Submitted:
22 May 2023
Posted:
23 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
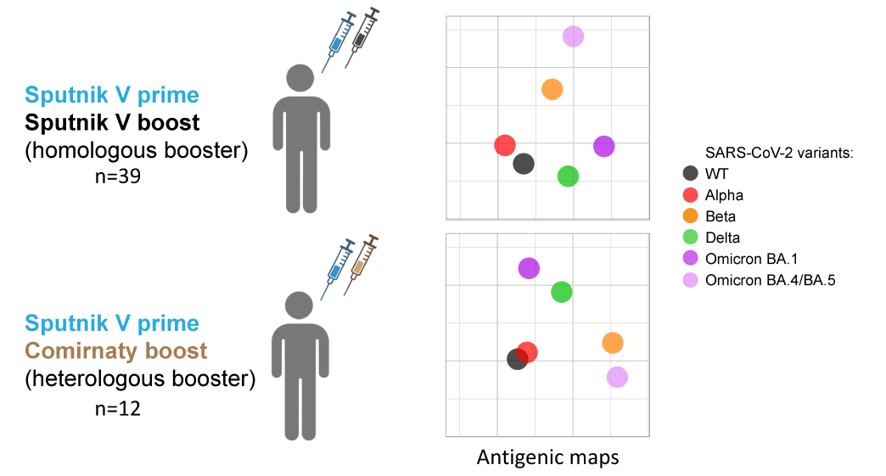
2.1. Study design
2.2. The level of post-boost SARS-CoV-2 NAbs depends on the type of booster vaccine and the history of exposure to SARS-CoV-2
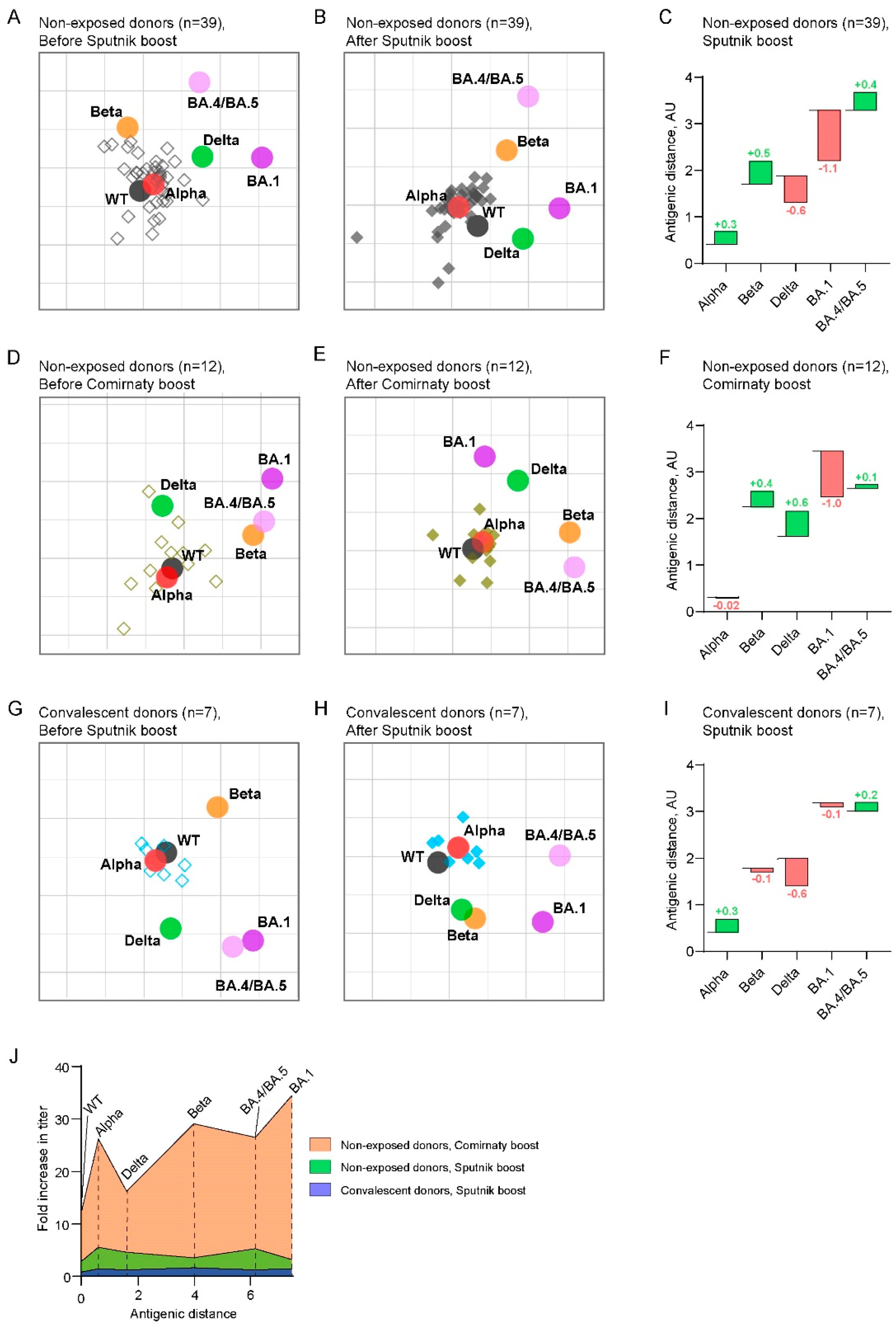
2.3. Antigenic maps reveal the emergence of booster-induced cross-neutralizing antibodies
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Volunteers and samples collection
4.3. SARS-CoV-2 pseudoviral particles production
4.4. Pseudotyped virus neutralization assay
4.5. Antigenic cartography
4.6. Statistical analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gattinger, P.; Ohradanova-Repic, A.; Valenta, R. Importance, Applications and Features of Assays Measuring SARS-CoV-2 Neutralizing Antibodies. Int J Mol Sci 2023, 24, 5352. [Google Scholar] [CrossRef] [PubMed]
- Sholukh, A.M.; Fiore-Gartland, A.; Ford, E.S.; Miner, M.D.; Hou, Y.J.; Tse, L. V.; Kaiser, H.; Zhu, H.; Lu, J.; Madarampalli, B.; et al. Evaluation of Cell-Based and Surrogate SARS-CoV-2 Neutralization Assays. J Clin Microbiol 2021, 59, e00527–21. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Lapedes, A.S.; De Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Mapping the Antigenic and Genetic Evolution of Influenza Virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef]
- Mykytyn, A.Z.; Rissmann, M.; Kok, A.; Rosu, M.E.; Schipper, D.; Breugem, T.I.; van den Doel, P.B.; Chandler, F.; Bestebroer, T.; de Wit, M.; et al. Antigenic Cartography of SARS-CoV-2 Reveals That Omicron BA.1 and BA.2 Are Antigenically Distinct. Sci Immunol 2022, 7, eabq4450. [Google Scholar] [CrossRef] [PubMed]
- Rössler, A.; Netzl, A.; Knabl, L.; Schäfer, H.; Wilks, S.H.; Bante, D.; Falkensammer, B.; Borena, W.; von Laer, D.; Smith, D.J.; et al. BA.2 and BA.5 Omicron Differ Immunologically from Both BA.1 Omicron and Pre-Omicron Variants. Nat Commun 2022, 13, 7701. [Google Scholar] [CrossRef]
- Wilks, S.H.; Mühlemann, B.; Shen, X.; Türeli, S.; LeGresley, E.B.; Netzl, A.; Caniza, M.A.; Chacaltana-Huarcaya, J.N.; Corman, V.M.; Daniell, X.; et al. Mapping SARS-CoV-2 Antigenic Relationships and Serological Responses. bioRxiv Prepr Serv Biol 2022, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Zou, J.; Kalveram, B.; Machado, R.R.G.; Ren, P.; Türeli, S.; Smith, D.J.; Weaver, S.C.; Xie, X.; et al. Cross-Neutralization and Cross-Protection among SARS-CoV-2 Viruses Bearing Different Variant Spikes. Signal Transduct Target Ther 2022, 7, 2020–2023. [Google Scholar] [CrossRef]
- Lusvarghi, S.; Pollett, S.D.; Neerukonda, S.N.; Wang, W.; Wang, R.; Vassell, R.; Epsi, N.J.; Fries, A.C.; Agan, B.K.; Lindholm, D.A.; et al. SARS-CoV-2 BA.1 Variant Is Neutralized by Vaccine Booster–Elicited Serum but Evades Most Convalescent Serum and Therapeutic Antibodies. Sci Transl Med 2022, 14, 8543. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lusvarghi, S.; Subramanian, R.; Epsi, N.J.; Wang, R.; Goguet, E.; Fries, A.C.; Echegaray, F.; Vassell, R.; Coggins, S.A.; et al. Antigenic Cartography of Well-Characterized Human Sera Shows SARS-CoV-2 Neutralization Differences Based on Infection and Vaccination History. Cell Host Microbe 2022, 30, 1745–1758.e7. [Google Scholar] [CrossRef] [PubMed]
- Neerukonda, S.N.; Vassell, R.; Lusvarghi, S.; Wang, R.; Echegaray, F.; Bentley, L.; Eakin, A.E.; Erlandson, K.J.; Katzelnick, L.C.; Weiss, C.D.; et al. SARS-CoV-2 Delta Variant Displays Moderate Resistance to Neutralizing Antibodies and Spike Protein Properties of Higher Soluble ACE2 Sensitivity, Enhanced Cleavage and Fusogenic Activity. Viruses 2021, 13, 2485. [Google Scholar] [CrossRef] [PubMed]
- van der Straten, K.; Guerra, D.; van Gils, M.J.; Bontjer, I.; Caniels, T.G.; van Willigen, H.D.G.; Wynberg, E.; Poniman, M.; Burger, J.A.; Bouhuijs, J.H.; et al. Antigenic Cartography Using Sera from Sequence-Confirmed SARS-CoV-2 Variants of Concern Infections Reveals Antigenic Divergence of Omicron. Immunity 2022, 55, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, V.A.; Pochtovyi, A.A.; Kustova, D.D.; Ogarkova, D.A.; Tarnovetskii, I.Y.; Belyaeva, E.D.; Divisenko, E. V.; Vasilchenko, L.A.; Shidlovskaya, E. V.; Kuznetsova, N.A.; et al. Dynamics of SARS-CoV-2 Major Genetic Lineages in Moscow in the Context of Vaccine Prophylaxis. Int J Mol Sci 2022, 23, 14670. [Google Scholar] [CrossRef] [PubMed]
- Horton, D.L.; McElhinney, L.M.; Marston, D.A.; Wood, J.L.N.; Russell, C.A.; Lewis, N.; Kuzmin, I. V.; Fouchier, R.A.M.; Osterhaus, A.D.M.E.; Fooks, A.R.; et al. Quantifying Antigenic Relationships among the Lyssaviruses. J Virol 2010, 84, 11841–11848. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, A.; Marini, V.; Di Gennaro, A.; Ronchi, G.F.; Casaccia, C.; Carelli, G.; Passantino, G.; D’Alterio, N.; D’Innocenzo, V.; Savini, G.; et al. Antigenic Relationship among Zoonotic Flaviviruses from Italy. Infect Genet Evol 2019, 68, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Katzelnick, L.C.; Fonville, J.M.; Gromowski, G.D.; Arriaga, J.B.; Green, A.; James, S.L.; Lau, L.; Montoya, M.; Wang, C.; Vanblargan, L.A.; et al. Dengue Viruses Cluster Antigenically but Not as Discrete Serotypes. Science 2015, 349, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Schaefer-Babajew, D.; Wang, Z.; Muecksch, F.; Cho, A.; Loewe, M.; Cipolla, M.; Raspe, R.; Johnson, B.; Canis, M.; DaSilva, J.; et al. Antibody Feedback Regulates Immune Memory after SARS-CoV-2 mRNA Vaccination. Nature 2023, 613, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Bekliz, M.; Adea, K.; Vetter, P.; Eberhardt, C.S.; Hosszu-Fellous, K.; Vu, D.L.; Puhach, O.; Essaidi-Laziosi, M.; Waldvogel-Abramowski, S.; Stephan, C.; et al. Neutralization Capacity of Antibodies Elicited through Homologous or Heterologous Infection or Vaccination against SARS-CoV-2 VOCs. Nat Commun 2022, 13, 1–10. [Google Scholar] [CrossRef]
- Fabricius, D.; Ludwig, C.; Scholz, J.; Rode, I.; Tsamadou, C.; Jacobsen, E.M.; Winkelmann, M.; Grempels, A.; Lotfi, R.; Janda, A.; et al. mRNA Vaccines Enhance Neutralizing Immunity against SARS-CoV-2 Variants in Convalescent and ChAdOx1-Primed Subjects. Vaccines 2021, 9, 918. [Google Scholar] [CrossRef]
- Hillus, D.; Schwarz, T.; Tober-Lau, P.; Vanshylla, K.; Hastor, H.; Thibeault, C.; Jentzsch, S.; Helbig, E.T.; Lippert, L.J.; Tscheak, P.; et al. Safety, Reactogenicity, and Immunogenicity of Homologous and Heterologous Prime-Boost Immunisation with ChAdOx1 NCoV-19 and BNT162b2: A Prospective Cohort Study. Lancet Respir Med 2021, 9, 1255–1265. [Google Scholar] [CrossRef]
- Godkov, M.A.; Ogarkova, D.A.; Gushchin, V.A.; Kleymenov, D.A.; Mazunina, E.P.; Bykonia, E.N.; Pochtovyi, A.A.; Shustov, V. V.; Shcheblyakov, D. V.; Komarov, A.G.; et al. Revaccination in Age-Risk Groups with Sputnik V Is Immunologically Effective and Depends on the Initial Neutralizing SARS-CoV-2 IgG Antibodies Level. Vaccines 2023, 11, 90. [Google Scholar] [CrossRef]
- Bates, T.A.; Leier, H.C.; McBride, S.K.; Schoen, D.; Lyski, Z.L.; Lee, D.X.; Messer, W.B.; Curlin, M.E.; Tafesse, F.G. An Extended Interval between Vaccination and Infection Enhances Hybrid Immunity against SARS-CoV-2 Variants. JCI Insight 2023, 8, e165265. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N Engl J Med 2020, 383, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N Engl J Med 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Logunov, D.Y.; Dolzhikova, I. V.; Zubkova, O. V.; Tukhvatullin, A.I.; Shcheblyakov, D. V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A. V.; Botikov, A.G.; et al. Safety and Immunogenicity of an rAd26 and rAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine in Two Formulations: Two Open, Non-Randomised Phase 1/2 Studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- Byazrova, M.G.; Kulemzin, S. V.; Astakhova, E.A.; Belovezhets, T.N.; Efimov, G.A.; Chikaev, A.N.; Kolotygin, I.O.; Gorchakov, A.A.; Taranin, A. V.; Filatov, A. V. Memory B Cells Induced by Sputnik V Vaccination Produce SARS-CoV-2 Neutralizing Antibodies Upon Ex Vivo Restimulation. Front Immunol 2022, 13, 840707. [Google Scholar] [CrossRef]
- Astakhova, E.A.; Byazrova, M.G.; Yusubalieva, G.M.; Kulemzin, S. V.; Kruglova, N.A.; Prilipov, A.G.; Baklaushev, V.P.; Gorchakov, A.A.; Taranin, A. V.; Filatov, A. V. Functional Profiling of In Vitro Reactivated Memory B Cells Following Natural SARS-CoV-2 Infection and Gam-COVID-Vac Vaccination. Cells 2022, 11. [Google Scholar] [CrossRef]

| Name of subgroup | Non-exposed donors, Sputnik V booster |
Non-exposed donors, Comirnaty booster |
Convalescent donors, Sputnik V booster |
|
|---|---|---|---|---|
| Number of participants | 39 | 12 | 7 | |
| Age | Years, median (range) | data data |
data data |
|
| Sex | Female | 19 | 6 | 3 |
| Male | 20 | 6 | 4 | |
| Booster vaccine | Spuntik V | Comirnaty | Spuntik V | |
| Probe sampling after second dose of the first vaccination | 195 (176 - 270) | 275 (261 - 335) | 207 (183 - 282) | |
| Probe sampling after boost | 33 (29 - 39) | 38 (30 - 46) | 31 (30 - 36) | |
| Anti-N IgG antibodies before boost | 0/39 | 0/12 | 5/7 | |
| Anti-S IgG antibodies before boost | 6.0 (0 – 10.4) | 7.3 (0 – 11.8) | 12.7 (11.2 – 14.4) | |
| PCR-confirmed COVID-19 | ||||
| Time between recovery and boost | 0/39 | 0/12 | 7/7 | |
| Infection period | - | - | May – August 2021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





