1. Introduction
In recent decades, there has been a strong demand to gradually replace intensive agriculture with sustainable cultivation practices, which are based on the use of organic amendments able to preserve the integrity of the soil and the healthiness of crops [
1,
2,
3]. Plants need not only light and water for their metabolism, but also a complete mineral nutrition whose availability is governed by soil properties and efficiency of water uptake by roots. Currently the functionality and productivity of the soil are seriously compromised by the intensive use for crops and by the degradation of fertile soils [
4]; the use of biofertilizers in agriculture help plants to enhance absorption of nutrient elements from soil and improve growth [
5]. Production of organic fertilizers from the urban organic waste is expected to increase and the challenge of the moment is to tackle these problems together by transforming food waste into a sustainable resource to be used to enrich the soils and enhance their nutritional quality and regeneration. Among the waste sources, those of the food residues and the municipal biosolids seems to be the most promising in the urban sustainable recycling process [
6]. Unfortunately, current recycling processes take months to degrade organic wastes, thus representing the main limiting factor for a large-scale application. Alternative processes, based on waste drying or high-temperature aerobic fermentation, were advanced to overcome this limit [
7,
8]. Nowadays, the classical Fenton reaction [
9] is considered as one of a set of advanced oxidation processes (AOPs) and is widely used for the chemical treatment of wastewater, industrial sludge, landfill leachate, soils, and sediments, which are contaminated with biorefractory organic compounds such as phenols, dyes, pesticides, organic solvents, pharmaceuticals, domestic chemicals [
10]. Specifically, fast degradation of urban solid waste by using the Fenton reaction has been already obtained [
11]; authors reported that, after few hours incubation, Fe (II) ions catalytic oxidation pathway was able to degrade dry organic matrices in C/N ratio lower than 12 with a high degree of oxidative decomposition into low-molecular-weight compounds at high oxidation state.
The aim of this research was to test the effects of the soil amendment formulation reported as Fast-Composted soil Amendment (FCA), obtained by urban organic solid waste fractions following the Fenton composting reaction on the growth of Lactuca sativa L. seedlings. Lettuce is one of the most intensively cultivated vegetables in Mediterranean countries. Millions of tons of lettuce for fresh consumption are produced annually in the EU, mainly in Mediterranean countries (FAO, 2022). Intensive lettuce cultivation practices are large consumers of fertilizers, thus entailing significant costs in terms of energy and the environment. Providing alternatives that improve the sustainability of these agricultural ecosystems without reducing productivity is therefore of great interest. Under our experimental conditions L. sativa was grown in a short-day period to prevent flowering and thus evaluating plant growth in its vegetative phase. Morphological, biochemical, and molecular analyses have been performed to elucidate the metabolic behavior of leaves and roots of plants grown on soil enriched with the FCA.
3. Discussion
Application of organic fertilizers can significantly improve the growth, yield, and quality of lettuce without negatively impacting the environment and human health [
12]. Authors reported that, when composted urban waste was used, there were inhibitory or growth-inducing effects of lettuce seedlings, possibly due to a combination of the high electrical conductivity, ammonia toxicity and degree of stabilization of this compost [
13,
14]. Further study on the effects of types of organic amendments on lettuce seedlings reported an enhanced plant biomass, a higher content of Rubisco large subunit and soluble proteins; on the contrary, an antagonistic effect was observed on the chlorophyll content [
12].
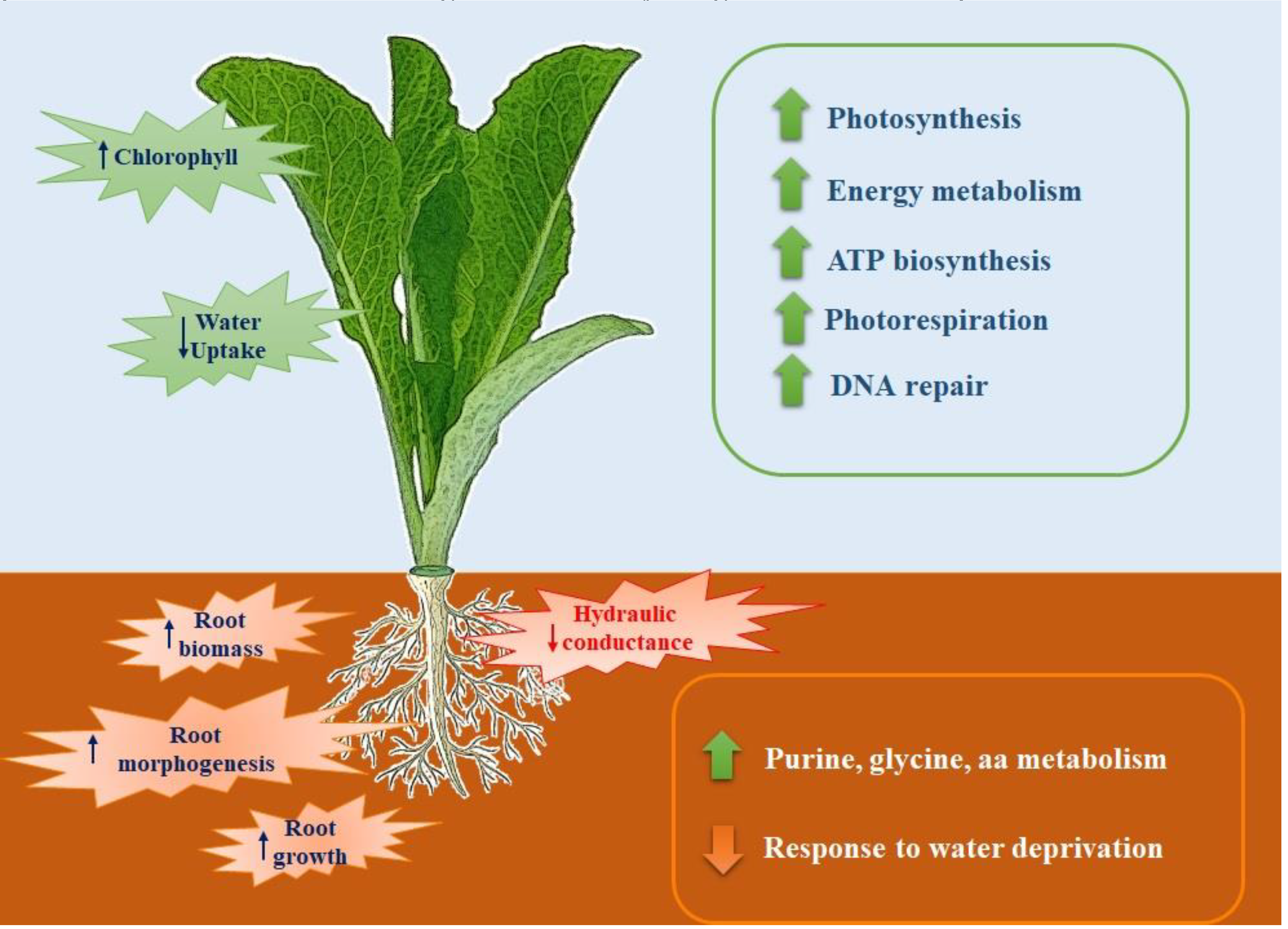
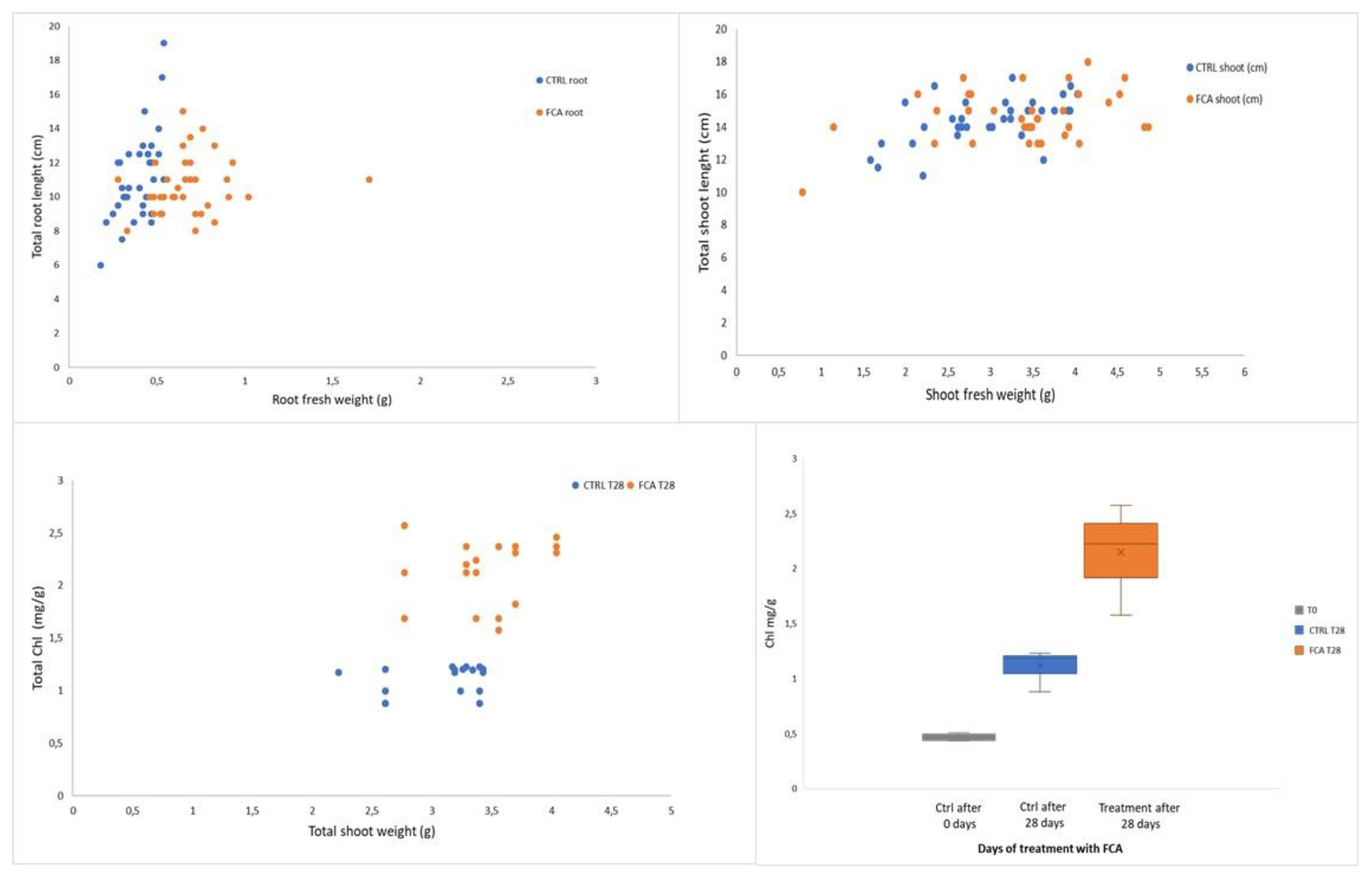
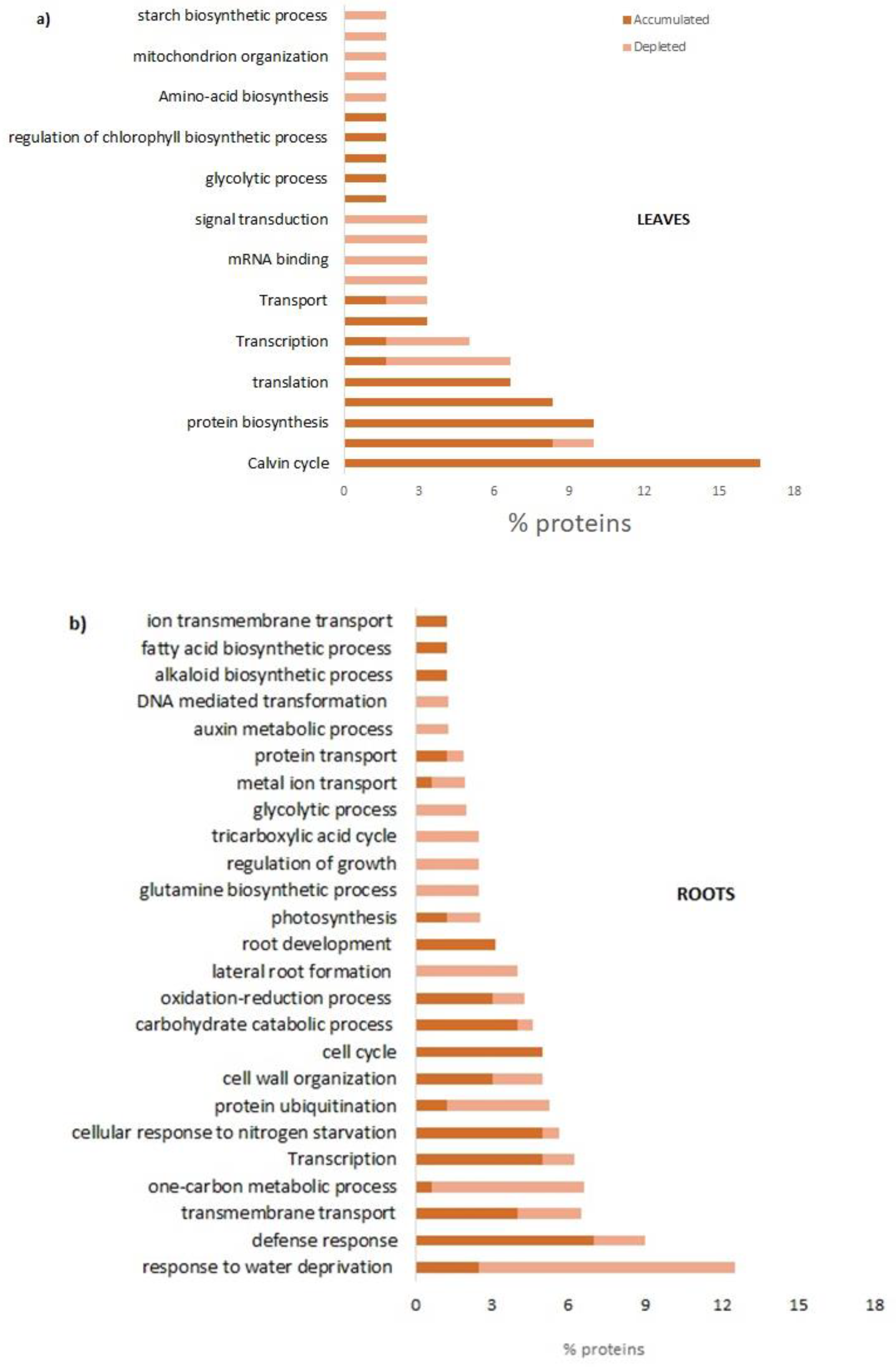
Fast-Composted soil Amendment (FCA) produced by the urban organic solid waste fractions stabilized by the Fenton’s reaction procedure, after 8 hours of treatment, resulted in a good degree of oxidative decomposition with C/N ratios as 22, compared to an initial value of 26. In addition, the FCA exhibited dark-brown color, odorless and had humidity of 15%. The product cannot be considered as a conventional compost, but it claimed to be a stabilized organic matrix. In this case the material does not go, at least within 6 months, in further biochemical transformation when stored under dry conditions. Property of the material as a soil fertilizer was tested on Lactuca sativa seedlings that underwent modification of the biochemical and molecular behavior; the FCA treatment, in fact, positively affected root growth and photosynthesis. After 28 days treatment of L. sativa seedlings root biomass and chlorophyll content significantly increased in respect to the control seedlings. The chlorophyll physiological adjustments induced by the FCA treatment appeared to be linked to an increase in photosynthesis, as the proteins related to this metabolism accumulated in shoots. Under our conditions, however, the shoot biomass was not affected by the treatment, indicating that the amendment exerted contrasting effects on shoot metabolism.
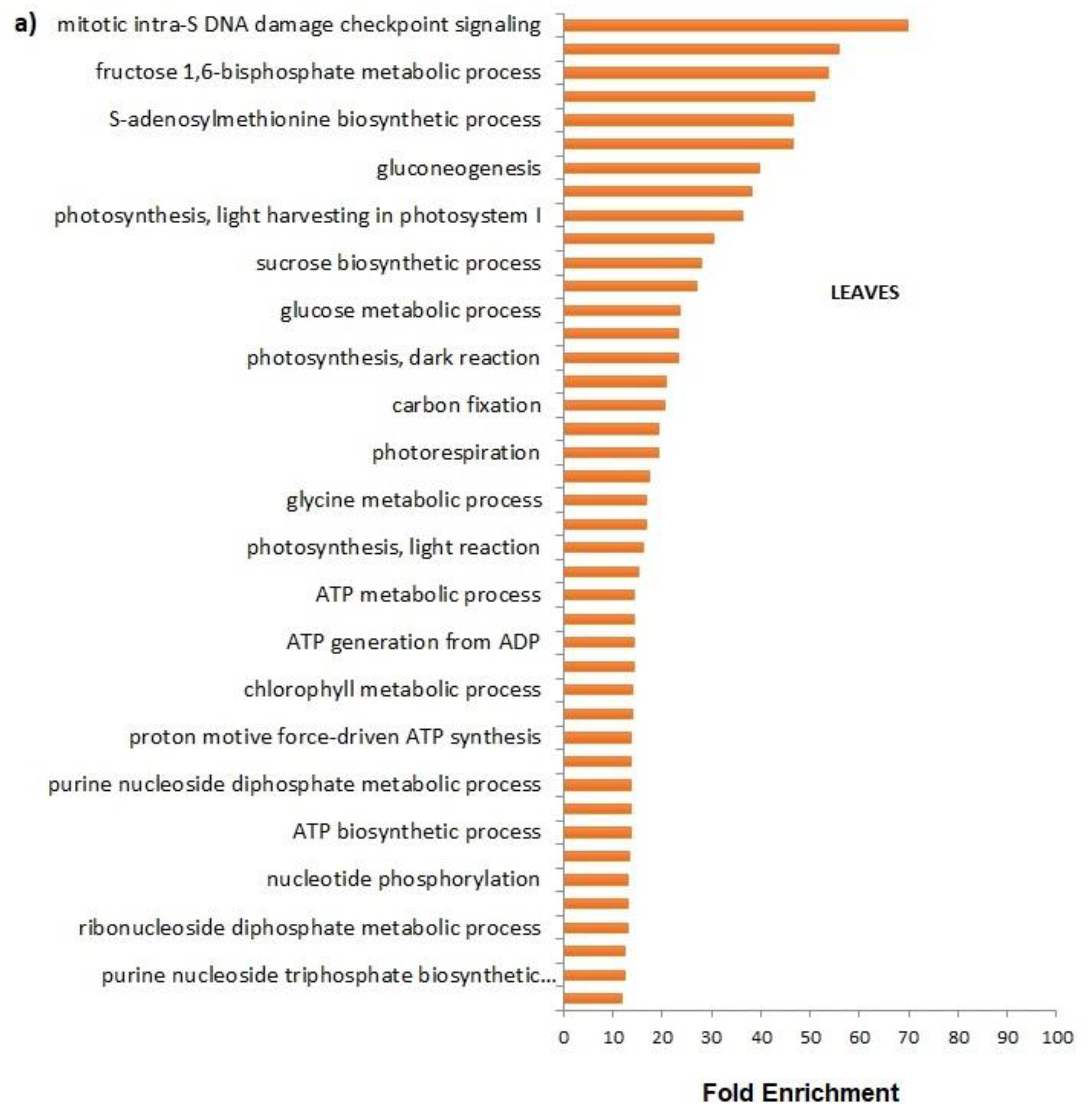
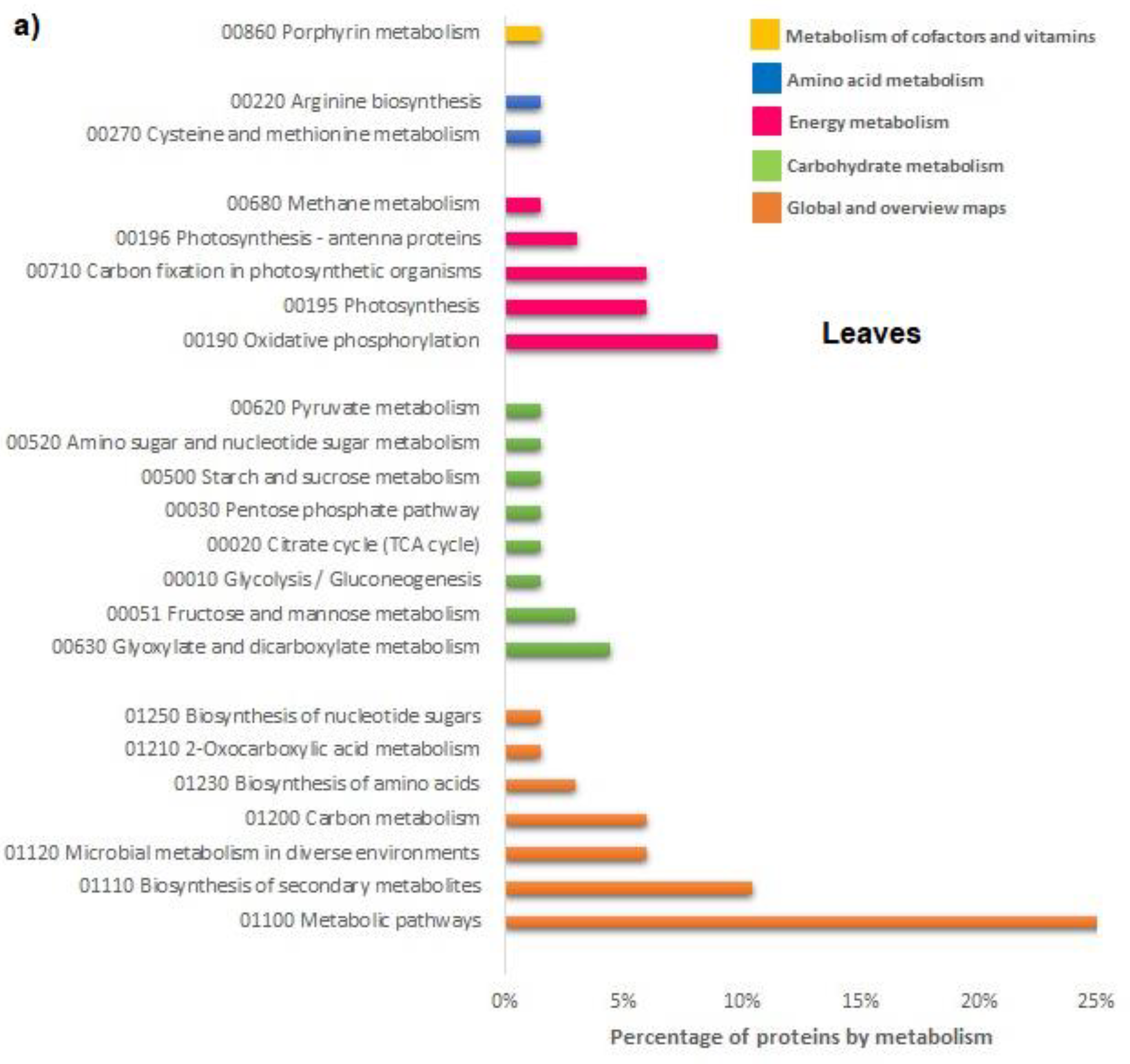
The FCA treatment promoted also the primary and secondary metabolisms in leaves; the mitotic intra-S DNA damage checkpoint signaling process has been enriched seventy times in respect to untreated leaves; during the S-phase of the cell cycle, in the event of problems during the replication process, DNA integrity checkpoints are activated slowing down the cell cycle to grant the cell time to repair the damage [
15]; evidences suggested also that this well-known mechanism has a role as growth regulator process in plants [
16]. On this view, the FCA imposed a stress condition that, however, the leaf cells coped with by activating the mechanisms that slow down the mitosis to repair the DNA, and this might lead to a slowdown in tissue growth.
Amendment also induced the enrichment of the S-adenosylmethionine (SAM) cycle; SAM is a key enzyme involved in many important biological processes, such as ethylene and polyamine biosynthesis, transmethylation, and transulfuration; SAMs genes showed differential expression in response to abiotic stresses and exogenous hormone treatments [
17]. Polyamines, for example, are important hormones that regulate cell growth during stress responses, pollen and flower development and protection of photosystem II [
18]. The cell demand for SAM compounds, of course, may change very markedly under different growth conditions with a metabolic cost of ATP consumption. In this regard, the metabolic processes of ATP production, mainly through photophosphorylation, have been enriched to cope with the high energy demand under treatment with FCA; the energy metabolism that uses carbohydrates was induced through the processes of photosynthetic gluconeogenesis, the biosynthesis of sucrose and glycolysis. To complete the effects on the metabolic pattern induced by FCA treatment, upregulation of processes involving the biosynthesis and metabolisms of the glycine have been found. It is well known that endogenous glycine accumulation mediates abiotic stress tolerance in plants involving the osmotic regulation [
19]. In response to FCA treatment, glycine should be synthesized in excess to adjust the osmotic stress-induced by stress, to maintain the sub-cellular structures and reduce the oxidative damages. In addition, catabolic process of amino acids and dicarboxylic acids, could be related to the high energy demand of treated leaves; catabolic pathways for some amino acids as alanine or glutamine are very short, it can be directly converted to pyruvate by alanine aminotransferases and to glutamate respectively contributing substantially to the energy state of plant cells under certain physiological conditions [
20].
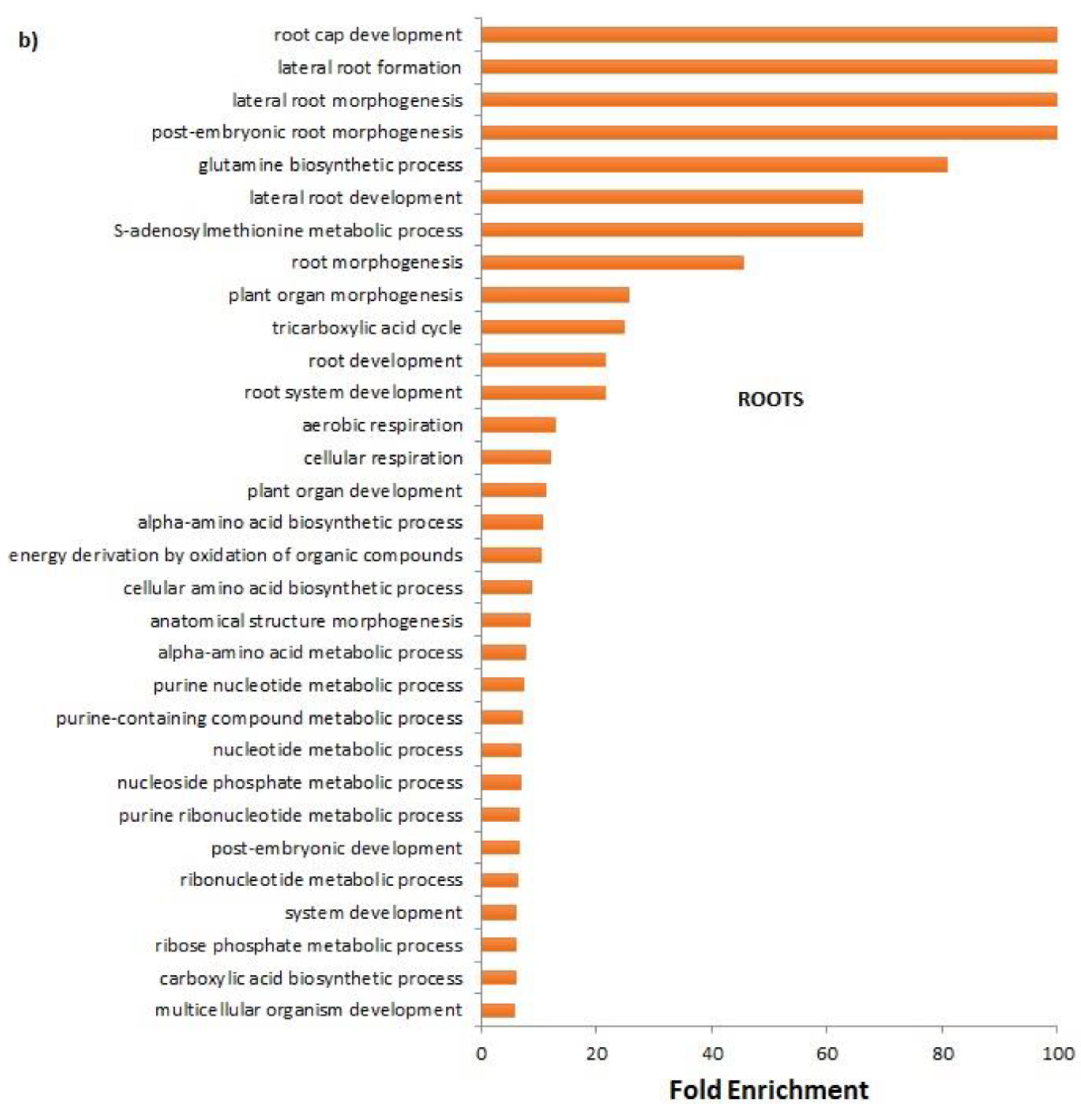
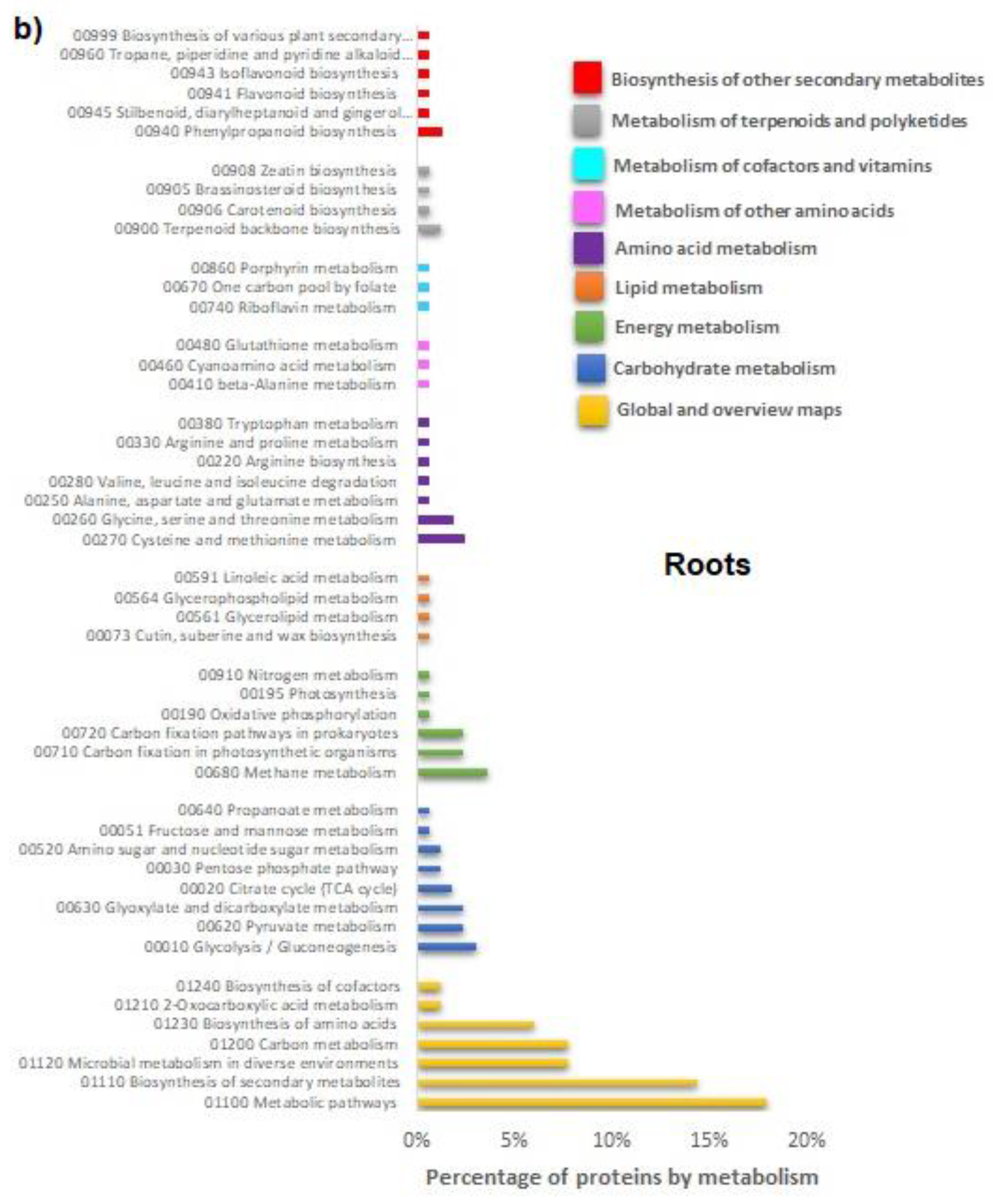
In roots the FCA strongly enriched the organs morphogenesis and developments; root cap development, lateral root formation, post-embryonic root morphogenesis were the main biological processes enriched by the treatment. It is well known that morphogenetic processes are the basis of new organ formation; lateral roots morphogenesis is a decisive process during root system formation [
21]. Regarding the root cap, it is the terminal tissue of the root of most plants. Historical evidence has shown that the root cap has not only the role of protecting proximal root meristem, but also to direct root growth in response to stimuli such gravity, light, gradients in temperature, humidity, ions, and other chemicals [
22]. Responses to water deprivation were strongly inhibited by the FCA treatment. The highly conserved plant aquaporins, known as Plasma membrane Intrinsic Proteins (PIPs), are the main gateways for cell membrane water exchange; in Arabidopsis, inhibition of aquaporin expression in roots induced an increase in root growth [
23]. Authors reported that, if plants lack aquaporins, thus having an increased resistance for the water movement in leaf and root cells, they compensate for this effect by increasing their root surface; this finding completes the evidence that FCA strongly altered the plant water transport and root growth dynamic. Additionally, biosynthetic processes of amino acids, the biosynthesis of ribonucleotides, and the biosynthetic process of carboxylic acids have also been enriched. Morphological, biochemical, and molecular findings were, then, completely consistent with a significant root growth promotion of lettuce seedlings cultivated on FCA-amended soil. FCA induced, in fact, the enrichment of the primary metabolism of purine, amino acids, and carbohydrates. Biological processes of the S-adenosylmethionine (SAM) cycle were strongly enriched. SAM, as reported above, is the main methyl group donor useful for methylation of DNA, RNA, protein, lignin, flavonoid and it also plays important roles in regulating plant development, in abiotic or biotic stress [
17] and heavy metal tolerance [
24].
4. Materials and Methods
4.1. Fast-Composted soil Amendment (FCA) production
The Fast-Composted soil Amendment (FCA) was produced starting from samples of municipal solid waste. Waste was dried under vacuum and finely ground; 200 gr of dried powder were placed in the glass reactor, Fe
2+ (as catalyst) at concentration of 0.01% FeSO
4 (VEBI Istituto Biochimico s.r.l., Italy) and 0.6 x 10-3 H2O2 (as oxidant) (Panreac Applichem, Spain) were added. The Fenton’s reaction was performed at pH 3.0, 60°C, under a pressure of 0.96 bar for 8 hours, during which the processes of chemical stabilization of the biomass took place. Further details of the reaction are reported in Roccotelli [
25]. For the elemental analysis of FCA, the stabilized biomasses were subjected to high-temperature combustion (T ≥ 900◦C) in a bomb containing oxygen under pressure and following the method described in Nelms [
26]; residues were analyzed by ICP/MS iCapQ (Thermofischer, Waltham, Massachusetts, USA) to evaluate the concentrations of C, H and N in terms of their relative percentages. Results of the elemental analysis are reported in the
Supplementary Table S3.
4.2. Plant acclimation, treatments, and growth
Seedlings of
Latuca sativa L. var. longifolia (n=70) with four leaves each, were transplanted in a soft and porous commercial soil, (Humifly, Humiflora, Italy) mixed with pine bark, light peat, and volcanic lapilli in a proportion of 2:1 (v/v). Seedlings of
L. sativa were acclimated under controlled temperature and short-day 10/14 h light/dark cycle. After seven days each seedling was weighed and measured in length, then divided into three sub-cultivations of 35 seedlings each, on soils whose compositions were as follows: i) soil without amendment (control), ii) soil with 2% FCA [
27]. After 30 days cultivation, all seedlings were collected; for each sub-cultivation, whole plant weight, weight of roots and leaves, number of total leaves for each seedling and number of healthy leaves were measured. Leaves and roots were, then, frozen in liquid nitrogen and stored at -80 °C to be used for the physiological and molecular analyses.
4.3. Chlorophyll extraction and measurement
1.0 g of frozen leaf tissue was ground in liquid nitrogen in a mortar to obtain a fine powder; 5 ml of 80% cold acetone was added to the tissue powder and incubated at 4 °C for 3 hours under weak shaking; then the samples were centrifuged at 800 g for 15 min. 1 ml of crude supernatant was transferred in a cuvette and the absorbance was measured at 663 nm and 645 nm with the 7310 Jenway spectrophotometer. The concentrations of chlorophylls a and b and of the total chlorophyll were determined by the following equations [
28]:
Chlorophylls content as mg/g fresh tissue were measured in six biological replicates.
4.4. Protein extraction and purification
4.4.1. Protein extraction and purification from leaves
Proteins from leaves of three biological replicates of control and FCA treatments, were extracted by the multistep procedures [
29]. For each extraction 1.4 g of tissues were crushed in a mortar in liquid nitrogen until a fine powder was obtained. Ground plant tissue was homogenized with a volume of 10% TCA in acetone and centrifuged at 14000 g for 5 min; a volume of 10% TCA in water was added and centrifuged at 14000 g for 5 min. Subsequently, four washes were performed using 80% acetone in water. After centrifugation, the pellet containing the precipitated proteins was dried at room temperature. Approximately 100 mg of powdered tissue was dissolved in 0.8 ml of phenol (buffered with Tris HCL, pH8.0, Sigma, St.Louis, MO, USA) and 0.8 ml of SDS buffer (30% sucrose, 2% SDS, 0.1M Tris-HCl, pH8.0, 5%2-mercaptoethanol) in a 2 ml microfuge tube. The samples were vortexed for 30 s and centrifuged at 14000 g for 5 min to allow proteins to solubilize in the phenol phase. The phenol phase was mixed with five volumes of 0.1 M ammonium acetate in cold methanol, and the mixture was stored at −20°C for 30 min to precipitate proteins. Proteins were collected by centrifugation at 14000 g for 5 min. Two washes were performed with 0.1M ammonium acetate in cold methanol, and two with cold 80% acetone, and centrifuged at 14000 g for 7 min. The final pellet containing purified protein was dried and dissolved in the Laemmli 1DE separation buffer overnight [
30]. Proteins were then quantified by measuring the absorbance at 595 nm according to the Bradford assay [
31]. Protein yield was calculated as mg of protein for g fresh tissue weight of each biological replicate.
4.4.2. Protein extraction and purification from roots
Proteins from roots of three biological replicates of control and FCA treatments, were extracted by 1 gram of root tissue, weighed and pulverized in liquid nitrogen. 1 mL of extraction buffer (0.7 M sucrose, 0.5 M Tris, 30 mM HCl, 50 mM EDTA, 0.1 M KCl, 2% 2-mercaptoethanol, 2 mM PMSF) was added to the pulverized tissue. The phenolic phase was performed by adding to the sample 500 µl of SDS Buffer (30% Sucrose, 2% SDS to be dissolved in 0.1 M Tris-HCl pH 8, 5% of 2-mercaptoethanol) and 500 µl of Sigma- Aldrich (equilibrated with 10 mM Tris HCl pH8 and 1Mm EDTA) [
32]. The sample was shaken and centrifuged at 13,000 rpm for 8 minutes. The supernatant phenolic phase was recovered to which 0.1 M of ammonium acetate in cold methanol was added. The sample was placed at -20°C for 30 minutes, then centrifuged at 13,000 rpm for 5 minutes. A second wash in ammonium acetate was performed, followed by two washes with 80% acetone. The final pellet was dried and then dissolved in the Laemmli 1DE separation buffer overnight [
30]. Proteins were then quantified by measuring the absorbance at 595 nm according to the Bradford assay [
31]. Protein yield was calculated as mg of protein for g fresh tissue weight of each biological replicate.
4.5. SDS-PAGE electrophoresis of proteins, in gel digestion and mass spectrometry
A gel was prepared at a concentration of 12.5% in the running gel and 6% in the stacking gel of acrylamide/bisacrylamide, according to the method of Laemmli [
30]. The samples were heated for 4 min at 100°C before being loaded on the gel. The electrophoretic run was carried out at 60 mA for the stacking gel and 120 mA in the running gel at constant power of 200 V.
The electrophoresis ran for an average of 1 h and 30 min. The gels were stained with Coomassie Blue overnight and subsequently destained with several changes of destaining solution (45% methanol, 10% acetic acid). Digitalized images of the SDS-PAGEs were analyzed by the Quantity One 1-D Analysis Software (Bio-Rad, Hercules, California, USA) to measure the optical densities at each lane of all biological replicates. Each lane of the same SDS-PAGE was divided in six slices from 200 to10 kDa and manually excised from the gel.
The CBB-stained gel slices were destained and then processed with reduction (DTT) and alkylation (IAA) steps [
33]. Gel pieces were digested by trypsin (Promega, Madison WI, USA) overnight at 37 °C adding an ammonium bicarbonate buffer to cover the gel matrix. The extracted peptides were immediately processed for mass spectrometry analysis.
4.6. Mass spectrometric analysis
LC−MS/MS analysis was performed on a EASY-LC 1000 (Thermo Fisher Scientific, Denmark) coupled to a hybrid quadrupole/Orbitrap Q-Exactive mass spectrometer (Thermo Fisher Scientific, Germany). An in-house made analytical column (length 14 cm, inner diameter 75 μm) packed with 3 μm C 18 silica particles (Dr. Maisch, Entringen, Germany) was used. Samples were diluted 5-fold in 0.1% formic acid; then, 2 μL of the resulting peptide mix was injected for LC-MS/MS analysis. Mobile phase A was 2% acetonitrile, 0.1% formic acid; mobile phase B was 80% acetonitrile, 0.1% formic acid. The LC mobile phase composition went from 0% mobile phase B to 3% mobile phase B in 1 s, then from 3 to 40% B in 120 min, then to 100% B in additional 8 min; after 5 min at 100% B, mobile phase composition was brought back to 0% B in 2 min, for a total run time of 135 min at a flow rate of 230 nL/min. The column effluent was subjected to nano-electrospray ionization (1600 V of nESI potential), and resulting charged species were detected by the Q-Exactive hybrid mass spectrometer operating in positive ion mode. A full MS scan was acquired in the Orbitrap analyzer at resolution 70,000, m/z range of 350−1800, and target AGC value of 1.00 × 106, Data-dependent MS/MS acquisition (DDA) procedure was performed by selecting the 12 most abundant peaks with more than two charges after each full scan analysis (top 12 method). Precursor ions were fragmented by HCD (high-energy collisional dissociation); HCD normalized collision energy was 25%. MS/MS analysis was carried in the Orbitrap analyzer at resolution 35,000 and target AGC value of 1.0 × 105, an intensity threshold of 5.0 × 10 4 ; the isolation window was set to 1.6 m/z. A maximum injection time of 50 ms was set for the full MS scan event, while 120 ms was the maximum injection time allowed for tandem MS/MS scans. Dynamic exclusion time was set to 30 s.
4.7. Bioinformatic analysis and proteins identification
From the MS/MS spectra, protein inference and validation were performed with the Scaffold software 4.8 (Proteome Software, Inc., Portland, Oregon, USA). MS/MS spectra were extracted from raw data by accepting one minimum sequence of eight amino acids and fusion scans with the same precursor within one mass window of ± 0.4 m/z, over a time interval of ± 30 s. The key parameters of research are Scored Peak Intensity, (SPI) ≥ 50%, precursor mass tolerance of ± 10 ppm and mass tolerance of product ions of ± 20 ppm. The carbamidomethylation of cysteine was fixed as a modification and trypsin was selected as the enzyme for the digestion, accepting two missing cleavages per peptide.
The Automatic thresholds were used for peptide identification in the software Scaffold. Generally, peptide probabilities are evaluated using a Bayesian approach for the estimation of the local FDR (LFDR) up to a value of 1%. The peptide sequences using Scaffold 4.8 Q+S system software, were interfaced with both the database of proteins deduced from generalist protein sequences of
Lactuca sativa deposited in the NCBI database (downloaded in June 2022) and in the bank UniProt data (downloaded in June 2022). Identified peptides assigned to each protein in all samples and the related statistical parameters for significant identification are reported in the
Supplementary Tables S2 and S3.
4.8. Semi-Quantitative Analysis of Identified Proteins
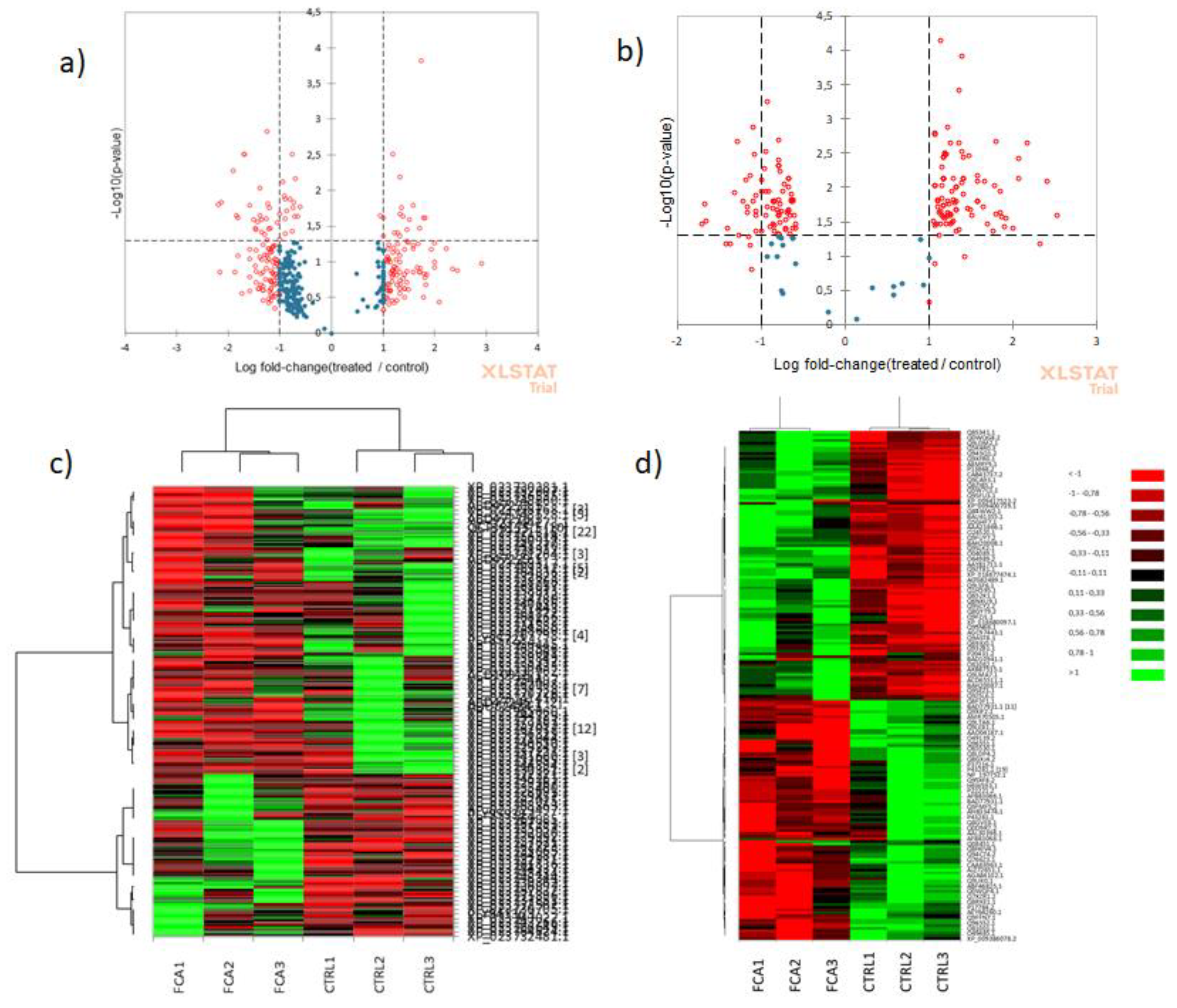
Three biological replicates of leaves and three of roots for each test were used for quantitative analyses. The relative abundance of proteins among samples was performed by choosing the label free quantitative method “Total Spectra” from the Quantitative menu of Scaffold software (Proteome Software, Inc., Portland, OR; version 5.1). This method uses the sum of all weighted spectra associated with a specific protein and within a sample, where the weight is a measure of how much a spectrum is shared by other proteins. Spectral count was undertaken only on statistically validated spectra to increase its accuracy. Consequently, it was used for quantitation comparisons. A peptide with less than two matches was discarded. The missing values were considered undetectable and assumed they were under the limit of detection, but present. Thus, when they were undetectable, a zero value was attributed, and they were considered in the statistical calculation. To identify proteins which show different quantitative abundances in two or more categories the test “Fold change by category” was used. Fold change (FC) is expressed as the ratio of the quantitative value in one sample (or category) over the quantitative value in a second sample. Values of FC > 1 indicate a high quantitative profile and protein results as accumulated, while values of FC < 1 indicate low quantitative profile and protein results as depleted. Because the specified minimum value replaces any missing values, if a zero appears in the denominator an INF will appear in the FC column. FC has been also log2 normalized and analysis was carried out using a threshold of 2 for the significance of the quantitative profile.
4.9. Gene Ontology categories and PANTHER and KEGG pathway enrichment analysis
Gene ontology (GO) categories of all differential accumulated proteins were assigned by means of Panther Classification System [
34]; then, a statistical assessment of differences in functional classes between two groups of sequences based on the Fisher test analysis were executed against the complete dataset of sequences of
Lactuca sativa. Three different significance parameters are given for false-positive control: false discovery rate (FDR), family-wise error rate (FWER), and single test P-value (Fisher P-value). By taking an FDR significance threshold of 0.05, we obtain those functionalities that are specific for the organs and significant for proteins in treated leaves and roots. Relative fold change of each GO term has been represented for “Biological process” categories.
4.10. Statistics
Comparison of differences among groups of values for biomass and photosynthetic pigment, were analyzed using t-test with p <0.05 threshold for statistical significance. All the statistical analyses were performed using Excel XLSTAT (©Addinsoft, Paris, France, released at 2022.6.1.1187). Significance was defined as p ≤ 0.05.
For the proteomics results, comparison of differences among the groups was carried out using the Differentially Expression and Heat map tools available at XLSTAT. Bonferroni test was used to test the assumption of homogeneity of variances. Threshold for significance was p ≤ 0.05.
Author Contributions
Conceptualization, S.M. and G.C.; methodology, A.P., D.O.and D.M.N.; software, D.M.N.; validation, A.P., D.O. and S.M.; formal analysis, A.P., D.O., I.L. and M.R.B.; investigation, A.P.; resources, G.C.; data curation, A.P. and S.M.; writing—original draft preparation, A.P., D.O. and S.M.; writing—review and editing, A.P. and S.M.; visualization, A.P. and D.O.; supervision, S.M.; project administration, G.C.; funding acquisition, G.C. All authors have read and agreed to the published version of the manuscript.