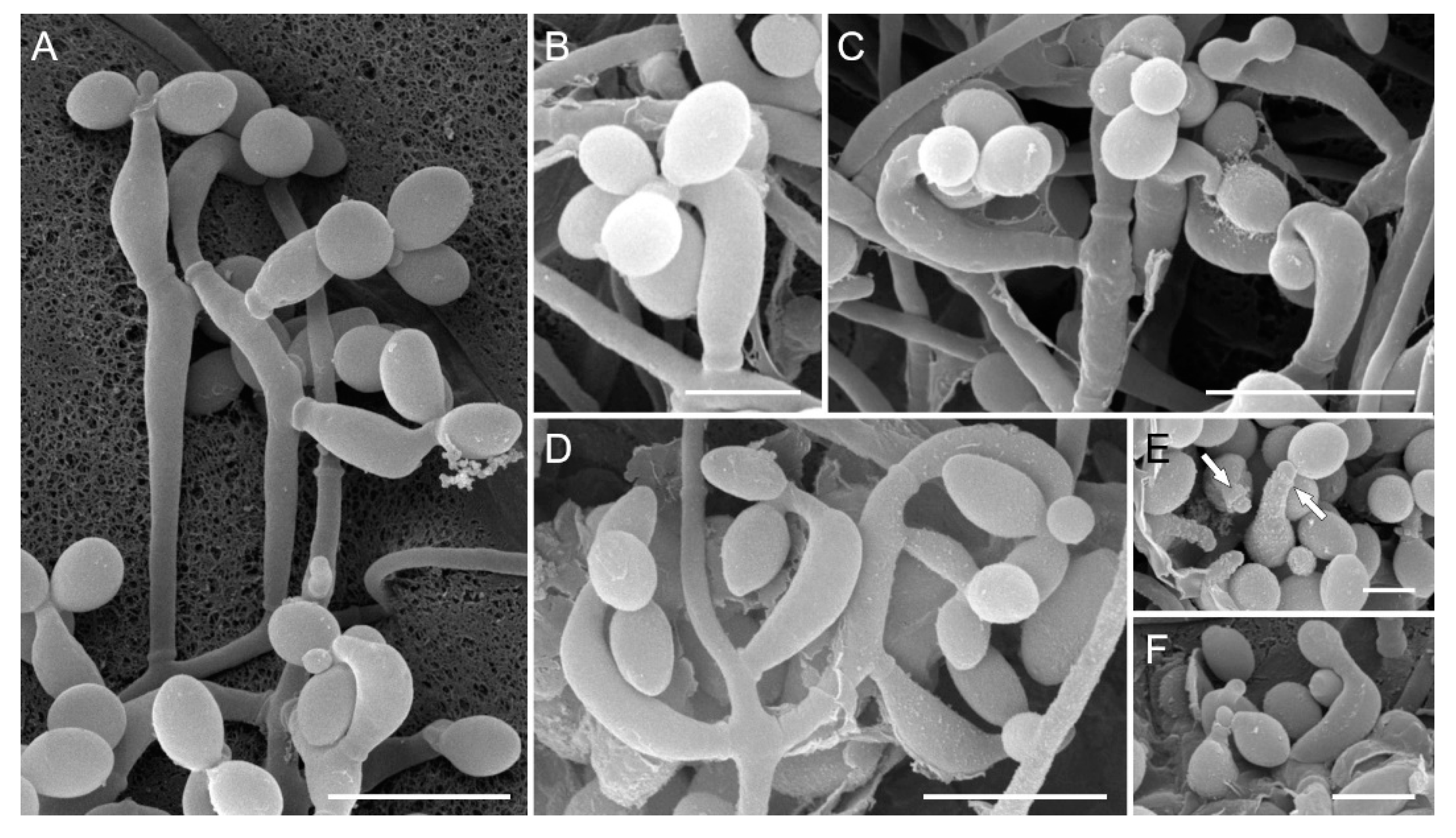
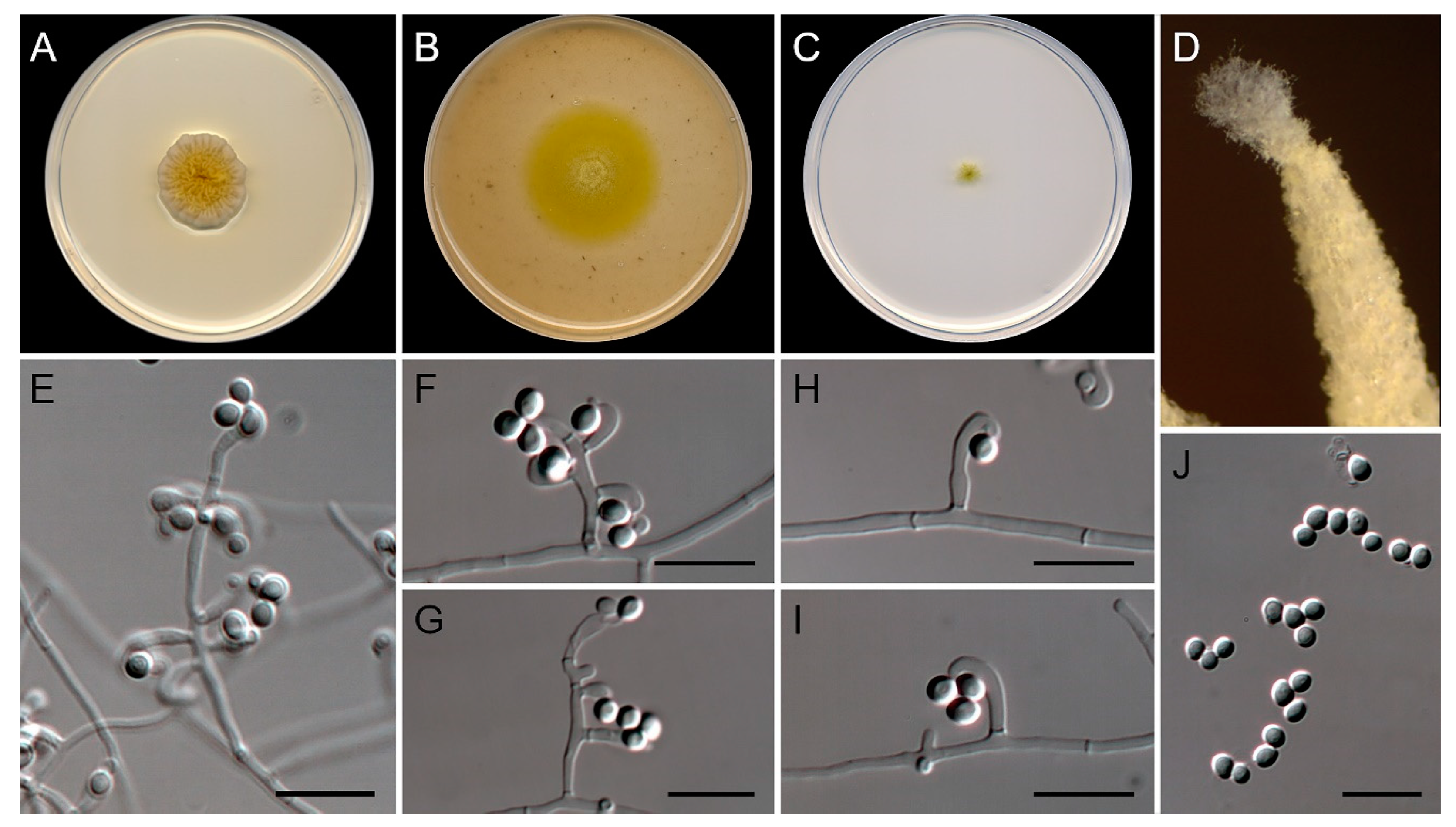
3.2. Morphological analysis
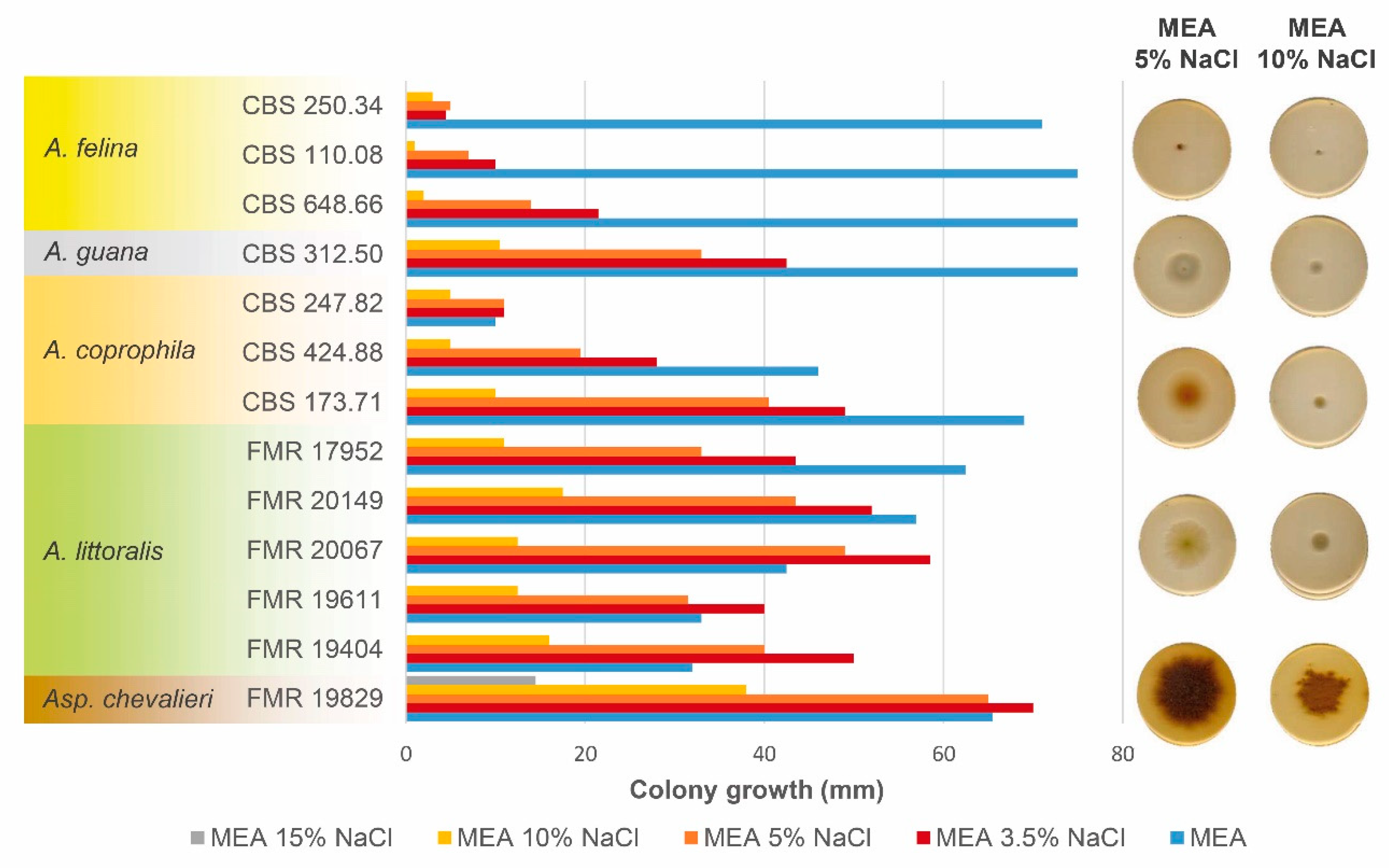
3.3. Salt tolerance test
3.4. Taxonomy
4. Discussion
5. Concluding remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined Families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Perera, R.H.; Hyde, K.D.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Bundhun, D.; Camporesi, E.; Akulov, A.; Liu, J.K.; Liu, Z.Y. Profile of Bionectriaceae, Calcarisporiaceae, Hypocreaceae, Nectriaceae, Tilachlidiaceae, Ijuhyaceae fam. nov., Stromatonectriaceae fam. nov. and Xanthonectriaceae fam. nov. Fungal Divers. 2023, 118, 95–271. [Google Scholar] [CrossRef]
- Kepler, R.M.; Luangsa-ard, J.J.; Hywel-Jones, N.L.; Quandt, C.A.; Sung, G.H.; Rehner, S.A.; Aime, M.C.; Henkel, T.W.; Sanjuan, T.; Zare, R.; et al. A Phylogenetically-Based Nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 2017, 8, 335–353. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.B.; Wang, Y.; Fan, Q.; Duan, D.E.; Zhang, G.-D.; Dai, R.-Q.; Dai, Y.-D.; Zeng, W.B.; Chen, Z.-H.; Li, D.D.; et al. Multigene Phylogeny of the Family Cordycipitaceae (Hypocreales): New Taxa and the New Systematic Position of the Chinese Cordycipitoid Fungus Paecilomyces hepiali. Fungal Divers. 2020, 103, 1–46. [Google Scholar] [CrossRef]
- Lamarck, J.B. De Flore françoise ou descriptions succinctes de toutes les plantes qui croissent naturellement en France, disposées selon une nouvelle méthode d’analyse, et précédées par un exposé des principes élémentaires de la botanique; Paris, 1815; Volume 6. [Google Scholar]
- Fries, E. Systema orbis vegetabilis: Primas lineas novae constructionis periclitatur; e Typographia academica: Lunde, 1825. [Google Scholar]
- Fries, E.M. Systema mycologicum: sistens fungorum ordines, genera et species, huc usque cognitas, quas ad normam methodi naturalis determinavit; Sumtibus Ernesti Mauritii; 1832; Volume 3. [Google Scholar]
- Hodge, K.T.; Gams, W.; Samson, R.A.; Korf, R.P.; Seifert, K.A. Lectotypification and Status of Isaria Pers. : Fr. Taxon 2005, 54, 485–489. [Google Scholar] [CrossRef]
- De Hoog, G.S. The Genera Beauveria, Isaria, Tritirachium and Acrodontium gen. nov. Stud. in Mycol. 1972, 1, 1–41. [Google Scholar]
- Clements, F.F.; Shear, C.L. The Genera of Fungi.; The H. W. Wilson Company: New York, 1931. [Google Scholar]
- Zhang, Z.F.; Liu, F.; Zhou, X.; Liu, X.Z.; Liu, S.J.; Cai, L. Culturable Mycobiota from Karst Caves in China, with Descriptions of 20 New Species. Persoonia 2017, 39, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-F.; Zhou, S.-Y.; Eurwilaichitr, L.; Ingsriswang, S.; Raza, M.; Chen, Q.; Zhao, P.; Liu, F.; Cai, L. Culturable Mycobiota from Karst Caves in China II, with Descriptions of 33 New Species. Fungal Divers. 2021, 106, 29–136. [Google Scholar] [CrossRef]
- Beyma, F.H. van. Beschreibung Einiger Neuer Pilzarten Aus Dem Centraalbureau Voor Schimmelcultures Baarn (Holland). Zentralblatt für Bakteriologie und Parasitenkunde 1935, 2, 345–355. [Google Scholar]
- Lendemer, J.C. Epitypes Are Forever: Best Practices for an Increasingly Misused Nomenclatural Action. Taxon 2020, 69, 849–850. [Google Scholar] [CrossRef]
- Gams, W.; Fisher, P.J.; Webster, J. Onychophora, a New Genus of Phialidic Hyphomycetes from Dung. TBMS 1984, 82, 174–177. [Google Scholar] [CrossRef]
- Expósito, N.; Rovira, J.; Sierra, J.; Folch, J.; Schuhmacher, M. Microplastics Levels, Size, Morphology and Composition in Marine Water, Sediments and Sand Beaches. Case Study of Tarragona Coast (Western Mediterranean). Sci. Total Environ. 2021, 786, 147453. [Google Scholar] [CrossRef] [PubMed]
- Kossuga, M.H.; Romminger, S.; Xavier, C.; Milanetto, M.C.; Valle, M.Z. do; Pimenta, E.F.; Morais, R.P.; Carvalho, E. de; Mizuno, C.M.; Coradello, L.F.C.; et al. Evaluating Methods for the Isolation of Marine-Derived Fungal Strains and Production of Bioactive Secondary Metabolites. Rev. Bras. Farmacogn. 2012, 22, 257–267. [Google Scholar] [CrossRef]
- Wingfield, B.D.; Wingfield, M.J.; Duong, T.A. Molecular Basis of Cycloheximide Resistance in the Ophiostomatales Revealed. Curr. Genet. 2022, 68, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Figueras, M.J.; Guarro, J. A Scanning Electron Microscopic Study of Ascoma Development in Chaetomium malaysiense. Mycol. 1988, 80, 298–306. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Methuen: London, UK, 1978. [Google Scholar]
- Müller, F.M.C.; Werner, K.E.; Kasai, M.; Francesconi, A.; Chanock, S.J.; Walsh, T.J. Rapid Extraction of Genomic DNA from Medically Important Yeasts and Filamentous Fungi by High-Speed Cell Disruption. J. Clin. Microbiol. 1998, 36, 1625–1629. [Google Scholar] [CrossRef] [PubMed]
- Vilgalys, R.; Hester, M. Rapid Genetic Identification and Mapping of Enzymatically Amplified Ribosomal DNA from Several Cryptococcus Species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; New York, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of Primer Sets Designed for Use with the PCR to Amplify Conserved Genes from Filamentous Ascomycetes. AEM 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria Phylogeny Inferred from Nuclear ITS and EF1-α Sequences: Evidence for Cryptic Diversification and Links to Cordyceps Teleomorphs. Mycol. 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES Science Gateway: Enabling High-Impact Science for Phylogenetics Researchers with Limited Resources. In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the Campus and Beyond, Chicago, IL, USA, 16–20 July 2012; Association for Computing Machinery: New York, NY, USA, 2012; pp. 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat Methods 2012, 9, 772–772. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Hespanhol, L.; Vallio, C.S.; Costa, L.M.; Saragiotto, B.T. Understanding and Interpreting Confidence and Credible Intervals around Effect Estimates. BJPT 2019, 23, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-Scale Generation and Analysis of Filamentous Fungal DNA Barcodes Boosts Coverage for Kingdom Fungi and Reveals Thresholds for Fungal Species and Higher Taxon Delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Z.-Y.; Ren, Y.-L.; Chen, W.-H.; Liang, J.-D.; Pan, J.-M.; Huang, J.-Z.; Liang, Z.-Q.; Han, Y.-F. Morphological characteristics and phylogenetic evidence reveal two new species of Acremonium (Hypocreales, Sordariomycetes). MycoKeys 2022, 91, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.-F.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Perera, R.H.; Thiyagaraja, V.; Hongsanan, S.; Wanasinghe, D.N.; Shen, H.-W.; Tian, X.; Yang, L.-Q.; et al. Taxonomy, Phylogeny and Evolution of Freshwater Hypocreomycetidae (Sordariomycetes), In Review. 2023.
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.E.St.J.; Gené, J.; Guarro, J.; Baseia, I.G.; García, D.; Gusmão, L.F.P.; Souza-Motta, C.M.; et al. Fungal Planet Description Sheets: 716–784. Persoonia 2018, 40, 240–393. [Google Scholar] [CrossRef]
- Lombard, L.; Houbraken, J.; Decock, C.; Samson, R.A.; Meijer, M.; Réblová, M.; Groenewald, J.Z.; Crous, P.W. Generic Hyper-Diversity in Stachybotriaceae. Persoonia 2016, 36, 156–246. [Google Scholar] [CrossRef]
- Huang, S.-K.; Hyde, K.; Maharachchikumbura, S.; Mckenzie, E.; Wen, T.-C. Taxonomic Studies of Coronophorales and Niessliaceae (Hypocreomycetidae). Mycosphere 2021, 12, 875–992. [Google Scholar] [CrossRef]
- Quandt, C.A.; Kepler, R.M.; Gams, W.; Araújo, J.P.M.; Ban, S.; Evans, H.C.; Hughes, D.; Humber, R.; Hywel-Jones, N.; Li, Z.; et al. Phylogenetic-Based Nomenclatural Proposals for Ophiocordycipitaceae (Hypocreales) with New Combinations in Tolypocladium. IMA Fungus 2014, 5, 121–134. [Google Scholar] [CrossRef]
- Rehner, S.A.; Minnis, A.M.; Sung, G.-H.; Luangsa-ard, J.J.; Devotto, L.; Humber, R.A. Phylogeny and Systematics of the Anamorphic, Entomopathogenic Genus Beauveria. Mycol. 2011, 103, 1055–1073. [Google Scholar] [CrossRef]
- Pascoe, I.G. Fusarium Morphology. I. Identification and Characterization of a Third Conidial Type, the Mesoconidium. Mycotaxon 1990, 37, 121–160. [Google Scholar]
- De Hoog, G.S.; Guarro, J.; Gené, J.; Ahmed, S.; Al-Hatmi, A.M.S.; Figueras, M.J.; Vitale, R.G. Atlas of Clinical Fungi: The Ultimate Benchtool for Diagnostics. Introductions, Lower Fungi, Basidiomicetes, Yeasts, Filamentous Ascomycetes, 4th ed.; Foundation Atlas: Hilversum, The Netherlands, 2020. [Google Scholar]
- Gonçalves, M.F.M.; Esteves, A.C.; Alves, A. Marine Fungi: Opportunities and Challenges. Encyclopedia 2022, 2, 559–577. [Google Scholar] [CrossRef]
- Ramanujam, B.; Poornesha, B.; Kandan, A.; Mohan, M.; Sivakumar, G. Natural Occurrence of Entomopathogenic Fungus Beauveria Felina (DC.) J.W. Carmich on Fall Armyworm, Spodoptera frugiperda (J. E. Smith). J. Entomol. Zool. Stud. 2021, 9, 140–143. [Google Scholar] [CrossRef]
| Species | Colony on PDA* | Colony on OA/SNA* | Microscopic features | Citation | ||
| Color | Diffusible pigment | Color | Conidiogenous cells size (µm) | Conidia size (µm) | ||
| A. cavernicola | Cream yellow to sea shell | Not observed | White | 4.5-8 x 2-3 | 2.5-4 x 2-3.5 | [12]; this study |
| A. coprophila | Orange to brownish orange | Greyish orange | Light yellow | 6-10 x 2-2.5 | 3.5-5.5 x 2-2.5 | [15]; this study |
| A. felina | White | Greyish orange | White | 3-8.5 x 2-2.5 | 2.5-4.5 x 2-3.5 | [9]; this study |
| A. guana | White to yellowish | Yellowish | White | 7-10 x 2-3 | 4.5-5.5 x 3.5-5 | [11]; this study |
| A. littoralis | Greenish yellow | Light yellow | Greenish yellow | 5.5-11.5 x 1.5-2.5 | 2.5-4 x 2.5-3 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





