Submitted:
01 June 2023
Posted:
02 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
- -
- First, viruses tend to have very few genes (fewer than 10 for most RNA viruses, for example, compared to over 20,000 in humans). Thus, a weak similarity between two viral proteins is much more meaningful than, say, a weak similarity between two human proteins;
- -
- Second, some proteins are found primarily in viruses, or even restricted only to a certain type of viruses. For example, movement proteins of the 30K superfamily are restricted to plant-infecting viruses (as well as to certain plants) [23]. Therefore, a protein which has only weak similarity to a 30K movement protein, but which comes from a plant-infecting virus, might reasonably be considered a candidate homolog that would have considerably diverged in sequence.
- -
- If HHpred detects a significant similarity, the two proteins are homologous;
- -
- If HHpred does not detect a significant similarity, either the two proteins are not homologous, or they are homologous but have diverged beyond recognition even by sequence profile methods. In such cases, only structure-based methods can confirm or infirm the homology (see below).
- -
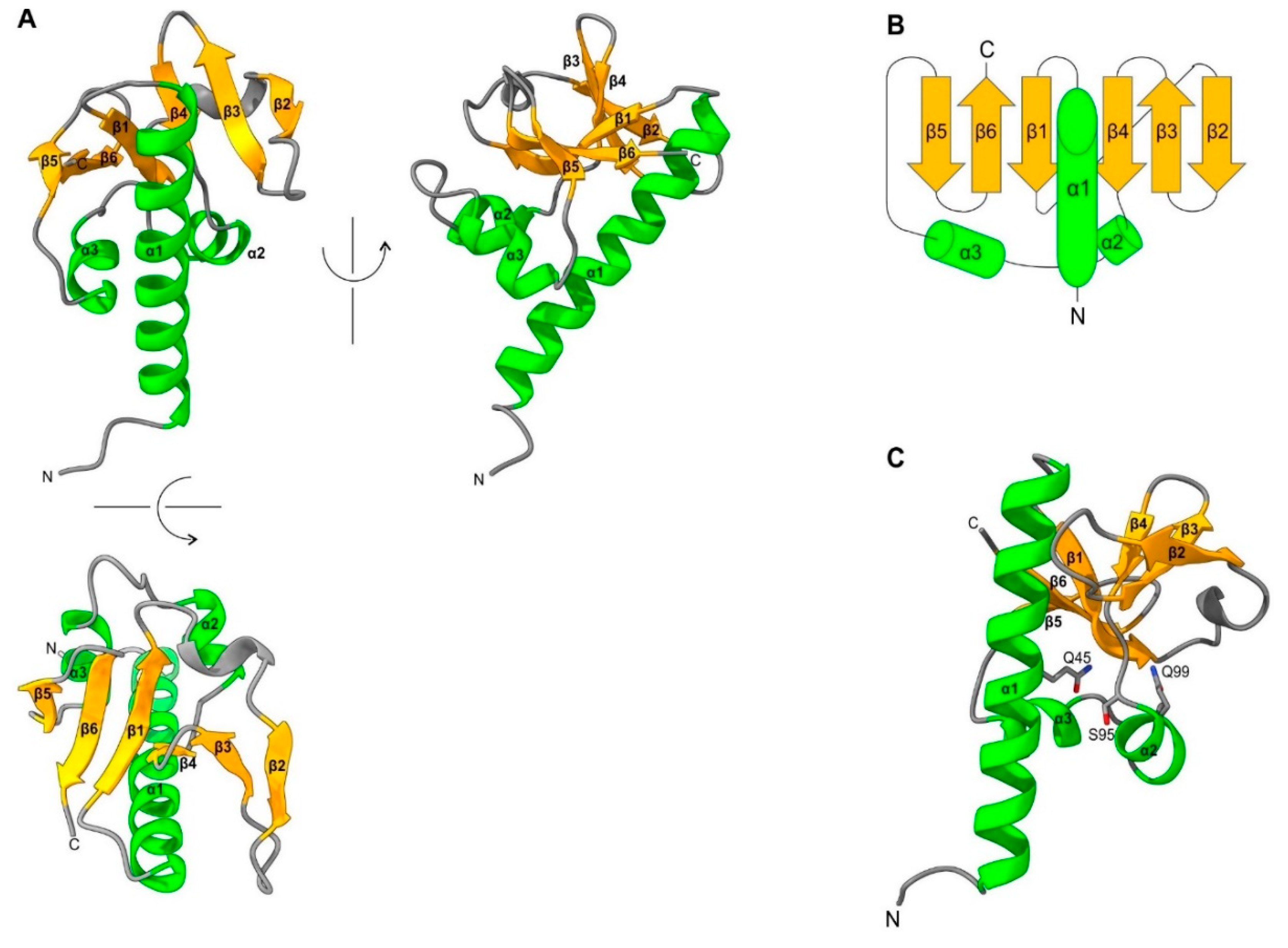
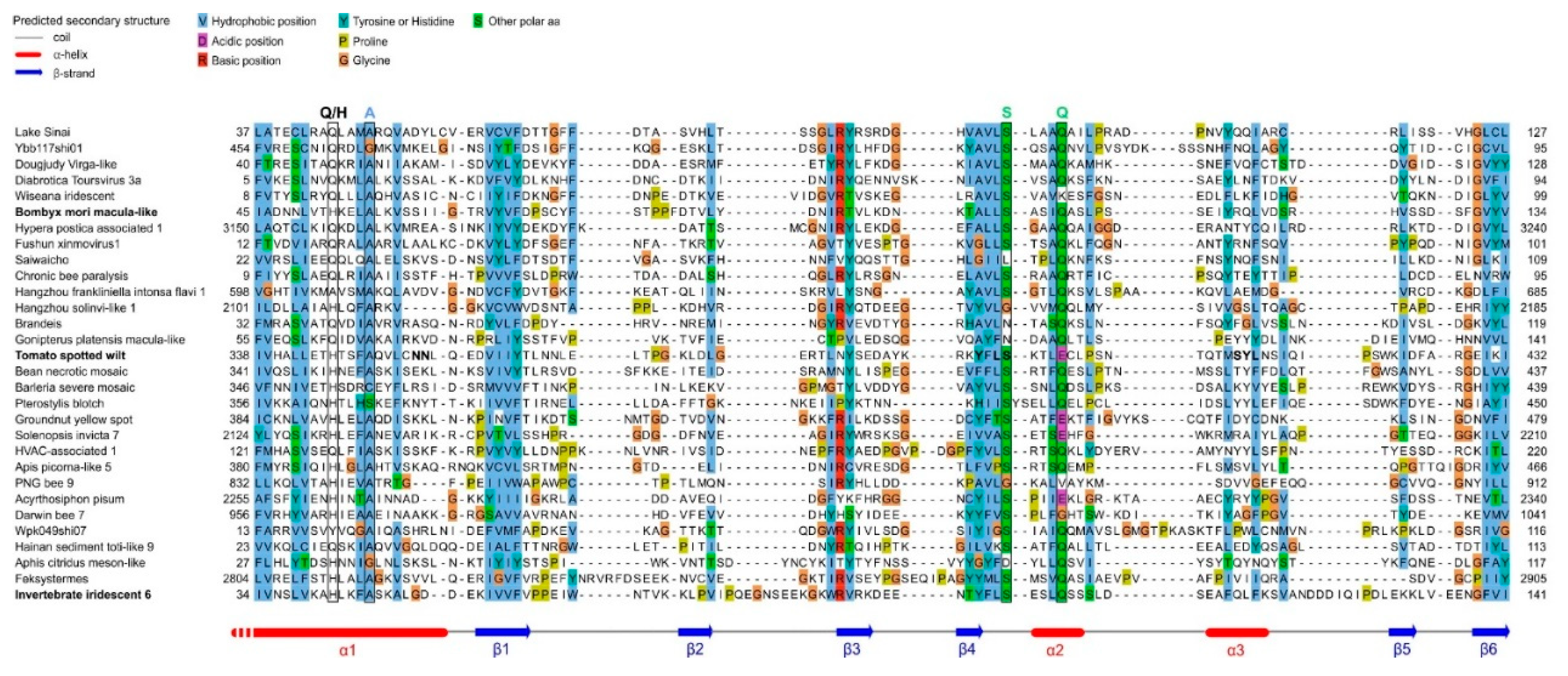
- As a standalone domain: WIV is found as a standalone domain in most positive-strand RNA viruses (Figure 2, panel A), except Picornavirales and an Amarillovirales; in a negative-strand RNA virus (Mononegavirales, panel B); and in some double-stranded DNA viruses (Pimascovirales, panel C). In some of these proteins, WIV is preceded by a signal peptide (e.g. in Lake Sinai virus ORF4).
- -
- Appended to a coiled-coil: In some species, WIV is appended to an N- or C-terminal coiled-coil, for example in Dougjudy virga-like virus RNA2 ORF1 (panel A), or in Wiseana iridescent virus gp049 (panel C);
- -
- Next to a double-stranded RNA-binding domain (dsRBD): WIV is wedged between a dsRBD domain and a capsid domain in some Picornavirales (panel A), and is located upstream of a dsRBD domain in a Ghabrivirales (panel D).
- -
- Downstream other types of domains: WIV is found at the very C-terminus of some proteins, such as Tospovirus NSs (panel B).
| Taxon | Virus species in the taxon |
|---|---|
| Brandeis virus group | Brandeis virus, Beult virus, Muthill virus, Bofa virus, Marsac virus, Buckhurst virus, Hubei virga-like virus 18, Broome virga-like virus, Hubei virga-like virus 19, Zeugodacus cucurbitae negev-like virus, Erysiphe necator associated virga-like virus 2 |
| Dougjudy virga-like virus group | Dougjudy virga-like virus, Hangzhou merodon fulcratus virga-like virus 1, Leuven Virga-like virus 1, Virga-like virus 21, Atrato virga-like virus 6, Atrato virga-like virus 7, Hammarskog virga-like virus, Erysiphe necator associated virga-like virus 1 |
| Family Acypiviridae (proposed in [32]) | Acyrthosiphon pisum virus, Darwin bee virus 7, Hangzhou solinvi-like virus 2, Grapevine-associated RNA virus 1, Hubei picorna-like virus 55, Hubei picorna-like virus 56, Aphis citridus picorna-like virus, Rosy apple aphid virus, Changjiang crawfish virus 6, Lasius picorna-like virus 7, Electric ant virus 1 |
|
Solenopsis invicta virus 7-like group, closely related to the proposed family Acypiviridae (see Figure S1h in [34]). |
Solenopsis invicta virus 7, Apis-Picorna-like virus 5, PNG bee virus 9, Hangzhou sesamia inferens solinvi-like virus 1, YCA-associated virus-like sequence 8 [34], HVAC-associated RNA virus 1, Apis picorna-like virus 3, Bundaberg bee virus 8, Milolii virus, Lasius picorna-like virus 9 |
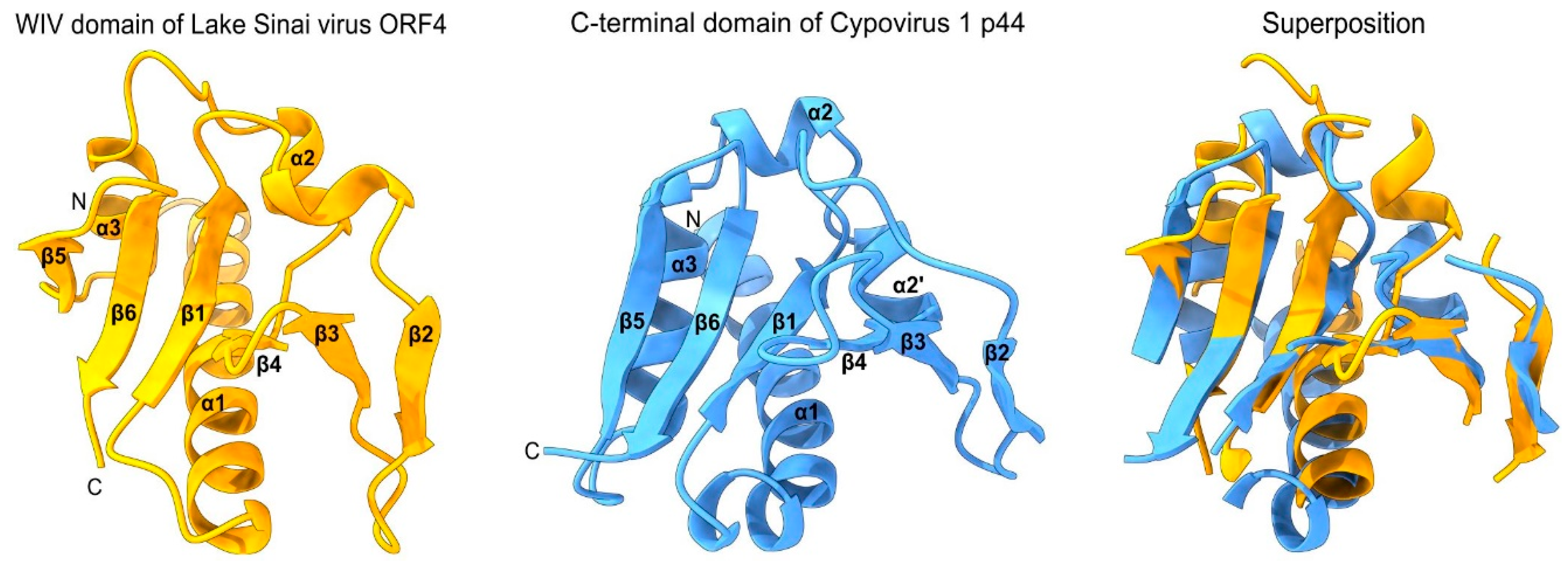
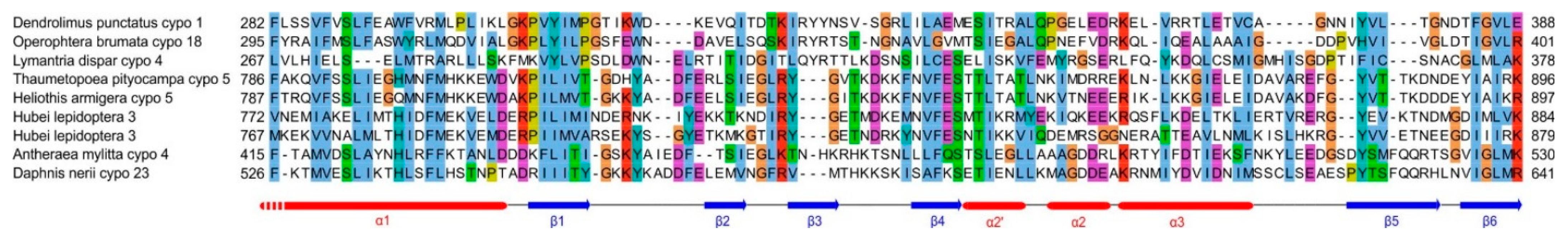
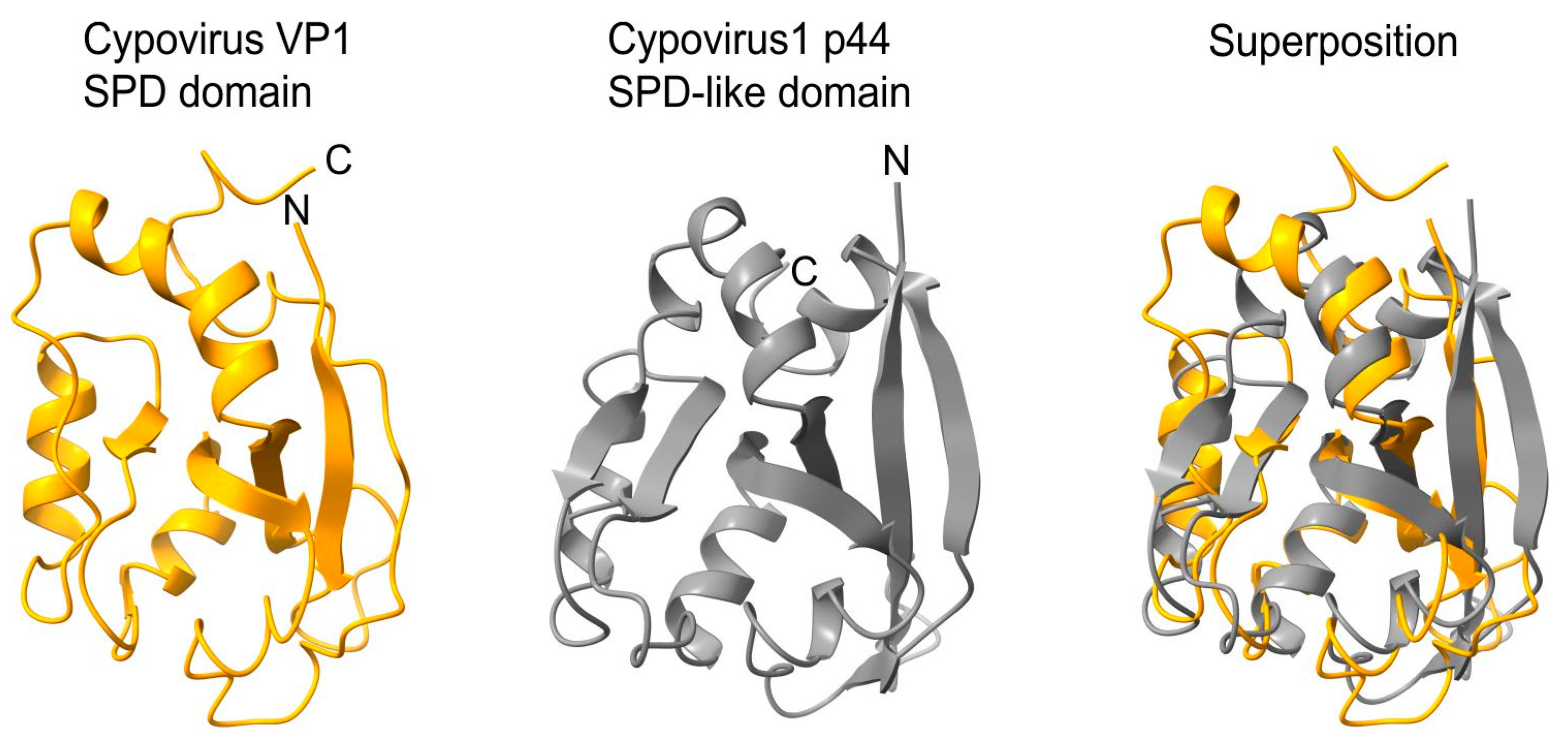
- -
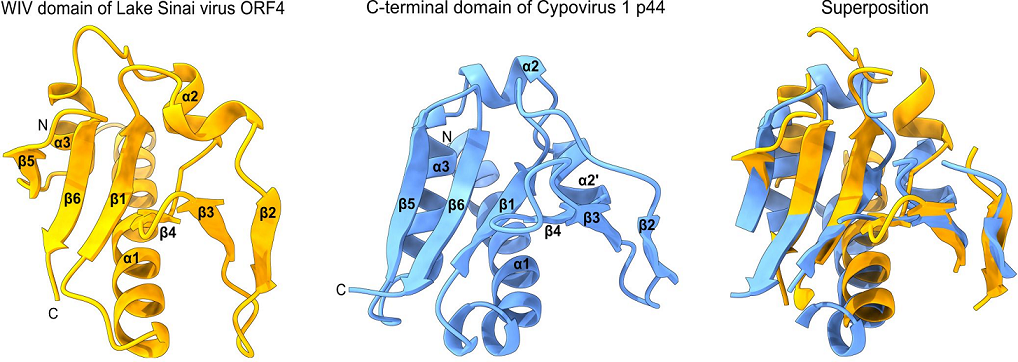
- an SPD-like domain in the p44 protein of cypovirus 1 and the related protein P8 of cypovirus 18;
- -
- an SPD-like domain followed by a Poly ADP-ribose glycohydrolase (PARG) domain in the P5 protein of cypovirus 5 and Hubei lepidoptera virus 3;
- -
- an unknown domain (s) in the P8 protein of cypovirus 4, 14 and 23.
3. Discussion
- -
- First, the reliability estimate (plDDT) provided by Alphafold2 has been proven to be accurate: a predicted structure with a plDDT≥0.90 is expected to be competitive with an experimentally determined structure [8]. The Alphafold2 structure for the WIV domain of Lake Sinai virus ORF4 has a plDDT of 0.95, and should therefore be close to the actual structure;
- -
- Second, the boundaries of the WIV domain frequently correspond exactly to an unassigned protein region between two known domains (or between a known domain and the extremity of a protein). For example, in the ORF1 of the virus wpk049shi07 [75] (Genbank accession QKE55054.1), related to Sinhaliviridae, the WIV domain, located between aa 1-113, is immediately followed by a 2A “StopGo” sequence (aa 127-139) (our observations; see Figure 2A, top). Such sequences (also called “Stop-Carryon”) mediate ribosome skipping during translation, which separates two proteins, akin to a cleavage, but without requiring a protease [76]. Their core motif is DxExNPGP, and the proteins are separated between the penultimate G and the final P (respectively G134 and P135 in the sequence of ORF1). Therefore, in this virus, the WIV domain should be found essentially as a standalone domain (with a short C-terminal extension, aa 114-134), providing strong biological support to our prediction.
4. Conclusion
5. Materials and Methods
4.1. Sequence Alignment and Sequence-BASED Homology Search
4.2. Prediction of Domain Organization
4.3. D structure Prediction
4.4. Structural Visualization and Alignment
Supplementary Materials
Acknowledgments
References
- Koonin EV, Krupovic M, Dolja VV. The global virome: How much diversity and how many independent origins? Environmental Microbiology. 2023;25: 40–44. [CrossRef]
- Zayed AA, Wainaina JM, Dominguez-Huerta G, Pelletier E, Guo J, Mohssen M, et al. Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science. 2022;376: 156–162. [CrossRef]
- Neri U, Wolf YI, Roux S, Camargo AP, Lee B, Kazlauskas D, et al. Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell. 2022;185: 4023-4037.e18. [CrossRef]
- Kuchibhatla DB, Sherman WA, Chung BYW, Cook S, Schneider G, Eisenhaber B, et al. Powerful sequence similarity search methods and in-depth manual analyses can identify remote homologs in many apparently “orphan” viral proteins. J Virol. 2014;88: 10–20. [CrossRef]
- Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2012;9: 173–175. [CrossRef]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50: D439–D444. [CrossRef]
- Marx V. Method of the Year: protein structure prediction. Nat Methods. 2022;19: 5–10. [CrossRef]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596: 583–589. [CrossRef]
- van Kempen M, Kim SS, Tumescheit C, Mirdita M, Söding J, Steinegger M. Foldseek: fast and accurate protein structure search. Bioinformatics; 2022 Feb. [CrossRef]
- Hildebrand A, Remmert M, Biegert A, Söding J. Fast and accurate automatic structure prediction with HHpred: Structure Prediction with HHpred. Proteins. 2009;77: 128–132. [CrossRef]
- Daughenbaugh K, Martin M, Brutscher L, Cavigli I, Garcia E, Lavin M, et al. Honey Bee Infecting Lake Sinai Viruses. Viruses. 2015;7: 3285–3309. [CrossRef]
- Runckel C, Flenniken ML, Engel JC, Ruby JG, Ganem D, Andino R, et al. Temporal Analysis of the Honey Bee Microbiome Reveals Four Novel Viruses and Seasonal Prevalence of Known Viruses, Nosema, and Crithidia. Moritz RFA, editor. PLoS ONE. 2011;6: e20656. [CrossRef]
- Parmentier L, Smagghe G, de Graaf DC, Meeus I. Varroa destructor Macula-like virus, Lake Sinai virus and other new RNA viruses in wild bumblebee hosts ( Bombus pascuorum , Bombus lapidarius and Bombus pratorum ). Journal of Invertebrate Pathology. 2016;134: 6–11. [CrossRef]
- Bigot D, Dalmon A, Roy B, Hou C, Germain M, Romary M, et al. The discovery of Halictivirus resolves the Sinaivirus phylogeny. Journal of General Virology. 2017;98: 2864–2875. [CrossRef]
- McMenamin AJ, Flenniken ML. Recently identified bee viruses and their impact on bee pollinators. Current Opinion in Insect Science. 2018;26: 120–129. [CrossRef]
- Ahola T, Karlin DG. Sequence analysis reveals a conserved extension in the capping enzyme of the alphavirus supergroup, and a homologous domain in nodaviruses. Biol Direct. 2015;10: 16. [CrossRef]
- Rehanek M, Karlin DG, Bandte M, Al Kubrusli R, Nourinejhad Zarghani S, Candresse T, et al. The Complex World of Emaraviruses—Challenges, Insights, and Prospects. Forests. 2022;13: 1868. [CrossRef]
- Hu G, Kurgan L. Sequence Similarity Searching. Current Protocols in Protein Science. 2019;95: e71. [CrossRef]
- Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430: 2237–2243. [CrossRef]
- Coin L, Bateman A, Durbin R. Enhanced protein domain discovery using taxonomy. BMC Bioinformatics. 2004;5: 56. [CrossRef]
- Aravind L. Guilt by Association: Contextual Information in Genome Analysis: Figure 1. Genome Res. 2000;10: 1074–1077. [CrossRef]
- Boekhorst J, Snel B. Identification of homologs in insignificant blast hits by exploiting extrinsic gene properties. BMC Bioinformatics. 2007;8: 356. [CrossRef]
- Mushegian AR, Elena SF. Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology. 2015;476: 304–315. [CrossRef]
- Dunbrack RL. Sequence comparison and protein structure prediction. Current Opinion in Structural Biology. 2006;16: 374–384. [CrossRef]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25: 3389–3402. [CrossRef]
- Paraskevopoulou S, Käfer S, Zirkel F, Donath A, Petersen M, Liu S, et al. Viromics of extant insect orders unveil the evolution of the flavi-like superfamily. Virus Evol. 2021;7: veab030. [CrossRef]
- Liu S, Sappington TW, Coates BS, Bonning BC. Sequences Encoding a Novel Toursvirus Identified from Southern and Northern Corn Rootworms (Coleoptera: Chrysomelidae). Viruses. 2022;14: 397. [CrossRef]
- Chen Y-M, Sadiq S, Tian J-H, Chen X, Lin X-D, Shen J-J, et al. RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat Microbiol. 2022;7: 1312–1323. [CrossRef]
- Medd NC, Fellous S, Waldron FM, Xuéreb A, Nakai M, Cross JV, et al. The virome of Drosophila suzukii, an invasive pest of soft fruit. Virus Evol. 2018;4: vey009. [CrossRef]
- Kondo H, Chiba S, Maruyama K, Andika IB, Suzuki N. A novel insect-infecting virga/nege-like virus group and its pervasive endogenization into insect genomes. Virus Research. 2019;262: 37–47. [CrossRef]
- Mahar JE, Shi M, Hall RN, Strive T, Holmes EC. Comparative Analysis of RNA Virome Composition in Rabbits and Associated Ectoparasites. Pfeiffer JK, editor. J Virol. 2020;94: e02119-19. [CrossRef]
- Valles SM, Oi DH, Becnel JJ, Wetterer JK, LaPolla JS, Firth AE. Isolation and characterization of Nylanderia fulva virus 1, a positive-sense, single-stranded RNA virus infecting the tawny crazy ant, Nylanderia fulva. Virology. 2016;496: 244–254. [CrossRef]
- Valles SM, Rivers AR. Nine new RNA viruses associated with the fire ant Solenopsis invicta from its native range. Virus Genes. 2019;55: 368–380. [CrossRef]
- Lee C-C, Hsu H-W, Lin C-Y, Gustafson N, Matsuura K, Lee C-Y, et al. First Polycipivirus and Unmapped RNA Virus Diversity in the Yellow Crazy Ant, Anoplolepis gracilipes. Viruses. 2022;14: 2161. [CrossRef]
- Webster CL, Longdon B, Lewis SH, Obbard DJ. Twenty-Five New Viruses Associated with the Drosophilidae (Diptera). Evol Bioinform Online. 2016;12s2: EBO.S39454. [CrossRef]
- Hernández-Pelegrín L, Llopis-Giménez Á, Crava CM, Ortego F, Hernández-Crespo P, Ros VID, et al. Expanding the Medfly Virome: Viral Diversity, Prevalence, and sRNA Profiling in Mass-Reared and Field-Derived Medflies. Viruses. 2022;14: 623. [CrossRef]
- He Y-J, Ye Z-X, Zhang C-X, Li J-M, Chen J-P, Lu G. An RNA Virome Analysis of the Pink-Winged Grasshopper Atractomorpha sinensis. Insects. 2022;14: 9. [CrossRef]
- Charles J, Tangudu CS, Hurt SL, Tumescheit C, Firth AE, Garcia-Rejon JE, et al. Discovery of a novel Tymoviridae-like virus in mosquitoes from Mexico. Arch Virol. 2019;164: 649–652. [CrossRef]
- Pei J, Tang M, Grishin NV. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Research. 2008;36: W30–W34. [CrossRef]
- Krell PJ. Reoviruses of Invertebrates (Reoviridae). Encyclopedia of Virology. Elsevier; 2021. pp. 867–882. [CrossRef]
- Dong R, Pan S, Peng Z, Zhang Y, Yang J. mTM-align: a server for fast protein structure database search and multiple protein structure alignment. Nucleic Acids Research. 2018 [cited 13 May 2022]. [CrossRef]
- Takatsuka J. A new cypovirus from the Japanese peppered moth, Biston robustus. Journal of Invertebrate Pathology. 2020;174: 107417. [CrossRef]
- Xu C, Wang J, Yang J, Lei C, Hu J, Sun X. NSP2 forms viroplasms during Dendrolimus punctatus cypovirus infection. Virology. 2019;533: 68–76. [CrossRef]
- Chavali VRM, Ghosh AK. Molecular cloning, sequence analysis and expression of genome segment 7 (S7) of Antheraea mylitta cypovirus (AmCPV) that encodes a viral structural protein. Virus Genes. 2007;35: 433–441. [CrossRef]
- Yu X, Ge P, Jiang J, Atanasov I, Zhou ZH. Atomic Model of CPV Reveals the Mechanism Used by This Single-Shelled Virus to Economically Carry Out Functions Conserved in Multishelled Reoviruses. Structure. 2011;19: 652–661. [CrossRef]
- Ren F, Swevers L, Lu Q, Zhao Y, Yan J, Li H, et al. Effect of mutations in capsid shell protein on the assembly of BmCPV virus-like particles. Journal of General Virology. 2021;102. [CrossRef]
- Zhu F, Li D, Song D, Huo S, Ma S, Lü P, et al. Glycoproteome in silkworm Bombyx mori and alteration by BmCPV infection. Journal of Proteomics. 2020;222: 103802. [CrossRef]
- Katsuma S, Tanaka S, Omuro N, Takabuchi L, Daimon T, Imanishi S, et al. Novel macula-like virus identified in Bombyx mori cultured cells. J Virol. 2005;79: 5577–5584. [CrossRef]
- Bejerman N, Debat H. Exploring the tymovirales landscape through metatranscriptomics data. Arch Virol. 2022;167: 1785–1803. [CrossRef]
- Katsuma S, Kawamoto M, Shoji K, Aizawa T, Kiuchi T, Izumi N, et al. Transcriptome profiling reveals infection strategy of an insect maculavirus. DNA Research. 2018;25: 277–286. [CrossRef]
- Feng Y, Zhang X, Kumar D, Kuang S, Liu B, Hu X, et al. Transient propagation of BmLV and dysregulation of gene expression in nontarget cells following BmLV infection. Journal of Asia-Pacific Entomology. 2021;24: 893–902. [CrossRef]
- Gupta R, Kwon S-Y, Kim ST. An insight into the tomato spotted wilt virus (TSWV), tomato and thrips interaction. Plant Biotechnol Rep. 2018;12: 157–163. [CrossRef]
- Whitfield AE, Falk BW, Rotenberg D. Insect vector-mediated transmission of plant viruses. Virology. 2015;479–480: 278–289. [CrossRef]
- Margaria P, Bosco L, Vallino M, Ciuffo M, Mautino GC, Tavella L, et al. The NSs Protein of Tomato spotted wilt virus Is Required for Persistent Infection and Transmission by Frankliniella occidentalis. Simon A, editor. J Virol. 2014;88: 5788–5802. [CrossRef]
- Oliveira VC, Bartasson L, de Castro MEB, Corrêa JR, Ribeiro BM, Resende RO. A silencing suppressor protein (NSs) of a tospovirus enhances baculovirus replication in permissive and semipermissive insect cell lines. Virus Research. 2011;155: 259–267. [CrossRef]
- de Oliveira VC, da Silva Morgado F, Ardisson-Araújo DMP, Resende RO, Ribeiro BM. The silencing suppressor (NSs) protein of the plant virus Tomato spotted wilt virus enhances heterologous protein expression and baculovirus pathogenicity in cells and lepidopteran insects. Arch Virol. 2015;160: 2873–2879. [CrossRef]
- Garcia S, Billecocq A, Crance J-M, Prins M, Garin D, Bouloy M. Viral suppressors of RNA interference impair RNA silencing induced by a Semliki Forest virus replicon in tick cells. Journal of General Virology. 2006;87: 1985–1989. [CrossRef]
- Kim C, Kim Y. In vivo transient expression of a viral silencing suppressor, NSs, derived from tomato spotted wilt virus decreases insect RNAi efficiencies. Arch Insect Biochem Physiol. 2022 [cited 12 Jan 2023]. [CrossRef]
- Nagata T, Storms MM, Goldbach R, Peters D. Multiplication of tomato spotted wilt virus in primary cell cultures derived from two thrips species. Virus Res. 1997;49: 59–66. [CrossRef]
- Wijkamp I, van Lent J, Kormelink R, Goldbach R, Peters D. Multiplication of tomato spotted wilt virus in its insect vector, Frankliniella occidentalis. J Gen Virol. 1993;74 ( Pt 3): 341–349. [CrossRef]
- Zhu M, van Grinsven IL, Kormelink R, Tao X. Paving the Way to Tospovirus Infection: Multilined Interplays with Plant Innate Immunity. Annu Rev Phytopathol. 2019;57: 41–62. [CrossRef]
- Turina M, Kormelink R, Resende RO. Resistance to Tospoviruses in Vegetable Crops: Epidemiological and Molecular Aspects. Annu Rev Phytopathol. 2016;54: 347–371. [CrossRef]
- de Ronde D, Pasquier A, Ying S, Butterbach P, Lohuis D, Kormelink R. Analysis of Tomato spotted wilt virus NSs protein indicates the importance of the N-terminal domain for avirulence and RNA silencing suppression: Mapping TSWV NSs Avr and RSS activity. Molecular Plant Pathology. 2014;15: 185–195. [CrossRef]
- Zhai Y, Bag S, Mitter N, Turina M, Pappu HR. Mutational analysis of two highly conserved motifs in the silencing suppressor encoded by tomato spotted wilt virus (genus Tospovirus, family Bunyaviridae). Arch Virol. 2014;159: 1499–1504. [CrossRef]
- Huang C-H, Foo M-H, Raja JAJ, Tan Y-R, Lin T-T, Lin S-S, et al. A Conserved Helix in the C-Terminal Region of Watermelon Silver Mottle Virus Nonstructural Protein S Is Imperative For Protein Stability Affecting Self-Interaction, RNA Silencing Suppression, and Pathogenicity. MPMI. 2020;33: 637–652. [CrossRef]
- Huang C-H, Hsiao W-R, Huang C-W, Chen K-C, Lin S-S, Chen T-C, et al. Two Novel Motifs of Watermelon Silver Mottle Virus NSs Protein Are Responsible for RNA Silencing Suppression and Pathogenicity. Ikegami T, editor. PLoS ONE. 2015;10: e0126161. [CrossRef]
- Yesilyurt A, Demirbag Z, van Oers MM, Nalcacioglu R. Conserved motifs in the invertebrate iridescent virus 6 (IIV6) genome regulate virus transcription. Journal of Invertebrate Pathology. 2020;177: 107496. [CrossRef]
- de Faria IJS, Aguiar ERGR, Olmo RP, Alves da Silva J, Daeffler L, Carthew RW, et al. Invading viral DNA triggers dsRNA synthesis by RNA polymerase II to activate antiviral RNA interference in Drosophila. Cell Reports. 2022;39: 110976. [CrossRef]
- İnce İA, Boeren SA, van Oers MM, Vervoort JJM, Vlak JM. Proteomic analysis of Chilo iridescent virus. Virology. 2010;405: 253–258. [CrossRef]
- Jiang L, Peng Z, Guo Y, Cheng T, Guo H, Sun Q, et al. Transcriptome analysis of interactions between silkworm and cytoplasmic polyhedrosis virus. Sci Rep. 2016;6: 24894. [CrossRef]
- Zhao SL, Liang CY, Zhang WJ, Tang XC, Peng HY. Characterization of the RNA-binding domain in the Dendrolimus punctatus cytoplasmic polyhedrosis virus nonstructural protein p44. Virus Research. 2005;114: 80–88. [CrossRef]
- Oliver JE, Whitfield AE. The Genus Tospovirus: Emerging Bunyaviruses that Threaten Food Security. Annu Rev Virol. 2016;3: 101–124. [CrossRef]
- Ribière M, Olivier V, Blanchard P. Chronic bee paralysis: A disease and a virus like no other? Journal of Invertebrate Pathology. 2010;103: S120–S131. [CrossRef]
- Beaurepaire A, Piot N, Doublet V, Antunez K, Campbell E, Chantawannakul P, et al. Diversity and Global Distribution of Viruses of the Western Honey Bee, Apis mellifera. Insects. 2020;11: 239. [CrossRef]
- Shan T, Yang S, Wang H, Wang H, Zhang J, Gong G, et al. Virome in the cloaca of wild and breeding birds revealed a diversity of significant viruses. Microbiome. 2022;10: 60. [CrossRef]
- de Lima JGS, Lanza DCF. 2A and 2A-like Sequences: Distribution in Different Virus Species and Applications in Biotechnology. Viruses. 2021;13: 2160. [CrossRef]
- Olivier V, Blanchard P, Chaouch S, Lallemand P, Schurr F, Celle O, et al. Molecular characterisation and phylogenetic analysis of Chronic bee paralysis virus, a honey bee virus. Virus Res. 2008;132: 59–68. [CrossRef]
- Zhan H, Unchwaniwala N, Rebolledo-Viveros A, Pennington J, Horswill M, Broadberry R, et al. Nodavirus RNA Replication Crown Architecture Reveals Proto-Crown Precursor and Viral Protein A Conformational Switching. Microbiology; 2022 Dec. [CrossRef]
- Wagner I, Volkmer M, Sharan M, Villaveces JM, Oswald F, Surendranath V, et al. morFeus: a web-based program to detect remotely conserved orthologs using symmetrical best hits and orthology network scoring. BMC Bioinformatics. 2014;15: 263. [CrossRef]
- Le Lay C, Shi M, Buček A, Bourguignon T, Lo N, Holmes E. Unmapped RNA Virus Diversity in Termites and Their Symbionts. Viruses. 2020;12: 1145. [CrossRef]
- Perrakis A, Sixma TK. AI revolutions in biology: The joys and perils of AlphaFold. EMBO Reports. 2021;22. [CrossRef]
- Tokuriki N, Oldfield CJ, Uversky VN, Berezovsky IN, Tawfik DS. Do viral proteins possess unique biophysical features? Trends in Biochemical Sciences. 2009;34: 53–59. [CrossRef]
- Lin Z, Akin H, Rao R, Hie B, Zhu Z, Lu W, et al. Evolutionary-scale prediction of atomic level protein structure with a language model. Synthetic Biology; 2022 Jul. [CrossRef]
- Bruley A, Mornon J-P, Duprat E, Callebaut I. Digging into the 3D Structure Predictions of AlphaFold2 with Low Confidence: Disorder and Beyond. Biomolecules. 2022;12: 1467. [CrossRef]
- Gulyaeva AA, Sigorskih AI, Ocheredko ES, Samborskiy DV, Gorbalenya AE. LAMPA, LArge Multidomain Protein Annotator, and its application to RNA virus polyproteins. Ponty Y, editor. Bioinformatics. 2020;36: 2731–2739. [CrossRef]
- Zhao Y, Sun J, Labropoulou V, Swevers L. Beyond Baculoviruses: Additional Biotechnological Platforms Based on Insect RNA Viruses. Advances in Insect Physiology. Elsevier; 2018. pp. 123–162. [CrossRef]
- Floden EW, Tommaso PD, Chatzou M, Magis C, Notredame C, Chang J-M. PSI/TM-Coffee: a web server for fast and accurate multiple sequence alignments of regular and transmembrane proteins using homology extension on reduced databases. Nucleic Acids Res. 2016;44: W339-343. [CrossRef]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25: 1189–1191. [CrossRef]
- Procter JB, Thompson J, Letunic I, Creevey C, Jossinet F, Barton GJ. Visualization of multiple alignments, phylogenies and gene family evolution. Nat Methods. 2010;7: S16-25. [CrossRef]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, et al. Pfam: The protein families database in 2021. Nucleic Acids Research. 2021;49: D412–D419. [CrossRef]
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Research. 2021;49: D437–D451. [CrossRef]
- Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19: 679–682. [CrossRef]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis: UCSF ChimeraX Visualization System. Protein Science. 2018;27: 14–25. [CrossRef]
- Li Z, Jaroszewski L, Iyer M, Sedova M, Godzik A. FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Research. 2020;48: W60–W64. [CrossRef]
- Holm L. Dali server: structural unification of protein families. Nucleic Acids Research. 2022;50: W210–W215. [CrossRef]

| Species | Protein name | Genbank accession number | Genomic material | Phylum | Order | Family | Genus |
|---|---|---|---|---|---|---|---|
| Acyrthosiphon pisum virus | P1 | QAA78863.1 | +ssRNA | Pisuviricota | Picornavirales | Acypiviridae(1) | Acypivirus (proposed) |
| Aphis citricidus meson-like virus | ORF2a | QPD01783.1 | +ssRNA | Pisuviricota | Nidovirales | Mesoniviridae | Unclassified |
| Barleria severe mosaic virus | NSs | QVY47427.1 | -ssRNA | Negarnaviricota | Bunyavirales | Tospoviridae | Orthotospovirus |
| Bombyx mori Macula-like virus | p15 | YP_004464932 | +ssRNA | Kitrinoviricota | Tymovirales | Tymoviridae | Inmaculavirus (proposed) |
| Bean necrotic mosaic virus | NSs | YP_006468899.1 | -ssRNA | Negarnaviricota | Bunyavirales | Tospoviridae | Orthotospovirus |
| Brandeis virus | ORF5 | AVZ66287.1 | +ssRNA | Kitrinoviricata | Martellivirales | Brandeis virus group(1) | Unclassified |
| Chronic bee paralysis virus | ORF1 from RNA2 | YP_001911139.1 | +ssRNA | Kitrinoviricata | Nodamuvirales | Unclassified | Chroparavirus |
| Darwin bee virus 7 | Nonstructural polyprotein | AWK77849.1 | +ssRNA | Pisuviricota | Picornavirales | Acypiviridae-like(1) | Unclassified |
| Diabrotica toursvirus 3a | F4ORF11 | UOX61048.1 | dsDNA | Nucleocytoviricota | Pimascovirales | Ascoviridae(2) | Toursvirus |
| Dougjudy virga-like virus | ORF4 | QIJ70140.1 | +ssRNA | Kitrinoviricata | Martellivirales | Dougjudy virga-like virus group (1) | Virga-like?? |
| Feksystermes virus | Polyprotein | QRW42904.1 | +ssRNA | Pisuviricota | Picornavirales | Unclassified | Unclassified |
| Fushun monolepta lauta xinmovirus(1) | ORF3 | UHM27673.1 | -ssRNA | Negarnaviricota | Mononegavirales | Xinmoviridae | Unclassified |
| Gonipterus platensis Macula-Like virus | ORF3 | QWX94186.1 | +ssRNA | Kitrinoviricata | Tymovirales | Tymoviridae | Unclassified |
| Groundnut yellow Spot virus | NSs | YP_009665191.1 | -ssRNA | Negarnaviricota | Bunyavirales | Tospoviridae | Orthotospovirus |
| Hangzhou frankliniella intonsa flavivirus(1) | RNA-dependent DNA polymerase | UHK03321.1 | +ssRNA | Kitrinoviricata | Amarillovirales | Flaviviridae-like | Unclassified, “Large-genome flaviviruses” [26] |
| Hangzhou sesamia inferens solinvi-like virus(1) | RNA helicase | UHR49866 | +ssRNA | Pisuviricota | Picornavirales | Acypiviridae-like(1) | Unclassified, similar to Solenopsis invicta virus 7 |
| Hainan sediment toti-like virus 9 | Putative coat protein | UHS72513.1 | dsRNA | Duplornaviricota | Ghabrivirales | Totiviridae | Artivirus |
| Hubei picorna-like virus 36 | ORF2 | KX883970.1 (coding sequence: nt 9360-9647) | +ssRNA | Pisuviricota | Picornavirales | Iflaviridae | Iflavirus |
| Hypera postica associated virus(1) | Hypothetical protein | QUS52866.1 | +ssRNA | Pisuviricota | Picornavirales | Solinvirididae-like | Unclassified |
| HVAC-associated RNA virus(1) | Polyprotein | AVD69112 | +ssRNA | Pisuviricota | Picornavirales | Acypiviridae-like(1) | Unclassified, similar to Solenopsis invicta virus 7 |
| Invertebrate iridescent virus 6 | 206R | Q91FW4 | dsDNA | Nucleocytoviricota | Pimascovirales | Iridoviridae | Iridovirus |
| Lake Sinai virus | ORF4 | YP_009333196.1 | +ssRNA | Kitrinoviricata | Nodamuvirales | Sinhaliviridae | Sinaivirus |
| PNG bee virus 9 | Polyprotein | QKW94212.1 | +ssRNA | Pisuviricota | Picornavirales | Acypiviridae-like(1) | Unclassified, similar to Solenopsis invicta virus 7 |
| Pterostylis blotch virus | NSs | ULN99190.1 | -ssRNA | Negarnaviricota | Bunyavirales | Tospoviridae | Orthotospovirus |
| Saiwaicho virus | ORF4 | AWA82266.1 | +ssRNA | Kitrinoviricota | Martellivirales | Unclassified | Unclassified, Nelorpivirus-like |
| Solenopsis invicta virus 7 | Polyprotein | QBL75890.1 | +ssRNA | Pisuviricota | Picornavirales | Acypiviridae-like(1) | Unclassified |
| Solenopsis invicta virus 8 | ORF6 | MH727525.2 (coding sequence: nt 4189-4476) | +ssRNA | Pisuviricota | Picornavirales | Polycipiviridae | Sopolycivirus |
| Tomato spotted wilt virus (strain Br20) | NSs | ABI94070.1 | -ssRNA | Negarnaviricota | Bunyavirales | Tospoviridae | Orthotospovirus |
| Isolate H4_Bulk_46_scaffold_6139 | Hypothetical protein | MN034786 (coding sequence: nt 3-1841)(3) | +ssRNA | Pisuviricota | Picornavirales | Acypiviridae-like | Unclassified |
| Wiseana Iridescent virus | gp049 | YP_004732832 | dsDNA | Nucleocytoviricota | Pimascovirales | Iridoviridae | Chloriridovirus |
| Wpk049shi07 isolate(4) | ORF1 | QKE55054.1 | +ssRNA | Kitrinoviricata | Nodamuvirales | Sinhaliviridae-like | Sinaivirus-like |
| Ybb117shi01 isolate | ORF2 (hypothetical protein) | QJI53477 | +ssRNA | Pisuviricota | Picornavirales | Acypiviridae-like | Unclassified |
| Virus species | Protein name | Genomic RNA Segment | Genbank accession number | Bibliographical information |
|---|---|---|---|---|
|
Dendrolimus punctatus cypovirus 1 (a strain of Bombyx mori cypovirus 1) |
p44 (also called nsp2 or NS2 or NS) | S8 | NP_149153.1 | p44 is a non-structural protein that forms viroplasms during infection by Dendrolimus punctatus cypovirus [43]. |
| Hubei lepidoptera virus 3 (proposed as Lymantria dispar cypovirus 3) | P5 (also called VP5) | S5 | YP_009330260.1 | |
| Hubei lepidoptera virus 3 (isolate LdCPV3) | P5 (also called VP5) | S5 | QJB76100.1 | |
| Antheraea mylitta cypovirus 4 | p61 | S7 | ABF83587.1 | p61 is a structural protein, i.e. found in virions [44]. |
| Thaumetopoea pityocampa cypovirus 5 | P5 | S5 | AJC97792.1 | |
| Heliothis armigera cypovirus 5 | P5 | S5 | YP_001883319.1 | |
|
Lymantria dispar cypovirus 14 |
P8 | S8 | NP_149142.1 | |
| Operophtera brumata cypovirus 18 | P8 | S8 | ABB17218 | |
| Daphnis nerii cypovirus 23 | P5 | S5 | YP_009551580 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





