Introduction
Nearly one-third of the world’s population is classified as overweight or obese by Body Mass Index (BMI), which characterizes obesity as a global health crisis [
1,
2,
3]. The rates of obesity have been steadily climbing over time, with the number of overweight and obese individuals worldwide increasing from 857 million (20 %) in 1980 to 2.1 billion (30 %) in 2013 [
4], and even higher to ~60% of the global population {Boutari, 2022 #495}. Among countries higher-income nations characterized as first-world some of the most significant increases in adult obesity have occurred in the United States (one-third of adults), Australia (~ 30 %), and the United Kingdom (~ 25 %) [
4]. With this growing epidemic, there has been a vested interest in better understanding other health-related outcomes associated with obesity. Not surprisingly, obesity, especially morbid obesity (BMI > 40), has been identified as a risk factor for numerous medical conditions, including diabetes mellitus, hypertension, cardiovascular disease, metabolic syndrome, and some cancers [
5]. Obesity was associated with an increased risk of 13 types of cancer [
6], including, but not limited to: breast, prostate, gastrointestinal (GI) (including colorectal, esophageal, liver and pancreas), kidney, and multiple myeloma.
Recent investigations into understanding the impacts of obesity on the tumor activity, the tumor microenvironment, and related immune components have led to the finding that obesity may improve survival in some cases [
7]. This paradoxically protective action has subsequently been termed the “obesity paradox.” For example, we recently showed that obesity enhances PD-1 mediated T-cell dysfunction at least partly by leptin signaling. Interestingly, obese patients have been reported to respond better to treatment with anti-PD1 immune checkpoint inhibitors [
8]. However, in seeking to identify the underlying biological etiology of the obesity paradox, we have realized this phenomenon may differ by sex. Findings of a study particularly interested in metastatic melanoma and the “obesity paradox” found the existence of the paradox is restricted to overweight/Class-I obesity, which is predominated by males with increased serum creatinine levels, a surrogate for skeletal muscle mass [
9]. These same associations were not seen in females, who tend to have lower creatinine levels. Other findings have suggested that the protective features of the obesity paradox are rooted in alterations to different transcripts compared to normal-weight patients [
10,
11] or alterations to the immune milieu [
12]. Much of this literature highlights that obesity impacts males and females differently. This is important to explore, given that globally, a higher proportion of women are obese than men [
3], yet most obesity treatment and service options are not sex-informed [
13]. Recently, the cross-talk between systemic and tumor metabolism has been identified as a modulator of cancer immunotherapy efficacy in patients [
14]. Several clinical trials are now looking at ways to combine checkpoint inhibition with the modulation of various metabolic pathways to improve therapeutic outcomes. As sex affects both systemic and tumor metabolism, there is a demonstrated need to identify the influence of sex on tumor growth [
15].
In this study, we aim to identify the unique tumor associated transcriptional and metabolic milieu associated with obesity and with sex in the context of obesity. To examine this phenomenon, we specifically queried patients with GI cancers (Colon (COAD) and Esophageal (ESCA) Adenocarcinomas), as these disease sites are both known to be impacted by obesity and sex differences and, further, to be associated with the obesity paradox. Here, we highlight consistent alterations to transcriptional tumor metabolism of obese male and female patients, albeit to a different extent, in lipid and amino acid-associated metabolic pathways compared to their normal-weight counterparts. Additionally, there are some varying metabolic pathway differences between the male and female obese patient populations, transcriptionally, associated with different hormone metabolism and, further, how these translate to implications in immune-related pathways, like IL-6 signaling. To support these findings, we utilized our serum samples from obese and non-obese patients, as well as publicly available metabolomics data that corresponds with The Cancer Genome Atlas (TCGA) samples. We confirmed metabolic dysregulation that overlapped both transcriptionally and metabolically and model pathways of interest to better examine patterns of metabolic flux associated with both sex and obesity.
Results: Obesity drives differential transcriptomic signature associated with metabolic reprogramming in GI cancers
Patients from the ORIEN database with an adenocarcinoma diagnosis and associated BMI data (n=930, Table 1) were classified as obese (BMI
> 30) or normal weight (18
<BMI
< 25). Tumor RNA- sequencing data from the two divergent groups was compared and resulted in the identification of 2511 significantly down-regulated and 1518 significantly upregulated (adj p < 0.05 and |LogFC| > 0.58) transcripts (
Figure 1A) in obese patients compared to normal weight patients. Top down-regulated transcripts with decreased expression in obese populations include CTRB1, CTRC, and CELA2B, all of which are chymotrypsin- and trypsinogen- related genes which are responsible for digestion and digestive processes. When taken as a ratio of each other, these genes are associated with the resolution of inflammatory symptoms and promote speedier recovery of acute tissue injury [
16]. Top up-regulated transcripts with increased expression in obese populations include ACPP (phosphatases) [
17], SCGB1D2 (glycoproteins) [
18] and PGR (steroid hormone receptors) [
19], which are all associated with hormonal regulation and metabolic processing. Gene set enrichment analysis (Normalize Enrichment Score >2, q-value<0.05;
Figure 1B) enriched for several types of pathways, including translation, therapeutic resistance, metabolism, immune dysfunction, and cellular processes. Metabolic pipeline analysis (
Figure 1C) showed enrichment of several metabolic pathways associated with obesity in adenocarcinomas, including lipids (like steroid hormone metabolism) and amino acids (like tryptophan and kynurenine metabolism), both of which are associated with immune cell dysfunction.
Obesity drives differential transcriptomic signature in a disease-type specific manner
Demographic tables of patients in the Colon Adenocarcinoma, TCGA-COAD (top) and Esophageal Adenocarcinoma TCGA-ESCA (bottom) cohorts reveal relatively similar distributions of race, tumor stage and BMI by sex. Of note, the only statistically significant differences were in the number of males and females in TCGA ESCA, and differences in tumor stage (Table 2). RNA-sequencing data of obese and non-obese patient populations from TCGA Colon Adenocarcinoma (COAD) (
Figure 1D) and TCGA Esophageal Adenocarcinoma (ESCA) (
Figure 1E) were compared, and differential metabolites are reported in volcano plots. Venn diagrams demonstrate very little overlap of differentially expressed genes between the two GI datasets (
Figure 1F), representing a non-significant overlap (P>0.05, Hypergeometric Test) between the two datasets, indicating differential transcript level dysregulation. Further, gene set enrichment analysis of the differentially expressed genes in TCGA COAD (
Figure 1G) and TCGA ESCA (
Figure 1H) highlight enrichment of both metabolic and immune pathways in obese and non-obese samples. While transcriptional changes were different between the two populations, enrichment reveals similar types of pathway dysregulation. Further, pathway modeling shows differential transcription of metabolic pathways by disease site (
Figure 1I; ESCA, left; COAD, right). This is demonstrated by the transcriptional model of tryptophan-NAD Synthesis, where a majority of the downstream Nicotinamide Adenine Dinucleotide (NAD) components are downregulated in obese ESCA populations as compared to large transcriptional increases in the obese COAD populations.
Obesity associated with differential lipid and amino acid-related metabolic programs
We conducted metabolomics on patient serum samples to better understand the metabolic state of obese GI adenocarcinoma patients, given that metabolomics data is not available through the TCGA. For this study, we obtained pre-treatment serum samples from 6 colorectal cancer patients (4 normal weight and 2 overweight, by a BMI > 30). We conducted global metabolomics using the Biocrates MxP Quant 500 kit, which measures 630 metabolites spanning 26 biochemical classes. This revealed distinct metabolic profiles that segregated patients by weight via Principal Component Analysis (PCA,
Figure 2A). Unsurprisingly, we found this differential clustering to be driven by a large number of increased lipids in the obese population compared to the normal weight population, as can be seen on the right side of the volcano plot (
Figure 2B). In fact, there are clear, statistically significant differences in metabolite abundance when comparing obese (blue) to non-obese (pink) patients. This is demonstrated by the top 10 differential metabolites, many of which were heavily associated with altered lipid metabolism (
Figure 2C). In particular, metabolite set enrichment analysis (
Figure 2D) revealed a large amount lipid enrichment as compared to any other class of metabolites (e.g., amino acids, carbohydrates, etc.). This enrichment is largely attributed to variation in a multitude of lipids, including Ceramides, Triacylglycerols, Sphingomyelins, and Phosphatidylcholines. Interestingly, and concordantly with the TCGA RNA-sequencing data, many of these lipids are highly associated with immune reprogramming and dysfunction [
20,
21]. Given our previous enrichment for immune dysregulation comparing obese to non-obese patient populations, we felt it is necessary to additionally investigate the immune profiles of these patients.
Obesity drives differential CD8 and CD14 metabolic and immune transcriptional profiles regardless of sex
To study the immune cell populations in the same 6 patients (4 normal and 2 overweight), we conducted RNA-sequencing of the flow cytometry-sorted CD8 and CD14 immune populations from peripheral blood mononuclear cells (PBMCs). This data revealed distinct transcriptional profiles in non- obese and obese populations, for both the CD8 (
Figure 3A) and CD14 (
Figure 3B) populations, with more notable differences in the CD8 cells. Gene set enrichment analysis of the differentially expressed genes were highly associated with differences in immune pathways, not surprisingly, given these were sorted immune populations, but also metabolism/metabolic pathways. For example, prostaglandin metabolism (
Figure 3C, Supplementary Figure 1A), is highly dysregulated in CD14+ cells of obese patients as compared to non-obese patients, revealing increasing transcriptional production of Arachidonic Acid and Prostaglandin H2, and decreased production of HPETE metabolites and Leukotrienes in CD14 cells of obese patients. Further, in CD8 cells, we also preferentially enriched for prostanoid and thromboxane metabolism (Supplementary Figure 1B), among other amino acid related pathways, like urea cycle (
Figure 3D), which revealed modest increases in transcripts associated with ornithine generation (OAT), lactate and oxaloacetate production, and decreased argininosuccinate and arginine generation. Additionally, modeling of predicted immunotherapy response (PD-L1 and CTLA-4) pathways (
Figure 3E, Supplementary Figure 2), demonstrate that obesity may play a role in differential transcriptional reprogramming that may specifically relate to altered immunotherapy response.
Obese males and females dysregulate similar amino acid and lipid-related metabolic pathways to varying extents
While the differences between obese and non-obese populations are interesting, in terms of both transcriptional and metabolomic alterations, the sex differences associated with this phenomenon are largely of interest to us as well. It is well known that males have a higher incidence of GI cancers [
22], females have significantly better overall survival [
23], and in immunotherapy studies, women tend to mount more of an immune response [
24]. Therefore, utilizing RNA-sequencing data from The Cancer Genome Atlas (TCGA), esophageal adenocarcinoma (ESCA) patient populations were designated as either obese (BMI
> 30) or non-obese (18
< BMI
< 25), and then later split by sex, resulting in 4 groups. Similar analyses were conducted in TCGA colon adenocarcinoma (COAD) (Supplementary Figure 3). Differential expression analysis (adjusted p < 0.05; |log2 Fold Change| > 0.58) revealed 5558 differentially expressed genes (DEGs) between obese and non-obese males (
Figure 4A) and 480 DEGs between obese and non-obese females (
Figure 4B). While there is extensive overlap (40 upregulated genes, 150 downregulated genes) in the transcriptional dysregulation that occurs between males and females, there were notable differences transcriptionally. Consequently, there is also overlap in the types of pathways enriched in the obese populations, as compared to their non-obese counterparts for both males and females. Obese males vs. non-obese males resulted in enrichment of pathways associated with stemness, differentiation, epigenetics, and metabolism (
Figure 4C). Obese females vs. non-obese females resulted in enrichment of pathways associated with epithelial to mesenchymal transition, development, and metabolism (
Figure 4D). Given that gene set enrichment analysis revealed both male and female obese patients dysregulated metabolism when compared to their non-obese counterparts, we applied our metabolic pipeline to understand which of 114 metabolic pathways had the most highly and statistically-significant transcriptional dysregulation in each of the two groups (
Figure 4E). This analysis revealed alterations to both lipid and amino acid metabolism, some of which were similarly dysregulated in both sexes, and some that were dysregulated uniquely in a single sex. One such pathway that is commonly significantly dysregulated in both sexes, is tryptophan/kynurenine metabolism (
Figure 4F; male, left; female, right). However, this immune-related pathway is dysregulated differently in each of the sexes. It is significantly transcriptionally downregulated in females, with predicted decreases in production of kynurenine from tryptophan. Conversely, in males, the earlier transcripts within this pathway are upregulated, indicating increased production of kynurenine from tryptophan. Taken together, it is important to not only consider obesity, but also sex, when assessing novel points of therapeutic leverage in GI cancer patients.
Discussion
Overall, this assessment of the obese tumor transcriptome compared to the non-obese transcriptome demonstrates a large amount of transcriptional metabolic dysregulation, which largely corresponds with serum-level microenvironmental metabolic dysregulation. More specifically, obesity is associated with differential transcriptome profiles in obese GI patient populations (
Figure 1) regardless of sex, many of which were immune- or metabolically altered. Obesity corresponds with differential metabolomics in the serum microenvironment, largely differentiated from normal-weight patient serum by lipids and amino acids, which are heavily associated with immune reprogramming (
Figure 2). Further, obesity is associated with differential metabolic and immune transcriptional profiles in immune cells, which may correspond with response to immunotherapy (
Figure 3). Our findings are similar to the recent study performed by Ringel et al. that showed tumor-infiltrating lymphocytes obtained from high-fat-diet (HFD) induced obese murine tumors were enriched for several pathways related to fat and cholesterol metabolism including glycosphingolipid biosynthesis, steroid biosynthesis, and fatty acid metabolism [
25]. Our study, in addition to the Ringel et al. [
25] study provides evidence that systemic metabolism influences tumor metabolism.
In our study, metabolic dysregulation trends in males and females were similar, with a much more prominent effect in females than in males (
Figure 4). Clinically, it has been demonstrated that immune checkpoint inhibitors work better in obese cancer patients compared to normal-weight patients, however, this association is mainly seen in obese male patients [
26]. Our prior collaborative work has confirmed the clinical phenomenon and further showed that HFD-induced obese male mice had significantly increased benefit from anti-PD-1 checkpoint inhibitors compared to diet-induced obese female mice or the lean male and female mice [
27]. This, therefore, highlights the need to study this phenomenon in greater depth, especially with the integration of additional levels of data (e.g., paired tumor transcriptomics and metabolomics, circulating cytokines).
While we believe this study provides vital information and evidence that obesity is associated with a differential metabolic and immune profile in both the tumor and systemic microenvironment of patients, we recognize that this study has flaws, the largest of which being BMI as a measure of adiposity. BMI fails to distinguish fat adequately from fat-free mass, such as muscle and bone and other bodily tissue {Humphreys, 2010 #496}. Therefore, for more direct measures of fat, DEXA scans, and visceral adiposity measurements from imaging, would better quantitate obesity. However, we are currently leveraging the wealth of publicly available data, where the most commonly used measure of obesity is BMI. Additionally, we drew on transcriptomic data, conducting metabolism-specific transcriptional analysis as a surrogate for metabolomics. While we have evidence that the pipeline is accurate in predicting metabolic dysregulation [
28,
29], we know that metabolomics is the best possible read-out for metabolic dysregulation. Unfortunately, the publicly available metabolomics data is limited and, further, is unavailable for most TCGA patients. Therefore, we utilized patient serum samples available to us, with available patient BMI information to ensure that we accurately captured obesity-associated effects and found that serum metabolomics from our patients corresponded with TCGA tumor transcriptomics. However, it is essential to note that we did have a limited sample size to draw on for this study. In order to enact clinical change, we plan to incorporate more patient samples in future studies.
This is a very clinically significant issue, given the current global obesity epidemic. Future studies should, additionally, focus on other immune cell populations (beyond CD8+ and CD14+), which may also play a role in carcinogenesis and therapeutic response, like macrophages [
31], which are impacted by visceral adiposity [
32]. Understanding how obesity influences tumor metabolism and immunity may have therapeutic implications. Tumor metabolism has been targeted as a mode of cancer therapy for a very long time, starting from chemotherapeutic agents like 5-fluorouracil, however, there is a new interest these days in harnessing metabolic pathways to make immunotherapy more efficacious [
33]. Therefore, in the future, we plan to assess not only the transcriptional and metabolic alterations that occur in the tumor and specific immune cell populations but also within the entirety of the immune milieu, both internal to and external to the tumor, in the context of both sex and obesity.
Methods
The Oncology Research Information Exchange Network (ORIEN) is an alliance of 18 U.S. cancer centers established in 2014. All ORIEN alliance members utilize a standard Total Cancer Care
® (TCC) protocol. As part of the TCC study, participants agree to have their clinical data followed over time, to undergo germline and tumor sequencing, and to be contacted in the future by their provider if an appropriate clinical trial or other study becomes available [
34]. TCC is a prospective cohort study with a subset of patients enrolled to the ORIEN Avatar program, which includes research use only (RUO) grade whole-exome tumor sequencing, RNA sequencing, germline sequencing, and collection of deep longitudinal clinical data with lifetime follow up. Nationally, over 325,000 participants have enrolled to TCC. ASTER INSIGHTS, the commercial and operational partner of ORIEN, harmonizes all abstracted clinical data elements and molecular sequencing files into a standardized, structured format to enable aggregation of de-identified data for sharing across the Network. ORIEN Avatar patients were utilized if they were diagnosed with an adenocarcinoma and consented to the TCC protocol from the participating members of ORIEN.
ORIEN Avatar specimens undergo nucleic acid extraction and sequencing at HudsonAlpha (Huntsville, AL) or Fulgent Genetics (Temple City, CA). For frozen and OCT tissue DNA extraction, Qiagen QIASymphony DNA purification is performed, generating 213 bp average insert size. For frozen and OCT tissue RNA extraction, Qiagen RNAeasy plus mini kit is performed, generating 216 bp average insert size. For FFPE tissue, Covaris Ultrasonication FFPE DNA/RNA kit is utilized to extract both DNA and RNA, generating 165b bp average insert size. For DNA sequencing, preparation of ASTER INSIGHTS Whole Exome Sequencing (WES) libraries involves hybrid capture using an enhanced IDT WES kit (38.7 Mb) with additional custom designed probes for double coverage of 440 cancer genes. Library hybridization is performed at either single or 8-plex, and sequenced on an Illumina NovaSeq 6000 instrument generating 100 bp paired reads. WES is performed on tumor/normal matched samples with the normal covered at 100X and the tumor covered at 300X (additional 440 cancer genes covered at double coverage; 200X for normal and 600X for tumor). Both tumor/normal concordance and gender identity QC checks are performed. Minimum threshold for hybrid selection is >80% of bases with >100X fold coverage for tumor and >50X fold coverage for normal. RNA sequencing (RNAseq) is performed using the Illumina TruSeq RNA Exome with single library hybridization, cDNA synthesis, library preparation, sequencing (100 bp paired reads at Hudson Alpha, 150 bp paired reads at Fulgent) to a coverage of 100M total reads / 50M paired reads.
Raw feature counts were downloaded from the TCGA, normalized and differential expression analysis was conducted using DESeq2 [
35]. Differential expression rank order was utilized for subsequent Gene Set Enrichment Analysis (GSEA), performed using the clusterProfiler package in R. Gene sets queried included the Hallmark, Canonical pathways, and GO Biological Processes Ontology collections available through the Molecular Signatures Database (MSigDB) [
36].
The metabolic pipeline utilizes Differentially Expressed Gene output. For differential gene expression analysis of raw counts, RNA-seq counts were processed to remove genes lacking expression in more than 80% of samples, and the DESeq2 package was utilized for differential analysis. However, most datasets were downloaded as normalized counts. In these cases, the “contrast matrix” function of the Limma package in R, was utilized to obtain differential analysis. Euclidian distances and hierarchal clustering were utilized to determine sample similarity. Scores for each gene were produced by multiplying the -log (adjusted p value)*logFC on a gene level. Further, absolute value scores were produced by taking the absolute value of the scores for each gene. Using the previously published pipeline [
28], we assessed transcriptional metabolic pathway dysregulation in several datasets.
Serum samples were prepared and analyzed in the Roswell Park Comprehensive Cancer Center Bioanalytics, Metabolomics and Pharmacokinetics Shared Resource, using the MxP Quant 500 kit (Biocrates Life Sciences AG, Innsbruck, Austria) in accordance with the user manual. 10 μl of each supernatant, quality control (QC) samples, blank, zero sample, or calibration standard were added on the filterspot (already containing internal standard) in the appropriate wells of the 96-well plate. The plate was then dried under a gentle stream of nitrogen. The samples were derivatized with phenyl isothiocyanate (PITC) for the amino acids and biogenic amines and dried again. Sample extract elution was performed with 5 mM ammonium acetate in methanol. Sample extracts were diluted with either water for the HPLC-MS/MS analysis (1:1) or kit running solvent (Biocrates Life Sciences AG) for flow injection analysis (FIA)-MS/MS (50:1), using a Sciex 5500 mass spectrometer. Data was processed using MetIDQ software (Biocrates Life Sciences AG), and Limma [
37] for differential metabolite analysis.
Pathway maps were generated using the Cytoscape software [
38], and specifically the VizMapper functions. Pathway maps were adapted from existing pathway maps in WikiPathways [
39]. DESeq2 output for DEG analysis, and Limma output for metabolomics analysis was utilized to direct shading of genes and metabolites within the pathway: red (positive fold change, statistically significant), blue (negative fold change, statistically significant), or gray (non-statistically significant), for individual cancer sites transcripts (triangles) and metabolites (rounded rectangles).
Authors’ contributions
S.R.R., B.D, Y.Z., H.H.H., E.I., J.W., S.L., P.K., and S.M. planned the experiments. S.R.R., Y.Z., H.H.H., E.I., and J.W. carried out RNA-sequencing and Metabolic Pipeline Analysis. S.R.R. carried out Metabolomics analysis. S.R.R., B.D, Y.Z., H.H.H., E.I., J.W., S.L., P.K., and S.M., assisted in the interpretation of data and biological assessment. S.R.R, and S.M. contributed to the writing of the manuscript.
Availability of data and materials
All data generated and/or analyzed in this the current study are either available on the Gene Expression Omnibus (GSE provided) or are available from the corresponding author upon reasonable request.
Acknowledgements
We would like to thank staff scientists Krystin Mantione, M.S., and Sarah Burke at the Roswell Park Comprehensive Cancer Center (RP) Bioanalytics, Metabolomics and Pharmacokinetics (BMPK) Shared Resource for guidance and assistance with our metabolomics studies. We thank Dr. Prashant Singh, Ph.D. and his research associates in the RP Genomics Shared Resource (GSR) for performing RNA-sequencing. We would also like to thank the RP Flow Cytometry Shared Resource. This work was supported by the IOTN: Data Management and Resource-Sharing Center (U24CA232979, S.R.R.), the TREC Fellowship Training Workshop (R25CA203650 (PI: Melinda Irwin); S.R.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Computational resources were maintained by the Center for Computational Research at the University at Buffalo.
Competing interests
The authors have no conflicts of interest to declare at this time.
References
- Jackson, S.E.; Llewellyn, C.H.; Smith, L. The obesity epidemic – Nature via nurture: A narrative review of high–income countries. SAGE Open Med 2020, 8, 2050312120918265. [Google Scholar] [PubMed]
- Kelly, T.; et al. Global burden of obesity in 2005 and projections to 2030. Int J Obes (Lond) 2008, 32, 1431–7. [Google Scholar] [CrossRef] [PubMed]
- Low, S.; Chin, M.C.; Deurenberg–Yap, M. Review on epidemic of obesity. Ann Acad Med Singap 2009, 38, 57–9. [Google Scholar] [PubMed]
- Ng, M.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–81. [Google Scholar] [CrossRef]
- Mokdad, A.H.; et al. Prevalence of obesity, diabetes, and obesity–related health risk factors, 2001. JAMA 2003, 289, 76–9. [Google Scholar] [PubMed]
- Lauby–Secretan, B.; et al. Body Fatness and Cancer––Viewpoint of the IARC Working Group. N Engl J Med 2016, 375, 794–8. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.C.; et al. Obesity paradox in cancer: new insights provided by body composition. Am J Clin Nutr 2014, 99, 999–1005. [Google Scholar]
- Wang, Z.; et al. Paradoxical effects of obesity on T cell function during tumor progression and PD–1 checkpoint blockade. Nat Med 2019, 25, 141–151. [Google Scholar]
- Naik, G.S.; et al. Complex inter–relationship of body mass index, gender and serum creatinine on survival: exploring the obesity paradox in melanoma patients treated with checkpoint inhibition. J Immunother Cancer 2019, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, M.; et al. Renal cell carcinoma survival and body mass index: a dose–response meta–analysis reveals another potential paradox within a paradox. Int J Obes (Lond) 2016, 40, 1817–1822. [Google Scholar]
- Toro, A.L.; et al. Effect of obesity on molecular characteristics of invasive breast tumors: gene expression analysis in a large cohort of female patients. BMC Obes 2016, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Naik, A.; Monjazeb, A.M.; Decock, J. The Obesity Paradox in Cancer, Tumor Immunology, and Immunotherapy: Potential Therapeutic Implications in Triple Negative Breast Cancer. Front Immunol 2019, 10, 1940. [Google Scholar] [CrossRef]
- Kanter, R.; Caballero, B. Global gender disparities in obesity: a review. Adv Nutr 2012, 3, 491–8. [Google Scholar] [PubMed]
- Wang, Y.; et al. Metabolic modulation of immune checkpoints and novel therapeutic strategies in cancer. Semin Cancer Biol 2022, 86 Pt 3, 542–565. [Google Scholar]
- Cheung, O.K.; Cheng, A.S. Gender Differences in Adipocyte Metabolism and Liver Cancer Progression. Front Genet 2016, 7, 168. [Google Scholar]
- Rosendahl, J.; et al. Genome–wide association study identifies inversion in the CTRB1–CTRB2 locus to modify risk for alcoholic and non–alcoholic chronic pancreatitis. Gut 2018, 67, 1855–1863. [Google Scholar] [CrossRef] [PubMed]
- Li, L.O.; Klett, E.L.; Coleman, R.A. Acyl–CoA synthesis, lipid metabolism and lipotoxicity. Biochim Biophys Acta 2010, 1801, 246–51. [Google Scholar]
- Vijayakumar, A.; Yakar, S.; Leroith, D. The intricate role of growth hormone in metabolism. Front Endocrinol (Lausanne) 2011, 2, 32. [Google Scholar] [CrossRef]
- Rudolph, A.; et al. Modification of menopausal hormone therapy–associated colorectal cancer risk by polymorphisms in sex steroid signaling, metabolism and transport related genes. Endocr Relat Cancer 2011, 18, 371–84. [Google Scholar]
- Mabrouk, N.; et al. Impact of Lipid Metabolism on Antitumor Immune Response. Cancers (Basel) 2022, 14. [Google Scholar]
- Yu, W.; et al. Contradictory roles of lipid metabolism in immune response within the tumor microenvironment. J Hematol Oncol 2021, 14, 187. [Google Scholar] [CrossRef]
- Rong, J.; et al. The prognostic value of gender in gastric gastrointestinal stromal tumors: a propensity score matching analysis. Biol Sex Differ 2020, 11, 43. [Google Scholar] [CrossRef]
- Abancens, M.; et al. Sexual Dimorphism in Colon Cancer. Front Oncol 2020, 10, 607909. [Google Scholar] [PubMed]
- Wang, S.; Cowley, L.A.; Liu, X.S. Sex Differences in Cancer Immunotherapy Efficacy, Biomarkers, and Therapeutic Strategy. Molecules 2019, 24. [Google Scholar] [CrossRef] [PubMed]
- Ringel, A.E.; et al. Obesity Shapes Metabolism in the Tumor Microenvironment to Suppress Anti–Tumor Immunity. Cell 2020, 183, 1848–1866. [Google Scholar] [CrossRef]
- McQuade, J.L.; et al. Association of body–mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol 2018, 19, 310–322. [Google Scholar]
- Vick, L.; Wang, Z.; Collins, C.; Khuat, L.; Dunai, C.; Stoffel, K.; Yendamuri, S.; Mukherjee, S.; Rosario, S.; Canter, R.; Monjazeb, A.; Murphy, W. Sex–linked differencecs in obesity markedly impact the anti–tumor efficacy of PD–1 blockade. J Immunother Cancer.
- Rosario, S.R.; et al. Pan–cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas. Nat Commun 2018, 9, 5330. [Google Scholar]
- Rosario, S.R.; et al. Altered acetyl–CoA metabolism presents a new potential immunotherapy target in the obese lung microenvironment. Cancer Metab 2022, 10, 17. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud 2015, 1, a000588. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar]
- Castoldi, A.; et al. The Macrophage Switch in Obesity Development. Front Immunol 2015, 6, 637. [Google Scholar] [CrossRef]
- Stine, Z.E.; et al. Nat Rev Drug Discov 2022, 21, 141–162. [PubMed]
- Dalton, W.S.; Sullivan, D.; Ecsedy, J.; Caligiuri, M.A. Patient Enrichment for Precision–Based Cancer Clinical Trials: Using Prospective Cohort Surveillance as an Approach to Improve Clinical Trials. Clin Pharmacol Ther. 2018, 104, 23–6. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA–seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–40. [Google Scholar] [CrossRef]
- Ritchie, M.E.; et al. limma powers differential expression analyses for RNA–sequencing and microarray studies. Nucleic Acids Res 2015, 43, e47. [Google Scholar] [CrossRef]
- Killcoyne, S.; et al. Cytoscape: a community–based framework for network modeling. Methods Mol Biol 2009, 563, 219–39. [Google Scholar]
- Slenter, D.N.; et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res 2018, 46, D661–D667. [Google Scholar] [CrossRef]
Figure 1.
Obesity corresponds with differential tumor transcriptomic signatures associated with metabolic reprogramming. A) Volcano plot showing significantly down-regulated (blue, adj. p < 0.05 and |logFC|>1.5) and up-regulated (pink, adj. p < 0.05 and |logFC|>1.5) genes in ORIEN Adenocarcinoma patients that were obese (BMI>30), as compared to normal weight (BMI<25) patients. B) Gene set enrichment analysis (GSEA) of these differentially expressed genes from ORIEN analysis show pathways down-regulated (blue, left) and up-regulated (red, right) in obese patients, as compared to normal patients, and C) further metabolic transcriptional pathway analysis specific to metabolic pathways in these same patients. Similar differential gene expression analysis comparing obese patients to non-obese patients were conducted and displayed as volcano plots for the D) TCGA colon adenocarcinoma (COAD) and E) TCGA esophageal carcinoma (ESCA). To demonstrate these transcriptional alterations, occur in a site-specific manner, F) we looked for overlap between the significantly down-regulated (top) and up-regulated (bottom) genes between the COAD and ESCA datasets, and found minimal overlap. Gene set enrichment analysis of G) COAD and H) ESCA revealed transcriptional enrichment of cellular processes decreased in obese patients (blue) like G2M checkpoint, and methylation, and up-regulated processes (red) like immune complement activation and therapeutic resistance. Further, metabolic analysis revealed several metabolic pathways to be differentially regulated in both ESCA and COAD, albeit to a different extent. For example, I) Modeling of the tryptophan/kynurenine/NAD pathway reveals a majority of transcripts are down-regulated (blue triangles, left), in ESCA obese as compared to normal patients. Conversely, a majority of transcripts are up-regulated (red triangles, right), in COAD obese as compared to normal patients.
Figure 1.
Obesity corresponds with differential tumor transcriptomic signatures associated with metabolic reprogramming. A) Volcano plot showing significantly down-regulated (blue, adj. p < 0.05 and |logFC|>1.5) and up-regulated (pink, adj. p < 0.05 and |logFC|>1.5) genes in ORIEN Adenocarcinoma patients that were obese (BMI>30), as compared to normal weight (BMI<25) patients. B) Gene set enrichment analysis (GSEA) of these differentially expressed genes from ORIEN analysis show pathways down-regulated (blue, left) and up-regulated (red, right) in obese patients, as compared to normal patients, and C) further metabolic transcriptional pathway analysis specific to metabolic pathways in these same patients. Similar differential gene expression analysis comparing obese patients to non-obese patients were conducted and displayed as volcano plots for the D) TCGA colon adenocarcinoma (COAD) and E) TCGA esophageal carcinoma (ESCA). To demonstrate these transcriptional alterations, occur in a site-specific manner, F) we looked for overlap between the significantly down-regulated (top) and up-regulated (bottom) genes between the COAD and ESCA datasets, and found minimal overlap. Gene set enrichment analysis of G) COAD and H) ESCA revealed transcriptional enrichment of cellular processes decreased in obese patients (blue) like G2M checkpoint, and methylation, and up-regulated processes (red) like immune complement activation and therapeutic resistance. Further, metabolic analysis revealed several metabolic pathways to be differentially regulated in both ESCA and COAD, albeit to a different extent. For example, I) Modeling of the tryptophan/kynurenine/NAD pathway reveals a majority of transcripts are down-regulated (blue triangles, left), in ESCA obese as compared to normal patients. Conversely, a majority of transcripts are up-regulated (red triangles, right), in COAD obese as compared to normal patients.
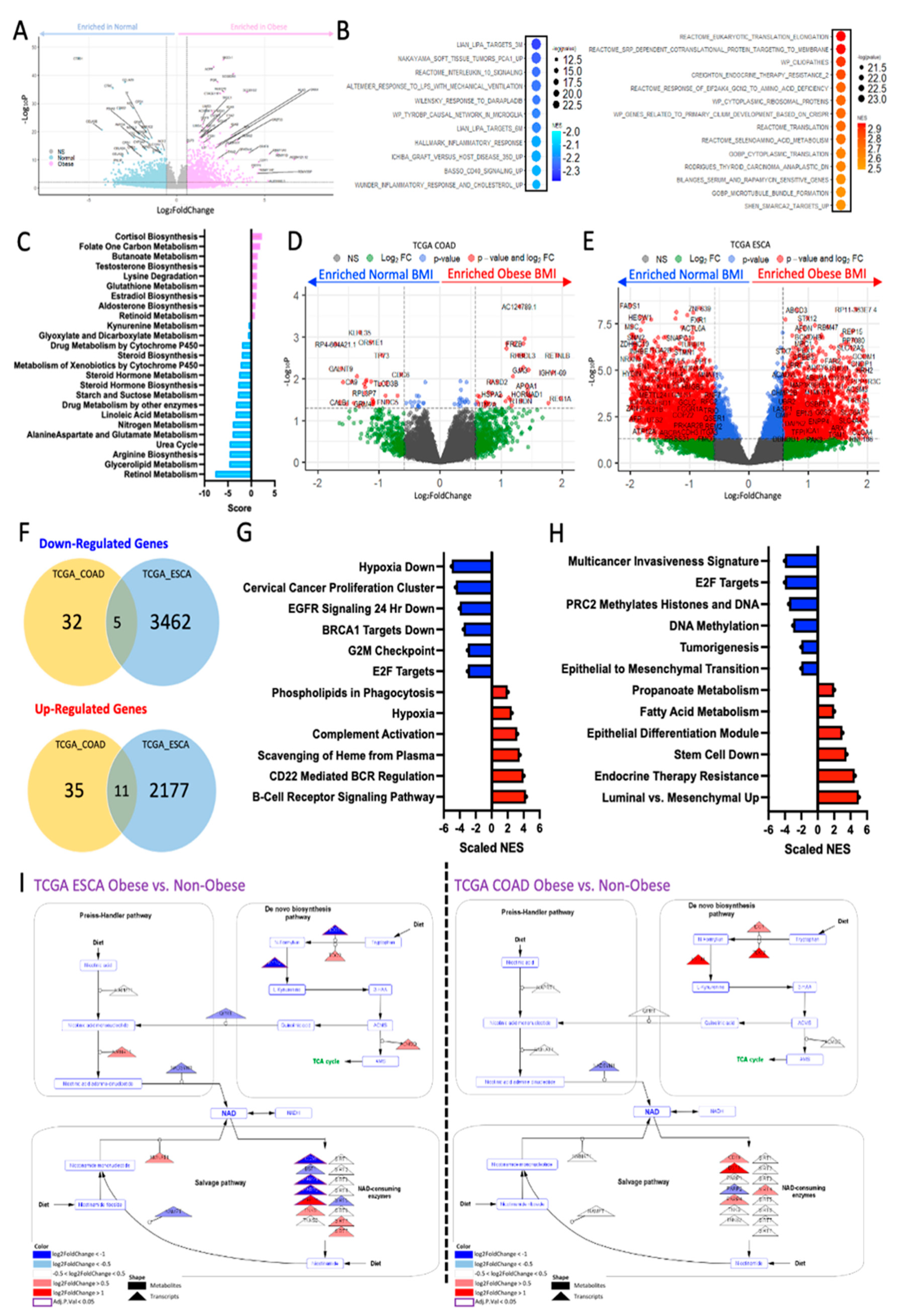
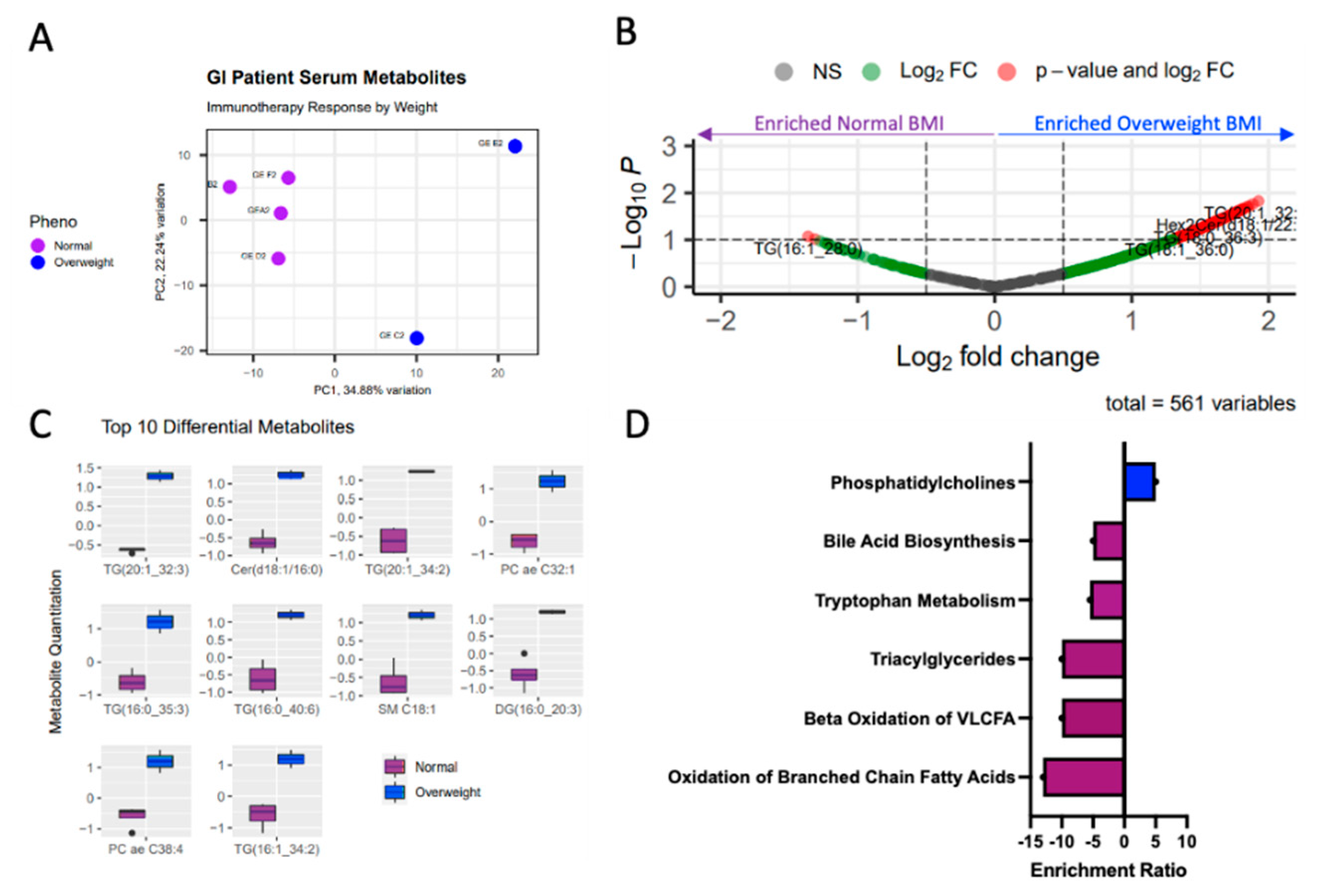
Figure 2.
Obesity associated with differential circulating serum metabolome. A) Principal component analysis reveals distinction between normal weight (purple) and overweight (blue) serum samples, based on 630 metabolite abundance. B) Volcano plot shows significantly differential metabolite abundances (p < 0.1, |logFC| > 1.5) between the normal BMI and overweight BMI groups. C) Demonstrates the top 10 significantly different metabolites, and D) the metabolite set enrichment associated with the differentially abundant metabolites highlighted decreased lipids, tryptophan metabolites and bile acid metabolites in the obese patients, as compared to the non-obese patients (purple), and phosphatidylcholines were increased in obese patients, as compared to normal weight patient serum samples.
Figure 2.
Obesity associated with differential circulating serum metabolome. A) Principal component analysis reveals distinction between normal weight (purple) and overweight (blue) serum samples, based on 630 metabolite abundance. B) Volcano plot shows significantly differential metabolite abundances (p < 0.1, |logFC| > 1.5) between the normal BMI and overweight BMI groups. C) Demonstrates the top 10 significantly different metabolites, and D) the metabolite set enrichment associated with the differentially abundant metabolites highlighted decreased lipids, tryptophan metabolites and bile acid metabolites in the obese patients, as compared to the non-obese patients (purple), and phosphatidylcholines were increased in obese patients, as compared to normal weight patient serum samples.
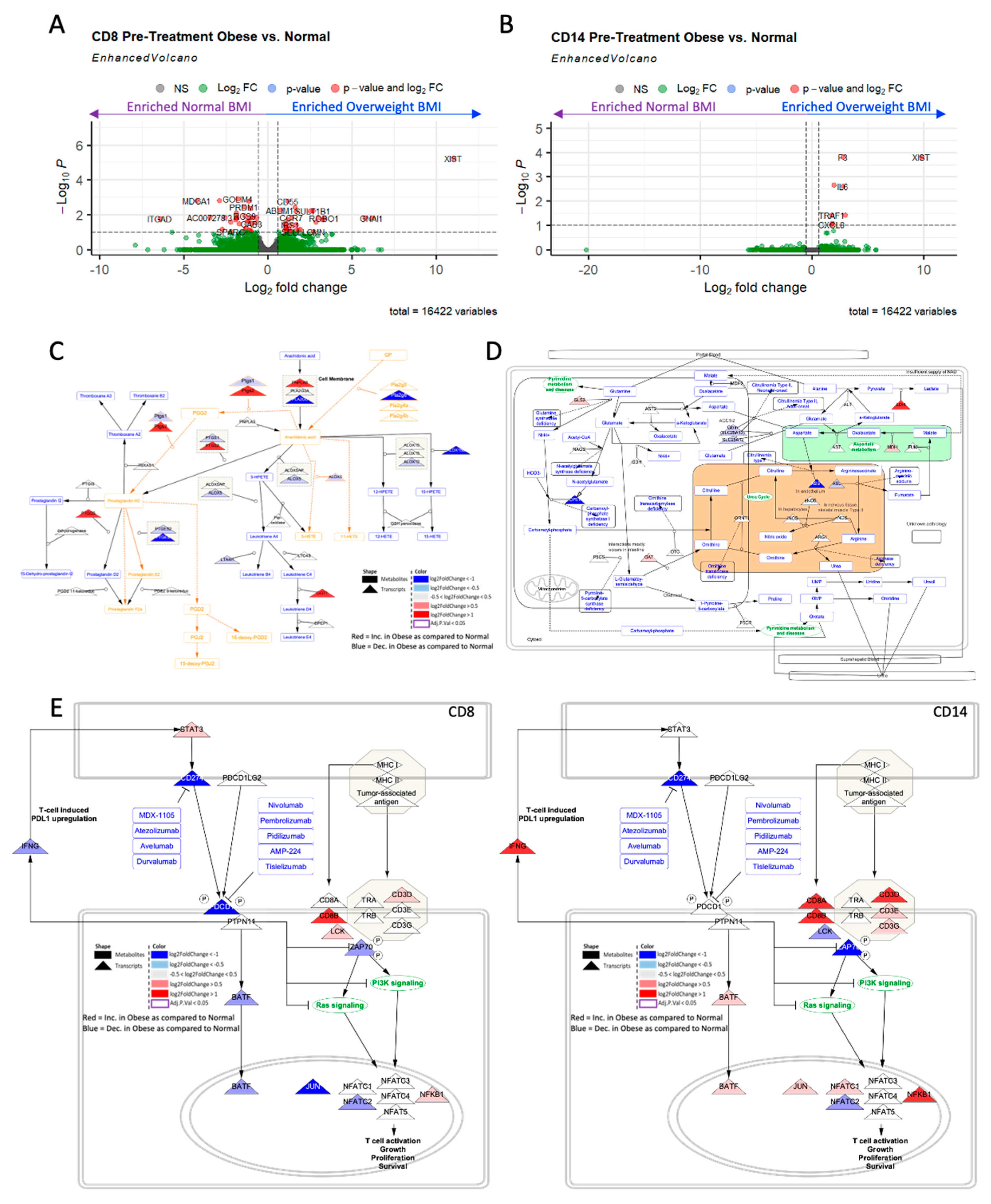
Figure 3.
Obesity associated with differential immune cell transcriptome. Obesity is associated with differential transcriptional profile in both A) CD8+ and B) CD14+ cells of patients with matched metabolomics. C) Prostaglandin pathway is highly upregulated transcriptionally in the Obese patients as compared to the non-obese patient’s CD14+ sorted T-cells. D) Urea pathway is highly upregulated transcriptionally in the Obese patients as compared to the non-obese patient’s CD8+ sorted T-cells. E) PD-L1 response pathway differentially dysregulated in CD8+ (left) and CD14+ (right) cells of obese as compared to non-obese patients.
Figure 3.
Obesity associated with differential immune cell transcriptome. Obesity is associated with differential transcriptional profile in both A) CD8+ and B) CD14+ cells of patients with matched metabolomics. C) Prostaglandin pathway is highly upregulated transcriptionally in the Obese patients as compared to the non-obese patient’s CD14+ sorted T-cells. D) Urea pathway is highly upregulated transcriptionally in the Obese patients as compared to the non-obese patient’s CD8+ sorted T-cells. E) PD-L1 response pathway differentially dysregulated in CD8+ (left) and CD14+ (right) cells of obese as compared to non-obese patients.
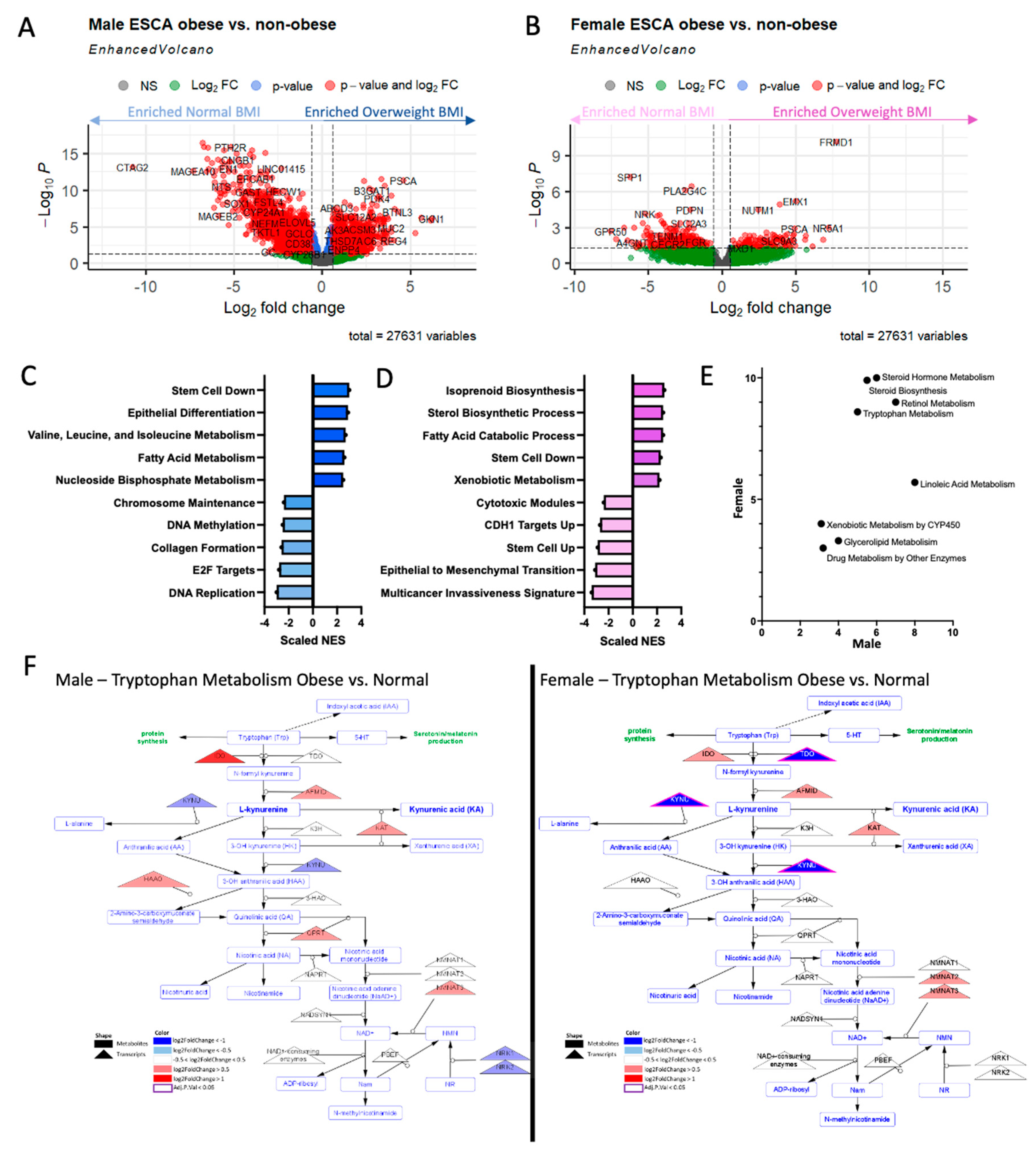
Figure 4.
Gender induces differential transcriptional metabolic profile. A) Male ESCA resulted in a large number of differentially expressed genes DEGs; adj. p < 0.05, |logFC|>1.5, red) when comparing patients with BMI considered normal (left, light blue) to those considered overweight (right, dark blue). B) Female ESCA resulted in a large number of differentially expressed genes (adj. p < 0.05, |logFC|>1.5, red) when comparing patients with BMI considered normal (left, light pink) to those considered overweight (right, dark pink). C) Gene set enrichment analysis (GSEA) of male ESCA DEGs resulted in enrichment of cellular processes, metabolism, and epigenetic associated pathways. D) Gene set enrichment analysis (GSEA) of female ESCA DEGs resulted in enrichment of similar cellular processes, and metabolism associated pathways. E) Metabolic pipeline analysis resulted in enrichment of steroid, tryptophan, and other lipid metabolic pathways in males and females. F) Comparison of tryptophan metabolic pathway in males (left) and females (right) demonstrated dysregulation of transcripts (triangles) in obese vs. normal weight patients, albeit to a different extent. Many of the transcripts were largely upregulated with obesity (red) in males and downregulated with obesity (blue) in females.
Figure 4.
Gender induces differential transcriptional metabolic profile. A) Male ESCA resulted in a large number of differentially expressed genes DEGs; adj. p < 0.05, |logFC|>1.5, red) when comparing patients with BMI considered normal (left, light blue) to those considered overweight (right, dark blue). B) Female ESCA resulted in a large number of differentially expressed genes (adj. p < 0.05, |logFC|>1.5, red) when comparing patients with BMI considered normal (left, light pink) to those considered overweight (right, dark pink). C) Gene set enrichment analysis (GSEA) of male ESCA DEGs resulted in enrichment of cellular processes, metabolism, and epigenetic associated pathways. D) Gene set enrichment analysis (GSEA) of female ESCA DEGs resulted in enrichment of similar cellular processes, and metabolism associated pathways. E) Metabolic pipeline analysis resulted in enrichment of steroid, tryptophan, and other lipid metabolic pathways in males and females. F) Comparison of tryptophan metabolic pathway in males (left) and females (right) demonstrated dysregulation of transcripts (triangles) in obese vs. normal weight patients, albeit to a different extent. Many of the transcripts were largely upregulated with obesity (red) in males and downregulated with obesity (blue) in females.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).