Submitted:
23 May 2023
Posted:
24 May 2023
You are already at the latest version
Abstract

Keywords:
1. Introduction
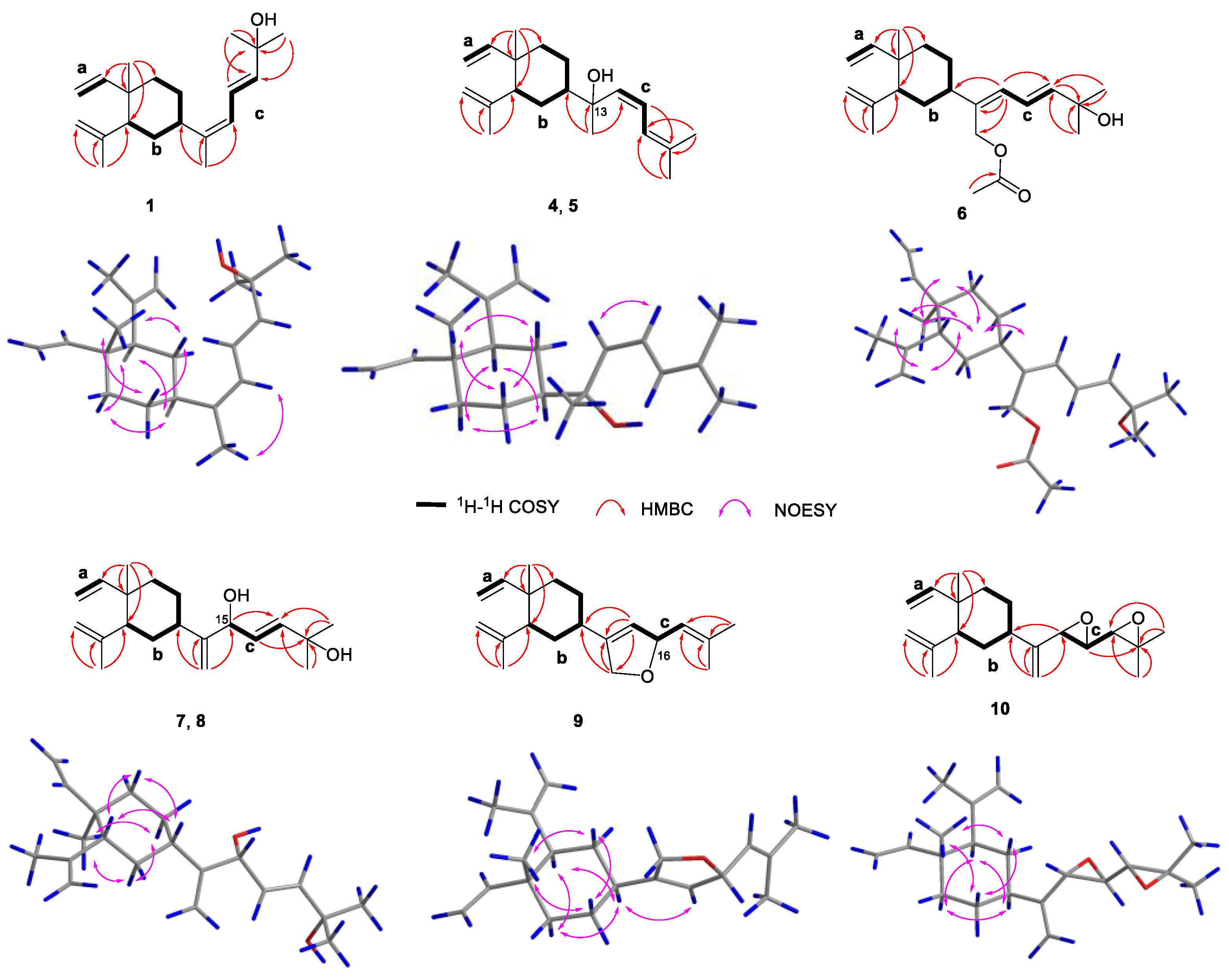
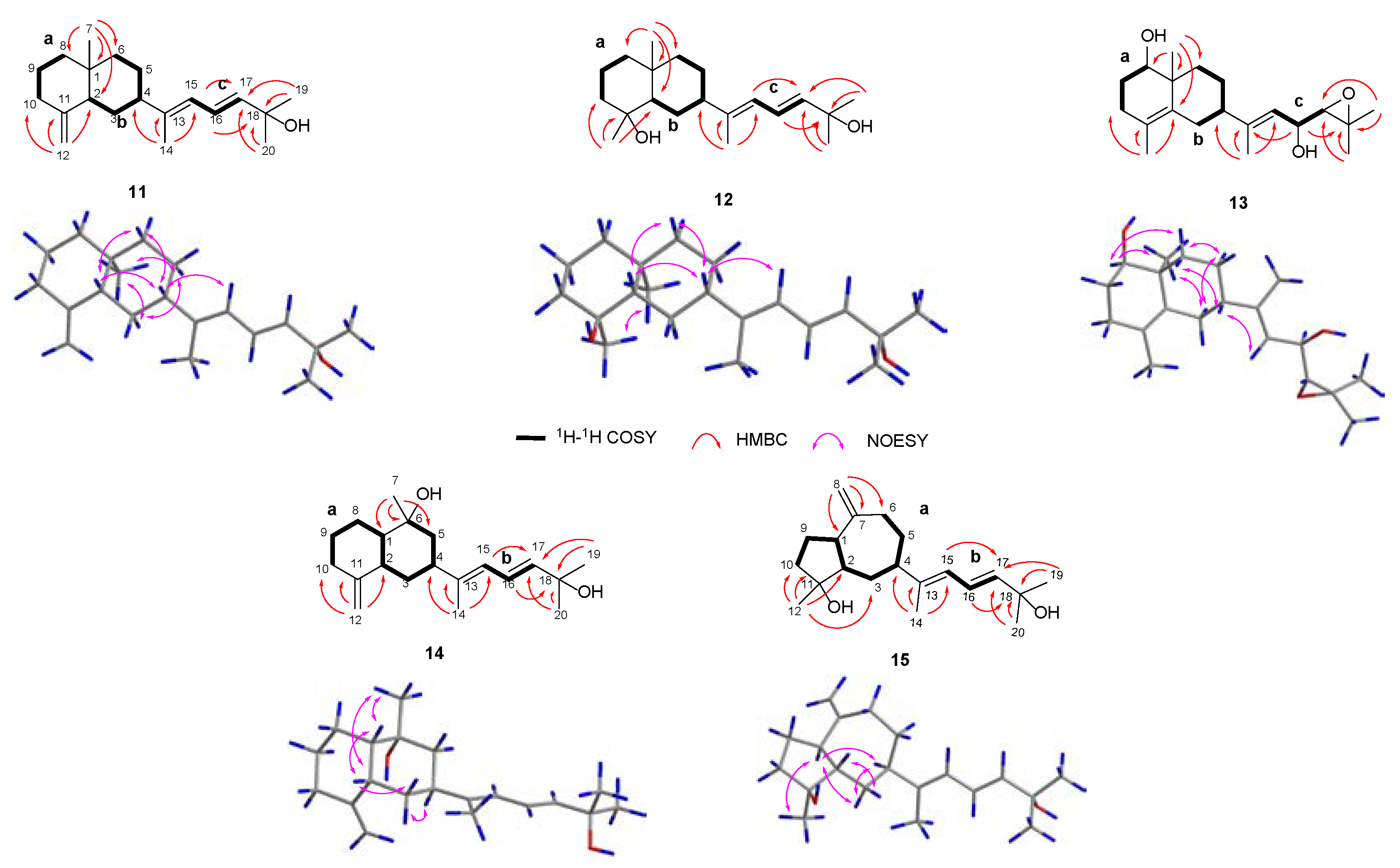
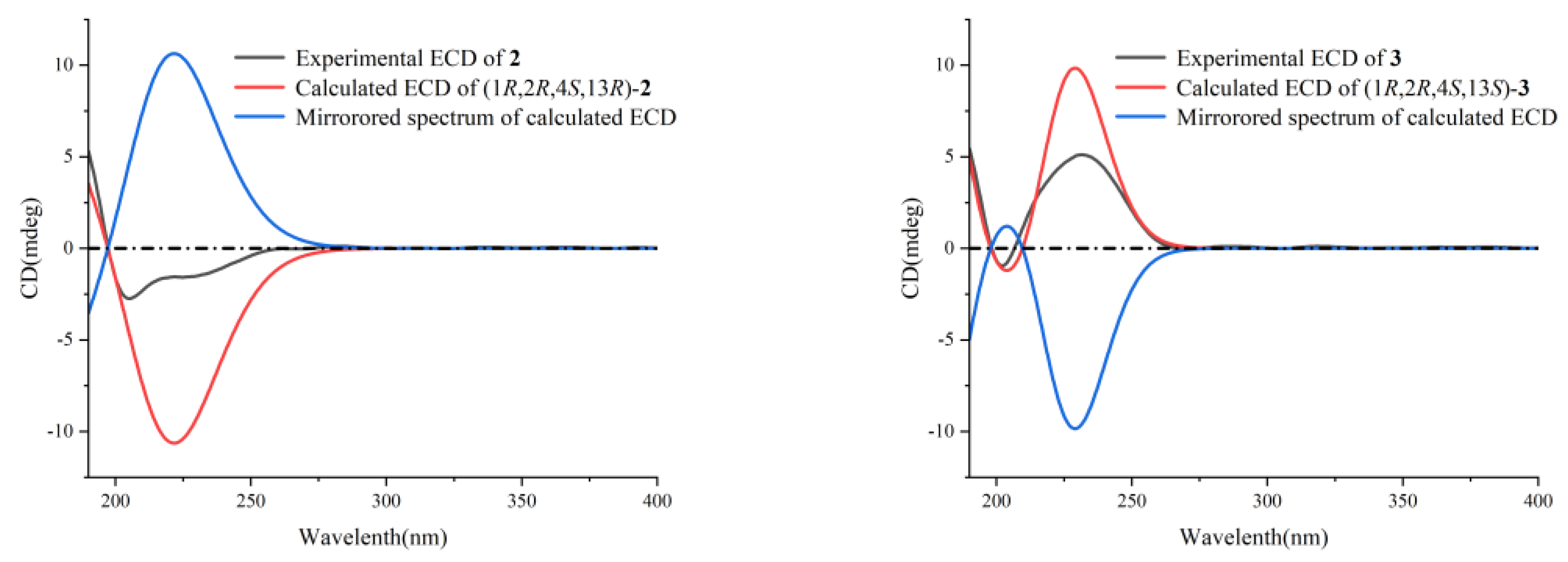
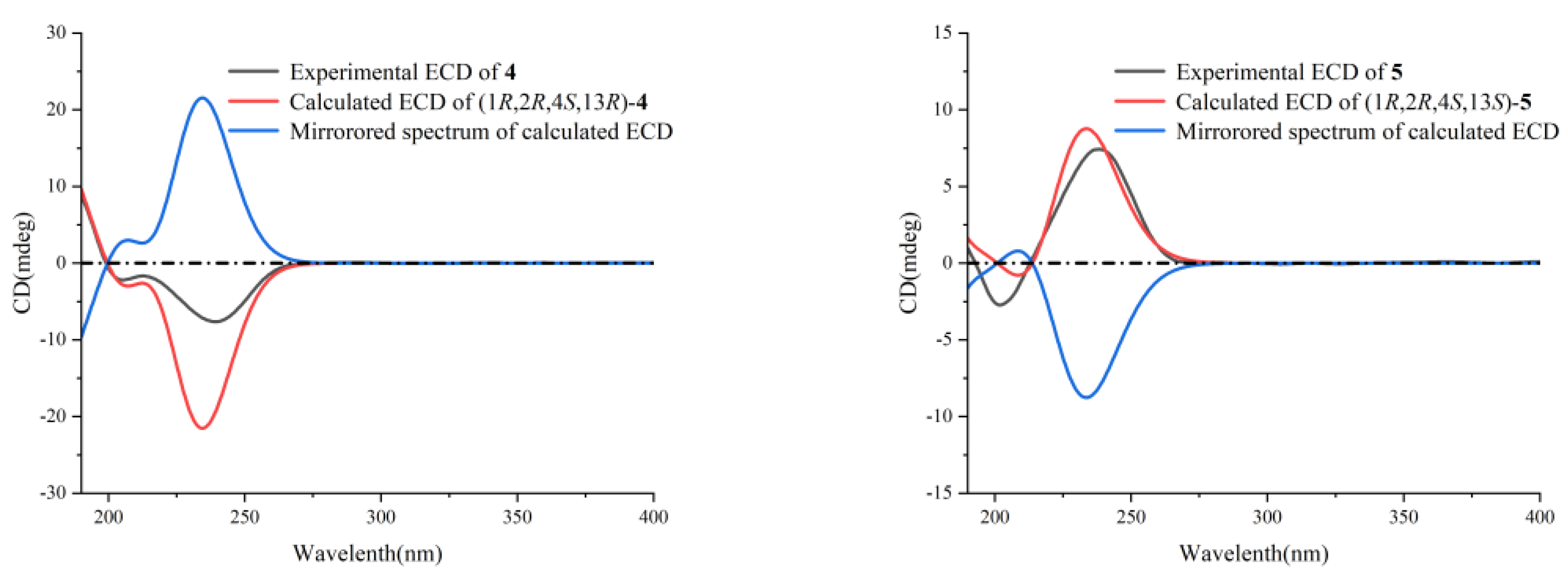
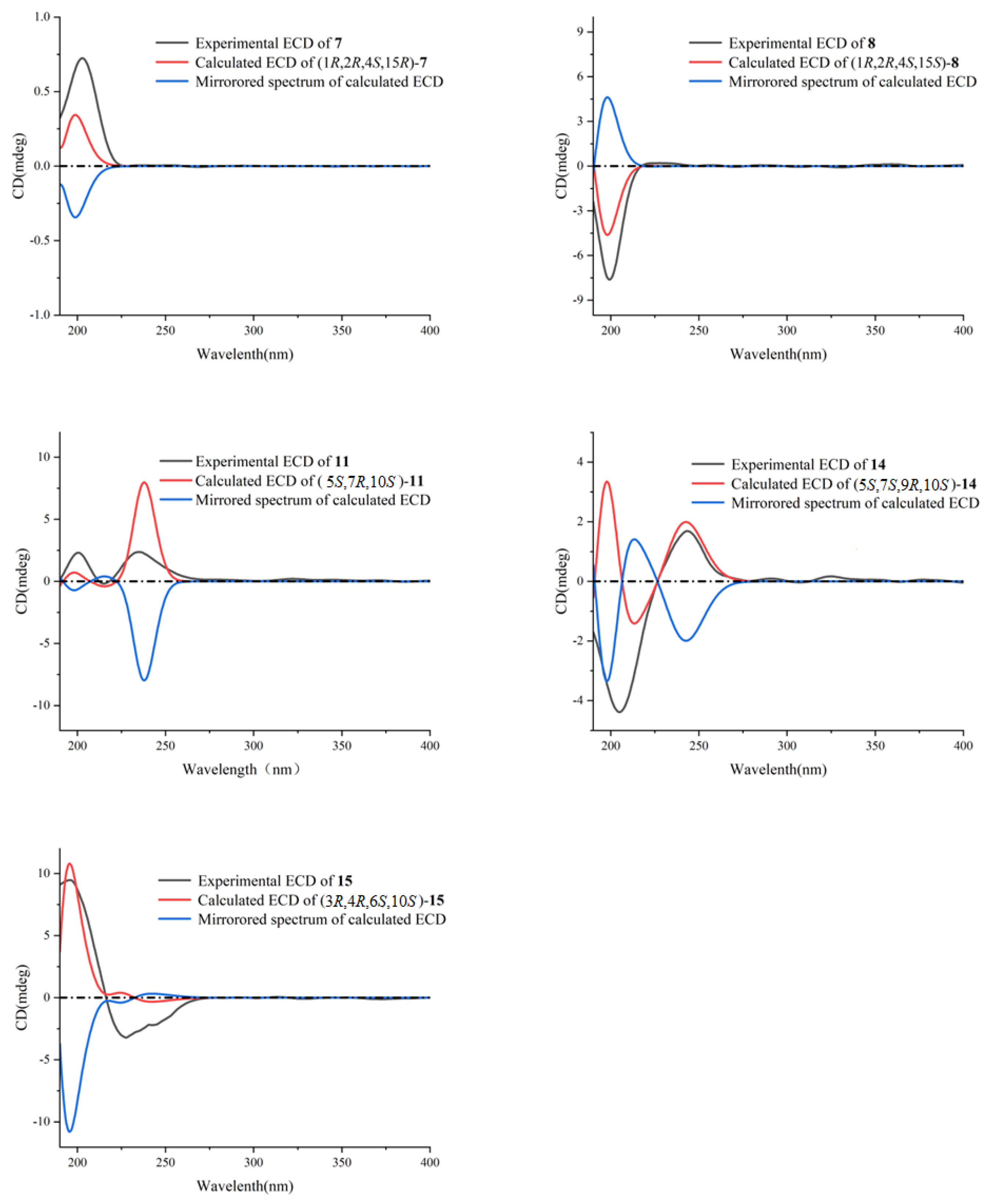
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.4. Spectroscopic Data of Compounds
3.5. Esterification of 3 and 5 with MTPA chlorides
3.6. QM-NMR Calculational Section
3.7. TDDFT-ECD Calculational Section
3.8. Antibacterial assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chill, L.; Berrer, N.; Benayahu, Y.; Kashman, Y. Eunicellin diterpenes from two Kenyan soft corals. J. Nat. Prod. 2005, 68, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liu, J.; Leng, X.; Ouyang, H. Chemical diversity and biological activity of secondary metabolites from soft coral genus Sinularia since 2013. Mar. Drugs 2021, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Elkhawas, Y.A.; Elissawy, A.M.; Elnaggar, M.S.; Mostafa, N.M.; Al-Sayed, E.; Bishr, M.M.; Singab, A.N.B.; Salama, O.M. Chemical diversity in species belonging to soft coral genus Sacrophyton and its impact on biological activity: a review. Mar. Drugs 2020, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-L.; Su, J.-H.; Huang, C.-Y.; Tai, C.-J.; Sung, P.-J.; Liaw, C.-C.; Sheu, J.-H. Simplexins P–S, eunicellin-based diterpenes from the soft coral Klyxum simplex. Mar. Drugs 2012, 10, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-W.; Wu, Y.-C.; Chiang, M.Y.; Su, J.-H.; Wang, W.-H.; Fan, T.-Y.; Sheu, J.-H. Eunicellin-based diterpenoids from the cultured soft coral Klyxum simplex. Tetrahedron 2009, 65, 7016–7022. [Google Scholar] [CrossRef]
- Chen, B.-W.; Chao, C.-H.; Su, J.-H.; Tsai, C.-W.; Wang, W.-H.; Wen, Z.-H.; Huang, C.-Y.; Sung, P.-J.; Wu, Y.-C.; Sheu, J.-H. Klysimplexins I–T, eunicellin-based diterpenoids from the cultured soft coral Klyxum simplex. Org. & Biomol. Chem. 2011, 9, 834–844. [Google Scholar] [CrossRef]
- Wu, S.-L.; Su, J.-H.; Wen, Z.-H.; Hsu, C.-H.; Chen, B.-W.; Dai, C.-F.; Kuo, Y.-H.; Sheu, J.-H. Simplexins A−I, eunicellin-based diterpenoids from the soft coral Klyxum simplex. J. Nat. Prod. 2009, 72, 994–1000. [Google Scholar] [CrossRef]
- Hsu, F.-J.; Chen, B.-W.; Wen, Z.-H.; Huang, C.-Y.; Dai, C.-F.; Su, J.-H.; Wu, Y.-C.; Sheu, J.-H. Klymollins A–H, bioactive eunicellin-based diterpenoids from the Formosan soft coral Klyxum molle. J. Nat. Prod. 2011, 74, 2467–2471. [Google Scholar] [CrossRef]
- Lin, M.-C.; Chen, B.-W.; Huang, C.-Y.; Dai, C.-F.; Hwang, T.-L.; Sheu, J.-H. Eunicellin-based diterpenoids from the Formosan sosft coral Klyxum molle with inhibitory activity on superoxide generation and elastase release by neutrophils. J. Nat. Prod. 2013, 76, 1661–1667. [Google Scholar] [CrossRef]
- Chang, F.-Y.; Hsu, F.-J.; Tai, C.-J.; Wei, W.-C.; Yang, N.-S.; Sheu, J.-H. Klymollins T–X, bioactive eunicellin-based diterpenoids from the soft coral Klyxum molle. Mar. Drugs 2014, 12, 3060–3071. [Google Scholar] [CrossRef]
- Chang, F.-Y.; Chokkalingam, U.; Tai, C.-J.; Huang, C.-Y.; Wei, W.-C.; Yang, N.-S.; Su, J.-H.; Sung, P.-J.; Sheu, J.-H. New eunicellin-derived diterpenoids from a Taiwanese soft coral Klyxum molle. Tetrahedron 2016, 72, 192–198. [Google Scholar] [CrossRef]
- Li, G.; Li, H.; Tang, W.; Guo, Y.-W.; Li, X.-W. Klyflaccilides A and B, diterpenoids with 6/5/8/3 fused tetracyclic carbon skeleton from the Hainan soft coral Klyxum flaccidum. Org. Lett. 2019, 21, 5660–5664. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Sun, L.-L.; Dickschat, J.S.; Guo, Y.-W. Klyflaccilins B-T, polyoxgenated eunicellins from the soft coral Klyxum flaccidum. Eur. J. Org. Chem. 2021, 9, 1402–1406. [Google Scholar] [CrossRef]
- Ahmed, A.F.; Tsai, C.-R.; Huang, C.-Y.; Wang, S.-Y.; Sheu, J.-H. Klyflaccicembranols A–I, new cembranoids from the soft coral Klyxum flaccidum. Mar. Drugs 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Tseng, W.-R.; Ahmed, A.F.; Huang, C.-Y.; Tsai, Y.-Y.; Tai, C.-J.; Orfali, R.S.; Hwang, T.-L.; Wang, Y.-H.; Dai, C.-F.; Sheu, J.-H. Bioactive capnosanes and cembranes from the soft coral Klyxum flaccidum. Mar. Drugs 2019, 17, 461. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Zhu, Z.-D.; Chen, J.-S.; Li, J.; Gu, Y.-C.; Zhu, W.-L.; Li, X.-W.; Guo, Y.-W. Xishacorenes A–C, diterpenes with bicyclo[3.3.1]nonane nucleus from the Xisha soft coral Sinularia polydactyla. Org. Lett. 2017, 19, 4183–4186. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Li, J.; Wu, Y.; Zhu, Z.-D.; Mollo, E.; Gavagnin, M.; Gu, Y.-C.; Zhu, W.-L.; Li, X.-W.; Guo, Y.-W. Sarinfacetamides A and B, nitrogenous diterpenoids with tricyclo[6.3.1.01,5]dodecane scaffold from the South China Sea soft coral Sarcophyton infundibuliforme. Org. Lett. 2018, 20, 2637–2640. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, X.-L.; Wang, J.-R.; Lei, X.; Tang, W.; Li, X.-W.; Sun, H.; Guo, Y.-W. Sarcomililate A, an unusual diterpenoid with tricyclo[11.3.0.02,16]hexadecane carbon skeleton, and its potential biogenetic precursors from the Hainan soft coral Sarcophyton mililatensis. J. Org. Chem. 2019, 84, 2568–2576. [Google Scholar] [CrossRef]
- Edrada, R. A.; Proksch, P.; Wray, V.; Witte, L.; Ofwegen, L. Four new bioactive lobane diterpenes of the soft coral Lobophytum pauciflorum from Mindoro, Philippines. J. Nat. Prod. 1998, 61, 358–361. [Google Scholar] [CrossRef]
- Chang, C.-H.; Ahmed, A.F.; Yang, T.-S.; Lin, Y.-C.; Huang, C.-Y.; Hwang, T.-L.; Sheu, J.-H. Isolation of lobane and prenyleudesmane diterpenoids from the soft coral Lobophytum varium. Mar. Drugs 2020, 18, 223. [Google Scholar] [CrossRef]
- Raju, B.L.; Subbaraju, G.V.; Rao, Ch.B.; Trimurtulu, G. Two new oxygenated lobanes from a soft coral of Lobophytum species of the Andaman and Nicobar Coasts. J. Nat. Prod. 1993, 56, 961–966. [Google Scholar] [CrossRef]
- Coll, J.C.; Bowden, B.F.; König, G.M.; Braslau, R.; Price, I.R. Studies of Australian soft corals. xxxx.1 The natural products chemistry of alcyonacean soft corals with special reference to the genus Lobophytum. Bull. Soc. Chim. Belg. 1986, 95, 815–834. [Google Scholar] [CrossRef]
- Li, X.-L.; Kurtán, T.; Hu, J.-C.; Mándi, A.; Li, J.; Li, X.-W.; Guo, Y.-W. Structural and stereochemical studies of Laurokamurols A–C, uncommon bis-sesquiterpenoids from the Chinese Red Alga Laurencia okamurai Yamada. J. Agric. Food Chem. 2017, 65, 1550–1555. [Google Scholar] [CrossRef] [PubMed]
- Pescitelli, G. ECD exciton chirality method today: a modern tool for determining absolute configurations. Chirality 2022, 34, 333–363. [Google Scholar] [CrossRef] [PubMed]
- Huo, Z.-Q.; Zhu, F.; Zhang, X.-W.; Zhang, X.; Liang, H.-B.; Yao, J.-C.; Liu, Z.; Zhang, G.- M.; Yao, Q.-Q.; Qin, G.-F. Approaches to configuration determinations of flexible marine natural products: advances and prospects. Mar. Drugs 2022, 20, 333. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-L.; Li, W.-S.; Li, J.; Zhang, H.-Y.; Yao, L.-G.; Luo, H.; Guo, Y.-W.; Li, X.-W. Uncommon diterpenoids from the South China Sea soft coral Sinularia humilis and their stereochemistry. J. Org. Chem. 2021, 86, 3367–3376. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-W.; Cuadrado, C.; Yao, L.-G.; Daranas, A.H.; Guo, Y.-W. Quantum mechanical–NMR-aided configuration and conformation of two unreported macrocycles isolated from the soft coral Lobophytum sp.: energy calculations versus coupling constants. Org. Lett. 2020, 22, 4093–4096. [Google Scholar] [CrossRef]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: an improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR Shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef]
- Grimblat, N.; Gavín, J.A.; Daranas, A.H.; Sarotti, A.M. Combining the power of J coupling and DP4 analysis on stereochemical assignments: The J-DP4 Methods. Org. Lett. 2019, 21, 4003–4007. [Google Scholar] [CrossRef]

| No. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | |
| 2 | 2.08, m | 1.94, m | 1.94, m | 1.97, m | 1.97, m |
| 3a | 1.68, m | 1.43, m | 1.43, m | 1.50, m | 1.50, m |
| 3b | 1.74, m | 1.59, m | 1.59, m | 1.62, m | 1.62, m |
| 4 | 2.70, m | 1.43, m | 1.43, m | 1.50, m | 1.50, m |
| 5a | 1.38, m | 1.44, m | 1.44, m | 1.33, m | 1.33, m |
| 5b | 1.59, m | 1.63, m | 1.63, m | 1.45, m | 1.45, m |
| 6a | 1.44, m | 1.44, m | 1.44, m | 1.46, m | 1.46, m |
| 6b | 1.55, m | ||||
| 7 | 1.02, s | 0.97, s | 0.97, s | 0.97, s | 0.97, s |
| 8 | 5.83, dd (17.2, 11.3) | 5.80, dd (15.3, 10.9) | 5.80, dd (15.3, 10.9) | 5.80, dd (15.3, 10.9) | 5.80, dd (15.3, 10.9) |
| 9a | 4.90, d (11.3) | 4.88, d (10.9) | 4.88, d (10.9) | 4.88, d (10.9) | 4.88, d (10.9) |
| 9b | 4.90, d (17.2) | 4.89, d (15.3) | 4.89, d (15.3) | 4.89, d (15.3) | 4.89, d (15.3) |
| 10a | 4.59, s | 4.58, s | 4.58, s | 4.58, s | 4.58, s |
| 10b | 4.81, s | 4.81, s | 4.81, s | 4.81, s | 4.81, s |
| 12 | 1.71, s | 1.70, s | 1.70, s | 1.70, s | 1.70, s |
| 14 | 1.76, s | 1.29, s | 1.29, s | 1.35, s | 1.35, s |
| 15 | 5.77, d (10.9) | 5.65, d (15.3) | 5.65, d (15.3) | 5.31, d (11.9) | 5.31, d (11.9) |
| 16 | 6.50, dd (15.2, 10.9) | 6.43, dd (15.3, 10.9) | 6.43, dd (15.3, 10.9) | 6.19, t (11.9) | 6.19, t (11.9) |
| 17 | 5.70, d (15.2) | 5.84, d (10.9) | 5.84, d (10.9) | 6.63, d (11.9) | 6.63, d (11.9) |
| 19 | 1.34, s | 1.78, s | 1.78, s | 1.74, s | 1.74, s |
| 20 | 1.34, s | 1.78, s | 1.78, s | 1.81, s | 1.81, s |
| No. | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|
| δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | |
| 2 | 2.02, m | 1.99, m | 1.99, m | 2.01, m | 2.02, m |
| 3a | 1.57, m | 1.57, m | 1.57, m | 1.61, m | 1.53, m |
| 3b | 1.60, m | ||||
| 4 | 2.11, m | 1.99, m | 1.99, m | 2.10, m | 1.99, m |
| 5a | 1.48, m | 1.47, m | 1.47, m | 1.44, m | 1.50, m |
| 5b | 1.63, m | 1.62, m | 1.62, m | 1.68, m | 1.62, m |
| 6a | 1.48, m | 1.45, m | 1.45, m | 1.46, m | 1.48, m |
| 6b | 1.51, m | 1.51, m | 1.51, m | 1.51, m | 1.51, m |
| 7 | 1.01, s | 1.02, s | 1.02, s | 1.01, s | 1.01, s |
| 8 | 5.82, dd (17.6, 10.6) | 5.81, dd (17.8, 10.5) | 5.81, dd (17.8, 10.5) | 5.81, dd (17.8, 10.5) | 5.81, dd (17.8, 10.5) |
| 9a | 4.90, d (10.6) | 4.90, d (10.5) | 4.90, d (10.5) | 4.90, d (10.5) | 4.90, d (10.5) |
| 9b | 4.90, d (17.6) | 4.91, d (17.8) | 4.91, d (17.8) | 4.90, d (17.8) | 4.92, d (17.8) |
| 10a | 4.58, s | 4.58, s | 4.58, s | 4.58, s | 4.57, s |
| 10b | 4.82, s | 4.82, s | 4.82, s | 4.83, s | 4.83, s |
| 12 | 1.70, s | 1.70, s | 1.70, s | 1.71, s | 1.71, s |
| 14a | 4.75, d (3.3) | 4.99, s | 4.99, s | 4.54, m | 4.95, s |
| 14b | 5.15, s | 5.15, s | 4.64, m | 5.09, s | |
| 15 | 6.10, d (11.0) | 4.62, m | 4.62, m | 5.31, m | 3.31, d (2.2) |
| 16 | 6.55, dd (15.2, 11.0) | 5.66, dd (15.6, 6.5) | 5.66, dd (15.6, 6.5) | 5.50, m | 2.73, ss (5.9, 2.2) |
| 17 | 5.89, d (15.2) | 5.90, d (15.6) | 5.90, d (15.6) | 5.15, m | 2.63, d (5.9) |
| 19 | 1.35, s | 1.33, s | 1.33, s | 1.73, s | 1.35, s |
| 20 | 1.35, s | 1.33, s | 1.33, s | 1.73, s | 1.39, s |
| 22 | 2.07 s |
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| δC mult. | δC mult. | δC mult. | δC mult. | δC mult. | δC mult. | δC mult. | δC mult. | δC mult. | δC mult. | |
| 1 | 39.7, C | 39.9, C | 39.9, C | 39.9, C | 39.9, C | 39.9, C | 39.9, C | 39.9, C | 39.9, C | 39.8, C |
| 2 | 52.6, CH | 52.8, CH | 52.8, CH | 52.7, CH | 52.8, CH | 52.9, CH | 53.0, CH | 53.0, CH | 52.6, CH | 52.8, CH |
| 3 | 31.9, CH2 | 28.2, CH2 | 28.5, CH2 | 28.1, CH2 | 28.5, CH2 | 33.3, CH2 | 34.3, CH2 | 34.7, CH2 | 33.1, CH2 | 33.9, CH2 |
| 4 | 40.5, CH | 48.8, CH | 48.9, CH | 49.5, CH | 49.6, CH | 41.1, CH | 40.9, CH | 41.0, CH | 37.2, CH | 41.1, CH |
| 5 | 26.0, CH2 | 22.7, CH2 | 22.4, CH2 | 22.6, CH2 | 22.2, CH2 | 27.2, CH2 | 28.7, CH2 | 28.2, CH2 | 27.2, CH2 | 27.1, CH2 |
| 6 | 39.8, CH2 | 40.0, CH2 | 39.9, CH2 | 40.0, CH2 | 40.0, CH2 | 40.0, CH2 | 40.2, CH2 | 40.1, CH2 | 39.7, CH2 | 39.8, CH2 |
| 7 | 16.8, CH3 | 16.7, CH3 | 16.7, CH3 | 16.7, CH3 | 16.7, CH3 | 16.8, CH3 | 16.8, CH3 | 16.8, CH3 | 16.7, CH3 | 16.7, CH3 |
| 8 | 150.4, CH | 150.4, CH | 150.4, CH | 150.5, CH | 150.5, CH | 150.2, CH | 150.3, CH | 150.3, CH | 150.1, CH | 150.1, CH |
| 9 | 110.1, CH2 | 110.0, CH2 | 110.0, CH2 | 110.0, CH2 | 110.0, CH2 | 110.2, CH2 | 110.1, CH2 | 110.1, CH2 | 110.2, CH2 | 110.3, CH2 |
| 10 | 112.3, CH2 | 112.2, CH2 | 112.2, CH2 | 112.2, CH2 | 112.2, CH2 | 112.4, CH2 | 112.3, CH2 | 112.4, CH2 | 112.4, CH2 | 112.4, CH2 |
| 11 | 147.7, C | 148.1, C | 148.0, C | 148.1, C | 148.0, C | 147.6, C | 147.6, C | 147.6, C | 147.5, C | 147.4, C |
| 12 | 25.0, CH3 | 24.9, CH3 | 24.9, CH3 | 24.8, CH3 | 24.9, CH3 | 24.9, CH3 | 24.9, CH3 | 24.9, CH3 | 24.9, CH3 | 25.0, CH3 |
| 13 | 143.1, C | 75.0, C | 75.1, C | 76.7, C | 76.7, C | 140.2, C | 156.0, C | 156.0, C | 145.7, C | 149.2, C |
| 14 | 20.2, CH3 | 26.2, CH3 | 26.2, CH3 | 27.4, CH3 | 27.5, CH3 | 61.6, CH2 | 109.1, CH2 | 108.9, CH2 | 75.3, CH2 | 109.4, CH2 |
| 15 | 124.6, CH | 135.3, CH | 135.2, CH | 133.5, CH | 133.4, CH | 128.4, CH | 75.0, CH | 74.9, CH | 120.9, CH | 56.0, CH |
| 16 | 122.0, CH | 124.4, CH | 124.4, CH | 125.5, CH | 125.5, CH | 122.0, CH | 128.1, CH | 128.2, CH | 83.1, CH | 58.5, CH |
| 17 | 139.3, CH | 124.8, CH | 124.8, CH | 121.4, CH | 121.5, CH | 143.0, CH | 139.6, CH | 139.6, CH | 125.7, CH | 63.0, CH |
| 18 | 71.1, C | 137.0, C | 137.0, C | 137.3, C | 137.3, C | 71.1, C | 70.8, C | 70.8, C | 135.5, C | 58.6, C |
| 19 | 30.2, CH3 | 18.5, CH3 | 18.5, CH3 | 17.8, CH3 | 17.8, CH3 | 29.9, CH3 | 29.9, CH3 | 29.9, CH3 | 18.2, CH3 | 19.7, CH3 |
| 20 | 30.2, CH3 | 26.0, CH3 | 26.1, CH3 | 26.7, CH3 | 26.7, CH3 | 29.9, CH3 | 30.0, CH3 | 29.9, CH3 | 26.0, CH3 | 24.7, CH3 |
| 21 | 171.3, C | |||||||||
| 22 | 21.2, CH3 |
| No. | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|
| δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | δH mult. (J Hz) | |
| 1 | 2.02, m | 2.27, m | |||
| 2 | 1.82, m | 1.25, m | 3.10, m | 1.79, m | |
| 3a | 1.34, m | 1.50, m1.50, m | 1.84, m | 1.71, m | 1.52, m |
| 3b | 1.52, m | 2.52, m | 1.86, m | 1.83, m | |
| 4 | 2.00, m | 1.99, m | 1.84, m | 2.30, m | 2.44, m |
| 5a | 1.50, m | 1.24, m | 1.53, m | 1.72, m | 1.64, m |
| 5b | 1.51, m | 1.82, m | 1.70, m | 1.66, m | |
| 6a | 1.28, m | 1.21, m | 1.21, m | 2.12, m | |
| 6b | 1.51, m | 1.44, m | 2.04, m | 2.49, m | |
| 7 | 0.73, s | 0.89, s | 1.03, s | 1.15, s | |
| 8a | 1.28, m | 1.10, m | 3.49, m | 1.52, m | 4.70, s |
| 8b | 1.44, m | 1.39, m | 1.57, m | 4.71. s | |
| 9a | 1.59, m | 1.55, m | 1.53, m | 1.55, m | 1.72, m |
| 9b | 1.64, m | 1.70, m | 1.82, m | 1.83, m | |
| 10a | 2.00, m | 1.37, m | 1.97, m | 2.30, m | 1.72, m |
| 10b | 2.31, m | 1.79, m | 2.17, m | 2.39, m | 1.76, m |
| 12a | 4.42 s | 1.11, s | 1.59, s | 4.75, s | 1.18, s |
| 12b | 4.70, s | 4.87, s | |||
| 14 | 1.80, s | 1.79, s | 1.73, s | 1.77, s | 1.75, s |
| 15 | 5.88, d (10.8) | 5.87, d (10.8) | 5.33, d (8.8) | 5.87, d (10.7) | 5.90, d (10.7) |
| 16 | 6.50, dd (15.3, 10.8) | 6.48, dd (15.3, 10.8) | 4.26, dd (8.8, 7.8) | 6.46, dd (15.3, 10.7) | 6.45, dd (15.3, 10.7) |
| 17 | 5.75, d (15.3) | 5.75, d (15.3) | 2.84, d (7.8) | 5.75, d (15.3) | 5.75, d (15.3) |
| 19 | 1.36, s | 1.35, s | 1.33, s | 1.36, s | 1.35, s |
| 20 | 1.36, s | 1.35, s | 1.35, s | 1.36, s | 1.35, s |
| No. | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|
| δC mult. | δC mult. | δC mult. | δC mult. | δC mult. | |
| 1 | 36.1, C | 34.7, C | 39.6, C | 48.6, CH | 48.4, CH |
| 2 | 50.0, CH | 55.0, CH | 133.4, C | 45.3, CH | 52.7, CH |
| 3 | 29.4, CH2 | 26.7, CH2 | 30.8, CH2 | 26.4, CH2 | 31.5, CH2 |
| 4 | 48.0, CH | 48.5, CH | 47.7, CH | 44.0, CH | 45.6, CH |
| 5 | 26.7, CH2 | 26.0, CH2 | 27.3, CH2 | 40.7, CH2 | 31.2, CH2 |
| 6 | 41.3, CH2 | 44.7, CH2 | 38.9, CH2 | 81.8, C | 37.0, CH2 |
| 7 | 16.5, CH3 | 18.9, CH3 | 17.5, CH3 | 24.1, CH3 | 153.4, C |
| 8 | 42.1, CH2 | 41.2, CH2 | 78.5, CH | 29.0, CH2 | 107.2, CH2 |
| 9 | 23.6, CH2 | 20.3, CH2 | 27.3, CH2 | 30.0, CH2 | 26.4, CH2 |
| 10 | 37.0, CH2 | 43.5, CH2 | 32.1, CH2 | 34.8, CH2 | 40.8, CH2 |
| 11 | 151.1, C | 72.4, C | 124.4, C | 151.5, C | 81.1, C |
| 12 | 105.6, CH2 | 22.9, CH3 | 19.2, CH3 | 109.3, CH2 | 24.0, CH3 |
| 13 | 144.1, C | 143.8, C | 145.9, C | 143.2, C | 143.9, C |
| 14 | 15.4, CH3 | 15.5, CH3 | 15.9, CH3 | 16.1, CH3 | 14.7, CH3 |
| 15 | 122.4, CH | 122.5, CH | 120.9, CH | 122.8, CH | 123.0, CH |
| 16 | 123.4, CH | 123.3, CH | 67.9, CH | 123.4, CH | 123.2, CH |
| 17 | 139.3, CH | 139.3, CH | 67.6, CH | 139.5, CH | 139.5, CH |
| 18 | 71.1, C | 71.1, C | 60.0, C | 71.1, C | 71.1, C |
| 19 | 30.1, CH3 | 30.1, CH3 | 19.7, CH3 | 30.1, CH3 | 30.1, CH3 |
| 20 | 30.1, CH3 | 30.1, CH3 | 25.1, CH3 | 30.1, CH3 | 30.1, CH3 |
| Compd. | MIC (μg/mL) | ||||
|---|---|---|---|---|---|
| Streptococcus parauberis | Lactococcus garvieae | Streptococcus parauberis | Phoyobacterium damselae | Aeromonas salmonicida | |
| FP KSP28 | MP5245 | SP0F3K | FP2244 | AS42 | |
| 1 | 14.4 | 28.8 | 7.2 | 7.2 | NA |
| 2 | 7.2 | NA | 28.8 | 14.4 | NA |
| 3 | 7.2 | NA | NA | 14.4 | 28.8 |
| 4 | 7.2 | 0.225 | NA | 28.8 | NA |
| 5 | 7.2 | 28.8 | NA | 28.8 | NA |
| 6 | 17.3 | NA | 17.3 | 17.3 | NA |
| 10 | 15.1 | NA | NA | NA | NA |
| 11 | 0.9 | 14.4 | NA | 7.2 | 28.8 |
| 16 | 7.2 | 14.4 | 3.6 | 7.2 | NA |
| 17 | NA | NA | NA | NA | NA |
| 18 | 7.6 | NA | NA | 15.2 | NA |
| Tetr | 3.01 | 0.38 | >24.05 | 0.02 | 6.11 |
| Oxy | 1.55 | 0.19 | 12.42 | 0.02 | 0.39 |
| Lev | 1.24 | 0.62 | 1.24 | 0.02 | 0.31 |
| AMP | 4.64 | 0.58 | 0.58 | 0.02 | >18.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





