1. Introduction
Following the advice during the 15
th meeting of the International Health Regulations (IHR) Emergency Committee regarding the coronavirus 2019 disease (COVID-19) pandemic, the World Health Organization (WHO) Director-General has recently determined that COVID-19 is now an established and ongoing health issue which no longer constitutes a public health emergency of international concern [
1]. However, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is still circulating among the population causing hospitalization and deaths mainly of frail patients. Currently, the prevalent SARS-CoV-2 circulating strains belong to the Omicron variant sub-lineage, called XBB.1.5. This variant is now considered a variant of interest (VOI) with no evidence of higher severity of associated disease, but increased transmissibility [
2].
A screenshot of the situation in Italy shows more than 60,000 hospitalized patients and 458 deaths in the last 30 days [
3], highlighting that COVID-19 still engages health care in an incisive way.
Of particular relevance are the patients in old age and with a compromised immune system, of which those with hematologic malignancy show a high mortality rate. The greater susceptibility is given by the lack of response to vaccination and the consequent insufficient antibody response against SARS-CoV-2 [
4]. Even if the correlation of humoral immunity with protection against SARS-CoV-2 is still debated [
5], the lack of a fully working immune system in hematologic patients leads to increased risk for severe COVID-19, prolonged positivity and persistent disease [
6,
7].
The current therapies available for COVID-19 mainly involve the use of antivirals. These include nucleoside analogues such as remdesivir and monlupiravir and a viral protease inhibitor nirmatrelvir given in combination with ritonavir [
8]. However, from March 2023 molnupiravir is no longer available for prescription, as “Agenzia Italiana del Farmaco” (AIFA) stated[
9]. Similar drugs were initially used against HIV and for this reason, it was possible to develop new ones in a very short time thanks to the knowledge already acquired and the means available for scientific research. Precisely in the fight against HIV, it has been seen that combined therapies are much more effective in containing the infection and guaranteeing total inhibition of viral replication.
The aim of this study was to test, for the first time, the combination of remdesivir and nirmatrelvir on a cell-based SARS-CoV-2 infection model to find a possible synergic effect against viral replication. Furthermore, we described a case report of the real-life use of this combination on a hematologic patient attending “Santa Maria” hospital of Terni, Italy.
2. Materials and Methods
2.1. SARS-CoV-2 Strains, Vero E6 Cell Cultures and Compounds
All the SARS-CoV-2 strains used in the experiments were isolated in Biosafety Level 3 (BSL3) Virology laboratory at “Santa Maria della Misericordia Hospital”, Perugia, Italy, as previously described [
10]. Vero E6 cells were cultured in Eagle’s minimum essential medium (EMEM) supplemented with 10% foetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C with 5% CO
2 and used for viral replication. The culture supernatants were titered by Half-maximal Tissue Culture Infectious Dose (TCID50) endpoint dilution assay [
11], aliquoted and stored at −80°C.
The full-length SARS-CoV-2 viral genome was obtained using the COVIDSeq Assay (Illumina MiSeq Instrument, San Diego, CA, USA) at the Virology laboratory of the Department of Medical Biotechnologies, University of Siena, Siena, Italy as previously described [
12]. Whole-genome sequencing was submitted to GISAID (
http://gisaid.org/) for variant assignment. The strain used for the experiments was a SARS-CoV-2 20A.EU1 (lineage B.1) clustered with viruses circulating in Europe from spring to the end of 2020, Omicron sub lineage BA.1 isolated on September 5
th, 2022, from the patient of the case report and sub-lineage BA.5 isolated from a symptomatic patient on June 4
th, 2022. Viral stock aliquots were thawed immediately before each experiment and discarded after use.
Remdesivir (Veklury®, Gilead, Foster City, CA, USA) and Nirmatrelvir (PF-07321332, MedChemExpress, USA) were suspended in sterile water and DMSO, respectively, following the manufacturer’s instructions. Stock aliquots were stored at −80°C. Before each experiment, the compounds were diluted to the desired concentration with EMEM supplemented with 10% of FBS.
2.2. Determination of Remdesivir Effective Concentrations 50 and 90 (EC50 and EC90)
The effect of remdesivir against SARS-CoV-2 strains was assessed through yield reduction assay as previously described [
13,
14]. Briefly, Vero E6 cells were seeded in 96-well clear flat-bottom plates and incubated at 37°C with 5% CO
2 for 24 h. After incubation, cells were infected using SARS-CoV-2 (multiplicity of infection 0.1) and subsequently treated with media containing 3-fold serial dilutions of remdesivir (0.62-50 µM). Negative controls (remdesivir alone), infected positive controls (SARS-CoV-2 alone) and mock-infected cells were included in each plate. Plates were incubated at 37°C with 5% CO
2 for 48 h and 72 h and then, cell viability was measured using the MTT reduction assay [
13].
Effective concentrations (EC50 and EC90) were calculated based on the analysis of the viability of infected cells by fitting drug dose–response curves using the variable-slope regression modelling.
The experiment was repeated thrice with three technical replicates.
2.3. Drug Combination Test
Nirmatrelvir EC50 and EC90 values were obtained from our previous paper [
14]. The synergistic effect of remdesivir and nirmatrelvir combinations was determined as previously described [
15]. The combinations were tested by yield reduction assay using 5 two-fold serial dilutions of each compound starting from EC90 previously obtained. After 48 and 72 h post-treatment, the supernatant was removed and stocked at -80°C for further tests (plaque assay). Viability was determined using the MTT reduction assay as described above. Viability recovery was calculated as follows:
All tests were conducted in triplicate in three independent experiments.
To test whether the drug combinations act synergistically, the observed responses were compared with expected combination responses. The expected responses were calculated based on the Highest Single Agent (HSA) reference model using SynergyFinder version 2 [
15].
2.4. Plaque-Reduction Assay
To better understand the effect on viral replication and viral particles release of treated cells after infection, SARS-CoV-2 titers were determined on selected supernatants through plaque assay as previously described [
16]. Briefly, Vero E6 cells (3 × 10
5 cells/well) were previously seeded in a 12-well plate and incubated at 37°C with 5% CO
2 for 24 h. The medium was removed, and 250 µL/well of ten-fold serial dilution of supernatant was inoculated and incubated for 1 h. Plates were rocked every fifteen minutes with front-to-back and side-to-side movements. The inoculum was removed and 1 mL of overlay medium (complete medium with agar 0.1%) was poured into each well and the plates were incubated for 72 h. After incubation, the overlay was removed, and cells were fixed for 30 min with 4% formalin and stained with 0.5% crystal violet. Viral titer was determined as plaque-forming units per mL, considering wells with plaques ranging from 2 to 50. For each concentration of compound, viral titration was performed in triplicate.
2.5. Statistical Analysis and Data Elaboration
Statistical analysis was performed using GraphPad 8.3 (GraphPad Software, San Diego, CA, USA). Data were tested for normality using the Kolmogorov-Smirnov test and were presented as mean with the respective standard deviation (SD) or median with interquartile range (IQR), as appropriate. EC50 and EC90 concentrations were calculated using four-parameter variable-slope regression modelling. All the experiments were performed at least twice with three technical replicates.
2.6. Case Report and In Vitro Infectivity
Here we described a case of a patient admitted for COVID-19 to the Clinic of Infectious Diseases of the “Santa Maria” hospital of Terni, Terni (TR), Italy. The patient provided informed oral consent to clinical data collection. The data were collected from electronic medical records (jpalio Hospital Information System, jHIS™).
Respiratory samples (nasopharyngeal swabs) were tested for SARS-CoV-2 RNA by the Xpert® Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA, USA) as previously described [
17].
The virus isolation and titration were conducted in the biosafety level-3 Virology laboratory at “Santa Maria della Misericordia Hospital”, Perugia, Italy, as previously described [
10].
3. Results
The antiviral effect and EC90 of nirmatrelvir were acquired from our previous article and used for the subsequent experiments. Nirmatrelvir EC50 and EC90 after 48 h treatment were 1.28 µM and 3.70 µM respectively. After 72 h, nirmatrelvir EC50 and EC90 were 1.75 µM and 4.46 µM, respectively [
18]. Remdesivir antiviral activity was tested
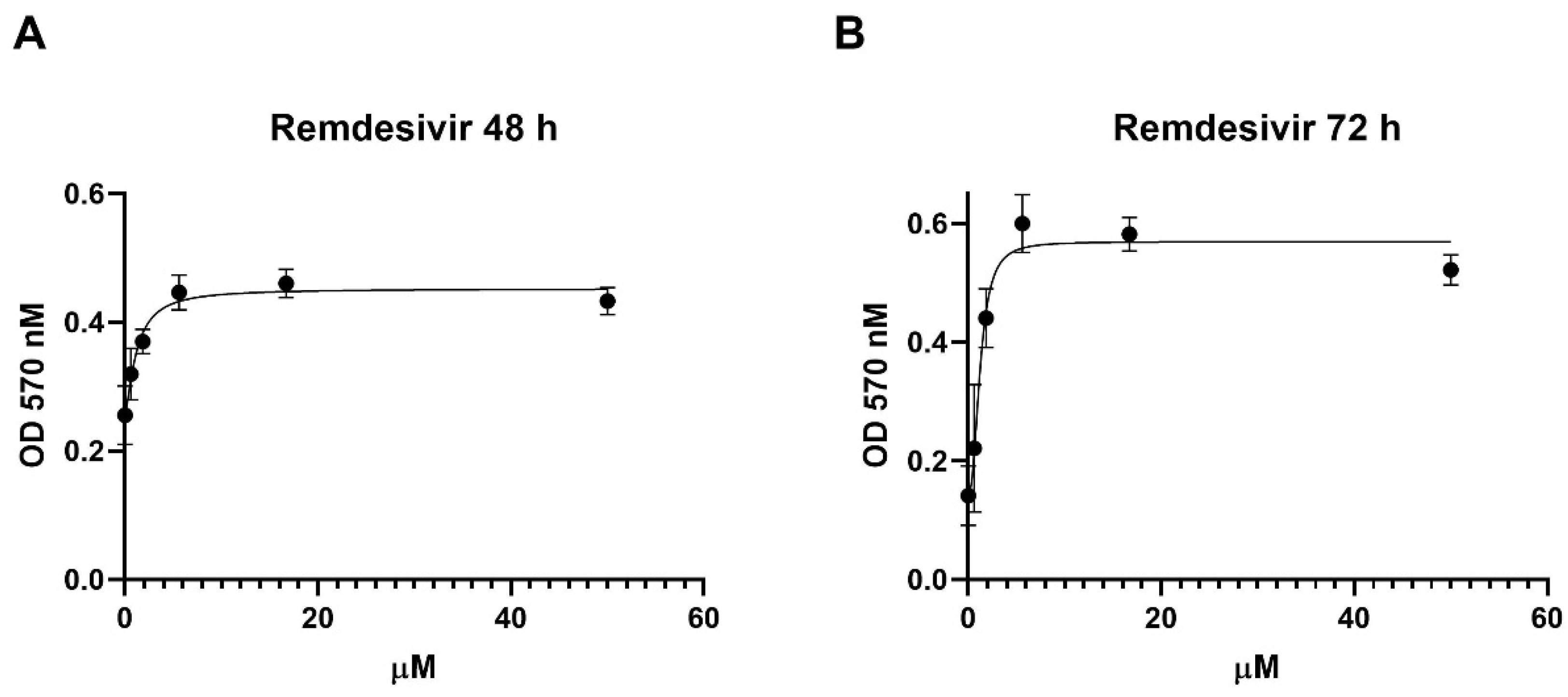
in vitro with a Vero E6 cell-based viability model. Four-parameter variable-slope regression modelling of remdesivir dose-response showed an EC50 of 1.2 µM (95% confidence interval, CI, 0.6-2.4) and an EC90 of 5.7 µM with a slope of 1.4 (95% CI 0.6-4.8) after 48 h of treatment (
Figure 1A). When the incubation was extended until 72 h, remdesivir EC50 and EC90 were 1.2 µM (95% CI 0.8-1.9) and 3.1 µM, respectively, with a slope of 2.4 (95% CI 1.3 to 3.8) (
Figure 1B).
The subsequent experiments with combination treatment were performed starting from the EC90s and proceeding with 2-fold serial dilutions. Vero E6 cells were previously infected with SARS-CoV-2 20A.EU1 strain, and then treated with the antiviral combination. After the incubations of 48 and 72 h viability was determined as above.
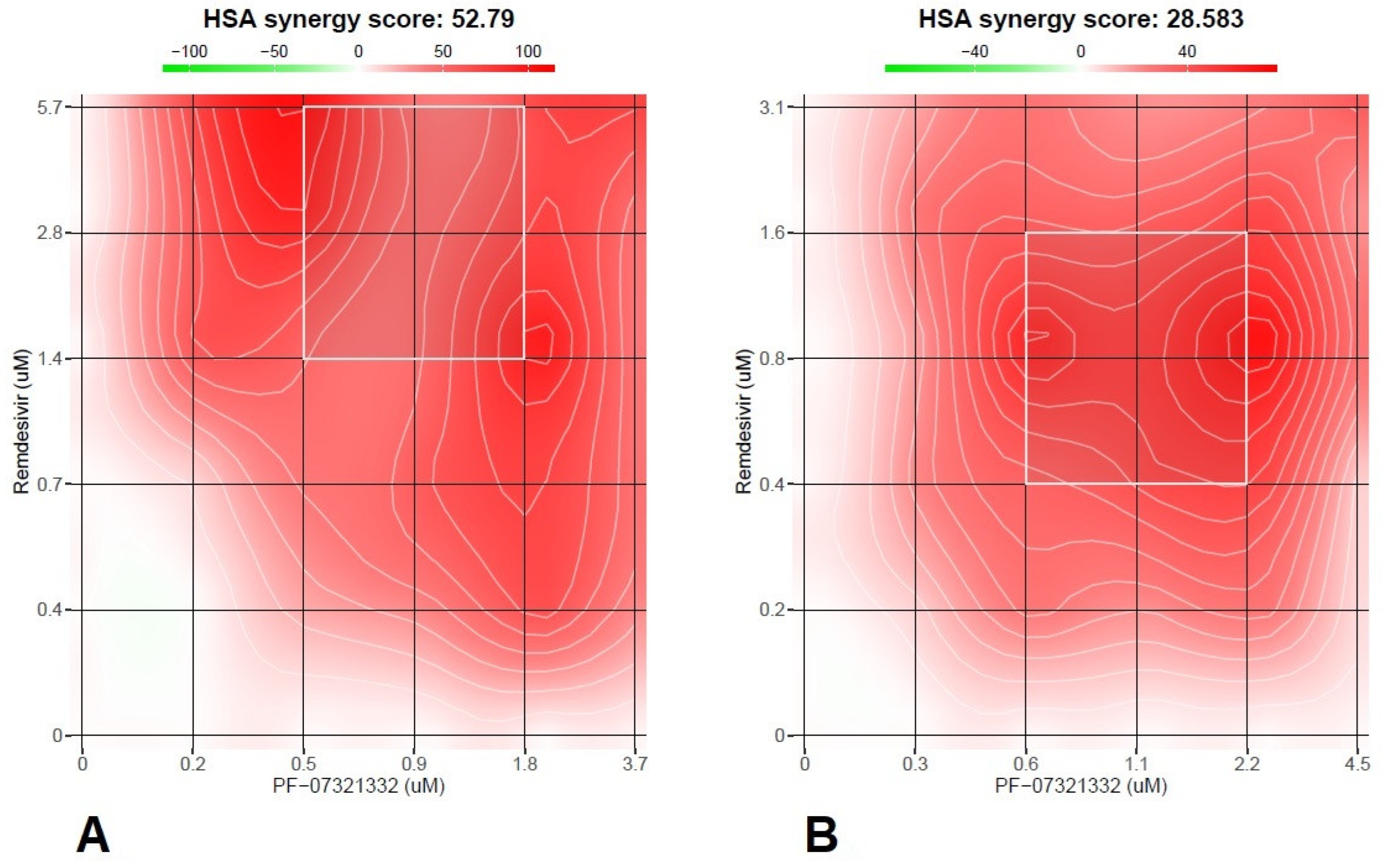
Remdesivir and nirmatrelvir showed a synergistic activity both at 48 h and 72 h with an HSA score of 52.8 and 28.6, respectively (p<0.0001,
Figure 2A,B).
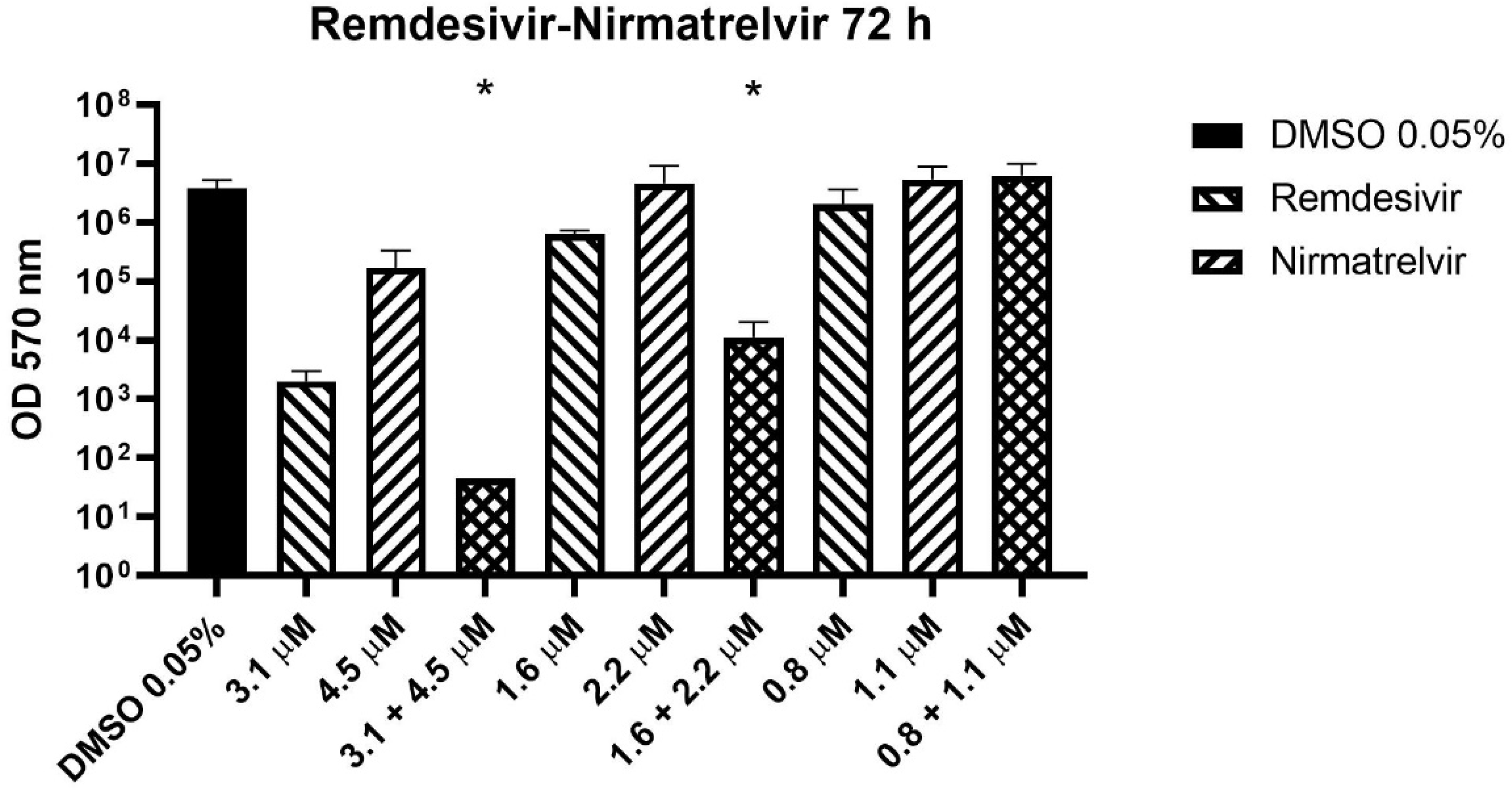
Before the MTT assay, the supernatants were collected and stored for subsequent determination of viral titer. Three concentrations from the 72 h experiment were selected to perform supernatant titration with plaque assay.
The combination of remdesivir and nirmatrelvir reduced the viral titer significantly better than remdesivir alone (the more active compound) at the two higher concentrations (
Figure 3, p = 0.0003). The compounds together caused a viral titer reduction by an extra 1.6-1.8 log compared to remdesivir alone.
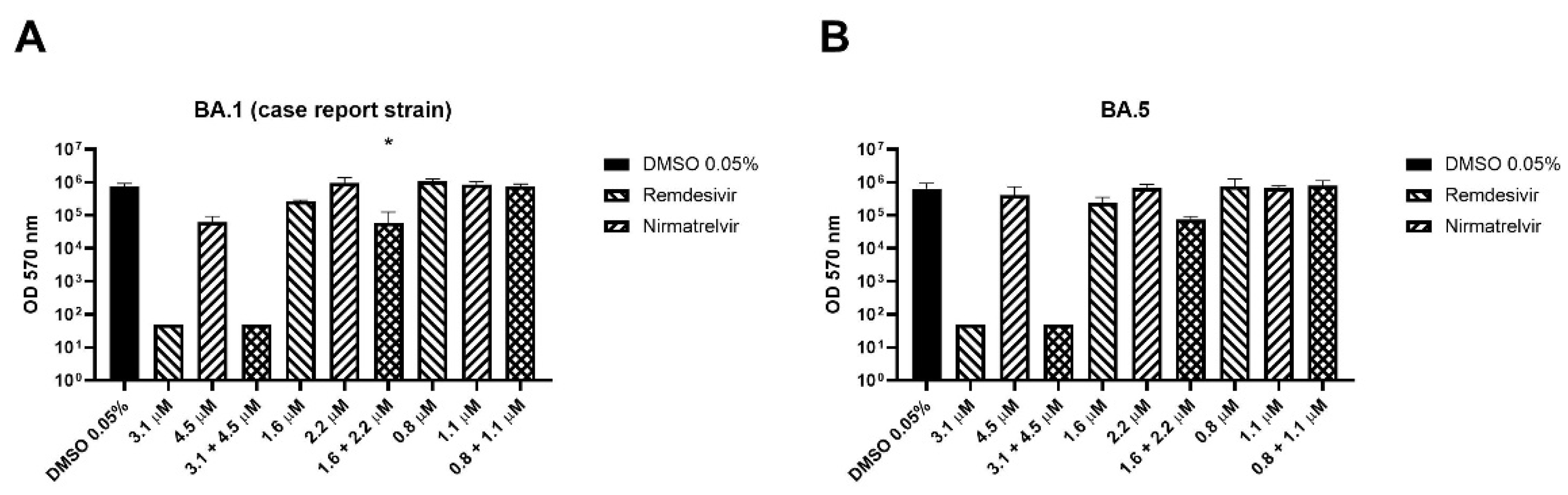
The combination has been also tested on BA.1 (isolated on September 5
th 2022 from the patient of the case report below) and BA.5 SARS-CoV-2 variants. As shown in
Figure 4A, remdesivir at the higher concentration reduced the viral titer below the detection limit, so it was not possible to establish if the combination is more effective. The remdesivir-nirmatrelvir combination at the respective concentrations of 1.6 and 2.2 μM reduced viral titer by 0.7 logs better than remdesivir alone (p = 0.048) while at the lower concentrations reduced the titer by 0.1 logs more than the remdesivir (p = 0.11).
The combination caused an extra viral titer reduction of 0.5 logs on the BA.5 strain compared to remdesivir alone, but the differences were not significant (p = 0.059,
Figure 4B). Similarly to the BA.1 strain, remdesivir at the higher concentration reduced the viral titer below the detection limit.
4. Case Report
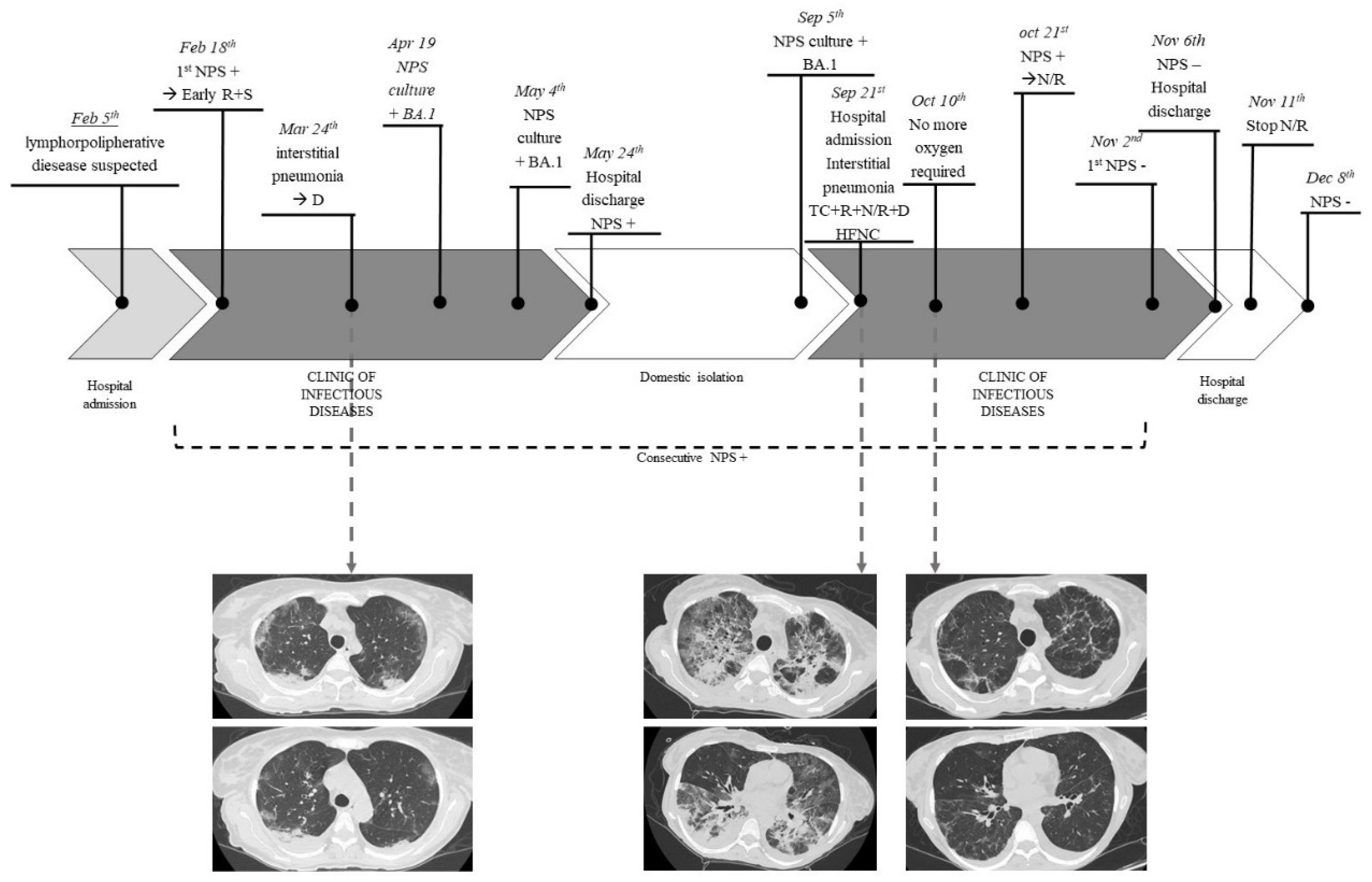
A 50-year-old woman was admitted to “Santa Maria” hospital of Terni for a suspected recurrence of lymphoproliferative disease on February 5th, 2022. She had a history of left adnexectomy for dermoid cyst at 19 years old, in 2015 left adrenalectomy for pheochromocytoma and simultaneous finding of non-Hodgkin (NH) follicular lymphoma. In 2018, she had a lymphoma recurrence. She was then treated with chemoimmunotherapy (rituximab as first-line and rituximab plus bendamustine at recurrence) and developed secondary liver damage as consequence of an autoimmune hepatitis. Therefore, she was treated with steroid therapy, leading to normal liver function values.
The patient previously received 3 doses of the Pfizer COVID-19 mRNA vaccine.
On February 18th, she underwent a nasopharyngeal swab that came positive (Xpert® Xpress SARS-CoV-2, Cepheid) and was transferred to the Clinic of Infectious Diseases of the same hospital. The patient had few COVID-19 symptoms (dry cough), so was treated with early antiviral therapy (remdesivir for three days course and sotrovimab).
She remained persistently asymptomatic until March 24th, when, following a progressive worsening of the respiratory picture, she performed chest CT angiography which was negative for pulmonary embolism and showed “the appearance of multiple and diffuse areas of parenchymal opacities with a ground glass appearance, with a tendency to consolidation in sloping areas.”
Therefore, she was treated with steroid therapy (dexamethasone 6 mg for 10 days) with the resolution of the clinical picture.
All exams performed to investigate the suspected lymphoproliferative disease recurrence were inconclusive.
However, during all this time, multiple nasopharyngeal swabs were collected and always resulted positive for SARS-CoV-2. Furthermore, on April 19th and May 4th, two nasopharyngeal swabs (resulted positive for SARS-CoV-2) were collected, and culture-based virus isolation in Vero E6 cells was performed to evaluate the potential infectivity. SARS-CoV-2 was isolated from these two specimens and both the viruses were sequenced as Omicron 1 variant (BA.1).
She was discharged on May 24, 2022, asymptomatic but still positive for SARS-CoV-2. During the following period, the patient remained in an isolation regimen at her home and performed multiple nasopharyngeal swabs, always resulting positive for SARS-CoV-2.
On September 5th, a new culture-based isolation test for SARS-CoV-2 from a swab specimen and also this resulted positive. This virus was sequenced as Omicron 1 (BA.1) variant, as in the previous specimens.
The patient was readmitted to the Clinic of Infectious Diseases to treat the infection with off-label antiviral therapy on 21st September 2022. The clinical conditions immediately appeared serious with acute progressive respiratory failure.
She performed a chest CT scan which showed “Bilateral extensive consolidative foci, “crazy paving” and “ground glass” opacities are evident in the lung parenchyma”. In addition, bronchoalveolar lavage was performed and it resulted positive for SARS-CoV-2, low-burden Haemophilus parainfluenzae, and Methicillin Sensitive Staphylococcus aureus (MSSA) 20000 CFU. It was also negative for Pneumocystis carinii and Cytomegalovirus. The specimen was also tested with BIOFIRE® FILMARRAY® Pneumonia Plus, which resulted positive for MSSA.
Due to severe respiratory insufficiency, she required high-flow nasal cannula (HFNC) oxygen support and had hemodynamic instability such as requiring inotropic support.
She was evaluated by the resuscitator specialist who judged the prognosis poor and defined the patient as not eligible for intensive care.
The following medical therapy was set as follows: targeted antibiotic therapy, off-label antiviral therapy with remdesivir (10 days course) plus nirmatrelvir/ritonavir (5 days course), tixagevimab/cilgavimab, and dexamethasone (6 mg/die for 10 days). No adverse effects were observed, except for mild bradycardia (50-60 beats per minute) on day 10 of therapy remdesivir, which regressed after the drug discontinuation. The patient presented an excellent clinical-radiological response with progressive reduction of oxygen requirement up to the suspension and improvement on control CT after 2 weeks. However, specifying that the patient was isolated in a single room without the risk of novel SARS-CoV-2 exposures, the nasopharyngeal swab was persistently positive for SARS-CoV-2, and she required further off-label prolonged therapy with nirmatrelvir/ritonavir to reach the negativization. She started this therapy on 21
st October until 11
th November, obtaining the first negative swab on 2
nd November 2022. No severe adverse events were reported, the patient complained of mild gastric intolerance but she completed the treatment. The patient was discharged on 6
th November, asymptomatic and without the need for oxygen therapy. She underwent a subsequent nasopharyngeal swab which resulted negative a month later (
Figure 5).
5. Discussion
After more than three years, World Health Organization (WHO) declared the end to COVID-19 as a health global emergency [
1]. This is a huge result and an important acknowledgement of the intense work of the scientific community that during the pandemic worked tirelessly to fight and contain the virus spread. Compared to the beginning of the pandemic, we have various weapons at our disposal to treat and prevent infection. Among these, vaccines and the three available antivirals (remdesivir, nirmatrelvir/ritonavir and molnupiravir) are certainly the key tools, while monoclonal antibodies and hyperimmune plasma are currently taking a back seat especially due to the emergencies of the variants of concern (VOCs) [
8].
However, SARS-CoV-2 is still spreading and killing, especially frail patients. Among frail patients, immunocompromised represented one of the most difficult challenges. It is known that both innate and adaptive immunity are involved in preventing, limiting and clearing SARS-CoV-2 infection [
7]. Firstly they showed poor response to the vaccine, with varying levels across different types of immunodepression [
19]. In particular, antibody responses to vaccines is lowest among solid organ transplant and anti-CD20 monoclonal recipients [
20]. They often showed prolonged SARS-CoV-2 infection with different levels of illness severity and consequent increased morbidity and mortality[
7,
21]. Furthermore, conventional therapeutic schemes are often not effective to treat COVID-19 in immunocompromised populations [
22,
23,
24]. However, the international guidelines do not provide specific therapeutic protocols for this kind of patient because there is insufficient evidence to guide clinical recommendations on using combinations of antiviral agents or on extending the duration of therapies[
8]. Indeed, poor data are available in the literature to support the need for specific protocols for immunocompromised patients.
The intuition that a combination of antivirals is more effective than a single drug is certainly not new. The efficacy of combination therapies has already been widely demonstrated for other viruses such as HIV and HCV, especially when the drugs act on different targets. Based on this, the scientific literature has begun to lay the foundations for carrying out similar treatments in fragile patients with difficult-to-treat COVID-19. There are few
in vitro and animal studies while there are no clinical trials but only retrospective clinical reports (case reports and a retrospective observational study) [
22,
23,
24,
25]. Our research group studied the combination of molnupiravir and nirmatrelvir, demonstrating the synergistic activity of this combination
in vitro [
18]. The biggest advantage of this combination is the possibility of administering as oral therapy, so without the need for hospitalization. Other studies have confirmed that this is a very promising combination also on Omicron variants both
in vitro and in animal models[
26,
27]. To the best of our knowledge, there is only a case report describing the outcome of a patient with persistent COVID-19 treated with this combination. The authors observed clinical, radiological, and microbiological success in an immunosuppressed patient [
28]. However, in Italy, molnupiravir is no longer available for prescription, so a different kind of drug needs to be tested in combination with nirmatrelvir[
9].
A wide number of drug combinations have been tested on SARS-CoV-2
in vitro. Remdesivir seems to be synergic with anti-HCV direct-acting antivirals (DAAS) in Vero E6
in vitro models, such as nelfinavir and amodaquine. SARS-CoV-2 polymerase is a good antiviral target and about 43 molecules showed synergistic activity with molnupiravir and remdesivir, especially if they have different targets. Indeed, molecules that act on the same target showed only additive effects [
29]. Other combinations with potential antiviral synergistic activity are nelfinavir with molnupiravir, nelfinavir with remdesivir in Calu-3
in vitro model or molnupiravir with favipiravir in hamster and Vero E6 models [
30,
31].
Poor data are available on in vitro and in vivo models about the combination of nirmatrelvir plus remdesivir, but paradoxically, compared to other combinations, it is the one on which we have the most clinical experience.
To the best of our knowledge, the only study that explored the synergistic activity of nirmatrelvir and remdesivir tested these compounds only on the feline coronavirus (FCoV) biotype but not on SARS-CoV-2. The authors found synergistic activity of this combination [
32].
The only
in vivo study available in the literature is inconclusive because remdesivir had been degraded in mice plasma by the enzyme carboxylesterase 1c (Ces1c) before it reaches tissues[
27].
To the best of our knowledge, this is the first in vitro study that tested the synergistic activity of nirmatrelvir and remdesivir. Both the compounds showed high activity on SARS-CoV-2, with micromolar effective concentrations. According to our data, the compounds showed a marked synergistic activity, independently from the time of incubation. Furthermore, the effect is also maintained on Omicron variants. Indeed, we tested BA.5 and BA.1 variants because the last one was isolated from the patient whose case report we have described. The combination was able to reduce the viral titer by an extra 1.6-1.8 logs more than the most effective single compound.
We hypothesize that the synergistic mechanism of this antiviral combination involves the inactivation of the exonuclease nsp14 that if active could contrast the activity of remdesivir. Nirmatrelvir inhibits the protease 3CL that is able to cleave 11 non-structural proteins including nsp14 inactivating it[
33]. Consequently, without the activity of the SARS-CoV-2 exonuclease, remdesivir is not inhibited. This hypothesis needs to be demonstrated.
Clinical data supporting the validity of this combination are in the early days. We have available only retrospective observational studies. The widest one is an Italian study published by Mikulska
et al. The authors enrolled 22 patients with relapsed/prolonged COVID-19 treated with nirmatrelvir/ritonavir, remdesivir and if available, anti-spike monoclonal antibodies (mAbs) [
25]. Of note, 68% of patients previously received anti-CD20 therapy, a condition they share with our patient and other cases described in the literature and that predisposes to an impaired immunological response[
19,
20,
22,
23,
24,
25]. Mikulska and colleagues observed that on the last follow-up day, 18/22 patients were negative and asymptomatic for SARS-CoV-2, 3 patients died, and 1 had improved symptoms but persistent antigen and PCR positivity. They also observed that the rate of virological response was higher in patients treated with a combination including mAbs and in patients with previous anti-CD20 treatment. Also, the number of vaccine doses influenced the rate of response [
25]. These data are in line with the other case reports described in the literature [
22,
23,
24] and also with the case here reported. Our patient has been treated with remdesivir, nirmatrelvir/ritonavir and tixagevimab/cilgavimab associated with antibiotic therapy and dexamethasone. This therapy led to impressive radiological and clinical improvement, but it was not enough to eradicate the infection after more than 7 months of positivity. Probably, a longer course of therapy could have been more effective to reach viral eradication. Indeed, she needed a prolonged course of nirmatrelvir/ritonavir to reach a stable negativity.
This study has a few limits such as the in vitro model that could overestimate the antiviral efficacy, the lack of other less common variants, and the use of non-human cell lines. Likewise, a single case report is not sufficient to validate a protocol therapy, but every experience could help physicians to manage the difficult-to-treat COVID-19.
Further studies are necessary to confirm the effectiveness of this combination, especially in vivo and hopefully in a clinical trial.
6. Conclusions
The remdesivir-nirmatrelvir combination showed good synergic activity in vitro. This combination may have a global impact on difficult-to-treat and severe COVID-19, especially in immunocompromised patients that often presented prolonged-relapsed COVID-19. Further studies are absolutely necessary to confirm these preliminary data.
Conflicts of Interest
The Vero E6 cell line was kindly provided by Istituto Zooprofilattico Sperimentale di Brescia, Brescia, Italy.
References
- World Health Organization WHO chief declares end to COVID-19 as a global health emergency. Available online: https://news.un.org/en/story/2023/05/1136367.
- Istituto Superiore di Sanità Prevalenza e distribuzione delle varianti di SARS-CoV-2 di interesse per la sanità pubblica in Italia. Available online: https://www.epicentro.iss.it/coronavirus/pdf/sars-cov-2-monitoraggio-varianti-rapporti-periodici-28-aprile-2023.pdf.
- Istituto Superiore di Sanità COVID-19 integrated surveillance data in Italy Available online:. Available online: https://www.epicentro.iss.it/en/coronavirus/sars-cov-2-dashboard (accessed on 24 May 2023).
- Langerbeins, P.; Hallek, M. COVID-19 in patients with hematologic malignancy. Blood 2022, 140. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Liu, B.; Wang, X.; Zhang, L. The humoral response and antibodies against SARS-CoV-2 infection. Nat. Immunol. 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Laracy, J.C.; Kamboj, M.; Vardhana, S.A. Long and persistent COVID-19 in patients with hematologic malignancies: From bench to bedside. Curr. Opin. Infect. Dis. 2022, 35. [Google Scholar] [CrossRef] [PubMed]
- DeWolf, S.; Laracy, J.C.; Perales, M.A.; Kamboj, M.; van den Brink, M.R.M.; Vardhana, S. SARS-CoV-2 in immunocompromised individuals. Immunity 2022, 55. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. National Institutes of Health. Available online: https://www.covid19treatmentguidelines.nih.gov/ (accessed on 23 May 2023).
- (AIFA), A.I.D.F. Sospensione di utilizzo del medicinale Lagevrio® (molnupiravir). Available online: https://www.aifa.gov.it/-/sospensione_utilizzo_lagevrio_molnupiravir.
- Gidari, A.; Sabbatini, S.; Bastianelli, S.; Pierucci, S.; Busti, C.; Bartolini, D.; Stabile, A.M.; Monari, C.; Galli, F.; Rende, M.; et al. SARS-CoV-2 Survival on Surfaces and the Effect of UV-C Light. Viruses 2021, 13, 408. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Gidari, A.; Sabbatini, S.; Bastianelli, S.; Pierucci, S.; Busti, C.; Monari, C.; Luciani Pasqua, B.; Dragoni, F.; Schiaroli, E.; Zazzi, M.; et al. Cross-neutralization of SARS-CoV-2 B.1.1.7 and P.1 variants in vaccinated, convalescent and P.1 infected. J. Infect. 2021, 83, 467–472. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Foo, C.S.; Jochmans, D.; Vangeel, L.; De Jonghe, S.; Augustijns, P.; Mols, R.; Weynand, B.; Wattanakul, T.; Hoglund, R.M.; et al. The oral protease inhibitor (PF-07321332) protects Syrian hamsters against infection with SARS-CoV-2 variants of concern. Nat. Commun. 2022, 13. [Google Scholar] [CrossRef]
- Gidari, A.; Sabbatini, S.; Schiaroli, E.; Bastianelli, S.; Pierucci, S.; Busti, C.; Comez, L.; Libera, V.; Macchiarulo, A.; Paciaroni, A.; et al. The Combination of Molnupiravir with Nirmatrelvir or GC376 Has a Synergic Role in the Inhibition of SARS-CoV-2 Replication In Vitro. Microorganisms 2022, 10, 1475. [Google Scholar] [CrossRef]
- Ianevski, A.; Giri, A.K.; Aittokallio, T. SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2021, 48. [Google Scholar] [CrossRef]
- Cox, R.M.; Wolf, J.D.; Plemper, R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021, 6, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Gidari, A.; Nofri, M.; Saccarelli, L.; Bastianelli, S.; Sabbatini, S.; Bozza, S.; Camilloni, B.; Fusco-Moffa, I.; Monari, C.; De Robertis, E.; et al. Is recurrence possible in coronavirus disease 2019 (COVID-19)? Case series and systematic review of literature. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gidari, A.; Sabbatini, S.; Schiaroli, E.; Bastianelli, S.; Pierucci, S.; Busti, C.; Comez, L.; Libera, V.; Macchiarulo, A.; Paciaroni, A.; et al. The Combination of Molnupiravir with Nirmatrelvir or GC376 Has a Synergic Role in the Inhibition of SARS-CoV-2 Replication In Vitro. Microorganisms 2022, 10. [Google Scholar] [CrossRef]
- Galmiche, S.; Luong Nguyen, L.B.; Tartour, E.; de Lamballerie, X.; Wittkop, L.; Loubet, P.; Launay, O. Immunological and clinical efficacy of COVID-19 vaccines in immunocompromised populations: A systematic review. Clin. Microbiol. Infect. 2022, 28. [Google Scholar] [CrossRef] [PubMed]
- Haidar, G.; Agha, M.; Bilderback, A.; Lukanski, A.; Linstrum, K.; Troyan, R.; Rothenberger, S.; McMahon, D.K.; Crandall, M.D.; Sobolewksi, M.D.; et al. Prospective Evaluation of Coronavirus Disease 2019 (COVID-19) Vaccine Responses Across a Broad Spectrum of Immunocompromising Conditions: The COVID-19 Vaccination in the Immunocompromised Study (COVICS). Clin. Infect. Dis. 2022, 75. [Google Scholar] [CrossRef] [PubMed]
- Malahe, S.R.K.; Hoek, R.A.S.; Dalm, V.A.S.H.; Broers, A.E.C.; den Hoed, C.M.; Manintveld, O.C.; Baan, C.C.; van Deuzen, C.M.; Papageorgiou, G.; Bax, H.I.; et al. Clinical Characteristics and Outcomes of Immunocompromised Patients With Coronavirus Disease 2019 Caused by the Omicron Variant: A Prospective, Observational Study. Clin. Infect. Dis. 2023, 76. [Google Scholar] [CrossRef] [PubMed]
- Baldi, F.; Dentone, C.; Mikulska, M.; Fenoglio, D.; Mirabella, M.; Magnè, F.; Portunato, F.; Altosole, T.; Sepulcri, C.; Giacobbe, D.R.; et al. Case report: Sotrovimab, remdesivir and nirmatrelvir/ritonavir combination as salvage treatment option in two immunocompromised patients hospitalized for COVID-19. Front. Med. 2023, 9. [Google Scholar] [CrossRef]
- Trottier, C.A.; Wong, B.; Kohli, R.; Boomsma, C.; Magro, F.; Kher, S.; Anderlind, C.; Golan, Y. Dual Antiviral Therapy for Persistent Coronavirus Disease 2019 and Associated Organizing Pneumonia in an Immunocompromised Host. Clin. Infect. Dis. 2023, 76. [Google Scholar] [CrossRef]
- Ford, E.S.; Simmons, W.; Karmarkar, E.N.; Yoke, L.H.; Braimah, A.B.; Orozco, J.J.; Ghiuzeli, C.M.; Barnhill, S.; Sack, C.L.; Benditt, J.O.; et al. Successful Treatment of Prolonged, Severe Coronavirus Disease 2019 Lower Respiratory Tract Disease in a B cell Acute Lymphoblastic Leukemia Patient With an Extended Course of Remdesivir and Nirmatrelvir/Ritonavir. Clin. Infect. Dis. 2023, 76. [Google Scholar] [CrossRef]
- Mikulska, M.; Sepulcri, C.; Denton, C.; Magne, F.; Balletto, E.; Baldi, F.B.; Labate, L.; Russo, C.; Mirabella, M.; Magnasco, L.; et al. Triple Combination Therapy With 2 Antivirals and Monoclonal Antibodies for Persistent or Relapsed Severe Acute Respiratory Syndrome Coronavirus 2 Infection in Immunocompromised Patients. Clin. Infect. Dis. 2023. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Lavrijsen, M.; Lamers, M.M.; de Vries, A.C.; Rottier, R.J.; Bruno, M.J.; Peppelenbosch, M.P.; Haagmans, B.L.; Pan, Q. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022, 32. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.H.; Chokkakula, S.; Min, S.C.; Kim, B.K.; Choi, W.S.; Oh, S.; Yun, Y.S.; Kang, D.H.; Lee, O.J.; Kim, E.G.; et al. Combination therapy with nirmatrelvir and molnupiravir improves the survival of SARS-CoV-2 infected mice. Antiviral Res. 2022, 208. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, D.; Antonello, R.M.; Coppi, Ma.; Palazzo, M.; Nassi, L.; Streva, N.; Povolo, L.; Malentacchi, F.; Zammarchi, L.; Rossolini, G.M.; et al. Combination regimen of nirmatrelvir ritonavir and molnupiravir for the treatment of persistent SARS-CoV-2 infection A case report and a scoping review of the literature. Int. J. Infect. Dis. 2023, May 5, 53–56. [Google Scholar] [CrossRef]
- White, J.M.; Schiffer, J.T.; Bender Ignacio, R.A.; Xu, S.; Kainov, D.; Ianevski, A.; Aittokallio, T.; Frieman, M.; Olinger, G.G.; Polyak, S.J. Drug Combinations as a First Line of Defense against Coronaviruses and Other Emerging Viruses. MBio 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Ianevski, A.; Yao, R.; Biza, S.; Zusinaite, E.; Männik, A.; Kivi, G.; Planken, A.; Kurg, K.; Tombak, E.M.; Ustav, M.; et al. Identification and tracking of antiviral drug combinations. Viruses 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Abdelnabi, R.; Foo, C.S.; Kaptein, S.J.F.; Zhang, X.; Do, T.N.D.; Langendries, L.; Vangeel, L.; Breuer, J.; Pang, J.; Williams, R.; et al. The combined treatment of Molnupiravir and Favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model. EBioMedicine 2021, 72. [Google Scholar] [CrossRef]
- Cook, S.; Wittenburg, L.; Yan, V.C.; Theil, J.H.; Castillo, D.; Reagan, K.L.; Williams, S.; Pham, C.D.; Li, C.; Muller, F.L.; et al. An Optimized Bioassay for Screening Combined Anticoronaviral Compounds for Efficacy against Feline Infectious Peritonitis Virus with Pharmacokinetic Analyses of GS-441524, Remdesivir, and Molnupiravir in Cats. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Wang, X.; Sacramento, C.Q.; Jockusch, S.; Chaves, O.A.; Tao, C.; Fintelman-Rodrigues, N.; Chien, M.; Temerozo, J.R.; Li, X.; Kumar, S.; et al. Combination of antiviral drugs inhibits SARS-CoV-2 polymerase and exonuclease and demonstrates COVID-19 therapeutic potential in viral cell culture. Commun. Biol. 2022, 5, 154. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).