1. Introduction
Breast cancer (BC) is a widespread form of cancer with millions of new diagnoses and deaths each year [
1]. In 2020 alone, there were 2.3 million new breast cancer diagnoses and 685,000 deaths [
2]. Although mortality rates have declined due to the implementation of regular mammography screening, early detection and treatment remain important for reducing cancer fatalities [
3]. Currently, early detection of BC from radiology images requires the expertise of highly trained radiologists. A looming shortage of radiologists in several countries will likely worsen this problem [
4]. Mammography screening also leads to a high incidence of false positive results. This can result in unnecessary anxiety, inconvenient follow-up care, extra imaging tests, and sometimes a need for tissue sampling (often a needle biopsy) [
5,
6]. Additionally, machine learning techniques have the potential to improve the process of evaluating screening mammograms by radiologists [
7]. Deep learning as a subset of machine learning in recent years has revolutionized the interpretation of diagnostic imaging studies [
8]. A convolutional neural network (CNN) is one of the most significant networks in the deep learning field [
9]. Compared to traditional screening techniques, computer aided diagnosis (CAD) systems utilizing convolutional neural networks (CNN) offer faster, more reliable, and more robust screening. CNNs have emerged as a prominent method for pattern recognition in image analysis [
10]. CNN has been extensively used for breast cancer detection in different types of breast cancer images such as ultrasound (US), magnatic resonance imaging (MRI), and X-ray as follows:
US Images: Eroğlu Y [
11] proposed a hybrid-based CNN system based on ultrasonography images for diagnosing BC by extracting features from Alexnet, MobilenetV2, and Resnet50 then, after concatenating them, mRMR features selection method were used to select the best features. This system used machine learning algorithms support vector machine (SVM) and k-nearest neighbors (k-NN) as a classifier. As a result,
an accuracy rate of 95.6% was achieved. Reference [
12] An image segmentation method is applied to split the breast US images into sub-regions, followed by an object recognition method that employs feature extraction, selection, and classification techniques to automatically detect the sub-regions related to BC. In [
13] a method was suggested to segment BCs via semantic classification and patch merging. The approach involves cropping a region of interest, enhancing it using filters and clustering techniques, extracting features, and performing classification with a neural network and a k-NN classifier.
MRI Images: Zhou J et al. [
14] proposed a 3D deep CNN for the detection and localization of BC in dynamic contrast-enhanced MRI data using a weakly supervised approach and achieved 83.7% accuracy. In [
15], a multi-layer CNN was designed to classify MRI images as malignant or benign tumors using pixel information and online data augmentation. The network achieved high accuracy as 98.33%.
X-ray Images: Authors in [
16] used pre-trained CNN models, InceptionV3 and ResNet50, on the DDSM dataset to differentiate benign and malignant mammogram tumors. Transfer learning, pre-processing, and data augmentation techniques were used due to limited data. ResNet50 achieved 85.7%, and InceptionV3 achieved 79.6% accuracies. In [
17], authors used a CNN model that combines features from multiple views of mediolateral oblique (MLO) and craniocaudal (CC). Multi-scale features and a penalty term were used and achieved 82.02% accuracy on the DDSM dataset. Ridhi Hela et al. in [
18] proposed a methodology for BC detection using the CBIS-DDSM image dataset. Image pre-processing was done, followed by feature extraction using multiple CNN models (AlexNet, VGG16, ResNet, GoogLeNet, and InceptionResNet). The extracted features were evaluated using a neural network classifier, achieving an accuracy of 88%.
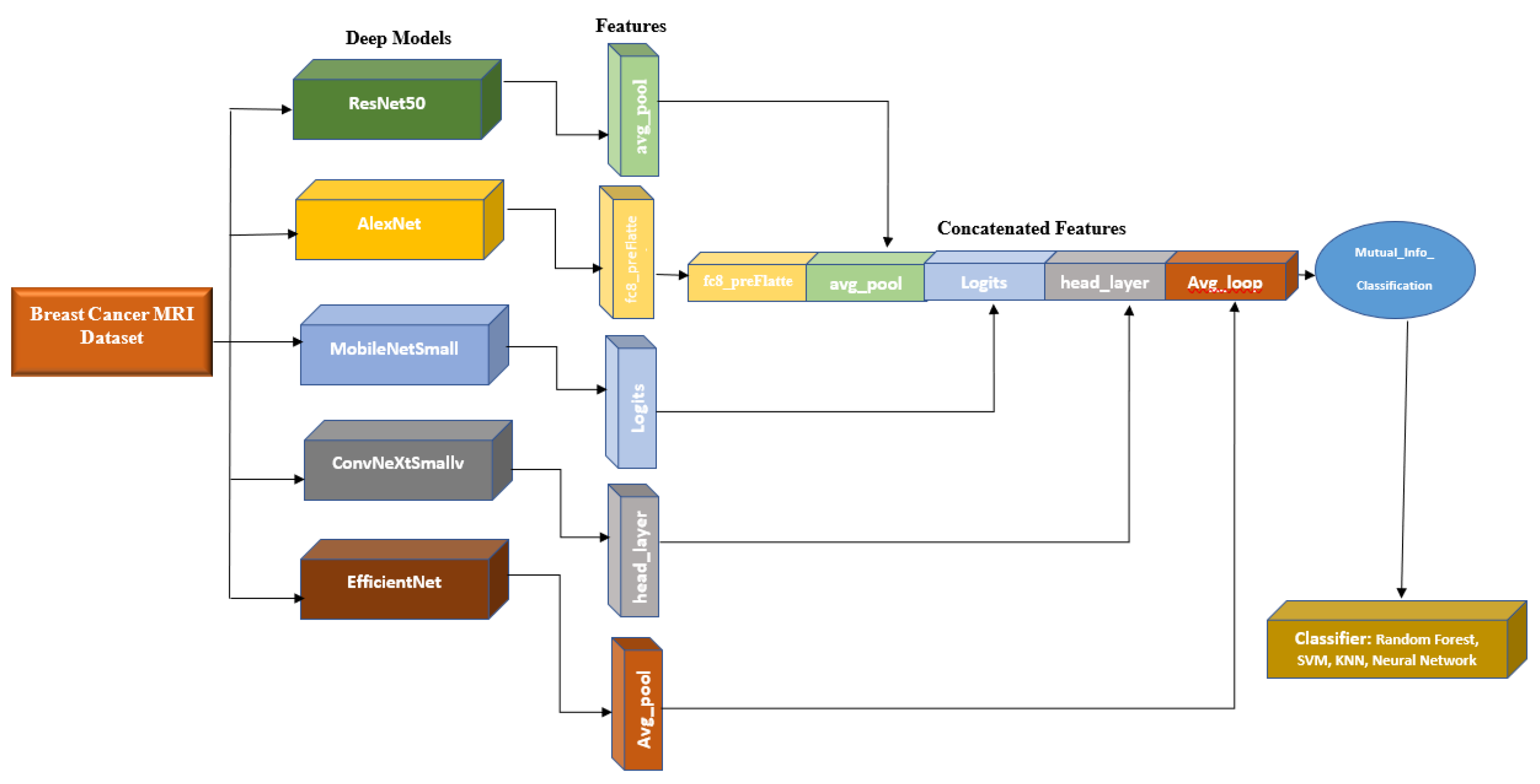
This paper provides two significant contributions to the existing literature. Firstly, it extracts a comprehensive set of features from diverse pre-trained convolutional neural networks (CNNs) for different perspectives. Additionally, it incorporates additional features like age to create a feature vector. Secondly, it employs a methodology to reduce feature vector dimensionality by eliminating weak features based on their mutual information with image labels.
The proposed system uses five base models, namely Alexnet, Resnet50, MobileNetSmall, ConvNeXtSmall, and EfficienNet, whose features are concatenated and extracted for optimal classification with a neural network (NN) model. This approach demonstrated its capability to enhance the accuracy of BC classification.
The rest of the paper is structured as follows:
Section 2 outlines the materials and models employed in the study, while
Section 3 presents the proposed model.
Section 4 discusses the results obtained for various datasets. The paper is concluded in
Section 5.
2. Materials and Methods
2.1 Datasets
A. The main dataset for this project is RSNA Screening Mammography BC from a recent Kaggle competition [
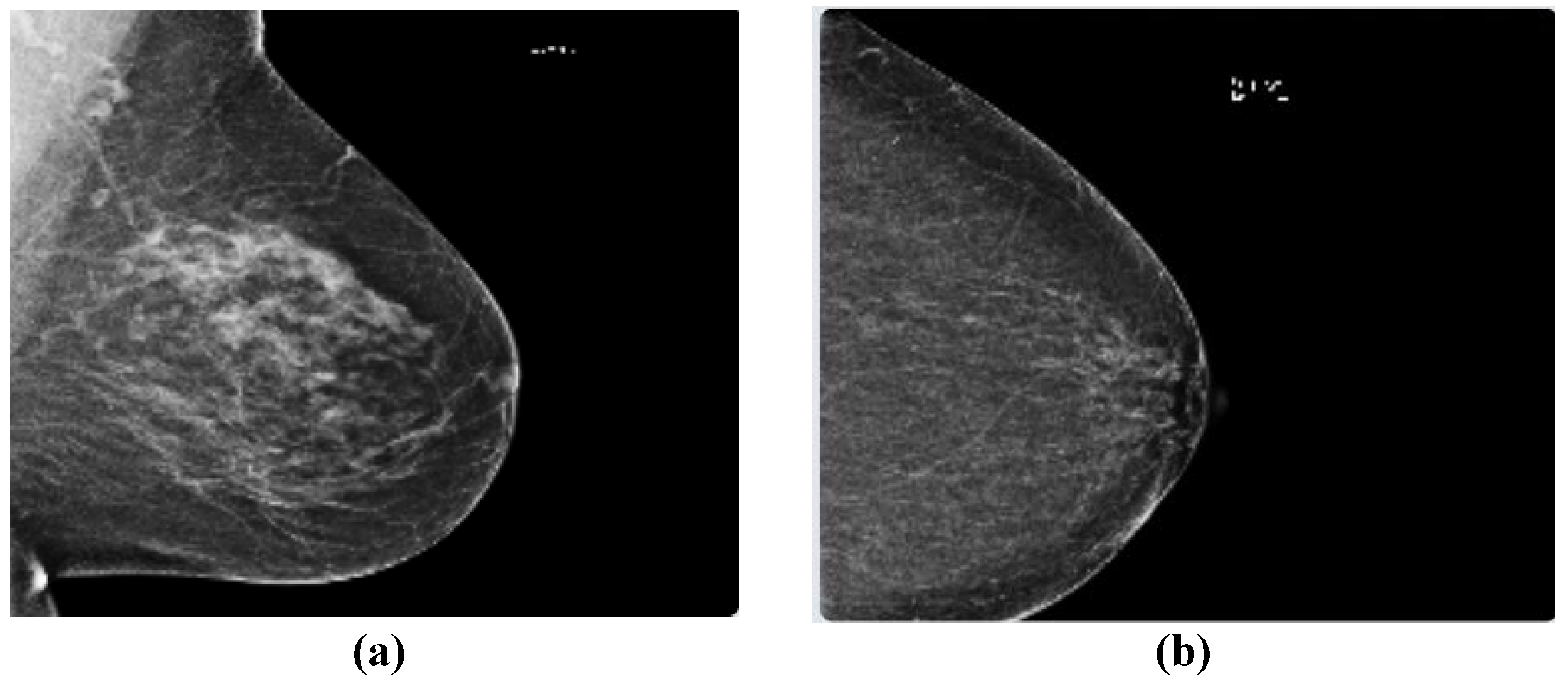
19]. The dataset contains 54713 images in dicom format, from roughly 11,000 patients. For each patient, there are at least four images from different laterality and views. For each subject, two different views CC and MLO, and images from left and right laterality were provided. The images are of various sizes and formats including jpeg and jpeg 2000 and different types such as monochrome-1 and monochrome-2. The dataset provides additional features which some of them can be used for classification purposes: age, implant, BIRADS, and density. We based our work on this dataset, but since this dataset is new, it has not been used in any published research yet. Hence, for comparison purposes, we used two other well-known datasets MIAS and DDSM. This dataset was imbalanced as only 2 percent of the images were from cancer patients, which makes any classification method biased. To compensate for this, we used all positive images and only 2320 images from negative cases.
Figure 1 depicts two sample images from this dataset for cancer and normal cases.
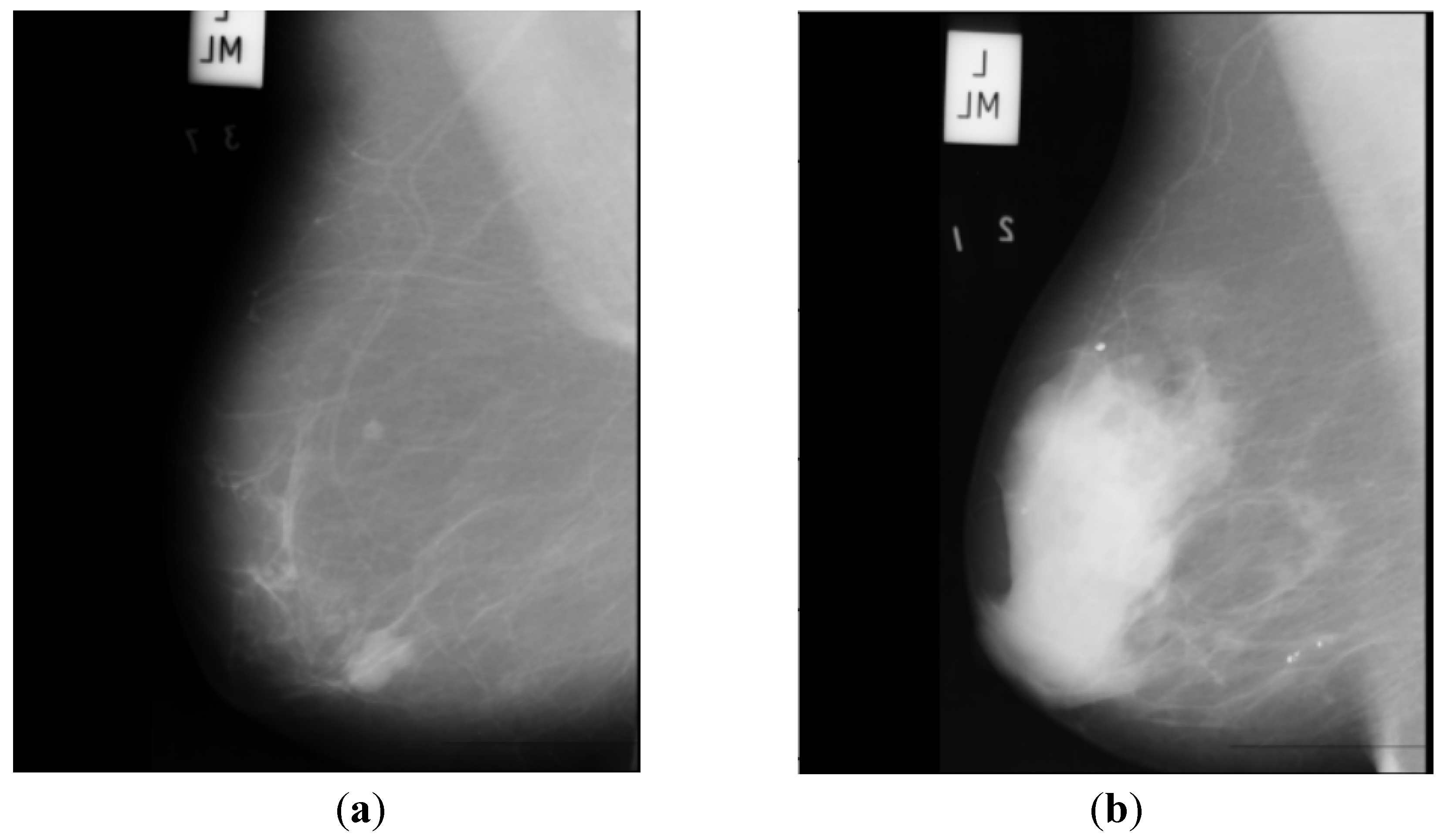
B. The Mammographic Image Analysis Society (MIAS) [
20] dataset is a well-known and widely used dataset for the development and evaluation of CAD systems for BC detection. It consists of 322 mammographic images, with each image accompanied by a corresponding ground truth classification of benign or malignant tumors. The dataset is particularly valuable for researchers interested in developing machine learning algorithms for BC detection, as it includes examples of both normal and abnormal mammograms, as well as a range of breast densities and lesion types.
Figure 2 depicts two sample images from this dataset for cancer and normal cases.
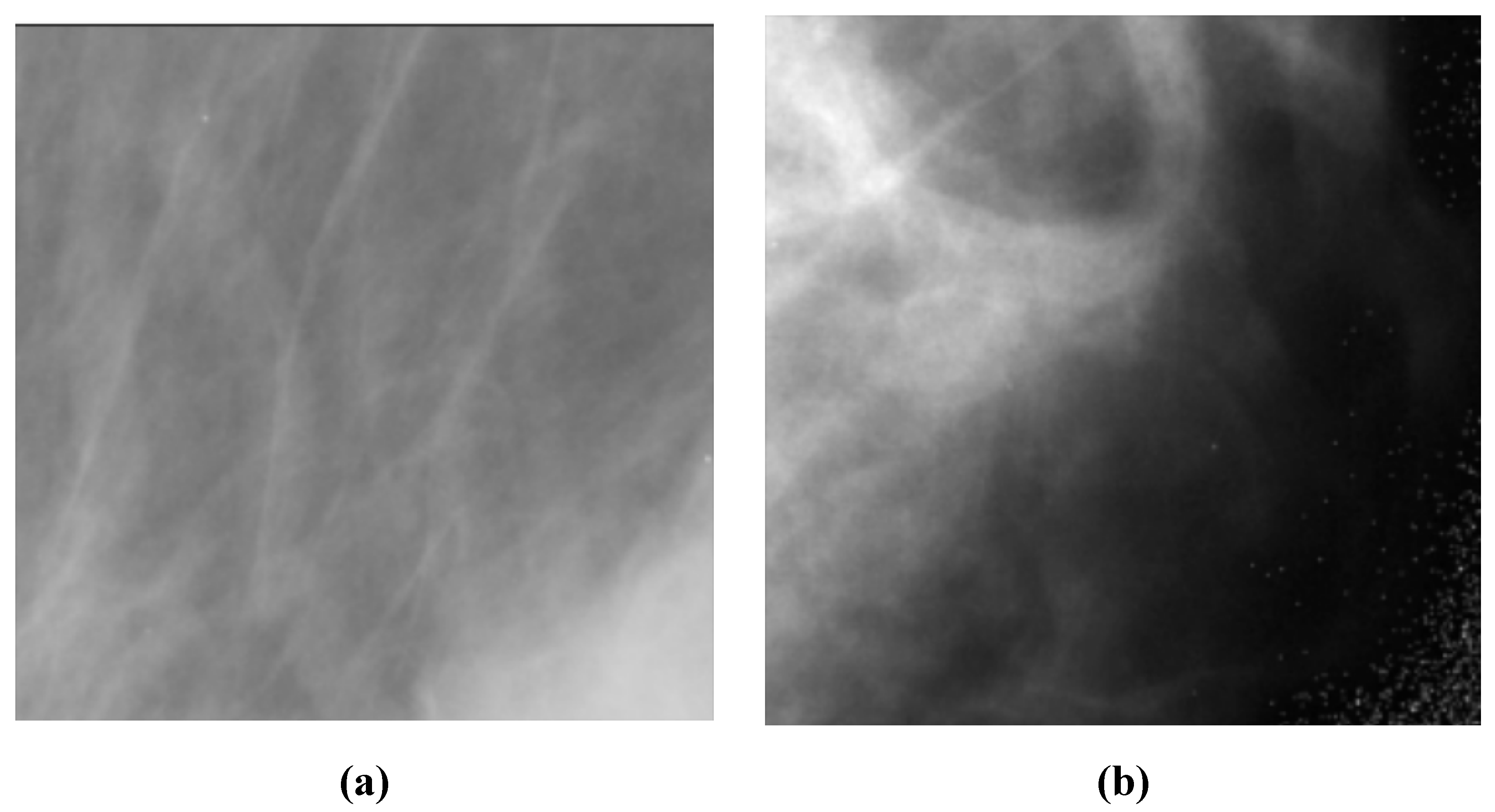
C. The Digital Database for Screening Mammography (DDSM) [
21] includes 55,890 images, of which 14% are positive, and the remaining 86% are negative. Images were tiled into 598x598 tiles, which were then resized to 299x299. A subset of this dataset which is for positive cases and is called CBIS-DDSM, has been annotated and the region of interest has been extracted by experts. In this research, we do not use the CBIS-DDSM and use the original DDSM dataset as we are classifying the images from normal subjects and cancer patients.
Figure 3 depicts two sample images from this dataset for cancer and normal cases.
2.2. Models
A. AlexNet [
22] is a deep CNN architecture that was introduced in 2012 and achieved a breakthrough in computer vision tasks such as image classification. It consists of eight layers, including five convolutional layers and three fully connected layers. The first convolutional layer uses a large receptive field to capture low-level features such as edges and textures, while subsequent layers use smaller receptive fields to capture increasingly complex and abstract features. AlexNet was the first deep network to successfully use Rectified Linear Unit (ReLU) activation functions, which have since become a standard activation function in deep learning. It also used dropout regularization to prevent overfitting during training. AlexNet’s success on the ImageNet dataset, which contains over one million images, demonstrated the potential of deep neural networks for image recognition tasks and paved the way for further advances in the field of computer vision.
B. ResNet50 [
23] is a deep CNN architecture that uses residual connections to enable learning from very deep architectures without suffering from the vanishing gradient problem. It consists of 50 layers, including convolutional layers, batch normalization layers, ReLU activation functions, and fully connected layers. ResNet50 also uses a skip connection that bypasses several layers in the network, allowing it to effectively learn both low-level and high-level features.
C. EfficientNet [
24] is a family of deep CNN architectures that were introduced in 2019 and have achieved state-of-the-art performance on a range of computer vision tasks. EfficientNet uses a compound scaling method to simultaneously optimize the depth, width, and resolution of the network, allowing it to achieve high accuracy while maintaining computational efficiency. EfficientNet consists of a backbone network that extracts features from input images and a head network that performs the final classification. The backbone network uses a combination of mobile inverted bottleneck convolutional layers and squeeze-and-excitation (SE) blocks to capture both spatial and channel-wise correlations in the input. The head network uses a combination of global average pooling and fully connected layers to perform the final classification.
D. MobileNet [
25] is a deep learning architecture suitable for efficient and accurate analysis of medical images, specifically in the context of BC diagnosis. With its emphasis on computational efficiency, MobileNet can effectively extract features from mammography images, enabling the detection of subtle patterns or abnormalities associated with breast cancer. By utilizing depthwise separable convolutions, MobileNet optimizes memory consumption and computational load, making it ideal for resource-constrained environments. The integration of the ReLU6 activation function further enhances efficiency and compatibility with medical imaging devices. Overall, MobileNet offers a valuable solution for BC analysis, providing accurate results while operating efficiently on limited computational resources.
E. ConvNeXt [
26] is an architecture that enhances the representational capacity of CNNs by leveraging parallel branches to capture diverse and complementary features, leading to improved performance on challenging visual recognition tasks. It has demonstrated excellent performance on various computer vision tasks, including image classification, object detection, and semantic segmentation. Its ability to capture complex relationships between features has made it a popular choice for tasks requiring a high-level understanding of visual data.