Submitted:
04 June 2023
Posted:
05 June 2023
You are already at the latest version
Abstract
Keywords:
Introduction
Method
2.1. Literature Review
2.2. Data and Data Source
2.3. Data Analysis
- Initialization: done randomly to determine the initial particle.
- Fitness: a measure in each particle in the population.
- Update: calculate the velocity of each particle with the equation below.
- 1)
- Construction with the equation below.
- 2)
- Termination: stop the process, if the termination criteria are met, and return to step 2 (fitness) if not met.
- 1)
- Determination of the variables to be tested.
- 2)
- Determining the number of sorters per variable according to the type of independent variable using the equation below.
- 3)
- Calculate the ‘Gini index’ value for each sorter according to the equation below.
- 4)
- Repeat step 2-3 to perform sorting until it is no longer possible.
- 5)
- Marking of terminal node class labels is based on the rules for the highest number of members using the equation below.
- 1)
- Insert testing result on the confusion matrix Table 1.
- 2)
- Calculate the accuracy value using the equation.
Result and Discussion
4.1. Data Mining
| No | Attribute | Variable Name |
|---|---|---|
| 1 | Diabetes Disease | DIABETE3 |
| 2 | High Blood Pressure | _RFHYPE5 |
| 3 | High Cholesterol | TOLDHI2 |
| 4 | Cholesterol Check | _CHOLCHK |
| 5 | BMI (Body Mass Index) | _BMI5 |
| 6 | Smoker | SMOKE100 |
| 7 | Physical Activity | _TOTINDA |
| 8 | Consume Fruit | _FRTLT1 |
| 9 | Consume Vegetables | _VEGLT1 |
| 10 | Alcohol Consumption | _RFDRHV5 |
| 11 | Stroke | CVDSTRK3 |
| 12 | Heart Disease | _MICHD |
| 13 | Health Care Coverage | HLTHPLN1 |
| 14 | Medical Checkup | MEDCOST |
| 15 | General Health | GENHLTH |
| 16 | Mental Health | MENTHLTH |
| 17 | Physical Health | PHYSHLTH |
| 18 | Difficult Walking Or Climbing Stairs | DIFFWALK |
| 19 | Sex | SEX |
| 20 | Age Category | _AGEG5YR |
| 21 | Highest Grade School | EDUCA |
| 22 | Income | INCOME2 |
4.2. Data Cleaning
4.3. Formatting Data
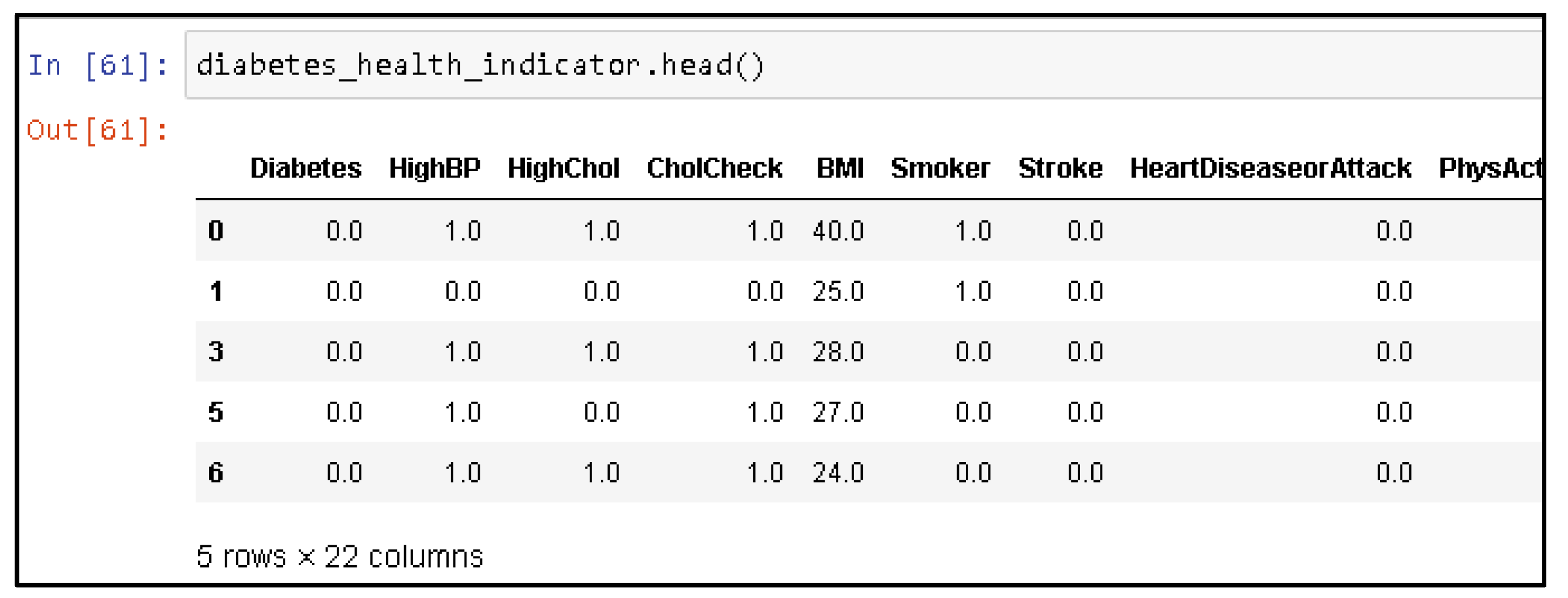
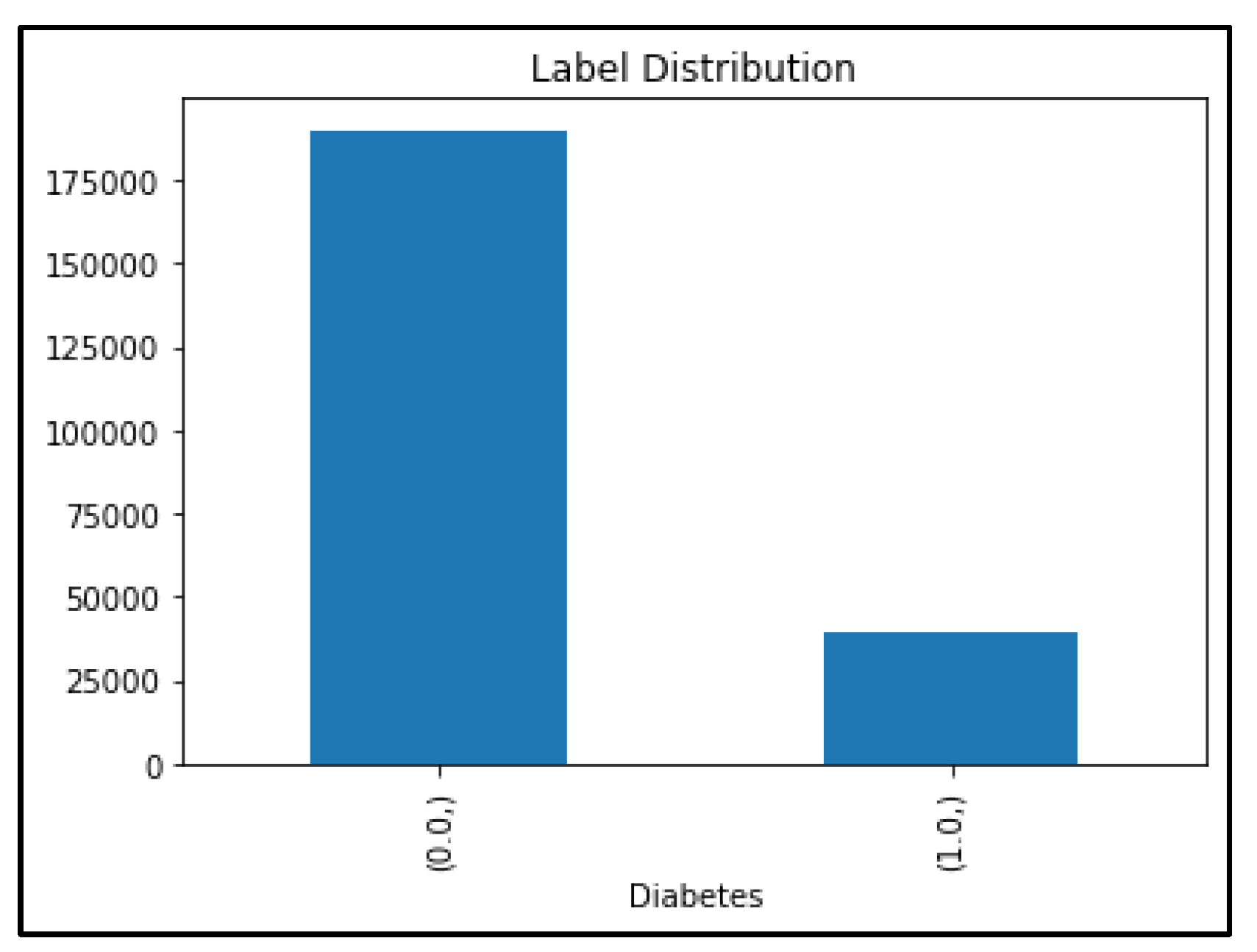
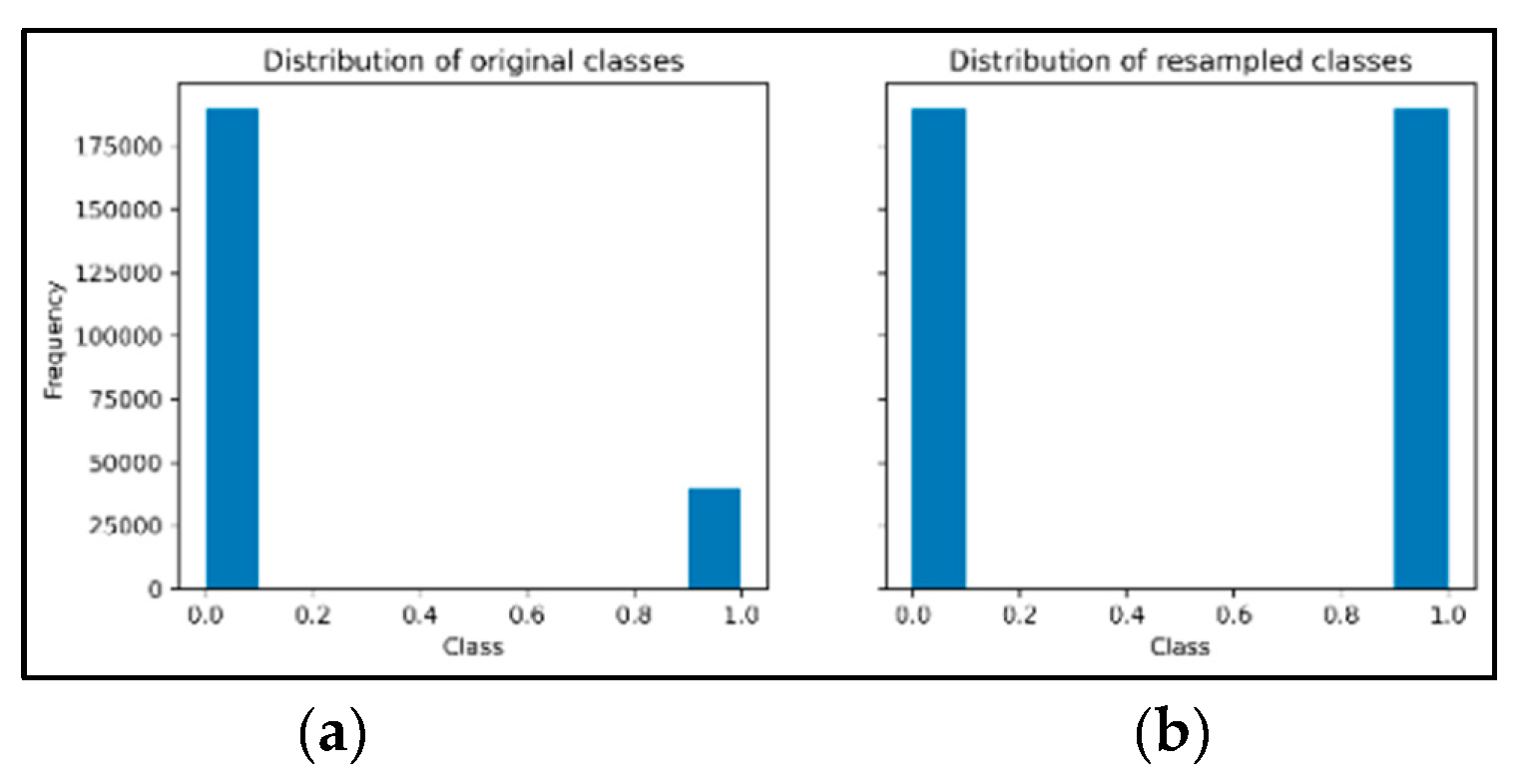
4.4. Oversampling Data Using SMOTE Technique
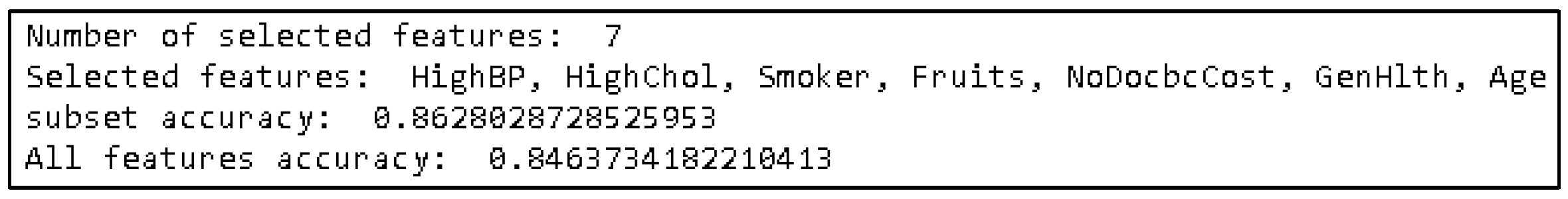
4.5. Feature Selection Algoritma PSO
4.6. Data Mining Result
- a.
- CART Classification Result
- The results of the first CART algorithm trial
| K-fold |
Accurary (%) |
Precission (%) |
Recall (%) |
F1-Score (%) |
APER (%) |
MAE |
|---|---|---|---|---|---|---|
| 1 | 75,54 | 31,39 | 34,12 | 32,69 | 24,46 | 0.2446 |
| 2 | 75,61 | 30,55 | 33,39 | 31,91 | 24,39 | 0.2439 |
| 3 | 75,43 | 30,31 | 33,70 | 31,91 | 24,57 | 0.2457 |
| 4 | 75,24 | 30,70 | 33,51 | 32,04 | 24,76 | 0.2476 |
| 5 | 75,10 | 30,72 | 34,01 | 32,28 | 24,90 | 0.2490 |
| 6 | 75,48 | 30,68 | 33,53 | 32,04 | 24,52 | 0.2452 |
| 7 | 74,70 | 30,78 | 33,72 | 32,19 | 25,30 | 0.2530 |
| 8 | 75,35 | 31,37 | 35,71 | 33,40 | 24,65 | 0.2465 |
| 9 | 75,34 | 29,32 | 32,31 | 30,74 | 24,66 | 0.2466 |
| 10 | 75,40 | 29,75 | 33,79 | 31,64 | 24,60 | 0.2460 |
| Median | 75,32 | 30,56 | 33,78 | 32,08 | 24,68 | 0.2468 |
- 2.
- The results of the second CART algorithms trial
| K-fold |
Accurary (%) |
Precission (%) |
Recall (%) |
F1-Score (%) |
APER (%) |
MAE |
|---|---|---|---|---|---|---|
| 1 | 75,58 | 31,54 | 34,34 | 32,88 | 24,42 | 0.2442 |
| 2 | 75,82 | 30,95 | 33,52 | 32,18 | 24,18 | 0.2418 |
| 3 | 75,32 | 30,04 | 33,42 | 31,64 | 24,68 | 0.2468 |
| 4 | 75,35 | 30,83 | 33,38 | 32,06 | 24,65 | 0.2465 |
| 5 | 75,26 | 31,10 | 34,38 | 32,66 | 24,74 | 0.2474 |
| 6 | 75,21 | 30,00 | 32,82 | 31,34 | 24,79 | 0.2479 |
| 7 | 74,71 | 30,70 | 33,46 | 32,02 | 25,29 | 0.2529 |
| 8 | 75,33 | 31,15 | 35,11 | 33,01 | 24,67 | 0.2467 |
| 9 | 75,38 | 29,60 | 32,90 | 31,16 | 24,62 | 0.2462 |
| 10 | 75,60 | 30,13 | 33,97 | 31,94 | 24,40 | 0.2440 |
| Median | 75,34 | 30,58 | 33,75 | 32,09 | 24,66 | 0.2466 |
- 3.
- The results of the third CART algorithm trial.
| K-fold |
Accurary (%) |
Precission (%) |
Recall (%) |
F1-Score (%) |
APER (%) |
MAE |
|---|---|---|---|---|---|---|
| 1 | 75,41 | 31,10 | 33,87 | 32,42 | 24,59 | 0.2459 |
| 2 | 75,58 | 30,73 | 34,00 | 32,28 | 24,42 | 0.2442 |
| 3 | 75,33 | 30,14 | 33,67 | 31,81 | 24,67 | 0.2467 |
| 4 | 75,22 | 30,49 | 33,01 | 31,70 | 24,78 | 0.2478 |
| 5 | 75,02 | 30,64 | 34,16 | 32,30 | 24,98 | 0.2498 |
| 6 | 75,50 | 30,81 | 33,78 | 32,23 | 24,50 | 0.2450 |
| 7 | 74,77 | 30,88 | 33,70 | 32,23 | 25,23 | 0.2523 |
| 8 | 75,63 | 31,83 | 35,76 | 33,68 | 24,37 | 0.2437 |
| 9 | 75,27 | 29,34 | 32,64 | 30,90 | 24,73 | 0.2473 |
| 10 | 75,40 | 29,82 | 34,00 | 31,77 | 24,60 | 0.2460 |
| Median | 75,33 | 30,58 | 33,79 | 32,10 | 24,67 | 0.2467 |
- b.
- SMOTE+PSO+CART Classification Results
- The results of the first CART+SMOTE+PSO algorithms.
| K-fold |
Accurary (%) |
Precission (%) |
Recall (%) |
F1-Score (%) |
APER (%) |
MAE |
|---|---|---|---|---|---|---|
| 1 | 86,09 | 92,33 | 78,73 | 84,99 | 13,91 | 0.1391 |
| 2 | 86,10 | 92,17 | 78,91 | 85,03 | 13,90 | 0.1390 |
| 3 | 86,37 | 93,14 | 78,74 | 85,34 | 13,63 | 0.1363 |
| 4 | 86,31 | 92,50 | 78,94 | 85,18 | 13,69 | 0.1369 |
| 5 | 86,09 | 91,98 | 79,00 | 85,00 | 13,91 | 0.1391 |
| 6 | 86,32 | 92,74 | 78,93 | 85,28 | 13,68 | 0.1368 |
| 7 | 86,26 | 92,66 | 78,88 | 85,22 | 13,74 | 0.1374 |
| 8 | 86,50 | 92,99 | 78,99 | 85,42 | 13,50 | 0.1350 |
| 9 | 86,17 | 92,75 | 78,52 | 85,05 | 13,83 | 0.1383 |
| 10 | 86,59 | 92,17 | 79,56 | 85,40 | 13,40 | 0.1341 |
| Median | 86,28 | 92,54 | 78,92 | 85,19 | 13,72 | 0.1372 |
- 2.
- The results of the second CART+SMOTE+PSO algorithms.
| K-fold |
Accurary (%) |
Precission (%) |
Recall (%) |
F1-Score (%) |
APER (%) |
MAE |
|---|---|---|---|---|---|---|
| 1 | 86,09 | 92,33 | 78,73 | 84,99 | 13,91 | 0.1391 |
| 2 | 86,10 | 92,17 | 78,91 | 85,03 | 13,90 | 0.1390 |
| 3 | 86,37 | 93,14 | 78,74 | 85,34 | 13,63 | 0.1363 |
| 4 | 86,31 | 92,50 | 78,94 | 85,18 | 13,69 | 0.1369 |
| 5 | 86,09 | 91,98 | 79,00 | 85,00 | 13,91 | 0.1391 |
| 6 | 86,32 | 92,74 | 78,93 | 85,28 | 13,68 | 0.1368 |
| 7 | 86,26 | 92,66 | 78,88 | 85,22 | 13,73 | 0.1373 |
| 8 | 86,50 | 92,99 | 78,99 | 85,42 | 13,50 | 0.1350 |
| 9 | 86,17 | 92,75 | 78,52 | 85,05 | 13,83 | 0.1383 |
| 10 | 86,59 | 92,17 | 79,56 | 85,40 | 13,41 | 0.1341 |
| Median | 86,28 | 92,54 | 78,92 | 85,19 | 13,72 | 0.1372 |
- 3.
- The results of the third CART+SMOTE+PSO algorithms.
| K-fold |
Accurary (%) |
Precission (%) |
Recall (%) |
F1-Score (%) |
APER (%) |
MAE |
|---|---|---|---|---|---|---|
| 1 | 86,09 | 92,33 | 78,73 | 84,99 | 13,91 | 0.1391 |
| 2 | 86,10 | 92,17 | 78,91 | 85,03 | 13,90 | 0.1390 |
| 3 | 86,37 | 93,14 | 78,74 | 85,34 | 13,63 | 0.1363 |
| 4 | 86,31 | 92,50 | 78,94 | 85,18 | 13,69 | 0.1369 |
| 5 | 86,09 | 91,98 | 79,00 | 85,00 | 13,91 | 0.1391 |
| 6 | 86,32 | 92,74 | 78,93 | 85,28 | 13,68 | 0.1368 |
| 7 | 86,26 | 92,66 | 78,88 | 85,22 | 13,73 | 0.1373 |
| 8 | 86,50 | 92,99 | 78,99 | 85,42 | 13,50 | 0.1350 |
| 9 | 86,17 | 92,75 | 78,52 | 85,05 | 13,83 | 0.1383 |
| 10 | 86,59 | 92,17 | 79,56 | 85,40 | 13,41 | 0.1341 |
| Median | 86,28 | 92,54 | 78,92 | 85,19 | 13,72 | 0.1372 |
- 1.
- Oversampling Data with SMOTE technique for Diabetes Health Indicators Dataset
- 2.
- Feature Selection on the Diabetes Health Indicators Dataset attribute using the PSO algorithm
- 3.
- Accuracy of the CART model Decision Tree algorithm using SMOTE and PSO
Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- IDF, IDF Diabetes Atlas 2021 – 10th edition, 10th ed., vol. 10. 2021. [CrossRef]
- W. C. Hsu, M. R. G. Araneta, A. M. Kanaya, J. L. Chiang, and W. Fujimoto, “BMI cut points to identify at-Risk asian americans for type 2 diabetes screening,” Diabetes Care, vol. 38, no. 1, pp. 150–158, 2015. [CrossRef]
- Asidik, Kusrini, and Henderi, “Decision Support System Model of Teacher Recruitment Using Algorithm C4 . 5 and Fuzzy Tahani Decision Support System Model of Teacher Recruitment Using Algorithm C4 . 5 and Fuzzy Tahani,” J. Phys. Conf. Ser. Pap., 2018. [CrossRef]
- I. H. Witten, E. Frank, and M. A. Hall, Data Mining Practical Machine Learning Tools and Techniques, 3rd ed. United atates, 2011. [CrossRef]
- P. Shella, “Sistem Pendukung Keputusan Dengan Menggunakan Decission Tree Dalam Pemberian Beasiswa Di Sekolah Menengah Pertama (Studi Kasus di SMPN 2 Rembang),” Universitas Negeri Semarang, 2015.
- T. Setiyorini and R. T. Asmono, “Komparasi Metode Decision Tree, Naive Bayes Dan K-Nearest Neighbor Pada Klasifikasi Kinerja Siswa,” J. Techno Nusa Mandiri, vol. 15, no. 2, pp. 85–92, 2018. [CrossRef]
- J. Han and M. Kamber, Data Mining: Concepts and Techniques : Concepts and Techniques. San Frasisco, 2012.
- B. Xue, M. Zhang, and W. N. Browne, “Particle swarm optimisation for feature selection in classification: Novel initialisation and updating mechanisms,” Appl. Soft Comput. J., vol. 18, pp. 261–276, 2014. [CrossRef]
- A. Puri and M. K. Gupta, “Comparative Analysis of Resampling Techniques under Noisy Imbalanced Datasets,” IEEE Int. Conf. Issues Challenges Intell. Comput. Tech. ICICT 2019, 2019. [CrossRef]
- N. V Chawla, K. W. Bowyer, L. O. Hall, and W. P. Kegelmeyer, “SMOTE: Synthetic Minority Over-sampling Technique,” J. Artif. Intell. Res., vol. 16, no. 2, pp. 321–357, 2002. [CrossRef]
- B. Krawczyk, “Learning from imbalanced data: open challenges and future directions,” Prog. Artif. Intell., vol. 5, no. 4, pp. 221–232, 2016. [CrossRef]
- A. N. Noercholis, “Comparative Analysis of 5 Algorithm Based Particle Swarm Optimization (Pso) for Prediction of Graduate Time Graduation,” Matics, vol. 12, no. 1, p. 1, 2020. [CrossRef]
- K. Ramandana and I. Carolina, “Seleksi Fitur Algoritma Neural Network Menggunakan Particle Swarm Optimization Untuk Memprediksi Kelahiran Prematur,” Kilat, vol. 6, no. 2, pp. 106–111, 2017. [CrossRef]
- V. F. Rodriguez-Galiano, J. A. Luque-Espinar, M. Chica-Olmo, and M. P. Mendes, “Feature selection approaches for predictive modelling of groundwater nitrate pollution: An evaluation of filters, embedded and wrapper methods,” Sci. Total Environ., vol. 624, pp. 661–672, 2018. [CrossRef]
- T. Dudkina, I. Meniailov, K. Bazilevych, S. Krivtsov, and A. Tkachenko, “Classification and prediction of diabetes disease using decision tree method,” CEUR Workshop Proc., vol. 2824, pp. 163–172, 2021.
- Z. Xie, O. Nikolayeva, J. Luo, and D. Li, “Building risk prediction models for type 2 diabetes using machine learning techniques,” Prev. Chronic Dis., vol. 16, no. 9, pp. 1–9, 2019. [CrossRef]
- V. Chang, M. A. Ganatra, K. Hall, L. Golightly, and Q. A. Xu, “An assessment of machine learning models and algorithms for early prediction and diagnosis of diabetes using health indicators,” Healthc. Anal., vol. 2, no. September, p. 100118, 2022. [CrossRef]
- N. Maulidah et al., “Seleksi Fitur Klasifikasi Penyakit Diabetes Menggunakan Particle Swarm Optimization (PSO) Pada Algoritma Naive Bayes,” Drh. Khusus Ibuk. Jakarta, vol. 13, no. 2, p. 21231170, 2020, [Online]. Available: http://journal.stekom.ac.id/index.php/elkom■page40.
- A. N. Rachman, S. Supratman, and E. N. F. Dewi, “Decision Tree and K-Nearest Neighbor (K-NN) Algorithm Based on Particle Swarm Optimization (PSO) for Diabetes Mellitus Prediction Accuracy Analysis,” CESS (Journal Comput. Eng. Syst. Sci., vol. 7, no. 2, p. 315, 2022. [CrossRef]
- I. Muqiit WS and R. Nooraeni, “Penerapan Metode Resampling Dalam Mengatasi Imbalanced Data Pada Determinan Kasus Diare Pada Balita Di Indonesia (Analisis Data Sdki 2017),” J. MSA ( Mat. dan Stat. serta Apl. ), vol. 8, no. 1, p. 19, 2020. [CrossRef]
- M. A. Muslim et al., Data Mining algoritma c 4.5 Disertai contoh kasus dan penerapannya dengan program computer. 2019.
- A. Subasi, Practical Machine Learning for Data Analysis Using Python. 2020. [CrossRef]
- M. Qois Syafi, “Increasing Accuracy of Heart Disease Classification on C4.5 Algorithm Based on Information Gain Ratio and Particle Swarm Optimization Using Adaboost Ensemble,” J. Adv. Inf. Syst. Technol., vol. 4, no. 1, pp. 100–112, 2022, [Online]. Available: https://journal.unnes.ac.id/sju/index.php/jaist.
| Two Class Classification | Actual Class | |||
| 0 | 1 | |||
| Predicted Class | 0 | TN | FP | |
| 1 | FN | TP | ||
| No | Attribute | Empty Data Amount |
|---|---|---|
| 1 | DIABETE3 | 7 |
| 2 | TOLDHI2 | 59154 |
| 3 | _BMI5 | 36398 |
| 4 | SMOKE100 | 14255 |
| 5 | “_MICHD | 3942 |
| 6 | DIFFWALK | 12334 |
| 7 | INCOME2 | 3301 |
| Variable Name | Formatting |
| DIABETE3 | 0 = Have no diabetes symptom 1 = Have diabetes symptoms type 1 or 2 |
| _RFHYPE5 | 0 = Have no blood pressure 1 = Have blood pressure |
| TOLDHI2 | 0 = Have no high cholesterol 1 = Have high cholesterol |
| c | 0 = Have no cholesterol check history within 5 years. 1 = Have cholesterol check history within 5 years. |
| _BMI5 | Integer value BMI |
| SMOKE100 | 0 = Does not smoke at least 100 cigarettes before. 1 = Does smoke at least 100 cigarettes before. |
| _TOTINDA | 0 = Have not done any form of workout within 30 days, besides work. 1 = Have done any form of workout within 30 days, besides work. |
| _FRTLT1 | 0 = Does not consume at least 1 kind of fruit a day. 1 = Consume at least 1 kind of fruit a day. |
| _VEGLT1 | 0 = Does not consume at least 1 kind of vegetable a day. 1 = Consume at least 1 kind of vegetable a day. |
| _RFDRHV5 | 0 = Does not consume over 14 liquor for adult male or 7 for female. 1 = Does consume over 14 liquor for adult male or 7 for female. |
| CVDSTRK3 | 0 = Have no history of having a stroke. 1 = Have history of having a stroke. |
| _MICHD | 0 = Have no history of having a myocardial infarction (MI). 1 = Have history of having a myocardial infarction (MI). |
| HLTHPLN1 | 0 = Does not have health care coverage. 1= Does have health care coverage. |
| MEDCOST | 0 = Does not have a medical chekup within the last 12 months cause of cost. 1 = Does have a medical chekup within the last 12 months. |
| GENHLTH | 0 = Does not state health condition in general. 1 = Excellent 2 = Very good 3 = Good 4 = Pretty good 5 = Bad |
| MENTHLTH | 0 = Have no stress, depretion, and or emotional problem within the last 30 days. 1-30 = Amount of day (s) having stress, depretion, and or emotional problem within the last 30 days. |
| PHYSHLTH | 0 = Have no physical injury within the last 30 days. 1-30 = Amount of days having physical injury within the last 30 days. |
| DIFFWALK | 0 = Have no problem in walking or climbing up stairs. 1 = Have problems in walking or climbing up stairs. |
| SEX | 0 = Female respondent 1 = Male respondent |
| _AGEG5YR | Respondent age category 1 = 18-24 y.o 2 = 25-29 y.o 3 = 30-34 y.o 4 = 35-39 y.o 5 = 40-44 y.o 6 = 45-49 y.o 7 = 50-54 y.o 8 = 55-59 y.o 9 = 60-64 y.o 10 = 65-69 y.o 11 = 70-74 y.o 12 = 75-79 y.o 13 = 80 y.o or above |
| EDUCA | Respondent last educational level category 1 = Have not or never kindergarten 2 = Pass between elementary school 3 = Pass between highschool 4 = Pass highschool 5 = Pass between college or technical school 6 = Pass college |
| INCOME2 | Respondent income categories for a year in dollars 1 = Lower than USD 10.000 2 = USD 10.000 – USD 15.000 3 = USD 15.000 – USD 20.000 4 = USD 20.000 – USD 25.000 5 = USD 25.000 – USD 35.000 6 = USD 35.000 – USD 50.000 7 = USD 50.000 – USD 75.000 8 = Over USD 75.000 |
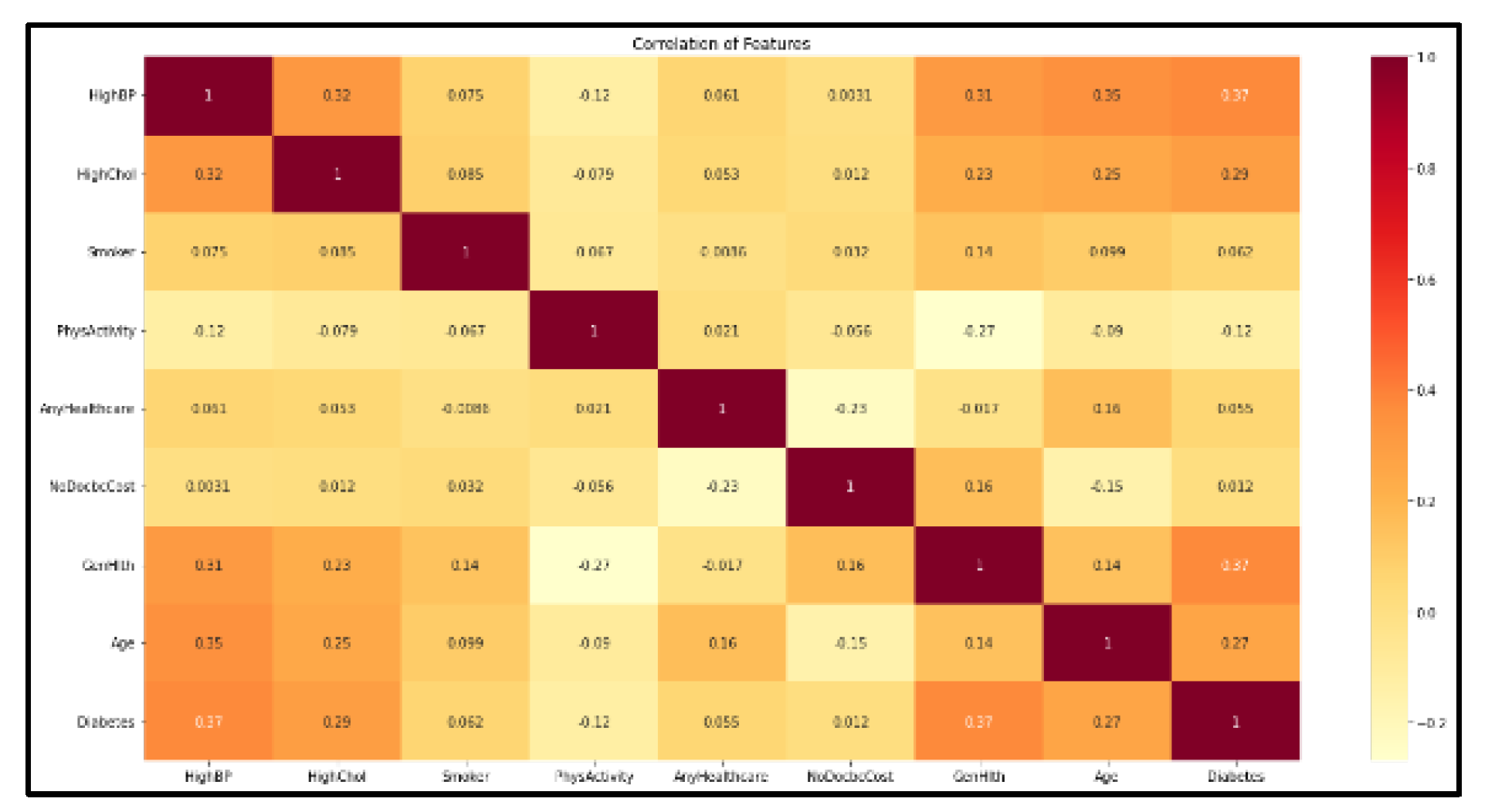
| Attribute Name | Correlation Value |
|---|---|
| HighBP | 0.37 |
| HighChol | 0.29 |
| Smoker | 0.062 |
| Fruits | 0.12 |
| NoDocbcCost | 0.012 |
| GenHlth | 0.37 |
| Age | 0.27 |
| Trial |
Accurary (%) |
Precission (%) |
Recall (%) |
F1-Score (%) |
APER (%) |
MAE |
|---|---|---|---|---|---|---|
| 1 | 75,32 | 30,56 | 33,78 | 32,08 | 24,68 | 0.2468 |
| 2 | 75,34 | 30,58 | 33,75 | 32,09 | 24,66 | 0.2466 |
| 3 | 75,33 | 30,58 | 33,79 | 32,10 | 24,67 | 0.2467 |
| Trial |
Accurary (%) |
Precission (%) |
Recall (%) |
F1-Score (%) |
APER (%) |
MAE |
|---|---|---|---|---|---|---|
| 1 | 86,28 | 92,54 | 78,92 | 85,19 | 13,72 | 0.1372 |
| 2 | 86,28 | 92,54 | 78,92 | 85,19 | 13,72 | 0.1372 |
| 3 | 86,28 | 92,54 | 78,92 | 85,19 | 13,72 | 0.1372 |
| Algorithm | Accuracy Result |
|---|---|
| CART | 75,34% |
| CART + SMOTE + PSO | 86,28% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




