Submitted:
03 June 2023
Posted:
05 June 2023
You are already at the latest version
Abstract
Keywords:
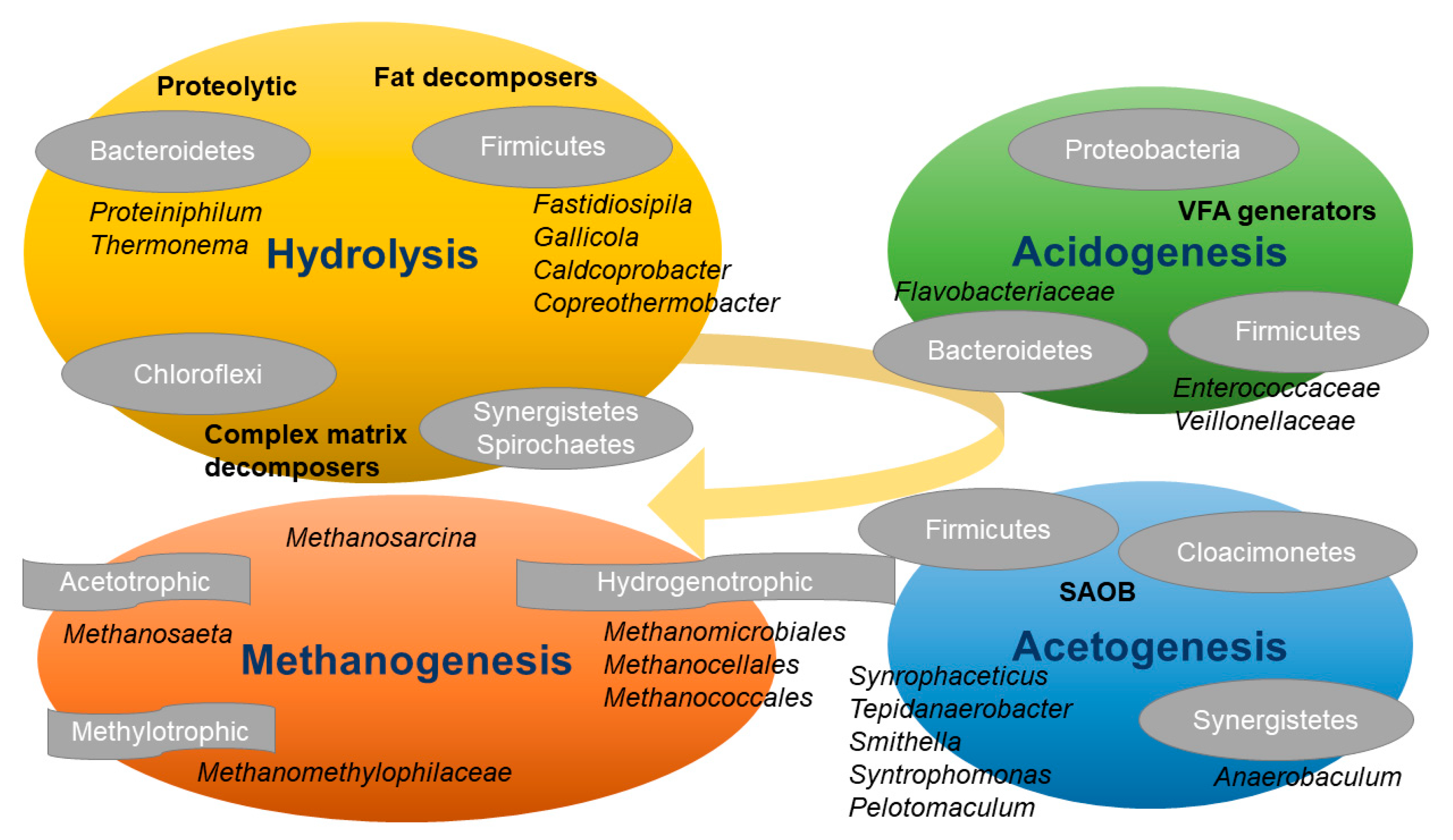
1. Introduction
2. Feedstock
2.1. Feedstock characteristics
2.2. Co-digestion of feedstocks
2.3. Microorganisms present in different types of feedstocks
2.4. Effect of the inoculum
3. Total solids
4. Ammonia concentration
4.1. Effect of elevated ammonia on microbial communities
5. Temperature
5.1. Different temperature regimes
5.2. Changes in microbial composition due to temperature variation
5.3. Effect of temperature combined with OLR changes
5.4. Microbial adaptation upon temperature alteration
6. Volatile Fatty Acids (VFAs)
6.1. Factors causing VFA accumulation
6.2. Effect of VFAs on microbial communities
7.1. The role of pH and its variation
7.2. Influence of pH on microbial communities
8. Hydraulic Retention Time (HRT) & Organic Loading Rate (OLR)
8.1. HRT and OLR in anaerobic digestion systems
8.2. HRT & OLR impact on microbial communities
8.2.1. HRT
8.2.2. OLR
8.2.3. Comparison of HRT versus OLR changes
9. Functional redundancy
10. Current status
11. Future perspectives
12. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, R.Z.; Fang, S.Y.; Zhang, L.; Huang, W.X.; Shao, Q.Q.; Fang, F.; Feng, Q.; Cao, J.S.; Luo, J.Y. Distribution patterns of functional microbial community in anaerobic digesters under different operational circumstances: A review. Bioresource Technology 2021, 341. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Muller, B.; Isaksson, S.; Schnurer, A. Trace element and temperature effects on microbial communities and links to biogas digester performance at high ammonia levels. Biotechnology for Biofuels 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; Patrone, V.; Puglisi, E.; Morelli, L.; Bassi, D.; Garuti, M.; Rossi, L.; Cappa, F. Effects of geographic area, feedstock, temperature, and operating time on microbial communities of six full-scale biogas plants. Bioresource Technology 2016, 218, 980–990. [Google Scholar] [CrossRef]
- Panigrahi, S.; Dubey, B.K. A critical review on operating parameters and strategies to improve the biogas yield from anaerobic digestion of organic fraction of municipal solid waste. Renewable Energy 2019, 143, 779–797. [Google Scholar] [CrossRef]
- Ziganshina, E.E.; Belostotskiy, D.E.; Ilinskaya, O.N.; Boulygina, E.A.; Grigoryeva, T.V.; Ziganshin, A.M. Effect of the Organic Loading Rate Increase and the Presence of Zeolite on Microbial Community Composition and Process Stability During Anaerobic Digestion of Chicken Wastes. Microbial Ecology 2015, 70, 948–960. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.K.; Lee, S.H.; Kim, Y.; Park, H.D. Current understanding and perspectives in anaerobic digestion based on genome-resolved metagenomic approaches. Bioresource Technology 2022, 344. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.P.; Wu, X.Y.; Zhou, B.Q.; Wang, Y.; Sun, Y.; Wang, Y.F.; Chen, Z.B.; Zhang, J.F. Effect of one step temperature increment from mesophilic to thermophilic anaerobic digestion on the linked pattern between bacterial and methanogenic communities. Bioresource Technology 2019, 292. [Google Scholar] [CrossRef]
- Gaby, J.C.; Zamanzadeh, M.; Horn, S.J. The effect of temperature and retention time on methane production and microbial community composition in staged anaerobic digesters fed with food waste. Biotechnology for Biofuels 2017, 10. [Google Scholar] [CrossRef]
- de Jonge, N.; Davidsson, A.; Jansen, J.L.; Nielsen, J.L. Characterisation of microbial communities for improved management of anaerobic digestion of food waste. Waste Management 2020, 117, 124–135. [Google Scholar] [CrossRef]
- Lim, J.W.; Park, T.; Tong, Y.W.; Yu, Z. Chapter One - The microbiome driving anaerobic digestion and microbial analysis. In Advances in Bioenergy; Li, Y., Khanal, S.K., Eds.; Elsevier, 2020; Volume 5, pp. 1–61. [Google Scholar]
- Langer, S.G.; Gabris, C.; Einfalt, D.; Wemheuer, B.; Kazda, M.; Bengelsdorf, F.R. Different response of bacteria, archaea and fungi to process parameters in nine full-scale anaerobic digesters. Microbial Biotechnology 2019, 12, 1210–1225. [Google Scholar] [CrossRef]
- Carballa, M.; Regueiro, L.; Lema, J.M. Microbial management of anaerobic digestion: exploiting the microbiome-functionality nexus. Current Opinion in Biotechnology 2015, 33, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Fenchel, T.; King, G.M.; Blackburn, T.H. Chapter 1 - Bacterial Metabolism. In Bacterial Biogeochemistry (Third Edition); Fenchel, T., King, G.M., Blackburn, T.H., Eds.; Academic Press: Boston, 2012; pp. 1–34. [Google Scholar]
- Sarmiento, B.F.; Leigh, J.A.; Whitrnan, W.B. GENETIC SYSTEMS FOR HYDROGENOTROPHIC METHANOGENS. Methods in Enzymology: Methods in Methane Metabolism, Pt A 2011, 494, 43–73. [Google Scholar] [CrossRef]
- Han, G.; Shin, S.G.; Cho, K.; Lee, J.; Kim, W.; Hwang, S. Temporal variation in bacterial and methanogenic communities of three full-scale anaerobic digesters treating swine wastewater. Environmental Science and Pollution Research 2019, 26, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, M.; Cydzik-Kwiatkowska, A.; Zielinski, M.; Debowski, M. Impact of temperature, microwave radiation and organic loading rate on methanogenic community and biogas production during fermentation of dairy wastewater. Bioresource Technology 2013, 129, 308–314. [Google Scholar] [CrossRef]
- Calusinska, M.; Goux, X.; Fossepre, M.; Muller, E.E.L.; Wilmes, P.; Delfosse, P. A year of monitoring 20 mesophilic full-scale bioreactors reveals the existence of stable but different core microbiomes in bio-waste and wastewater anaerobic digestion systems. Biotechnology for Biofuels 2018, 11. [Google Scholar] [CrossRef]
- Achinas, S.; Achinas, V.; Euverink, G.J.W. A Technological Overview of Biogas Production from Biowaste. Engineering 2017, 3, 299–307. [Google Scholar] [CrossRef]
- Saha, S.; Basak, B.; Hwang, J.H.; Salama, E.; Chatterjee, P.K.; Jeon, B.H. Microbial Symbiosis: A network towards Biomethanation. Trends in Microbiology 2020, 28, 968–984. [Google Scholar] [CrossRef]
- Puig-Castellvi, F.; Cardona, L.; Bouveresse, D.J.R.; Cordella, C.B.Y.; Mazeas, L.; Rutledge, D.N.; Chapleur, O. Assessment of the microbial interplay during anaerobic co-digestion of wastewater sludge using common components analysis. Plos One 2020, 15. [Google Scholar] [CrossRef]
- Yun, Y.M.; Cho, S.K.; Kim, H.W.; Jung, K.W.; Shin, H.S.; Kim, D.H. Elucidating a synergistic effect of food waste addition on the enhanced anaerobic digestion of waste activated sludge. Korean Journal of Chemical Engineering 2015, 32, 1542–1546. [Google Scholar] [CrossRef]
- Shin, S.G.; Han, G.; Lee, J.; Shin, J.; Hwang, S. A snapshot of microbial community structures in 20 different field-scale anaerobic bioreactors treating food waste. Journal of Environmental Management 2019, 248. [Google Scholar] [CrossRef]
- Lee, J.; Kim, E.; Han, G.; Tongco, J.V.; Shin, S.G.; Hwang, S. Microbial communities underpinning mesophilic anaerobic digesters treating food wastewater or sewage sludge: A full-scale study. Bioresource Technology 2018, 259, 388–397. [Google Scholar] [CrossRef]
- Lee, J.; Shin, S.G.; Han, G.; Koo, T.; Hwang, S. Bacteria and archaea communities in full-scale thermophilic and mesophilic anaerobic digesters treating food wastewater: Key process parameters and microbial indicators of process instability. Bioresource Technology 2017, 245, 689–697. [Google Scholar] [CrossRef]
- Sposob, M.; Moon, H.S.; Lee, D.; Kim, T.H.; Yun, Y.M. Comprehensive analysis of the microbial communities and operational parameters of two full-scale anaerobic digestion plants treating food waste in South Korea: Seasonal variation and effect of ammonia. Journal of Hazardous Materials 2020, 398. [Google Scholar] [CrossRef]
- Montanes, R.; Solera, R.; Perez, M. Anaerobic co-digestion of sewage sludge and sugar beet pulp lixiviation in batch reactors: Effect of temperature. Bioresource Technology 2015, 180, 177–184. [Google Scholar] [CrossRef]
- Fitamo, T.; Treu, L.; Boldrin, A.; Sartori, C.; Angelidaki, I.; Scheutz, C. Microbial population dynamics in urban organic waste anaerobic co-digestion with mixed sludge during a change in feedstock composition and different hydraulic retention times. Water Research 2017, 118, 261–271. [Google Scholar] [CrossRef]
- Lin, Q.; De Vrieze, J.; Fang, X.Y.; Li, L.J.; Li, X.Z. Labile carbon feedstocks trigger a priming effect in anaerobic digestion: An insight into microbial mechanisms. Bioresource Technology 2022, 344. [Google Scholar] [CrossRef]
- Kalamaras, S.D.; Vasileiadis, S.; Karas, P.; Angelidaki, I.; Kotsopoulos, T.A. Microbial adaptation to high ammonia concentrations during anaerobic digestion of manure-based feedstock: biomethanation and 16S rRNA gene sequencing. Journal of Chemical Technology and Biotechnology 2020, 95, 1970–1979. [Google Scholar] [CrossRef]
- Pereira, M.A.; Sousa, D.Z.; Mota, M.; Alves, M.M. Mineralization of LCFA associated with anaerobic sludge: Kinetics, enhancement of methanogenic activity, and effect of VFA. Biotechnology and Bioengineering 2004, 88, 502–511. [Google Scholar] [CrossRef]
- Kougias, P.G.; Campanaro, S.; Treu, L.; Zhu, X.Y.; Angelidaki, I. A novel archaeal species belonging to Methanoculleus genus identified via de-novo assembly and metagenomic binning process in biogas reactors. Anaerobe 2017, 46, 23–32. [Google Scholar] [CrossRef]
- Niu, Q.; Kobayashi, T.; Takemura, Y.; Kubota, K.; Li, Y.Y. Evaluation of functional microbial community's difference in full-scale and lab-scale anaerobic digesters feeding with different organic solid waste: Effects of substrate and operation factors. Bioresource Technology 2015, 193, 110–118. [Google Scholar] [CrossRef]
- Yi, J.; Dong, B.; Jin, J.W.; Dai, X.H. Effect of Increasing Total Solids Contents on Anaerobic Digestion of Food Waste under Mesophilic Conditions: Performance and Microbial Characteristics Analysis. Plos One 2014, 9. [Google Scholar] [CrossRef]
- Ziganshin, A.M.; Ziganshina, E.E.; Kleinsteuber, S.; Nikolausz, M. Comparative Analysis of Methanogenic Communities in Different Laboratory-Scale Anaerobic Digesters. Archaea-an International Microbiological Journal 2016, 2016. [Google Scholar] [CrossRef]
- Peces, M.; Astals, S.; Jensen, P.D.; Clarke, W.P. Deterministic mechanisms define the long-term anaerobic digestion microbiome and its functionality regardless of the initial microbial community. Water Research 2018, 141, 366–376. [Google Scholar] [CrossRef]
- Duan, N.; Kougias, P.G.; Campanaro, S.; Treu, L.; Angelidaki, I. Evolution of the microbial community structure in biogas reactors inoculated with seeds from different origin. Science of The Total Environment 2021, 773, 144981. [Google Scholar] [CrossRef]
- Demichelis, F.; Tommasi, T.; Deorsola, F.A.; Marchisio, D.; Fino, D. Effect of inoculum origin and substrate-inoculum ratio to enhance the anaerobic digestion of organic fraction municipal solid waste (OFMSW). Journal of Cleaner Production 2022, 351. [Google Scholar] [CrossRef]
- Quintero, M.; Castro, L.; Ortiz, C.; Guzman, C.; Escalante, H. Enhancement of starting up anaerobic digestion of lignocellulosic substrate: fique's bagasse as an example. Bioresource Technology 2012, 108, 8–13. [Google Scholar] [CrossRef]
- Bella, K.; Rao, P.V. Anaerobic co-digestion of cheese whey and septage: Effect of substrate and inoculum on biogas production. Journal of Environmental Management 2022, 308. [Google Scholar] [CrossRef]
- Li, Y.Y.; Wang, Y.Q.; Yu, Z.H.; Lu, J.X.; Li, D.Y.; Wang, G.Y.; Li, Y.; Wu, Y.; Li, S.Y.; Xu, F.Q.; et al. Effect of inoculum and substrate/inoculum ratio on the performance and methanogenic archaeal community structure in solid state anaerobic co-digestion of tomato residues with dairy manure and corn stover. Waste Management 2018, 81, 117–127. [Google Scholar] [CrossRef]
- Bao, R.; Wei, Y.F.; Guan, R.L.; Li, X.J.; Lu, X.B.; Rong, S.Y.; Zuo, X.Y.; Yuan, H.R. High-solids anaerobic co-digestion performances and microbial community dynamics in co-digestion of different mixing ratios with food waste and highland barley straw. Energy 2023, 262. [Google Scholar] [CrossRef]
- Liu, C.; Li, H.; Zhang, Y.Y.; Si, D.D.; Chen, Q.W. Evolution of microbial community along with increasing solid concentration during high-solids anaerobic digestion of sewage sludge. Bioresource Technology 2016, 216, 87–94. [Google Scholar] [CrossRef]
- Wang, Z.Z.; Jiang, Y.; Wang, S.; Zhang, Y.Z.; Hu, Y.S.; Hu, Z.H.; Wu, G.X.; Zhan, X.M. Impact of total solids content on anaerobic co-digestion of pig manure and food waste: Insights into shifting of the methanogenic pathway. Waste Management 2020, 114, 96–106. [Google Scholar] [CrossRef]
- Abbassi-Guendouz, A.; Brockmann, D.; Trably, E.; Dumas, C.; Delgenes, J.P.; Steyer, J.P.; Escudie, R. Total solids content drives high solid anaerobic digestion via mass transfer limitation. Bioresource Technology 2012, 111, 55–61. [Google Scholar] [CrossRef]
- Liu, C.; Li, H.; Zhang, Y.Y.; Chen, Q.W. Characterization of methanogenic activity during high-solids anaerobic digestion of sewage sludge. Biochemical Engineering Journal 2016, 109, 96–100. [Google Scholar] [CrossRef]
- Liao, X.C.; Li, H. Biogas production from low-organic-content sludge using a high-solids anaerobic digester with improved agitation. Applied Energy 2015, 148, 252–259. [Google Scholar] [CrossRef]
- Duan, N.N.; Dong, B.; Wu, B.; Dai, X.H. High-solid anaerobic digestion of sewage sludge under mesophilic conditions: Feasibility study. Bioresource Technology 2012, 104, 150–156. [Google Scholar] [CrossRef]
- Arelli, V.; Begum, S.; Anupoju, G.R.; Kuruti, K.; Shailaja, S. Dry anaerobic co-digestion of food waste and cattle manure: Impact of total solids, substrate ratio and thermal pre treatment on methane yield and quality of biomanure. Bioresource Technology 2018, 253, 273–280. [Google Scholar] [CrossRef]
- Han, G.; Shin, S.G.; Lee, J.; Shin, J.; Hwang, S. A comparative study on the process efficiencies and microbial community structures of six full-scale wet and semi-dry anaerobic digesters treating food wastes. Bioresource Technology 2017, 245, 869–875. [Google Scholar] [CrossRef]
- Choi, G.; Kim, H.; Lee, C. Long-term monitoring of a thermal hydrolysis-anaerobic co-digestion plant treating high-strength organic wastes: Process performance and microbial community dynamics. Bioresource Technology 2021, 319. [Google Scholar] [CrossRef]
- Duc, L.V.; Miyagawa, Y.; Inoue, D.; Ike, M. Identification of key steps and associated microbial populations for efficient anaerobic digestion under high ammonium or salinity conditions. Bioresource Technology 2022, 360. [Google Scholar] [CrossRef]
- Su, L.H.; Sun, X.; Liu, C.W.; Ji, R.T.; Zhen, G.Y.; Chen, M.; Zhang, L.J. Thermophilic Solid-State Anaerobic Digestion of Corn Straw, Cattle Manure, and Vegetable Waste: Effect of Temperature, Total Solid Content, and C/N Ratio. Archaea-an International Microbiological Journal 2020, 2020. [Google Scholar] [CrossRef]
- Fotidis, I.A.; Karakashev, D.; Kotsopoulos, T.A.; Martzopoulos, G.G.; Angelidaki, I. Effect of ammonium and acetate on methanogenic pathway and methanogenic community composition. Fems Microbiology Ecology 2013, 83, 38–48. [Google Scholar] [CrossRef]
- Zhang, H.; Yuan, W.D.; Dong, Q.; Wu, D.; Yang, P.J.; Peng, Y.; Li, L.; Peng, X.Y. Integrated multi-omics analyses reveal the key microbial phylotypes affecting anaerobic digestion performance under ammonia stress. Water Research 2022, 213. [Google Scholar] [CrossRef]
- Westerholm, M.; Calusinska, M.; Dolfing, J. Syntrophic propionate-oxidizing bacteria in methanogenic systems. Fems Microbiology Reviews 2022, 46. [Google Scholar] [CrossRef]
- Heyer, R.; Benndorf, D.; Kohrs, F.; De Vrieze, J.; Boon, N.; Hoffmann, M.; Rapp, E.; Schluter, A.; Sczyrba, A.; Reichl, U. Proteotyping of biogas plant microbiomes separates biogas plants according to process temperature and reactor type. Biotechnology for Biofuels 2016, 9. [Google Scholar] [CrossRef]
- Ziganshin, A.M.; Liebetrau, J.; Proter, J.; Kleinsteuber, S. Microbial community structure and dynamics during anaerobic digestion of various agricultural waste materials. Applied Microbiology and Biotechnology 2013, 97, 5161–5174. [Google Scholar] [CrossRef]
- Bardi, M.J.; Aminirad, H. Synergistic effects of co-trace elements on anaerobic co-digestion of food waste and sewage sludge at high organic load. Environmental Science and Pollution Research 2020, 27, 18129–18144. [Google Scholar] [CrossRef]
- Shaw, G.T.W.; Liu, A.C.; Weng, C.Y.; Chou, C.Y.; Wang, D.Y. Inferring microbial interactions in thermophilic and mesophilic anaerobic digestion of hog waste. Plos One 2017, 12. [Google Scholar] [CrossRef]
- Moestedt, J.; Ronnberg, J.; Nordell, E. The effect of different mesophilic temperatures during anaerobic digestion of sludge on the overall performance of a WWTP in Sweden. Water Science and Technology 2017, 76, 3213–3219. [Google Scholar] [CrossRef]
- Kabaivanova, L.; Petrova, P.; Hubenov, V.; Simeonov, I. Biogas Production Potential of Thermophilic Anaerobic Biodegradation of Organic Waste by a Microbial Consortium Identified with Metagenomics. Life-Basel 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Tukacs-Hajos, A.; Pap, B.; Maroti, G.; Szendefy, J.; Szabo, P.; Retfalvi, T. Monitoring of thermophilic adaptation of mesophilic anaerobe fermentation of sugar beet pressed pulp. Bioresource Technology 2014, 166, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Angelidaki, I.; Sanders, W. Assessment of the anaerobic biodegradability of macropollutants. Re/Views in Environmental Science & Bio/Technology 2004, 3, 117–129. [Google Scholar] [CrossRef]
- Guo, X.H.; Wang, C.; Sun, F.Q.; Zhu, W.J.; Wu, W.X. A comparison of microbial characteristics between the thermophilic and mesophilic anaerobic digesters exposed to elevated food waste loadings. Bioresource Technology 2014, 152, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Goux, X.; Calusinska, M.; Lemaigre, S.; Marynowska, M.; Klocke, M.; Udelhoven, T.; Benizri, E.; Delfosse, P. Microbial community dynamics in replicate anaerobic digesters exposed sequentially to increasing organic loading rate, acidosis, and process recovery. Biotechnology for Biofuels 2015, 8. [Google Scholar] [CrossRef]
- Shaw, G.T.W.; Weng, C.Y.; Chen, C.Y.; Weng, F.C.H.; Wang, D.Y. A systematic approach re-analyzing the effects of temperature disturbance on the microbial community of mesophilic anaerobic digestion. Scientific Reports 2019, 9. [Google Scholar] [CrossRef]
- Liu, J.; Zuo, X.Y.; Peng, K.; He, R.; Yang, L.Y.; Liu, R.F. Biogas and Volatile Fatty Acid Production During Anaerobic Digestion of Straw, Cellulose, and Hemicellulose with Analysis of Microbial Communities and Functions. Applied Biochemistry and Biotechnology 2022, 194, 762–782. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, C.X.; Yuan, Z.W.; Wang, R.M.; Angelidaki, I.; Zhu, G.F. Syntrophy mechanism, microbial population, and process optimization for volatile fatty acids metabolism in anaerobic digestion. Chemical Engineering Journal 2023, 452. [Google Scholar] [CrossRef]
- Franke-Whittle, I.H.; Walter, A.; Ebner, C.; Insam, H. Investigation into the effect of high concentrations of volatile fatty acids in anaerobic digestion on methanogenic communities. Waste Management 2014, 34, 2080–2089. [Google Scholar] [CrossRef]
- Mathai, P.P.; Nicholes, M.S.; Venkiteshwaran, K.; Brown, C.M.; Morris, R.L.; Zitomer, D.H.; Maki, J.S. Dynamic shifts within volatile fatty acid-degrading microbial communities indicate process imbalance in anaerobic digesters. Applied Microbiology and Biotechnology 2020, 104, 4563–4575. [Google Scholar] [CrossRef]
- Nguyen, D.; Wu, Z.Y.; Shrestha, S.; Lee, P.H.; Raskin, L.; Khanal, S.K. Intermittent micro-aeration: New strategy to control volatile fatty acid accumulation in high organic loading anaerobic digestion. Water Research 2019, 166. [Google Scholar] [CrossRef]
- Hori, T.; Haruta, S.; Ueno, Y.; Ishii, M.; Igarashi, Y. Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Applied and Environmental Microbiology 2006, 72, 1623–1630. [Google Scholar] [CrossRef]
- Wagner, A.O.; Malin, C.; Lins, P.; Illmer, P. Effects of various fatty acid amendments on a microbial digester community in batch culture. Waste Management 2011, 31, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.F.; Hu, K.; Zheng, Z.H.; Zhang, Y.; Guo, S.Y.; Zhao, X.L.; Cui, Z.J.; Wang, X.F. Effects of adding EDTA and Fe2+ on the performance of reactor and microbial community structure in two simulated phases of anaerobic digestion. Bioresource Technology 2019, 275, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Venkiteshwaran, K.; Bocher, B.; Maki, J.; Zitomer, D. Relating Anaerobic Digestion Microbial Community and Process Function : Supplementary Issue: Water Microbiology. Microbiology Insights 2015, 8s2, MBI–S33593. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.G.; Yang, X.; Yao, X.F. Effects of pH on the biodegradation characteristics of thermophilic micro-aerobic digestion for sludge stabilization. Rsc Advances 2019, 9, 8379–8388. [Google Scholar] [CrossRef]
- Zhou, X.N.; Lu, Y.; Huang, L.; Zhang, Q.; Wang, X.Y.; Zhu, J.Y. Effect of pH on volatile fatty acid production and the microbial community during anaerobic digestion of Chinese cabbage waste. Bioresource Technology 2021, 336. [Google Scholar] [CrossRef] [PubMed]
- Castro-Ramos, J.J.; Solis-Oba, A.; Solis-Oba, M.; Calderon-Vazquez, C.L.; Higuera-Rubio, J.M.; Castro-Rivera, R. Effect of the initial pH on the anaerobic digestion process of dairy cattle manure. Amb Express 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, R.; Liu, F.W.; Yong, X.Y.; Wu, X.Y.; Zheng, T.; Jiang, M.; Jia, H.H. Biogas production and microbial community shift through neutral pH control during the anaerobic digestion of pig manure. Bioresource Technology 2016, 217, 44–49. [Google Scholar] [CrossRef]
- Peces, M.; Astals, S.; Jensen, P.D.; Clarke, W.P. Transition of microbial communities and degradation pathways in anaerobic digestion at decreasing retention time. New Biotechnology 2021, 60, 52–61. [Google Scholar] [CrossRef]
- Tena, M.; Perez, M.; Solera, R. Effect of hydraulic retention time on the methanogenic step of a two-stage anaerobic digestion system from sewage sludge and wine vinasse: Microbial and kinetic evaluation. Fuel 2021, 296. [Google Scholar] [CrossRef]
- Shi, X.S.; Dong, J.J.; Yu, J.H.; Yin, H.; Hu, S.M.; Huang, S.X.; Yuan, X.Z. Effect of Hydraulic Retention Time on Anaerobic Digestion of Wheat Straw in the Semicontinuous Continuous Stirred-Tank Reactors. Biomed Research International 2017, 2017. [Google Scholar] [CrossRef]
- Xu, R.; Yang, Z.H.; Zheng, Y.; Liu, J.B.; Xiong, W.P.; Zhang, Y.R.; Lu, Y.; Xue, W.J.; Fan, C.Z. Organic loading rate and hydraulic retention time shape distinct ecological networks of anaerobic digestion related microbiome. Bioresource Technology 2018, 262, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Christou, M.L.; Vasileiadis, S.; Karpouzas, D.G.; Angelidaki, I.; Kotsopoulos, T.A. Effects of organic loading rate and hydraulic retention time on bioaugmentation performance to tackle ammonia inhibition in anaerobic digestion. Bioresource Technology 2021, 334. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Meng, X.S.; Zhou, G.N.; Zhou, Z.Z.; Zheng, T.; Bai, Y.E.; Yuan, H.R.; Huhe, T. Effects of organic loading rates on the anaerobic co-digestion of fresh vinegar residue and pig manure: Focus on the performance and microbial communities. Biochemical Engineering Journal 2022, 183. [Google Scholar] [CrossRef]
- Yang, S.M.; Luo, F.; Yan, J.; Zhang, T.L.; Xian, Z.Y.; Huang, W.Y.; Zhang, H.G.; Cao, Y.J.; Huang, L. Biogas production of food waste with in-situ sulfide control under high organic loading in two-stage anaerobic digestion process: Strategy and response of microbial community. Bioresource Technology 2023, 373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiao, P.; Zhang, M.; Wu, P.; Zhang, Y.; Wang, Y.; Xu, K.; Yu, J.; Ma, L. Impacts of organic loading rate and hydraulic retention time on organics degradation, interspecies interactions and functional traits in thermophilic anaerobic co-digestion of food waste and sewage sludge. Bioresource Technology 2023, 370, 128578. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, A.; Khadka, A.; Sapkota, L.; Ghimire, A. Effect of Hydraulic Retention Time and Organic-Loading Rate on Two-Staged, Semi-Continuous Mesophilic Anaerobic Digestion of Food Waste during Start-Up. Fermentation-Basel 2022, 8. [Google Scholar] [CrossRef]
- Pramanik, S.K.; Suja, F.B.; Pramanik, B.K. Effects of hydraulic retention time on the process performance and microbial community structure of an anaerobic single-stage semi-pilot scale reactor for the treatment of food waste. International Biodeterioration & Biodegradation 2020, 152. [Google Scholar] [CrossRef]
- Razaviarani, V.; Buchanan, I.D. Reactor performance and microbial community dynamics during anaerobic co-digestion of municipal wastewater sludge with restaurant grease waste at steady state and overloading stages. Bioresource Technology 2014, 172, 232–240. [Google Scholar] [CrossRef]
- Wu, Z.L.; Zhang, Q.; Xia, Z.Y.; Gou, M.; Sun, Z.Y.; Tang, Y.Q. The responses of mesophilic and thermophilic anaerobic digestion of municipal sludge to periodic fluctuation disturbance of organic loading rate. Environmental Research 2023, 218. [Google Scholar] [CrossRef]
- Braz, G.H.R.; Fernandez-Gonzalez, N.; Lema, J.M.; Carballa, M. The time response of anaerobic digestion microbiome during an organic loading rate shock. Applied Microbiology and Biotechnology 2018, 102, 10285–10297. [Google Scholar] [CrossRef]
- Yachi, S.; Loreau, M. Biodiversity and ecosystem productivity in a fluctuating environment: The insurance hypothesis. Proceedings of the National Academy of Sciences of the United States of America 1999, 96, 1463–1468. [Google Scholar] [CrossRef] [PubMed]
- Roopnarain, A.; Rama, H.; Ndaba, B.; Bello-Akinosho, M.; Bamuza-Pemu, E.; Adeleke, R. Unravelling the anaerobic digestion 'black box': Biotechnological approaches for process optimization. Renewable & Sustainable Energy Reviews 2021, 152. [Google Scholar] [CrossRef]
- Lv, Z.; Leite, A.F.; Harms, H.; Richnow, H.H.; Liebetrau, J.; Nikolausz, M. Influences of the substrate feeding regime on methanogenic activity in biogas reactors approached by molecular and stable isotope methods. Anaerobe 2014, 29, 91–99. [Google Scholar] [CrossRef] [PubMed]
| TS content | Biogas Yield | Favored | |
|---|---|---|---|
| Wet AD | ≤10% | Increased [45] |
Porphyromonadaceae Sphingobacteriaceae Syntrophomonadaceae [49] |
| HSAD | 10-20% | Declined [44] Increased [33,42] |
Clostridiaceae Patulibacteraceae Pseudonocardiaceae Lachnospiraceae Rikenellaceae Methanobacteriaceae [49] |
| Dry AD | >20% | Increased [41,48] Declined [43] |
Proteiniphilum Fastidiosipila, Gallicola Aminobacterium Syntrophaceticus W5053 [41] |
| Digester | Microbial communities favored | Role | Reference |
|---|---|---|---|
| Mesophilic sludge digesters |
Ca. Brevefilum (Chloroflexi) Ca. Cloacimonas Syntrophorhabdus |
SPOB | [51] |
|
Paraclostridium Enterococcus Romboutsia Proteiniphilum Turicibacter |
hydrogen-producing | [51] | |
| Methanosarcina | acetoclastic and hydrogenotrophic methanogenesis | [9,33,51] | |
| Mesophilic reactors co-digesting household waste and albumin |
Clostridium ultunense Syntrophaceticus schinkii Tepidanaerobacter acetatoxydans |
SAOB | [2,41] |
| Mesophilic reactors with chicken waste |
Marinilabiaceae Porphyromonadaceae |
SAOB | [5] |
| Methanosarcina | cooperating with Clostridium for hydrogenotrophic methanogenesis | [5,56] | |
| Mesophilic digesters fed with sewage sludge and FW | Methanoculleus receptaculi | [23] | |
| Cloacimonadales W27 | syntrophic partner to hydrogenotrophic methanogens | [23] | |
|
Caldicoprobacter algeriensis Clostridium senegalense Ercella succinigene Gelria glutamica |
[25] | ||
|
Gelria glutamica Defluviitoga tunisiensis |
positively correlated with high NH4+ | [25] | |
| Batch cultures and manure-based substrate |
Cellulosilyticum ruminicola (Cellulosilyticum) Alkaliflexus imshenetskii (Ruminofilibacter) |
syntrophic associations | [29] |
| Hydrogenispora ethanolica Acetomicrobium hydrogeniformans | [29] | ||
| Methanosarcina flavescens | [29] |
| pH | Microbial communities favoured | Reference |
|---|---|---|
| Acidic | Proteobacteria, Aminicenantes, Actinobacteria, and Nitrospirae | [52] |
| Bacteroidetes | [77] | |
|
Clostridium IV, Clostridium sensu stricto, unclassified_Ruminococcaceae |
[77] | |
| Caproiciproducens | [78] | |
| Turicibacter | [78] | |
| Methanosarcina | [79] | |
| Neutral | Sphaerochaeta (Spirochaetes) | [79] |
| Basic | Petrimonas and Proteiniphilum | [24] |
| Tissierella and Tepidimicrobium | [76] | |
| Porphyromonadaceae | [79] | |
| Solibacillus silvestris | [79] | |
| Methanosarcina | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





