1. Introduction
1.1. Biofouling
Organisms living on surfaces take advantage of the energetics of water movement to carry away wastes and propagules and obtain food. The term biofouling is usually used for the growth of organisms on man-made surfaces[
1]. Biofouling includes microorganisms (microfouling) and macroscopic organisms (macrofouling). Biofilms include layers of microorganisms in a matrix of organic molecules attached to surfaces [
2].
Microfouling includes bacteria, archaea, fungi, microalgae, sessile metazoans, and microalgae [
3]. Microfouling can include microbes living in biofilms attached to a hard substratum, the surface of the water, marine snow, and other particles [
4]. Microfouling results from the active or passive movement of microbes onto surfaces from the water column. Microfouling can be instantaneous or take several hours for transition of individual bacteria from the water column to a sessile existance [
5]. Microfouling is dynamic and much of it can be reversed.
Macrofouling includes metazoan organisms with at least one sessile life stage. As in the case of microfouling, macrofouling can be immediate upon submergence of a surface or after a while when the biofilm is transformed [
6]. Usually, macrofouling is due to attachment to a surface by a propagule/dispersal life stage. Upon attachment, propagules metamorphose into juvenile organisms [
7]. It can be instantaneous upon contact with the surface as in the case of many ascidian and barnacle larvae. Or it takes longer times as is the case for tubeworm larvae which require pre-existing biofilms. This process can take a few minutes to a few days. Some propagules require biofilms for their settlement and metamorphosis [
7,
8,
9]. Many macrofoulers are gregarious and gregariousness is mediated by cues and pheromones [
6]. Cues can originate from a variety of sources including symbiotic microalgae, conspecifics, generated by the action of exoproteases, molecules leaching from degrading biological adhesives, and anthropogenically derived synthetic mimics. Most macrofouing is not reversible.
Most sessile macrofoulers have planktonic larval stages that function in the dispersal and colonization of new substrates. These larval stages are complex in their physical, behavioral, and chemical response repertoire and have entertained biologists for tens of decades. Larval settlement is a logical control step for biofouling management [
8].
1.2. Antifouling techniques
Organisms living on man-made surfaces degrade performance. Degradation is by corrosion, the build-up of biomass that causes resistance to flow decreasing structural stability and increasing resistance [
10]. On hulls, speed, and fuel consumption are compromised. Biofouling costs money. There are over 2000 years of history of attempts to control biofouling [
11].
Historically biofouling is mediated by killing what grows on surfaces [
11]. Commercial solutions are broad-spectrum biocides ranging from chlorine gas which oxidizes organisms to heavy metal ions which disrupt metabolic processes [
10,
12]. As environmental damage is recognized, regulations on biocides are increased and the most toxic are banned. In the last several decades-long live organic biocides have been added to the commercial repertoire. These co-biocides can be more environmentally damaging than the biocides they augment. A recent twist on using toxic metals is nanomaterials like nano-silver, nano-copper, nano-titania, and nano-zinc [
13,
14,
15]. Nanomaterials generate reactive oxygen species and release toxic metal ions that kill biofouling organisms [
16].
Foul-release technology is now being developed by coatings companies and is a major improvement [
12,
17]. This technology is proposed as non-toxic and environmentally benign. However, due to loopholes in the regulations, these coatings routinely contain toxic compounds that kill organisms and alter their enzymes and adhesives [
18]. Thus, it is necessary to find new and non-toxic antifouling solutions.
1.3. Enhancement of biofouling
Biofouling has not only negative but also positive consequences. Coral reefs are the largest fouling communities in the world occupying an area as large as 284,000 km
2 and habitat for many other marine species [
3]. Mussel beds and oyster reefs are other examples of large fouling communities [
19].
Due to anthropogenic effects, coral reefs and marine ecosystems are under constant threat. However, restoration of damaged ecosystems, like coral and oyster reefs, is possible by enhancement of settlement of larvae of these species onto new substrata [
19]. Larvae of corals and oysters require specific chemical cues/pheromones that can be used in order to trigger the release of gametes and larval settlement in the laboratory [
20,
21,
22,
23,
24]. Later the substrates with young settlers can be moved to the desired habitats. The manipulation of spawning and larval settlement of endangered biofouling species requires extensive knowledge of their chemical ecology and species-species interactions.
Some biofouling species are cultivated and used for food production. These include mussel species like
Mytilus, Saccrostrea and
Perna. These species use chemical and biological cues to find suitable substratum and for metamorphosis [
7,
9,
20,
21]. In order to successfully cultivate them one needs to know how to deliver the chemical cues involved in the process as well as the mechanisms of larval settlement and metamorphosis.
1.4. Omics and biofouling studies
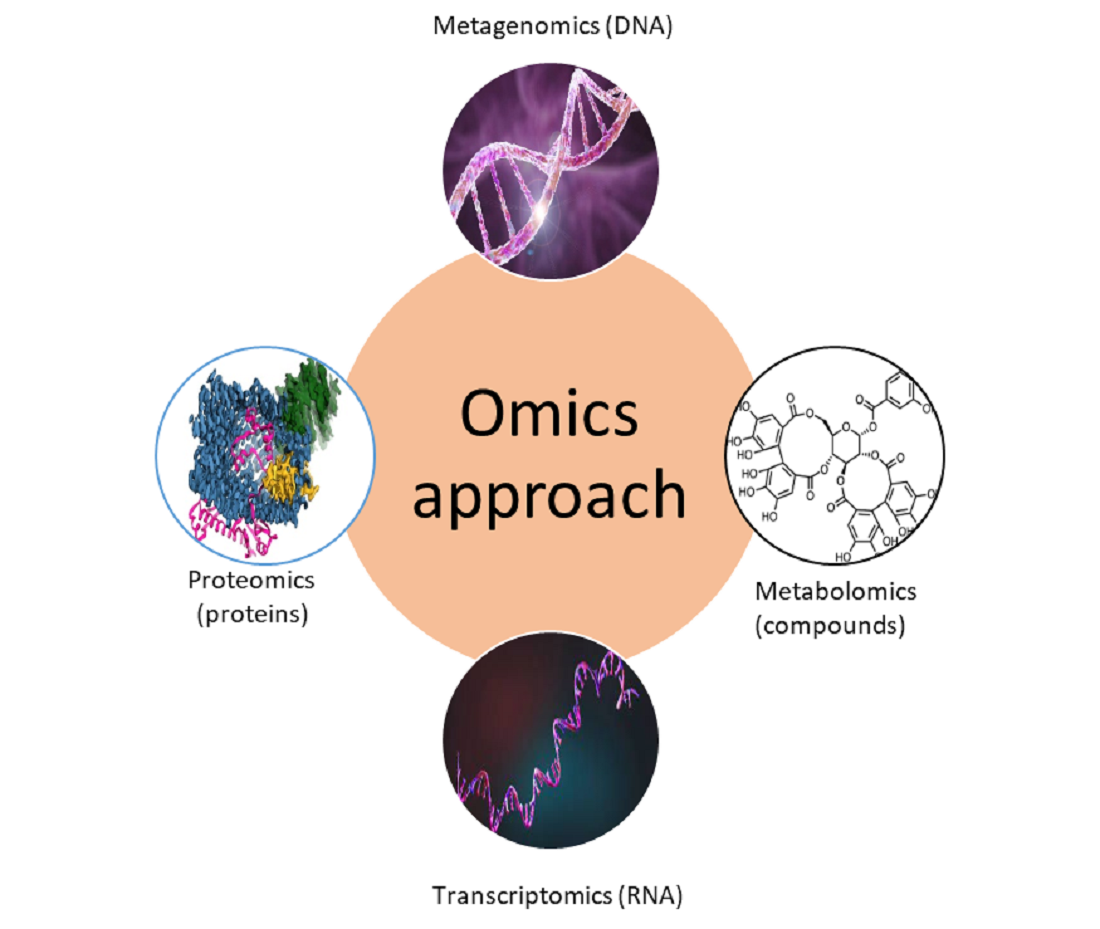
Traditionally, biofouling research was focused on the isolation and identification of single micro- and macrofouling organisms and the investigation of their biological properties (attachment, settlement behavior, metamorphosis, etc) and chemical metabolites. However, recently so called “omics” approaches have been used to study complex biofouling communities and their interactions with the environment. These “omics” methods include mainly metagenomics, metabolomics, transcriptomics, and proteomics approaches (
Table 1,
Figure 1). The main questions that novel approaches try to answer are “who is there?“, “what are they doing?”, “which compounds (proteins) are produced?“, and “how does the environment affect biofouling organisms and their metabolites?”.
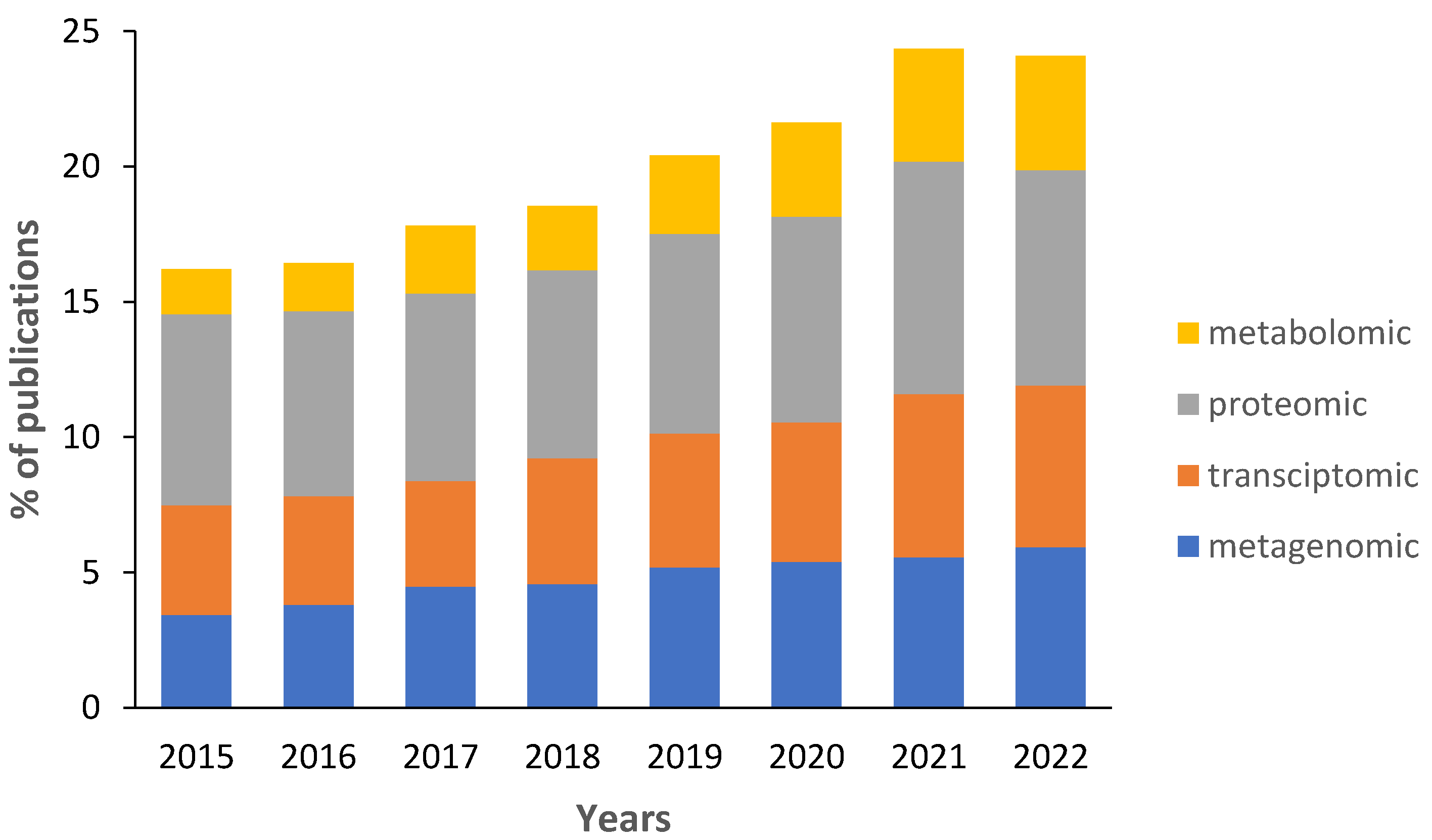
Data on the percentage of biofouling studies that use at least one “omics” technique remains low (
Figure 2). Since 2015, the percentage of publications has increased from 15% to 23%. Because the figure over counts studies that use multiple omics approaches, the actual percentage is lower. Most biofouling studies use proteomics to investigate the synthesis of proteins of biofouling organisms and to study their adhesive proteins (
Table 1). The second most common type of publication uses metagenomic and transcriptomic approaches (
Figure 2). Transcriptomics is used to investigate genes that are transcribed during biofilm formation or larval settlement and metamorphosis [
7,
25]. Most metagenomic studies use 16S or 18S genes to identify micro- and microorganisms in biofouling communities (
Table 1). However, there are some studies that investigate genes present in biofilms and larvae of macrofouling organisms. Studies that investigate metabolomes of biofouling organisms or complex communities are rare but their percentage is increasing every year (
Figure 2). In particular, biofilms associated with corals [
26,
27], seaweeds [
28,
29], sponges [
30,
31], and other marine organisms are reported.
Several reviews about “omics”-based studies have been published. The use of “omics” datasets for the identification of marine algal natural products is reviewed [
32] and the role of “omics” in the exploration of marine phytoplankton is evaluated [
33]. A recent review investigates the use of “omics” tools for assessing the biodeterioration of cultural heritage artifacts [
34]. The role of “omics” approaches in studying proteins of invertebrates was also reviewed [
35]. “Omics” research of an important aquaculture species, like abalone, is summarized [
36]. Other reviews investigate the role of biofilms in biofouling [
37] and the induction of larval settlement by marine biofilms [
7]. However, “omics” approaches in biofouling studies has not been reviewed.
The main aim of this review is to evaluate the use of different omic techniques in biofouling research. In particular, we review biofouling-related metagenomic, transcriptomic, proteomic, and metabolomic studies. We provide examples of marine biofilms on man-made surfaces and the settlement of commercially important invertebrate species and those that cause significant industrial damage. Studies of biofouling on living organisms (epibiosis) are omitted. Specific emphasis is given to metagenomic and proteomic studies. Finally, we highlight the future directions of using omic techniques and a combination of omic techniques in biofouling studies.
2. Omics in context
“Omics” techniques can be daunting for beginners. These techniques originate in the health sector and are dominated by for-profit businesses, proprietary kits, platforms, and a special language for each subdiscipline that includes acronyms and jargon. If you were involved in the initial planning and funding the development of “omics ”, you would know that there were many assumptions and limits placed on funding for this technology. An example is funding and planning for initial work usually stopped at the cellular level. One can see the consequences of this narrow approach today as the technology spreads to human health, microbial ecology, metazoans, biofouling and other communiteis as well as environmental health.
At the hands-on level, the kits and specialized tools make benchwork easy. A drawback to the kits approach is many users do not understand what they are doing. Lack of understanding makes it hard to problem solve and to understand what kind of kits might be correct for the interest you have. Many of the kits are optimized for specific research and final products. If ones work does not fit exactly, then there may be nuances that impact the research.
Finally, “omics projects” generate a lot of what is known as “big data”. So much data that new data analysis is essential and comes with its own jargon, like pipeline and trivial names for analysis techniques. For most users, the solution is to take advantage of services provided by centers at universities or private companies that accept samples at some level, process them to generate the data, and then do preliminary analysis that enables a user to answer their specific questions.
To summarize, beginning “omics research” is like going to a foreign country without being able to read or speak the language. Luckily, the “omics” centers are jammed with kind and helpful people who will help you with the basics. However, these people do not necessarily know about marine biology and biofouling. Thus, you have to learn the language, the culture, the power, and the limits of “omics” technology. Once you can understand, exciting new pathways for answering your questions will emerge. As a user, you can tweak what you are doing to help you understand the output and answer your specific research questions.
3. Use of metagenomics in biofouling research
Pioneering studies of Prof. Claude E. ZoBell and his colleagues from Scripps Institution of Oceanography (USA) provided the first information about the diversity of marine microbes in marine biofilms. Culture-dependent techniques were developed that for years allowed researchers to isolate and investigate the properties of marine bacteria. However, less than 1% of marine bacteria can be cultivated in the laboratory using traditional techniques [
38]. Thus, culture-dependent techniques limit our understanding of microbial diversity. Modern metagenomic techniques allow the identification of all microbes from biofilms and seawater and estimate their abundances (
Table 2).
The majority of metagenomic studies use amplicon sequencing of small pieces (300-1000 bp) of 16S ribosomal DNA for prokaryotes and pieces of 18S ribosomal DNA and pieces of COI (cytochrome oxidase I) for eukaryote identification in biofouling communities. Metagenomics is the study of DNA sequences from microbes and from metazoans [
39]. A key step is the use of the polymerase chain reaction (PCR) for making many copies of existing sequences of DNA that have been extracted from your environmental sample. For PCR you use specific DNA “primer” sequences that you add that enable you to amplify specific genes [
40]. The amplified DNA is called amplicons. These amplicons you use to identify organisms (microbes, animals, or plants) by comparison of obtained DNA sequences with ones in the database. There are many databases but the most used ones are NCBI (National Institutes of Health, USA) and RDP (Ribosomal Database Project from Michigan State University, USA).
The first marine metagenomic study was conducted in 1991 by Dr. Norman R. Pace and colleagues who cloned 16S DNA of the picoplankton community from the Pacific Ocean [
41]. A few years later, one of the first metagenomics studies of biofilms was done on sulfate-reducing bacteria from marine sediments [
42]. In this study, 16S DNA genes were cloned in
E.coli and identified by Sanger sequencing. The development of sequencing techniques (
Table 2) and the reduction of costs and labor time resulted in many biofouling studies that utilize metagenomic approaches, which are reviewed using selected examples below.
3.1. Metagenomics of biofilms on man-made substrata
Biofilms are a significant part of biofouling. Knowing the composition of the biofilm community is important for environmental toxicology, forensics, understanding surface microbe interaction, management, and ecology. For example, investigators using high throughput 454 pyrosequencing, the detection of light released during nucleotide incorporation during the polymerase chain reaction [
51] of 16S DNA genes. Analysis of pyrosequencing results demonstrated that bacterial communities developed on black and white panels exposed to fouling in the sea were different [
45]. However, classes Alphaproteobacteria and Firmicutes dominated in all biofilms. Another study that used the same next-generation sequencing (NGS) technique investigated the composition of microbial communities developed on a bioreactor [
52]. The investigators found that microbial diversity decreased under high aeration. Similarly, results of pyrosequencing demonstrate that aeration affected the diversity and species richness of bacterial and archaeal communities on osmosis membranes [
46].
Illumina is next-generation, high-throughput “sequencing by synthesis technology” based on tracking the addition of fluorescently labeled nucleotides during DNA polymerization [
51]. Illumina technology has frequently been used in biofouling metagenomic studies for analysis of species composition of microbial biofilms developed on man-made structures [
53,
54,
55]. Analysis of microbial communities developed in membrane bioreactors by Illumina sequencing showed that high salinity increased the proportion of
Flavobacterium, Aequorivita, Gelidibacter, Microbacterium, and
Algoriphagus genera [
47]. Another study investigated biofouling developed on sea gliders using 16S and 18S amplicon sequencing [
56]. The researchers observed differences in the number of OTUs (operational taxonomic units) between biofilms on different parts of the glider. Bacteria belonging to the classes Gamma- and Alpha-proteobacteria dominated prokaryotic communities, while hydrozoans and Chlorophyta dominated eukaryotic communities. Distinct bacterial communities were detected using MiSeq Illumina on stainless steel exposed to biofouling in a Northern Portugal port [
57]. In contrast, bacterial communities developed on stainless steel, polyethylene and titanium investigated by MiSeq Illumina shared some similarities [
58]. However, the communities changed over time. Most studies using NGS technology have found that microbial communities developed on submerged surfaces are different from those in seawater [
56,
59].
High throughput sequencing using the Illumina platform is used in studies of metabolic assembled genomes from biofouling communities. For example, a study by Walter et al. [
60] of microbial mats of a coastal lagoon showed that they are dominated by cyanobacteria responsible for photosynthesis, Chroococcales responsible for nitrogen and ammonia assimilation, and Desulfobacterales contributing to sulfate reduction. Taxonomic and functional metagenomic analysis of biofilms developed in different locations and their effect on larval settlement of the polychaete
Hydroides elegans was investigated [
61]. The investigators demonstrated that the microbial communities were significantly different in coastal waters as compared to off-shore waters. However, the functional genes were similar between sites and related to carbohydrates, amino acids, and protein metabolism.
The temporal shift of microbial communities on wood and concrete used for artificial reefs was investigated using Ion Torrent sequencing technology [
62]. This technology is based on the detection of hydrogen ions during DNA polymerization [
63]. The investigators found that the relative abundances of bacterial phyla decreased differently on different substrata over time [
62]. Similarly, microbial communities developed in brackish waters on different classes of stainless steel showed that one type of steel had different microbial communities dominated by Actinobacteria, while Proteobacteria dominated other types. Ion Torrent sequencing technology is also used to examine microorganisms on membranes [
48] and in bioreactors [
64].
MinION Oxford nanopore is a new sequencing technology (
Table 2). It can be used for amplicon sequencing as well as for the assembly of full genomes. The advantage of this technique is that it does not require a PCR step and can detect femtograms of DNA [
65]. Probably because of this technology is novel, only a few biofouling-related studies were found (
Table 2). Complete genomic sequences of biofouling bacteria
Vibrio cambelli [
50,
66] and
Pseudomonas putida [
67] were determined using this technology. Similarly, the genome of important fouling species was sequenced using Illumina and Oxford nanopore technologies [
49].
NGS technology can be used to assess the composition of biofilms and investigate their effect on the settlement of macrofoulers. Biofilms and their mussel-inducing activity were investigated by Illumina MiSeq [
68]. The Phylum Proteobacteria dominated all biofilms. The composition of biofilms inducing
Mytilus coruscus settlement was different from low inductive ones. The coral settlement-inducing activity of biofilms of crustose coralline alga was investigated by Illumina metagenomics and isolation of bacteria [
69]. From the data analysis, there was no correlation between inductive settlement capacities and species of bacteria. A recent study demonstrated that biofilms formed in different environmental conditions affect the formation of macrofouling communities [
70]. Biofilms were developed in areas of high and low anthropogenic impact and then were translocated. A low settlement rate of non-indigenous species
Watersipora subatra on biofilms developed in marine protected areas and moved to the area with high anthropogenic impact. This demonstrates that metagenomics can be a useful tool in marine conservation.
Many studies investigated biofouling communities developed on plastics floating in the oceans or deposited in sediments using metagenomic approaches. Due to limited space, we refer readers to reviews about this topic [
71,
72,
73]. In general, the studies confirmed the presence of diverse and specific microbial communities associated with different types of plastic which were different from microbes in the water column. Thus, microbial biofilms on the surface of plastic are called the “plastisphere” [
71]. Additionally, omic studies are used to identify species responsible for the degradation of plastics [
74].
3.2. Metagenomics of biofilms on antifouling coatings and biocides
Metagenomics of biofilms on antifouling coatings opened up a new world for researchers and industries. First, the studies indicated that microbial communities contain many species of Prokaryotic and Eukaryotic organisms. Diverse microbial communities were observed during a 1-year study of microbial biofilms on 11 biocidal antifouling coatings in Oman waters [
54]. Communities were dominated by Alpha- and Gamma-Proteobacteria, Cyanobacteria, and Flavobacteria. Similarly, clear differences between communities representing three different types of coatings were observed. 454 pyrosequencing of 16S rRNA demonstrated spatiotemporal changes in microbial communities on different antifouling coatings in French waters [
75].
Second, metagenomic studies enable the identification of biofouling species present on fouling management coatings. 454 pyrosequencing of 16S rRNA was used to evaluate microbes in biofilms on different coatings in French coastal waters [
75]. The study reports that the biocidal coating reduced the abundance of Bacteroidetes. Similarly, clear differences in microbial communities developed on ZnO nanorod coatings and copper-based coating on fishing nets were recorded using Illumina MiSeq technology in Oman coastal waters [
76]. 16S and 18S amplicon Illumina MiSeq sequencing were used to evaluate prokaryotic and eukaryotic communities on different substrata in New Zealand waters [
77]. The authors found that the orientation of the substrate, as well as the presence of antifouling coatings, impacted the composition of microbial communities. Similarly, clear differences in prokaryotic and eukaryotic communities on antifouling coatings were detected in another study [
55]. However, in a different study, only prokaryotic communities were different between toxic and non-toxic coatings on sea gliders [
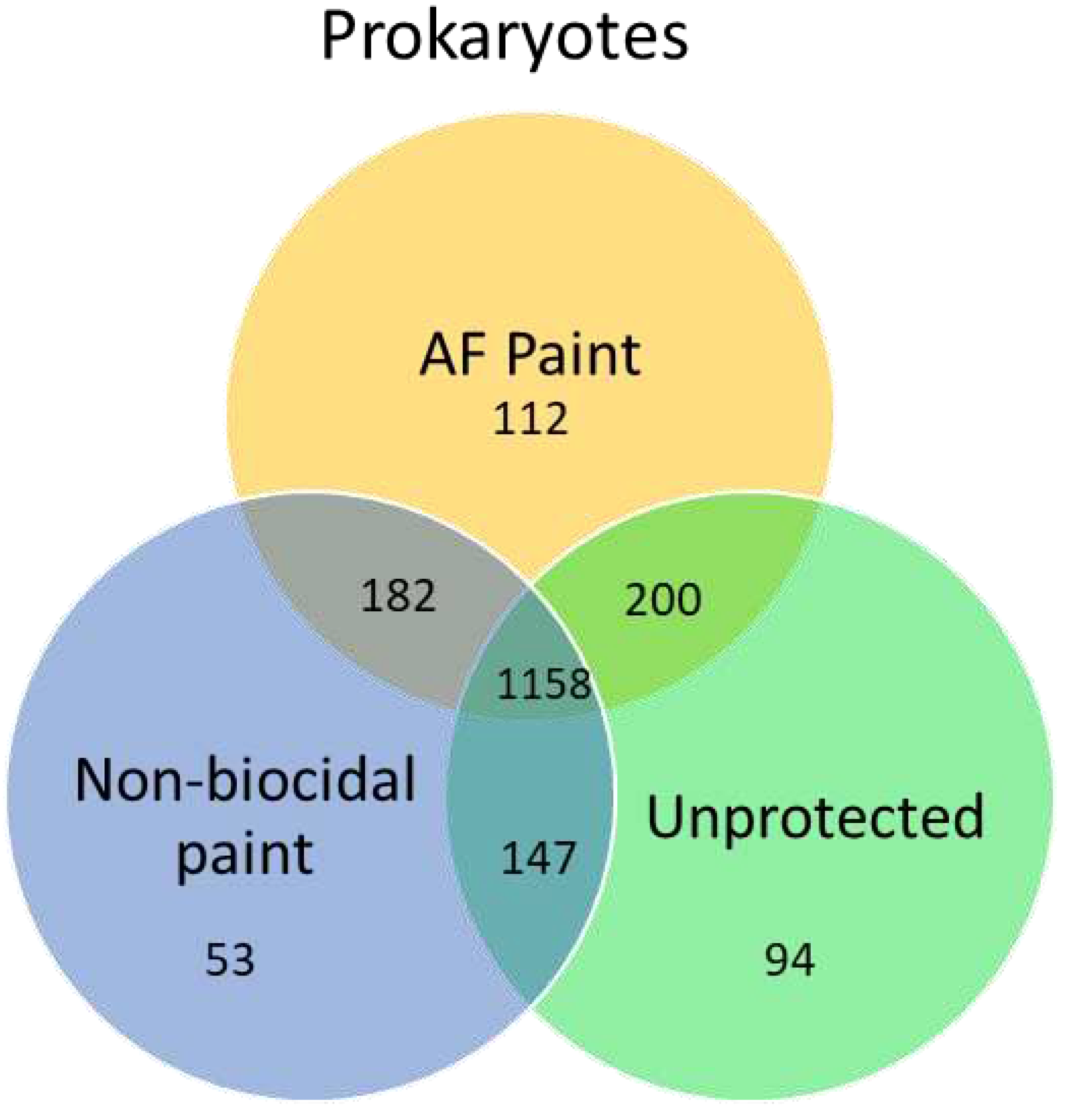
56]. In this study, the majority of bacteria species were shared between different surfaces (
Figure 3). The highest number of unique operational taxonomic units (OTUs) were observed on biocidal antifouling paint, while the lowest OTUs were found in non-biocidal paint. In another study, marine biofilms on biocidal and non-biocidal antifouling coatings were studied using 16S amplicon MiSeq sequencing [
53]. Genera
Loktanella, Sphingorhabdus, and
Erythrobacter dominated biocidal coatings, while
Portibacter dominated fouling release ones. Though for decades people working with roof shingle fouling focused on one cyanobacterium
Gloeocapsa spp., and when dust samples from shingles were analyzed
Gloeocapsa proved to be a minor biofouling component [
34]. Shingle manufacturers interested in understanding their biofouling problem could take advantage of microbiome analysis and use it to develop an effective antifouling defense.
Metagenomics is a powerful tool to detect the toxicity of coatings because of differences between microbial communities developed on toxic and non-toxic substrates. This can be used in forensic studies to detect compounds that are not listed in the recipes for industrial products. Ward et al. [
78] reported the results of microbial growth on seven different kinds of plastic preproduction pellets in seawater. The investigators report that biofilms on plastic pellets were initially different and remained different on pellets, especially on PVC which was different from all the rest of the microbiomes on pellets at every sample interval. After 70 days, there were four distinct microbiomes on the pellets, with the convergence of microbiomes on similar plastics. The authors found that groups of bacteria associated with toxic fouling management coatings were found on some plastics at all time intervals suggesting compounds leaching from the plastic had a role in biofilm community composition. The more we understand microbial communities using metagenomics the better we will be able to manage human health, ecosystem health, and food security and develop effective antifouling and antimicrobial defenses.
3. Environmental DNA (e-DNA)
One use of metagenomics for metazoans in the biofouling community is environmental DNA (eDNA) [
79]. The idea is that DNA in water from near a fouling community can be filtered and then one can selectively amplify particular genes, such as Cytochrome Oxidase I (COI) or 18S ribosomal DNA, to determine members of the fouling community including invasive and cryptic species [
80]. Obtained sequences can be compared with publicly available data and groups of interest can be identified. What is excellent about this approach is one needn't be an expert in systematics because there are extensive public databases containing systematic information that can be queried and used.
Several groups around the world have developed this approach to make lists of the species in their biofouling communities. For example, e-DNA was used to detect the invasive golden mussel
Limnoperna fortunei in Chinese waters [
80]. Another study used e-DNA from plastic litter to detect 4 invasive species [
81]. However, there are challenges associated with this technique. The e-DNA technique is too new to answer questions about the presence or absence of particular species and estimate their abundance at the time of sampling. No one knows e-DNA shedding rates for different kinds of organisms. The sequences of some biofouling species are either absent in the databases or primers (for COI and 18S) do not allow distinguishing them from other similar species. The DNA distribution in water is patchy [
82]. The methods of sampling and DNA extraction can have an impact on the results [
83]. Finally, all e-DNA is not equal and in complex communities, some dominant species can camouflage the presence of rare species.
During the development of this review, we realized that there are a lot of important unknowns in the development and use of “omic” techniques in the environment. For example, the half-life of arginine carboxy-terminal signal peptides in marine environments is approximately 2 hours. The half-life of eDNA is in a similar range. However, though eDNA might still be present, the pieces could be too short to be useful to be amplified as intact sequences of DNA hundreds of base pairs in length are required to identify organisms. More generally, the question of shedding rates and forms of shedding for different kinds of metazoans remains open. For example, do mollusks produce the same amount of eDNA per gram of a living animal as arthropods or polychaetes, or cnidaria? There is plenty to do to improve “omic” techniques in the future.
4. Transcriptomics
Transcriptomics is defined as “everything RNA” (
Figure 1). Transcriptomics allows us to detect DNA sequences that are transcribed in the current moment and used by biofouling organisms. Like other “omics” approaches, it has its jargon, incredible strengths, understated weaknesses, and the explosive growth of science on a rapidly expanding technological frontier (
Figure 2). In the RNA world, Deep Sequencing means the same thing as Next Generation Sequencing in the DNA world and refers to the output of a very powerful sequencing platform. Transcriptomics has its limitations. Compared to DNA, RNA is less stable, thus extra precautions need to be taken. Experts estimate that RNA amounts to as little as 1% of the total can be identified [
25]. The result is the equivalent of a fingerprint for a cell at one point in time or a disease state, or an individual, or in our dreams, a community. The field is in the rapid discovery of new kinds of RNA that are important to cellular function and disease states. 1% seems like a fine level of precision until one realizes that most enzymes occur at much lower percentages. One must also remember that like all “omic” techniques, processing and poorly understood amplification bias blurs the final picture. Nevertheless, transcriptomics is revolutionizing our understanding of how cells, organisms, and diseases work.
In biofouling research, transcriptomics has been used to investigate genes expressed during larval metamorphosis, and genes transcribed in biofilms. For example, the treatment of biofilms with a quorum sensing blocker (furanone) changed the expression of 61 genes [
84]. These genes were responsible for quorum sensing signal production, flagellar assembly, and aspartate kinase. Subsequently, changes in biofilms resulted in a lower larval settlement. The effect of climate change on larval development was studied using a transcriptomic approach [
85,
86,
87]. The experiment showed that ocean acidification suppressed the immune response pathways of pacific oyster
Crassostrea gigas larvae [
88]. Transcriptomics can provide additional information about the mechanism of action of antifouling compounds. The effect of Di(1H-indol-3-yl)methane on the transcription of DNA genes of the bryozoan larvae
Bugula neritina showed that this “environmentally friendly” compound down-regulates steroid hormone biosynthesis genes, which could have long-term effects on the bryozoan [
25]. Transcriptomes have been used in the analysis of genes responsible for adhesive proteins of barnacles [
89] and mussels [
90]. In most cases, transcriptomics is combined with proteomics for a more complete analysis of expressed genes and produced proteins [
91].
5. Proteomics
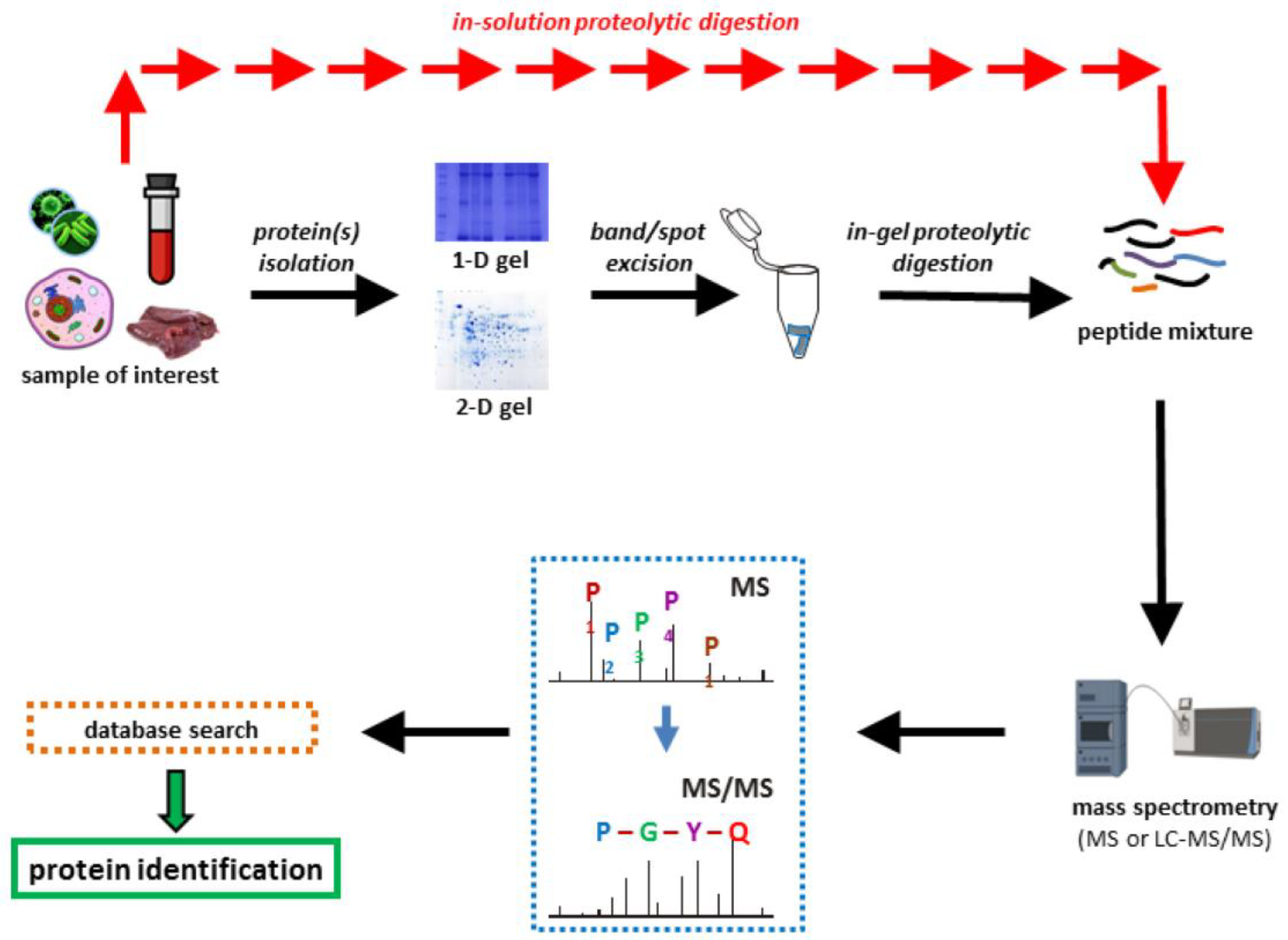
Proteomics is the study of peptides and their associated proteins (
Figure 1). Proteomics is based on the use of high-resolution mass spectrometry [
92,
93]. In the standard approach, a researcher isolates proteins from the organism or community, fractionates them, breaks the proteins into peptides with a pure enzyme (i.e. trypsin), and identifies the mass of resulting peptides (
Figure 4). Finally, extensive databases (such as NCBI ) that contain masses and amino acid sequences of known peptides and proteins are used to determine peptides and their origins. There are free user-friendly analysis tools (such as Mascot and Scaffold-TM) that enable a novice to understand what proteins were present in the mixture. The databases provide a rich context that enables one to deduce the families of proteins to which their peptides belong.
As with all the omics, there is a complex language that includes trivial names for analysis techniques i.e mascot, scaffold, etc., and very practical concepts like false discovery rate and the likelihood that your peptide fragments are from specific families of proteins.
With Darwin and Wallace, biologists' problem-solving skills increased because they can use principles of evolution, like relatedness to help solve biological puzzles. Proteins and their evolutionary relations in series like Protein Structure and Function provide insight into protein evolution and relatedness and help us understand biological phenomena. The literature is so vast, no researcher can know it. For example, there is a compilation of over 700 papers in a series that describes different kinds of proteolytic enzymes [
93]. However, modern databases of protein sequences provide powerful insight into protein structure and function. Pathways common to all metazoans provide particular insights.
Proteomics of biofouling organisms can be used to identify proteins expressed during metamorphosis or during inhibition of metamorphosis (
Table 3). Most of the studies conducted are with dominant biofouling species. Data for other fouling species are less common. In contrast to terrestrial species whose proteomes are well studied, marine species receive less attention.
One of the first studies analyzed the protein expression of a bryozoan
Bugula neritina and a barnacle
Balanus (= Amphibalanus) amphitrite during larval metamorphosis [
94]. Induction and inhibition of metamorphosis for both species changed the phosphorylation status of proteins. Similar results were observed for the phosphorylation of proteins of the polychaete
Hydroides elegans [
95]. More than 700 proteins were identified during the metamorphosis of
B. amphitrite [
96]. Another study with the bryozoan
B.neritina reported that proteins involved in energy metabolism were downregulated while proteins responsible for transcription and translation were upregulated during the metamorphosis of this species [
97].
Proteomics enabled the study of the effect of climate change on the proteins of marine organisms (
Table 3). The effect of multiple stressors associated with climate change on the proteome of oyster larvae
Crassostrea gigas was investigated [
98]. The interpretation of the data suggested that climate change will have a significant impact on proteins expressed in larval stages. Similar results were found for another species of oyster
Crassostrea hongkongensis [
99]. Up-regulated and down-regulated proteins have been found during the exposure of oyster larvae
Saccostrea glomerata to acidic conditions [
100]. Exposure of
Mytilus species to different salinities leads to changes in proteins involved in energy metabolism and scavenging of reactive oxygen species [
101].
Some proteomics studies focus on adhesive proteins (
Table 3). It was hypothesized that metazoan glue curing was related to other processes where proteins coagulated in water. Testing the hypothesis with barnacle glue provided a preliminary conclusion that barnacle glue curing is related to blood clotting [
102]. However, many biologists, biochemists, and structural protein chemists have not accepted the hypothesis. 14 years later, further work with collaborators from other disciplines confirms the relationship between adhesive glue and hemolymph proteins [
103]. In the study, researchers have shown that barnacle glue curing involves an ancient pathway, the innate immune response, that neutralizes pathogens. The researchers found evidence that in addition to neutralizing pathogens the innate immune response provided chemistries, like reactive oxygen species, involved oxidative crosslinking and the processes involved in reworking and calcifying the barnacle glue. This has changed our view of biological adhesives. In addition to complex structural protein self-assembly, the process also involves many enzymes, a variety of reactive species from several sources, and secondary processes that result in calcification and curing for weeks if not months [
103]. Additionally, peptides generated during and after this process of glue formation and glue curing mediate gregariousness in barnacles, oysters, and a large number of other processes that organize marine communities [
103].
Proteomics has several weaknesses. Many proteins of marine organisms, including biofouling organisms, are not well studied. Thus, their identification is hard. Complete genomes of most marine organisms are not available, which makes the prediction of proteins difficult. Thus, the function of novel proteins is difficult to predict. A large number of proteins, make it difficult to identify functionally important proteins especially enzymes, existing in lower numbers in comparison with dominant ones. This problem can be mediated by employing additional techniques such as antigen-antibody reporting which amplifies signals with reporter enzymes. As with all new techniques, time will enable a better understanding and interpretation of the proteomics data.
Figure 4.
The workflow of proteomics. Reproduced with permission from [
92].
Figure 4.
The workflow of proteomics. Reproduced with permission from [
92].
Table 3.
Relevant proteomic studies of marine biofouling organisms.
Table 3.
Relevant proteomic studies of marine biofouling organisms.
| Group |
Species |
Propose of study |
References |
| Prokaryotes |
Cyanobacteria |
Effect of hydrodynamics |
[104] |
| |
mixed communities |
Biocorrosion |
[105] |
| |
mixed communities |
Effect of contaminants |
[106] |
| Arthropoda |
Balanus amphitrite |
Proteins during larval metamorphosis |
[96] |
| |
Balanus amphitrite |
Larval response to AF compound |
[107] |
| |
Balanus amphitrite |
Glue proteins |
[103] |
| |
|
|
|
| Bryozoa |
Bugula neritina |
Proteins during larval metamorphosis |
[108] |
| |
Bugula neritina |
Impact of AF compound |
[109] |
| Polychaeta |
Hydroides elegans |
Proteins during larval metamorphosis |
[95] |
| Mollusca |
Crasosstrea gigas |
Effect of climate change |
[98] |
| |
Crassostrea hongkongensis |
Effect of climate change |
[99] |
| |
Saccostrea glomerata |
Effect of climate change |
[100] |
| |
Mytilus trossulus
M. galloprovincialis
|
Effect of salinity |
[101] |
6. Metabolomics
Metabolomics studies all the metabolites found in organisms and the environments around them [
110]. In a metabolomic project, metabolites are extracted first by polar and non-polar solvents. Then, crude extracts are fractionated using chromatography techniques (gas or liquid chromatography) and individual compounds are identified using high-resolution mass spectrometry (MS). Finally, the compounds are identified by comparison of their retention times, fragmentation patterns, and m/z values by comparison with compounds in libraries. However, due to the limitations of the methods at this point, only small molecules can be analyzed. Metabolomics can be targeted or untargeted [
111]. In the targeted method, researchers focus on the analysis of individual metabolites involved in a specific pathway. This method can be used for “fingerprint” analysis of samples. In untargeted metabolomics, thousands of metabolites from an organism or community are analyzed. This method is useful for the identification of biomarkers.
For researchers working with biofouling, metabolomics is intriguing but daunting. Metabolomics can be used as a snapshot to examine the physiological impacts of toxins or biocides on organisms or communities [
112,
113]. The effect of an antifouling compound on metabolites of
Vibrio sp. was studied [
113]. Researchers found that during treatment some metabolites were downregulated while others were upregulated. Overall, the number of metabolomic studies is limited in biofouling studies due to a lack of basic understanding of the metabolic pathways in biofouling animals.
Combinations of omics techniques can be useful. Biofouling was studied on protected and non-protected surfaces using 18S amplicon sequencing and metabolomic analysis [
112]. The results demonstrated that the metabolomes of biofouling communities developed on artificial substrata in different sites were different from each other. Metabolites of biofouling organisms found on different antifouling paints were also different. A combination of metabolomics with metagenomics approaches showed that different metabolomes were associated with different biofouling phyla [
112]. A study of metabolomics and proteomics of a marine bacterium
Pseudoltermonas liplytica in planktonic and attached cultures showed different metabolites were produced [
114]. Proteomics showed major differences in peptidases, oxidases, and membrane proteins.
Metabolomics can be used to determine biofouling species, especially in the case of microbes. Identification of bacteria by their metabolites is currently used by a MALDI-biotyper. However, the biotyper is not very effective for the identification of marine bacteria due to limited databases. However,
Persicivirga (
Nonlabens)
mediterranea,
Pseudoalteromonas lipolytica, and
Shewanella sp. were identified based on their metabolites using LC-MS [
115]. An MS/MS database for tested species of marine bacteria allowed the investigators to distinguish them based on their metabolites.
The main difficulties with metabolomics, as with many omics techniques, are the absence of standardized methods for the extraction and analysis of metabolites and difficulties in the quantification and identification of compounds. Lack of information on the metabolic processes of marine organisms limits the analysis of metabolites. For example, the major fouling species barnacles,
Amphibalanus (Balanus) amphitrite, polychaetes
Hydorides elegans, and bryozoans
Bugula neritna are missing in the metabolic pathway MetaCyc (
https://metacyc.org/) and BioCyc databases (
https://biocyc.org/). An additional difficulty is associated with the analysis of the metabolome of complex communities that contain many species. The combination of metabolomics with other omic techniques, for example, proteomics and metagenomics, can provide additional insight into biofouling processes. Standardization of extraction and analysis methods as well as the development of new analytical methods for the identification of compounds will help in the future development of metabolomics.
7. Conclusions and future outlook
Our review demonstrates that the proportion of biofouling publications that utilize “omics” techniques is quite low. However, this number is constantly increasing. “Omics” provide information about the composition of biofouling communities and allow the detection of biofouling and invasive species. “Omics” approaches allow us to investigate the response of biofouling species to environmental factors and biocides. “Omics” opens new paths to study and better understand biofouling species and communities and their impact on marine installations. However, “omics” approaches have their limitations that need to be considered during investigations. Some of these limitations are specific to each technique but others are general. The limitations and possible solutions were highlighted during this review.
There is extraordinary power in combining “omics” approaches. Most combination approaches are by multidisciplinary teams that are asking specific questions and have an interest in crossing the boundaries of each “omics” approach. Researchers in the “omics” silos have plenty of development to do within each technology and may find research at the boundaries less interesting.
Each “omics” technology has its strengths and weaknesses. All are limited by competition between targets, limits to the technologies, inherent biases in technological approaches, and lack of a larger biological perspective. Combining the techniques enables using one “omics” to overcome a shortcoming of another. As one can surmise, making this experimental stride requires a substantial understanding of the techniques involved and then the ability to communicate across sub-disciplinary boundaries in the context of the biological questions asked.
Specialized language makes it difficult to understand and interpret the ‘omics’ data for biologists and non-specialists. Help from colleagues and specialized agencies is essential. One solution is to simply look only for targeted genes or compounds or metabolic pathways. Large massive data require careful analysis and quality assessment. Additionally, researchers need to include informaticians or big data analytics in their research design. Finally, biological meaning can be lost during the analysis.
Using “omic” approaches could be beneficial in the development of the non-toxic or low-toxic antifouling defense. The toxicity of antifouling compounds and their mechanism of action can be tested using transcriptomics, metabolomics, and proteomics approaches. The presence of particular micro- and macro-fouling organisms on the surface of antifouling paints can be detected by metagenomic approaches. Additionally, one would be able to identify the role of microorganisms present on coatings or marine installations. Using these approaches will be easier and cheaper in the future. We believe that in the future even non-specialists would be able to identify the presence of invasive or biofouling species in industrial marine applications.
During the preparation of this review, we realized that most “omics” publications are dealing with dominant species of micro- and macroorganisms. The information about other species, their genomes, proteins, and metabolites are limited in the databases. For some biofouling species, we have no information about their genes, proteins, and metabolic pathways. For example, even the very common
Balanus genus has two sequences in the NCBI database. Only one complete shotgun genome sequence for
Amphibalanus (Balanus) amphitrite was found (
https://www.ncbi.nlm.nih.gov/nuccore/VIIS01000292.1). This makes it difficult to interpret the results even for dominant species. In contrast, the data show that biofouling communities are highly diverse and composed of many biofouling species. At this point, it is difficult to use “omics' approaches for biofouling communities. Thus, in the future, more studies should be done to solve this and other problems associated with the use of “omics” technologies.
Author Contributions
Conceptualization, S.D. and D.R.; data curation, S.D.; writing—original draft preparation, S.D. and D.R.; writing—review and editing, S.D. and D.R.; visualization, S.D.; project administration, S.D. and D.R.; funding acquisition, S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by Sultan Qaboos University, grant number IG/CEMB/ XXX IG/DVC/CEMB/21/01.
Data Availability Statement
Data are available upon request from the authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wahl, M. Marine Epibiosis. I. Fouling and Antifouling: Some Basic Aspects. Marine Ecology Progress Series 1989, 58, 175–189.
- Carvalho, D.; R, C.C.C. Marine Biofilms: A Successful Microbial Strategy With Economic Implications. Front. Mar. Sci. 2018, 5. [CrossRef]
- Railkin, A.I. Marine Biofouling: Colonization Processes and Defenses; CRC Press, 2003; ISBN 978-0-203-50323-2.
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS Matrix: The “House of Biofilm Cells.” Journal of Bacteriology 2007, 189, 7945–7947. [CrossRef]
- Flemming, H.-C.; Wingender, J. The Biofilm Matrix. Nature Reviews Microbiology 2010, 8, 623–633. [CrossRef]
- Rittschof, D. Body Odors and Neutral-Basic Peptide Mimics: A Review of Responses by Marine Organisms. Integr Comp Biol 1993, 33, 487–493. [CrossRef]
- Dobretsov, S.; Rittschof, D. Love at First Taste: Induction of Larval Settlement by Marine Microbes. International Journal of Molecular Sciences 2020, 21, 731. [CrossRef]
- Dobretsov, S.; Dahms, H.-U.; Qian, P.-Y. Inhibition of Biofouling by Marine Microorganisms and Their Metabolites. Biofouling 2006, 22, 43–54. [CrossRef]
- Hadfield, M.G. Biofilms and Marine Invertebrate Larvae: What Bacteria Produce That Larvae Use to Choose Settlement Sites. Annual Review of Marine Science 2011, 3, 453–470. [CrossRef]
- Yebra, D.M.; Kiil, S.; Dam-Johansen, K. Antifouling Technology—Past, Present and Future Steps towards Efficient and Environmentally Friendly Antifouling Coatings. Progress in Organic Coatings 2004, 50, 75–104. [CrossRef]
- Jones, G. 2 - The Battle against Marine Biofouling: A Historical Review. In Advances in Marine Antifouling Coatings and Technologies; Hellio, C., Yebra, D., Eds.; Woodhead Publishing Series in Metals and Surface Engineering; Woodhead Publishing, 2009; pp. 19–45 ISBN 978-1-84569-386-2.
- Maréchal, J.-P.; Hellio, C. Challenges for the Development of New Non-Toxic Antifouling Solutions. International Journal of Molecular Sciences 2009, 10, 4623–4637. [CrossRef]
- Pérez, H.; Vargas, G.; Silva, R. Use of Nanotechnology to Mitigate Biofouling in Stainless Steel Devices Used in Food Processing, Healthcare, and Marine Environments. Toxics 2022, 10, 35. [CrossRef]
- Kumar, S.; Ye, F.; Dobretsov, S.; Dutta, J. Nanocoating Is a New Way for Biofouling Prevention. Frontiers in Nanotechnology 2021, 3.
- Uskoković, V.; Bertassoni, L.E. Nanotechnology in Dental Sciences: Moving towards a Finer Way of Doing Dentistry. Materials (Basel) 2010, 3, 1674–1691. [CrossRef]
- Al-Fori, M.; Dobretsov, S.; Myint, M.T.Z.; Dutta, J. Antifouling Properties of Zinc Oxide Nanorod Coatings. Biofouling 2014, 30, 871–882. [CrossRef]
- Carl, C.; Poole, A.J.; Vucko, M.J.; Williams, M.R.; Whalan, S.; Nys, R. de Enhancing the Efficacy of Fouling-Release Coatings against Fouling by Mytilus Galloprovincialis Using Nanofillers. Biofouling 2012, 28, 1077–1091. [CrossRef]
- Rittschof, D.; Orihuela, B.; Genzer, J.; Efimenko, K. PDMS Networks Meet Barnacles: A Complex and Often Toxic Relationship. Biofouling 2022, 38, 876–888. [CrossRef]
- Rittschof, D.; Dobretsov, S. Ecosystem Restoration: Enhancing Ecosystem Services with Floating Aquaculture. Journal of Fisheries Science 2022, 4.
- Bonar, D.B.; Coon, S.L.; Walch, M.; Weiner, R.M.; Fitt, W. Control of Oyster Settlement and Metamorphosis by Endogenous and Exogenous Chemical Cues Available online: https://www.ingentaconnect.com/content/umrsmas/bullmar/1990/00000046/00000002/art00018 (accessed on 17 November 2019).
- Hidu, H. Gregarious Setting in the American OysterCrassostrea Virginica Gmelin. Chesapeake Science 1969, 10, 85–92. [CrossRef]
- Paul, V.J.; Ritson-Williams, R. Marine Chemical Ecology. Nat. Prod. Rep. 2008, 25, 662–695. [CrossRef]
- Webster, N.S.; Smith, L.D.; Heyward, A.J.; Watts, J.E.M.; Webb, R.I.; Blackall, L.L.; Negri, A.P. Metamorphosis of a Scleractinian Coral in Response to Microbial Biofilms. Appl. Environ. Microbiol. 2004, 70, 1213–1221. [CrossRef]
- Tran, C.; Hadfield, M.G. Larvae of Pocillopora Damicornis (Anthozoa) Settle and Metamorphose in Response to Surface-Biofilm Bacteria. Marine Ecology Progress Series 2011, 433, 85–96. [CrossRef]
- Xu, Y.; Zhang, L.; Wang, K.-L.; Zhang, Y.; Wong, Y.H. Transcriptomic Analysis of the Mode of Action of the Candidate Anti-Fouling Compound Di(1H-Indol-3-Yl)Methane (DIM) on a Marine Biofouling Species, the Bryozoan Bugula Neritina. Marine Pollution Bulletin 2020, 152, 110904. [CrossRef]
- Sánchez-Quinto, A.; Falcón, L.I. Metagenome of Acropora Palmata Coral Rubble: Potential Metabolic Pathways and Diversity in the Reef Ecosystem. PLOS ONE 2019, 14, e0220117. [CrossRef]
- Littman, R.; Willis, B.L.; Bourne, D.G. Metagenomic Analysis of the Coral Holobiont during a Natural Bleaching Event on the Great Barrier Reef. Environmental Microbiology Reports 2011, 3, 651–660. [CrossRef]
- Rothäusler, E.; Dobretsov, S.; Gómez, M.F.; Jofré-Madariaga, D.; Thiel, M.; Véliz, K.; Tala, F. Effect of UV-Radiation on the Physiology of the Invasive Green Seaweed Codium Fragile and Its Associated Bacteria. Marine Environmental Research 2022, 180, 105708. [CrossRef]
- Egan, S.; Harder, T.; Burke, C.; Steinberg, P.; Kjelleberg, S.; Thomas, T. The Seaweed Holobiont: Understanding Seaweed–Bacteria Interactions. FEMS Microbiol Rev 2013, 37, 462–476. [CrossRef]
- The Sponge Hologenome | MBio Available online: https://journals.asm.org/doi/full/10.1128/mBio.00135-16 (accessed on 6 May 2023).
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The Sponge Holobiont in a Changing Ocean: From Microbes to Ecosystems. Microbiome 2018, 6, 46. [CrossRef]
- Lauritano, C.; Ferrante, M.I.; Rogato, A. Marine Natural Products from Microalgae: An -Omics Overview. Marine Drugs 2019, 17, 269. [CrossRef]
- Pierella Karlusich, J.J.; Ibarbalz, F.M.; Bowler, C. Phytoplankton in the Tara Ocean. Annual Review of Marine Science 2020, 12, 233–265. [CrossRef]
- Beata, G. The Use of -Omics Tools for Assessing Biodeterioration of Cultural Heritage: A Review. Journal of Cultural Heritage 2020, 45, 351–361. [CrossRef]
- Chandramouli, K.H.; Qian, P.-Y.; Ravasi, T. Proteomics Insights: Proteins Related to Larval Attachment and Metamorphosis of Marine Invertebrates. Front. Mar. Sci. 2014, 1. [CrossRef]
- Nguyen, T.V.; Alfaro, A.C.; Mundy, C.; Petersen, J.; Ragg, N.L.C. Omics Research on Abalone (Haliotis Spp.): Current State and Perspectives. Aquaculture 2022, 547, 737438. [CrossRef]
- Qian, P.-Y.; Cheng, A.; Wang, R.; Zhang, R. Marine Biofilms: Diversity, Interactions and Biofouling. Nat Rev Microbiol 2022, 20, 671–684. [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic Identification and in Situ Detection of Individual Microbial Cells without Cultivation. Microbiol Rev 1995, 59, 143–169. [CrossRef]
- Escobar-Zepeda, A.; Vera-Ponce de León, A.; Sanchez-Flores, A. The Road to Metagenomics: From Microbiology to DNA Sequencing Technologies and Bioinformatics. Frontiers in Genetics 2015, 6.
- Machida, R.J.; Knowlton, N. PCR Primers for Metazoan Nuclear 18S and 28S Ribosomal DNA Sequences. PLOS ONE 2012, 7, e46180. [CrossRef]
- Schmidt, T.M.; DeLong, E.F.; Pace, N.R. Analysis of a Marine Picoplankton Community by 16S RRNA Gene Cloning and Sequencing. J Bacteriol 1991, 173, 4371–4378. [CrossRef]
- Devereux, R.; Mundfrom, G.W. A Phylogenetic Tree of 16S RRNA Sequences from Sulfate-Reducing Bacteria in a Sandy Marine Sediment. Applied and Environmental Microbiology 1994, 60, 3437–3439. [CrossRef]
- Huang, Y.-L.; Li, M.; Yu, Z.; Qian, P.-Y. Correlation between Pigmentation and Larval Settlement Deterrence by Pseudoalteromonas Sp. Sf57. Biofouling 2011, 27, 287–293. [CrossRef]
- Egan, S.; James, S.; Holmström, C.; Kjelleberg, S. Correlation between Pigmentation and Antifouling Compounds Produced by Pseudoalteromonas Tunicata. Environmental Microbiology 2002, 4, 433–442. [CrossRef]
- Dobretsov, S.; Abed, R.M.M.; Voolstra, C.R. The Effect of Surface Colour on the Formation of Marine Micro and Macrofouling Communities. Biofouling 2013, 29, 617–627. [CrossRef]
- Zhang, Q.; Jie, Y.W.; Loong, W.L.C.; Zhang, J.; Fane, A.G.; Kjelleberg, S.; Rice, S.A.; McDougald, D. Characterization of Biofouling in a Lab-Scale Forward Osmosis Membrane Bioreactor (FOMBR). Water Res 2014, 58, 141–151. [CrossRef]
- Guo, X.; Miao, Y.; Wu, B.; Ye, L.; Yu, H.; Liu, S.; Zhang, X.-X. Correlation between Microbial Community Structure and Biofouling as Determined by Analysis of Microbial Community Dynamics. Bioresour Technol 2015, 197, 99–105. [CrossRef]
- Zhang, L.; Xu, L.; Graham, N.; Yu, W. Unraveling Membrane Fouling Induced by Chlorinated Water Versus Surface Water: Biofouling Properties and Microbiological Investigation. Engineering 2022, 15, 154–164. [CrossRef]
- Yang, J.-L.; Feng, D.-D.; Liu, J.; Xu, J.-K.; Chen, K.; Li, Y.-F.; Zhu, Y.-T.; Liang, X.; Lu, Y. Chromosome-Level Genome Assembly of the Hard-Shelled Mussel Mytilus Coruscus, a Widely Distributed Species from the Temperate Areas of East Asia. GigaScience 2021, 10, giab024. [CrossRef]
- Colston, S.M.; Ellis, G.A.; Kim, S.; Wijesekera, H.W.; Leary, D.H.; Lin, B.; Kirkup, B.C.; Hervey, W.J.; Vora, G.J. Complete Genome Sequences of Two Bioluminescent Vibrio Campbellii Strains Isolated from Biofouling Communities in the Bay of Bengal. Genome Announcements 2018, 6, e00422-18. [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr Protoc Mol Biol 2018, 122, e59. [CrossRef]
- Ma, J.; Wang, Z.; Yang, Y.; Mei, X.; Wu, Z. Correlating Microbial Community Structure and Composition with Aeration Intensity in Submerged Membrane Bioreactors by 454 High-Throughput Pyrosequencing. Water Res 2013, 47, 859–869. [CrossRef]
- Papadatou, M.; Robson, S.C.; Dobretsov, S.; Watts, J.E.M.; Longyear, J.; Salta, M. Marine Biofilms on Different Fouling Control Coating Types Reveal Differences in Microbial Community Composition and Abundance. Microbiologyopen 2021, 10, e1231. [CrossRef]
- Muthukrishnan, T.; Abed, R.M.M.; Dobretsov, S.; Kidd, B.; Finnie, A.A. Long-Term Microfouling on Commercial Biocidal Fouling Control Coatings. Biofouling 2014, 30, 1155–1164. [CrossRef]
- Winfield, M.O.; Downer, A.; Longyear, J.; Dale, M.; Barker, G.L.A. Comparative Study of Biofilm Formation on Biocidal Antifouling and Fouling-Release Coatings Using next-Generation DNA Sequencing. Biofouling 2018, 34, 464–477. [CrossRef]
- Dobretsov, S.; Abed, R.M.M.; Muthukrishnan, T.; Sathe, P.; Al-Naamani, L.; Queste, B.Y.; Piontkovski, S. Living on the Edge: Biofilms Developing in Oscillating Environmental Conditions. Biofouling 2018, 34, 1064–1077. [CrossRef]
- Antunes, J.T.; Sousa, A.G.G.; Azevedo, J.; Rego, A.; Leão, P.N.; Vasconcelos, V. Distinct Temporal Succession of Bacterial Communities in Early Marine Biofilms in a Portuguese Atlantic Port. Front Microbiol 2020, 11, 1938. [CrossRef]
- Sushmitha, T.J.; Rajeev, M.; Murthy, P.S.; Ganesh, S.; Toleti, S.R.; Pandian, S.K. Bacterial Community Structure of Early-Stage Biofilms Is Dictated by Temporal Succession Rather than Substrate Types in the Southern Coastal Seawater of India. PLOS ONE 2021, 16, e0257961. [CrossRef]
- Vaksmaa, A.; Egger, M.; Lüke, C.; Martins, P.D.; Rosselli, R.; Asbun, A.A.; Niemann, H. Microbial Communities on Plastic Particles in Surface Waters Differ from Subsurface Waters of the North Pacific Subtropical Gyre. Marine Pollution Bulletin 2022, 182, 113949. [CrossRef]
- Walter, J.M.; de Oliveira, L.S.; Tschoeke, D.A.; Meirelles, P.M.; Neves, M.H.C.B.; Batista, D.; Carvalho, A.P.; Santos Costa, R.D.; Dobretsov, S.; Coutinho, R.; et al. Metagenomic Insights Into Ecosystem Function in the Microbial Mats of a Large Hypersaline Coastal Lagoon System. Frontiers in Marine Science 2021, 8.
- Lema, K.A.; Constancias, F.; Rice, S.A.; Hadfield, M.G. High Bacterial Diversity in Nearshore and Oceanic Biofilms and Their Influence on Larval Settlement by Hydroides Elegans (Polychaeta). Environmental Microbiology 0. [CrossRef]
- Guo, Z.; Wang, L.; Cong, W.; Jiang, Z.; Liang, Z. Comparative Analysis of the Ecological Succession of Microbial Communities on Two Artificial Reef Materials. Microorganisms 2021, 9, 120. [CrossRef]
- Ribani, A.; Schiavo, G.; Utzeri, V.J.; Bertolini, F.; Geraci, C.; Bovo, S.; Fontanesi, L. Application of next Generation Semiconductor Based Sequencing for Species Identification and Analysis of Within-Species Mitotypes Useful for Authentication of Meat Derived Products. Food Control 2018, 91, 58–67. [CrossRef]
- Harb, M.; Xiong, Y.; Guest, J.; Amy, G.; Hong, P.-Y. Differences in Microbial Communities and Performance between Suspended and Attached Growth Anaerobic Membrane Bioreactors Treating Synthetic Municipal Wastewater. Environmental Science: Water Research & Technology 2015, 1, 800–813. [CrossRef]
- Liu, Y.; Jeraldo, P.; Mendes-Soares, H.; Masters, T.; Asangba, A.E.; Nelson, H.; Patel, R.; Chia, N.; Walther-Antonio, M. Amplification of Femtograms of Bacterial DNA Within 3 h Using a Digital Microfluidics Platform for MinION Sequencing. ACS Omega 2021, 6, 25642–25651. [CrossRef]
- Colston, S.M.; Hervey, W.J.; Horne, W.C.; Haygood, M.G.; Petersen, B.D.; van Kessel, J.C.; Vora, G.J. Complete Genome Sequence of Vibrio Campbellii DS40M4. Microbiology Resource Announcements 2019, 8, e01187-18. [CrossRef]
- Hosoe, A.; Suenaga, T.; Sugi, T.; Iizumi, T.; Nagai, N.; Terada, A. Complete Genome Sequence of Pseudomonas Putida Strain TS312, Harboring an HdtS-Type N-Acyl-Homoserine Lactone Synthase, Isolated from a Paper Mill. Microbiology Resource Announcements 2020, 9, e00055-20. [CrossRef]
- Li, Y.-F.; Chen, Y.-R.; Yang, J.-L.; Bao, W.-Y.; Guo, X.-P.; Liang, X.; Shi, Z.-Y.; Li, J.-L.; Ding, D.-W. Effects of Substratum Type on Bacterial Community Structure in Biofilms in Relation to Settlement of Plantigrades of the Mussel Mytilus Coruscus. International Biodeterioration & Biodegradation 2014, 96, 41–49. [CrossRef]
- Petersen, L.-E.; Moeller, M.; Versluis, D.; Nietzer, S.; Kellermann, M.Y.; Schupp, P.J. Mono- and Multispecies Biofilms from a Crustose Coralline Alga Induce Settlement in the Scleractinian Coral Leptastrea Purpurea. Coral Reefs 2021, 40, 381–394. [CrossRef]
- Cacabelos, E.; Ramalhosa, P.; Canning-Clode, J.; Troncoso, J.S.; Olabarria, C.; Delgado, C.; Dobretsov, S.; Gestoso, I. The Role of Biofilms Developed under Different Anthropogenic Pressure on Recruitment of Macro-Invertebrates. International Journal of Molecular Sciences 2020, 21, 2030. [CrossRef]
- Dey, S.; Rout, A.K.; Behera, B.K.; Ghosh, K. Plastisphere Community Assemblage of Aquatic Environment: Plastic-Microbe Interaction, Role in Degradation and Characterization Technologies. Environmental Microbiome 2022, 17, 32. [CrossRef]
- Zhurina, M.V.; Bogdanov, K.I.; Gannesen, A.V.; Mart’yanov, S.V.; Plakunov, V.K. Microplastics as a New Ecological Niche For Multispecies Microbial Biofilms within the Plastisphere. Microbiology 2022, 91, 107–123. [CrossRef]
- Singh, S.P.; Sharma, P.; Bano, A.; Nadda, A.K.; Varjani, S. Microbial Communities in Plastisphere and Free-Living Microbes for Microplastic Degradation: A Comprehensive Review. Green Analytical Chemistry 2022, 3, 100030. [CrossRef]
- Wright, R.J.; Erni-Cassola, G.; Zadjelovic, V.; Latva, M.; Christie-Oleza, J.A. Marine Plastic Debris: A New Surface for Microbial Colonization. Environ. Sci. Technol. 2020, 54, 11657–11672. [CrossRef]
- Briand, J.-F.; Barani, A.; Garnier, C.; Réhel, K.; Urvois, F.; LePoupon, C.; Bouchez, A.; Debroas, D.; Bressy, C. Spatio-Temporal Variations of Marine Biofilm Communities Colonizing Artificial Substrata Including Antifouling Coatings in Contrasted French Coastal Environments. Microb Ecol 2017, 74, 585–598. [CrossRef]
- Sathe, P.; Laxman, K.; Myint, M.T.Z.; Dobretsov, S.; Richter, J.; Dutta, J. Bioinspired Nanocoatings for Biofouling Prevention by Photocatalytic Redox Reactions. Scientific Reports 2017, 7, 3624. [CrossRef]
- von Ammon, U.; Wood, S.A.; Laroche, O.; Zaiko, A.; Tait, L.; Lavery, S.; Inglis, G.; Pochon, X. The Impact of Artificial Surfaces on Marine Bacterial and Eukaryotic Biofouling Assemblages: A High-Throughput Sequencing Analysis. Marine Environmental Research 2018, 133, 57–66. [CrossRef]
- Ward, C.S.; Diana, Z.; Ke, K.M.; Orihuela, B.; Schultz, T.P.; Rittschof, D. Microbiome Development of Seawater-Incubated Pre-Production Plastic Pellets Reveals Distinct and Predictive Community Compositions. Frontiers in Marine Science 2022, 8.
- Joardar, I.; Dutta, S. A Selective Review on the Novel Approaches and Potential Control Agents of Anti-Biofouling and Anti-Biofilming. Appl Biochem Biotechnol 2022. [CrossRef]
- Xia, Z.; Gu, J.; Wen, Y.; Cao, X.; Gao, Y.; Li, S.; Haffner, G.D.; MacIsaac, H.J.; Zhan, A. EDNA-Based Detection Reveals Invasion Risks of a Biofouling Bivalve in the World’s Largest Water Diversion Project. Ecological Applications n/a, e2826. [CrossRef]
- Ibabe, A.; Rayón, F.; Martinez, J.L.; Garcia-Vazquez, E. Environmental DNA from Plastic and Textile Marine Litter Detects Exotic and Nuisance Species Nearby Ports. PLOS ONE 2020, 15, e0228811. [CrossRef]
- Bowers, H.A.; Pochon, X.; von Ammon, U.; Gemmell, N.; Stanton, J.-A.L.; Jeunen, G.-J.; Sherman, C.D.H.; Zaiko, A. Towards the Optimization of EDNA/ERNA Sampling Technologies for Marine Biosecurity Surveillance. Water 2021, 13, 1113. [CrossRef]
- Pawlowski, J.; Bruce, K.; Panksep, K.; Aguirre, F.I.; Amalfitano, S.; Apothéloz-Perret-Gentil, L.; Baussant, T.; Bouchez, A.; Carugati, L.; Cermakova, K.; et al. Environmental DNA Metabarcoding for Benthic Monitoring: A Review of Sediment Sampling and DNA Extraction Methods. Science of The Total Environment 2022, 818, 151783. [CrossRef]
- Li, Y.-F.; Zhu, X.; Cheng, Z.-Y.; Liang, X.; Zhu, Y.-T.; Feng, D.-D.; Dobretsov, S.; Yang, J.-L. 2(5H)-Furanone Disrupts Bacterial Biofilm Formation and Indirectly Reduces the Settlement of Plantigrades of the Mussel Mytilus Coruscus. Frontiers in Marine Science 2020, 7.
- Barbosa, M.; Schwaner, C.; Pales Espinosa, E.; Allam, B. A Transcriptomic Analysis of Phenotypic Plasticity in Crassostrea Virginica Larvae under Experimental Acidification. Genes 2022, 13, 1529. [CrossRef]
- Todgham, A.E.; Hofmann, G.E. Transcriptomic Response of Sea Urchin Larvae Strongylocentrotus Purpuratus to CO2-Driven Seawater Acidification. Journal of Experimental Biology 2009, 212, 2579–2594. [CrossRef]
- Griffith, A.W.; Harke, M.J.; DePasquale, E.; Berry, D.L.; Gobler, C.J. The Harmful Algae, Cochlodinium Polykrikoides and Aureococcus Anophagefferens, Elicit Stronger Transcriptomic and Mortality Response in Larval Bivalves (Argopecten Irradians) than Climate Change Stressors. Ecology and Evolution 2019, 9, 4931–4948. [CrossRef]
- Dineshram, R.; Xiao, S.; Ko, G.W.K.; Li, J.; Smrithi, K.; Thiyagarajan, V.; Zhang, Y.; Yu, Z. Ocean Acidification Triggers Cell Signaling, Suppress Immune and Calcification in the Pacific Oyster Larvae. Frontiers in Marine Science 2021, 8.
- Al-Aqeel, S.; Ryu, T.; Zhang, H.; Chandramouli, K.H.; Ravasi, T. Transcriptome and Proteome Studies Reveal Candidate Attachment Genes during the Development of the Barnacle Amphibalanus Amphitrite. Frontiers in Marine Science 2016, 3.
- Rees, D.J.; Hanifi, A.; Obille, A.; Alexander, R.; Sone, E.D. Fingerprinting of Proteins That Mediate Quagga Mussel Adhesion Using a De Novo Assembled Foot Transcriptome. Sci Rep 2019, 9, 6305. [CrossRef]
- Liu, C.; Zhang, R. Identification of Novel Adhesive Proteins in Pearl Oyster by Proteomic and Bioinformatic Analysis. Biofouling 2021, 37, 299–308. [CrossRef]
- Dupree, E.J.; Jayathirtha, M.; Yorkey, H.; Mihasan, M.; Petre, B.A.; Darie, C.C. A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of This Field. Proteomes 2020, 8, 14. [CrossRef]
- Slattery, M.; Ankisetty, S.; Corrales, J.; Marsh-Hunkin, K.E.; Gochfeld, D.J.; Willett, K.L.; Rimoldi, J.M. Marine Proteomics: A Critical Assessment of an Emerging Technology. J. Nat. Prod. 2012, 75, 1833–1877. [CrossRef]
- Thiyagarajan, V.; Wong, T.; Qian, P.-Y. 2D Gel-Based Proteome and Phosphoproteome Analysis During Larval Metamorphosis in Two Major Marine Biofouling Invertebrates. J. Proteome Res. 2009, 8, 2708–2719. [CrossRef]
- Zhang, Y.; Sun, J.; Xiao, K.; Arellano, S.M.; Thiyagarajan, V.; Qian, P.-Y. 2D Gel-Based Multiplexed Proteomic Analysis during Larval Development and Metamorphosis of the Biofouling Polychaete Tubeworm Hydroides Elegans. J. Proteome Res. 2010, 9, 4851–4860. [CrossRef]
- Chen, Z.-F.; Zhang, H.; Wang, H.; Matsumura, K.; Wong, Y.H.; Ravasi, T.; Qian, P.-Y. Quantitative Proteomics Study of Larval Settlement in the Barnacle Balanus Amphitrite. PLOS ONE 2014, 9, e88744. [CrossRef]
- Quantitative Proteomics Identify Molecular Targets That Are Crucial in Larval Settlement and Metamorphosis of Bugula Neritina | Journal of Proteome Research Available online: https://pubs.acs.org/doi/abs/10.1021/pr100817v (accessed on 15 May 2023).
- Dineshram, R.; Chandramouli, K.; Ko, G.W.K.; Zhang, H.; Qian, P.-Y.; Ravasi, T.; Thiyagarajan, V. Quantitative Analysis of Oyster Larval Proteome Provides New Insights into the Effects of Multiple Climate Change Stressors. Global Change Biology 2016, 22, 2054–2068. [CrossRef]
- Dineshram, R.; Q., Q.; Sharma, R.; Chandramouli, K.; Yalamanchili, H.K.; Chu, I.; Thiyagarajan, V. Comparative and Quantitative Proteomics Reveal the Adaptive Strategies of Oyster Larvae to Ocean Acidification. PROTEOMICS 2015, 15, 4120–4134. [CrossRef]
- Parker, L.; Ross, P.; Raftos, D.; Thompson, E.; O’Connor, W. The Proteomic Response of Larvae of the Sydney Rock Oyster, Saccostrea Glomerata to Elevated PCO2. Australian Zoologist 2012, 35, 1011–1023. [CrossRef]
- Tomanek, L.; Zuzow, M.J.; Hitt, L.; Serafini, L.; Valenzuela, J.J. Proteomics of Hyposaline Stress in Blue Mussel Congeners (Genus Mytilus): Implications for Biogeographic Range Limits in Response to Climate Change. Journal of Experimental Biology 2012, 215, 3905–3916. [CrossRef]
- Dickinson, G.H.; Vega, I.E.; Wahl, K.J.; Orihuela, B.; Beyley, V.; Rodriguez, E.N.; Everett, R.K.; Bonaventura, J.; Rittschof, D. Barnacle Cement: A Polymerization Model Based on Evolutionary Concepts. J. Exp. Biol. 2009, 212, 3499–3510. [CrossRef]
- Essock-Burns, T.; Soderblom, E.J.; Orihuela, B.; Moseley, M.A.; Rittschof, D. Hypothesis Testing With Proteomics: A Case Study Using Wound Healing Mechanisms in Fluids Associated With Barnacle Glue. Front. Mar. Sci. 2019, 6. [CrossRef]
- Romeu, M.J.; Domínguez-Pérez, D.; Almeida, D.; Morais, J.; Araújo, M.J.; Osório, H.; Campos, A.; Vasconcelos, V.; Mergulhão, F.J. Quantitative Proteomic Analysis of Marine Biofilms Formed by Filamentous Cyanobacterium. Environ Res 2021, 201, 111566. [CrossRef]
- Avelino-Jiménez, I.A.; Hernández-Maya, L.; Larios-Serrato, V.; Quej-Ake, L.; Castelán-Sánchez, H.; Herrera-Díaz, J.; Garibay-Febles, V.; Rivera-Olvera, J.N.; Zavala-Olivares, G.; Zapata-Peñasco, I. Biofouling and Biocorrosion by Microbiota from a Marine Oil Pipeline: A Metagenomic and Proteomic Approach. Journal of Environmental Chemical Engineering 2023, 11, 109413. [CrossRef]
- Ram, R.J.; VerBerkmoes, N.C.; Thelen, M.P.; Tyson, G.W.; Baker, B.J.; Blake, R.C.; Shah, M.; Hettich, R.L.; Banfield, J.F. Community Proteomics of a Natural Microbial Biofilm. Science 2005, 308, 1915–1920. [CrossRef]
- Han, Z.; Sun, J.; Zhang, Y.; He, F.; Xu, Y.; Matsumura, K.; He, L.-S.; Qiu, J.-W.; Qi, S.-H.; Qian, P.-Y. ITRAQ-Based Proteomic Profiling of the Barnacle Balanus Amphitrite in Response to the Antifouling Compound Meleagrin. J. Proteome Res. 2013, 12, 2090–2100. [CrossRef]
- Zhang, H.; Wong, Y.H.; Wang, H.; Chen, Z.; Arellano, S.M.; Ravasi, T.; Qian, P.-Y. Quantitative Proteomics Identify Molecular Targets That Are Crucial in Larval Settlement and Metamorphosis of Bugula Neritina. J. Proteome Res. 2011, 10, 349–360. [CrossRef]
- Qian, P.-Y.; Wong, Y.H.; Zhang, Y. Changes in the Proteome and Phosphoproteome Expression in the Bryozoan Bugula Neritina Larvae in Response to the Antifouling Agent Butenolide. PROTEOMICS 2010, 10, 3435–3446. [CrossRef]
- Liu, R.; Bao, Z.-X.; Zhao, P.-J.; Li, G.-H. Advances in the Study of Metabolomics and Metabolites in Some Species Interactions. Molecules 2021, 26, 3311. [CrossRef]
- Gutarowska, B.; Celikkol-Aydin, S.; Bonifay, V.; Otlewska, A.; Aydin, E.; Oldham, A.L.; Brauer, J.I.; Duncan, K.E.; Adamiak, J.; Sunner, J.A.; et al. Metabolomic and High-Throughput Sequencing Analysis—Modern Approach for the Assessment of Biodeterioration of Materials from Historic Buildings. Front. Microbiol. 2015, 6. [CrossRef]
- Briand, J.-F.; Pochon, X.; Wood, S.A.; Bressy, C.; Garnier, C.; Réhel, K.; Urvois, F.; Culioli, G.; Zaiko, A. Metabarcoding and Metabolomics Offer Complementarity in Deciphering Marine Eukaryotic Biofouling Community Shifts. Biofouling 2018, 34, 657–672. [CrossRef]
- Chandramouli, K.H.; Dash, S.; Zhang, Y.; Ravasi, T.; Qian, P.-Y. Proteomic and Metabolomic Profiles of Marine Vibrio Sp. 010 in Response to an Antifoulant Challenge. Biofouling 2013, 29, 789–802. [CrossRef]
- Favre, L.; Ortalo-Magné, A.; Pichereaux, C.; Gargaros, A.; Burlet-Schiltz, O.; Cotelle, V.; Culioli, G. Metabolome and Proteome Changes between Biofilm and Planktonic Phenotypes of the Marine Bacterium Pseudoalteromonas Lipolytica TC8. Biofouling 2018, 34, 132–148. [CrossRef]
- Favre, L.; Ortalo-Magné, A.; Greff, S.; Pérez, T.; Thomas, O.P.; Martin, J.-C.; Culioli, G. Discrimination of Four Marine Biofilm-Forming Bacteria by LC–MS Metabolomics and Influence of Culture Parameters. J. Proteome Res. 2017, 16, 1962–1975. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).