Submitted:
26 June 2023
Posted:
28 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Data collection and processing
2.2. Differential Gene Expression Analysis
2.3. Ortholog prediction
2.4. Signature analysis
2.5. Metabolites prediction
3. Results
3.1. Datasets
| Species | Sample size | Platform | PFAS compound |
Concentration | Setup | Tissue | Reference |
|---|---|---|---|---|---|---|---|
| H. sapiens | 607 | RNA-seq | PFOS PFOA PFBS PFDS |
0.02, 0.1, 0.2, 1, 2, 10, 20, 50, 100 µM | in vitro | Primary liver spheroids | Rowan-Carroll et al. 2021 [38] |
| H. sapiens | 1201 | RNA-seq | PFBA PFPeA PFHxA PFHpA PFOA PFNA PFDA PFUnA PFTeDA PFBS PFHxS PFHpS PFOS PFDS PFOSA 8:2MonoPAP 6:2MonoPAP 8:2 FtS 6:2 FtS 4:2 FtS 8:2 FtOH 6:2 FtOH 5:3 Acid |
Various concentrations in the range 0.2 – 100 µM |
in vitro | Primary liver spheroids | Reardon et al. 2021 [44] |
| H. sapiens | 23 | RNA-seq | PFOS | 10 mg/kg | in vivo | Prostate cancer cells xenograft |
Imir et al. 2021 [45] |
| M. musculus | 32 | RNA-seq | PFOA GenX |
0.05, 0.3 mg/kg body weight/day | in vivo | Liver | Attema et al. 2022 [39] |
| M. musculus | 37 | RNA-seq | HFPO-DA | 0.1, 0.5, 5 mg/kg | in vivo | Liver | Heintz et al. 2022 [46] |
| M. musculus | 18 | Microarray | PFOS PFNA |
0.0003% of low-fat diet or high-fat diet | in vivo | Liver | Pfohl et al. 2021 [47] |
| C. elegans | 60 | RNA-seq | HFPO-DA | 1.25x10-5, 6.25x10-5, 3.13x10-4, 1.56x10-3, 7.81x10-3, 1.56x10-2, 3.13x10-2, 6.25x10-2, 0.125, 0.25, 0.5, 1, 2, 4 g/L |
in vivo | Whole body | Feng et al. 2022 [40] |
| D. rerio | 16 | RNA-seq | PFOSA | 12.5 µM | in vivo | Embryo | Dasgupta et al. 2020 [41] |
| G. morhua | 48 | RNA-seq | PFOS PFOA PFNA |
Low, medium, high, 1x, 20x, 100x | in vitro | Ovary | Khan et al. 2021 [43] |
| M. salmoides | 72 | Microarray | PFDA PFUnA PFDoA PFOS | Different for each lake and each PFAS | in vivo | Liver & Testis |
Collí-Dulá et al. 2016 [48] |
| P. promelas | 30 | Microarray | PFOS PFBA PFHxA PFHpA PFOA PFNA PFDA |
0.5, 25 µg/L | in vivo | Liver & Whole blood | Rodríguez-Jorquera et al. 2019 [49] |
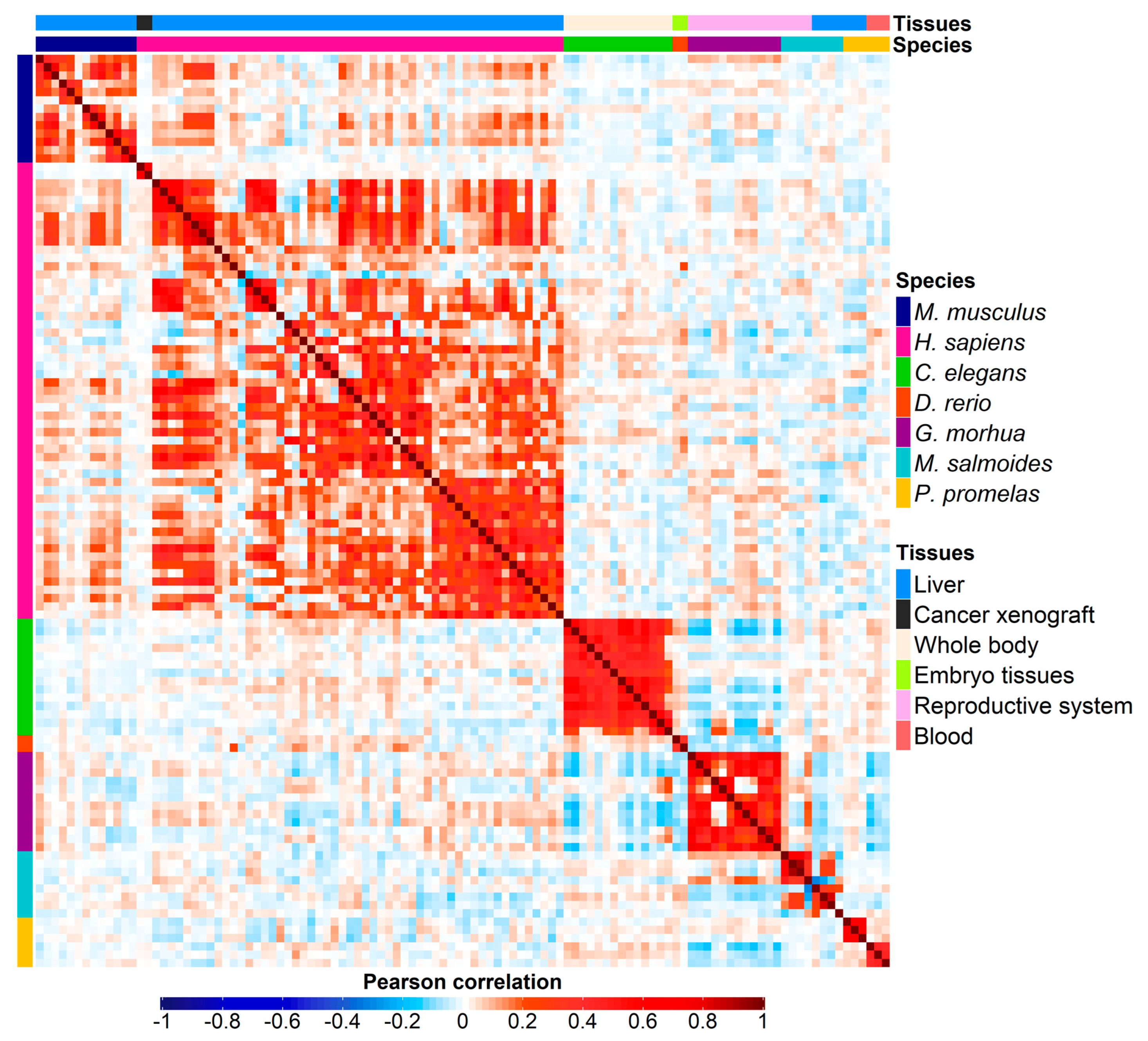
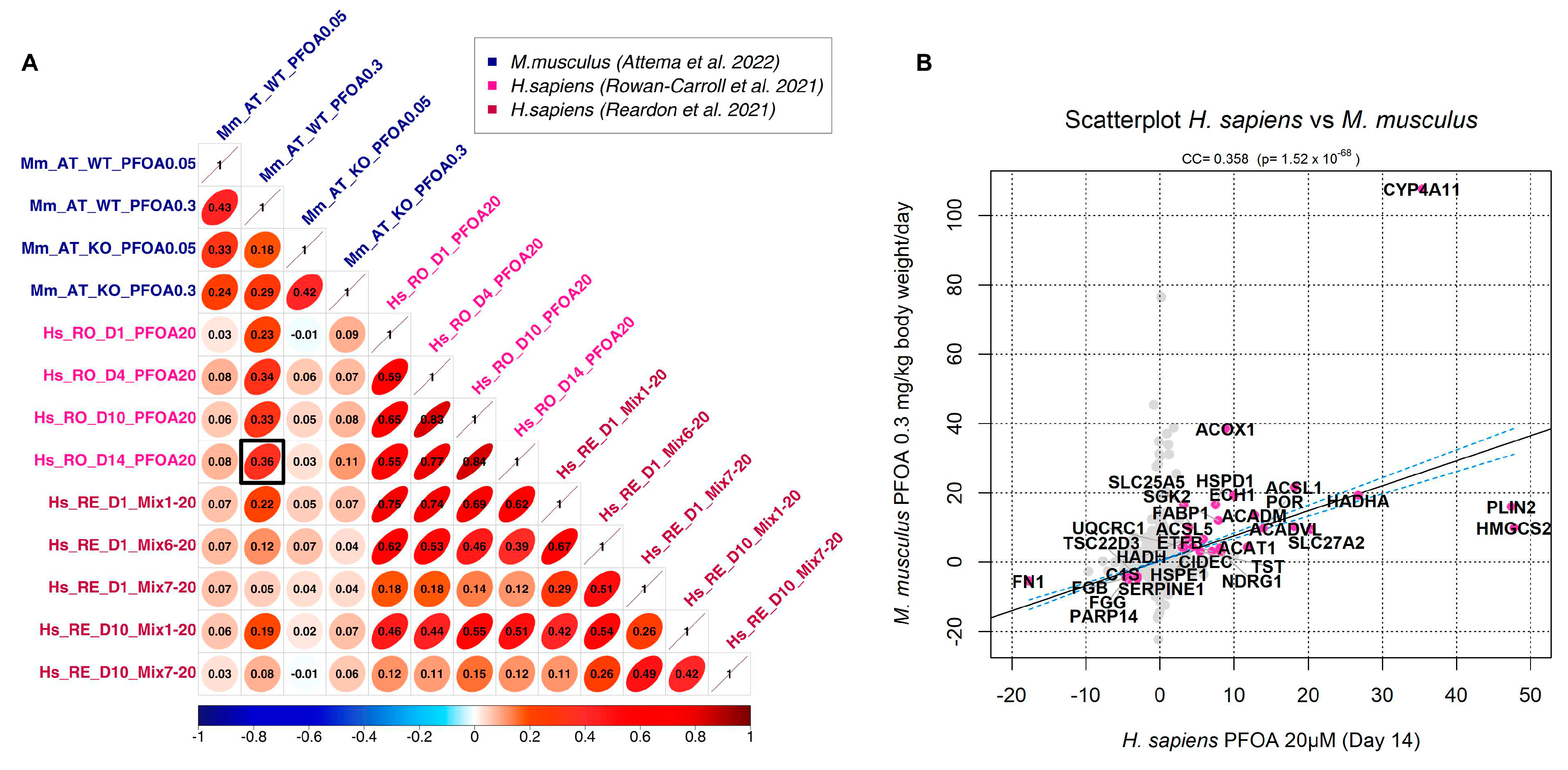
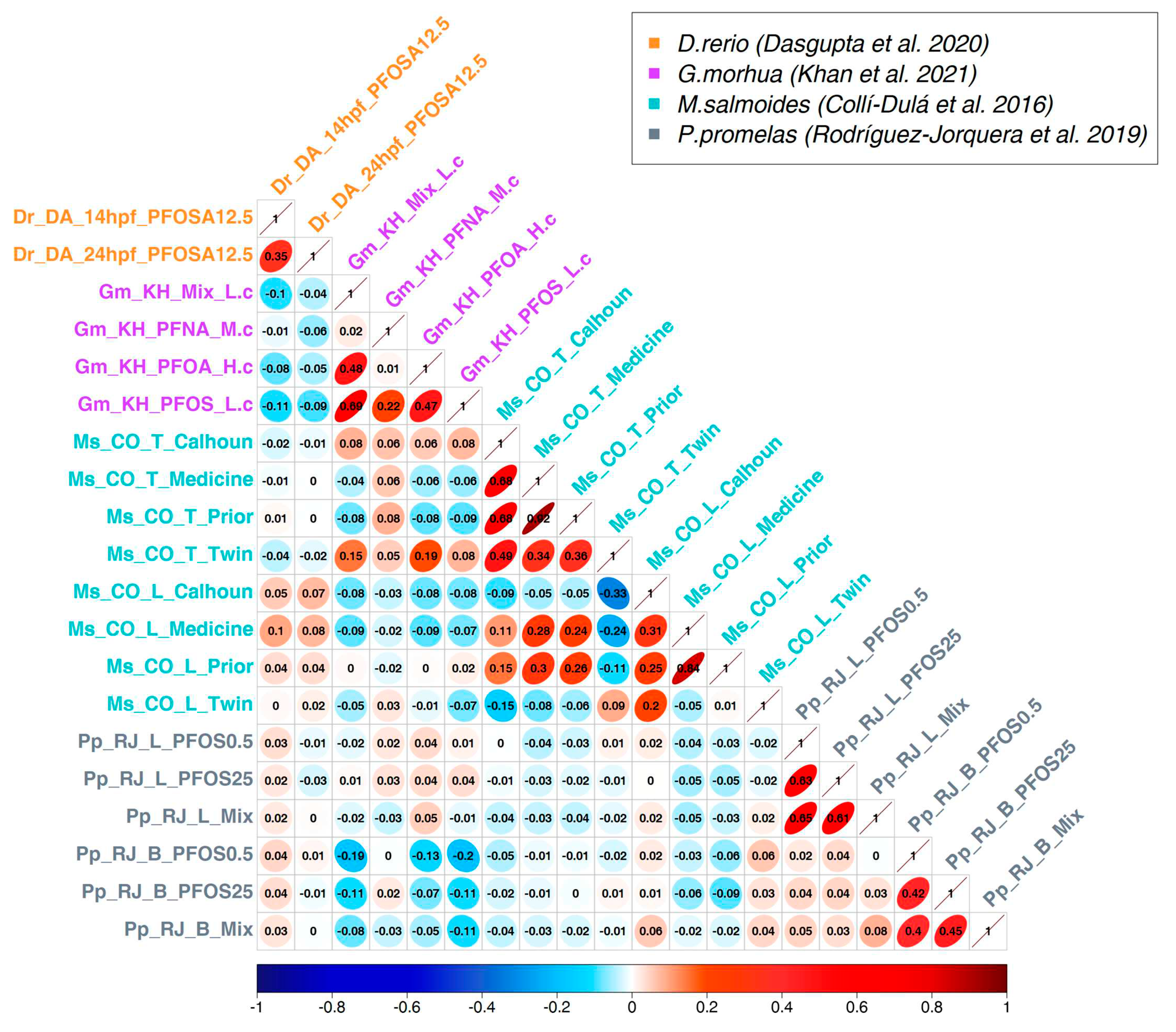
3.2. Correlation analysis
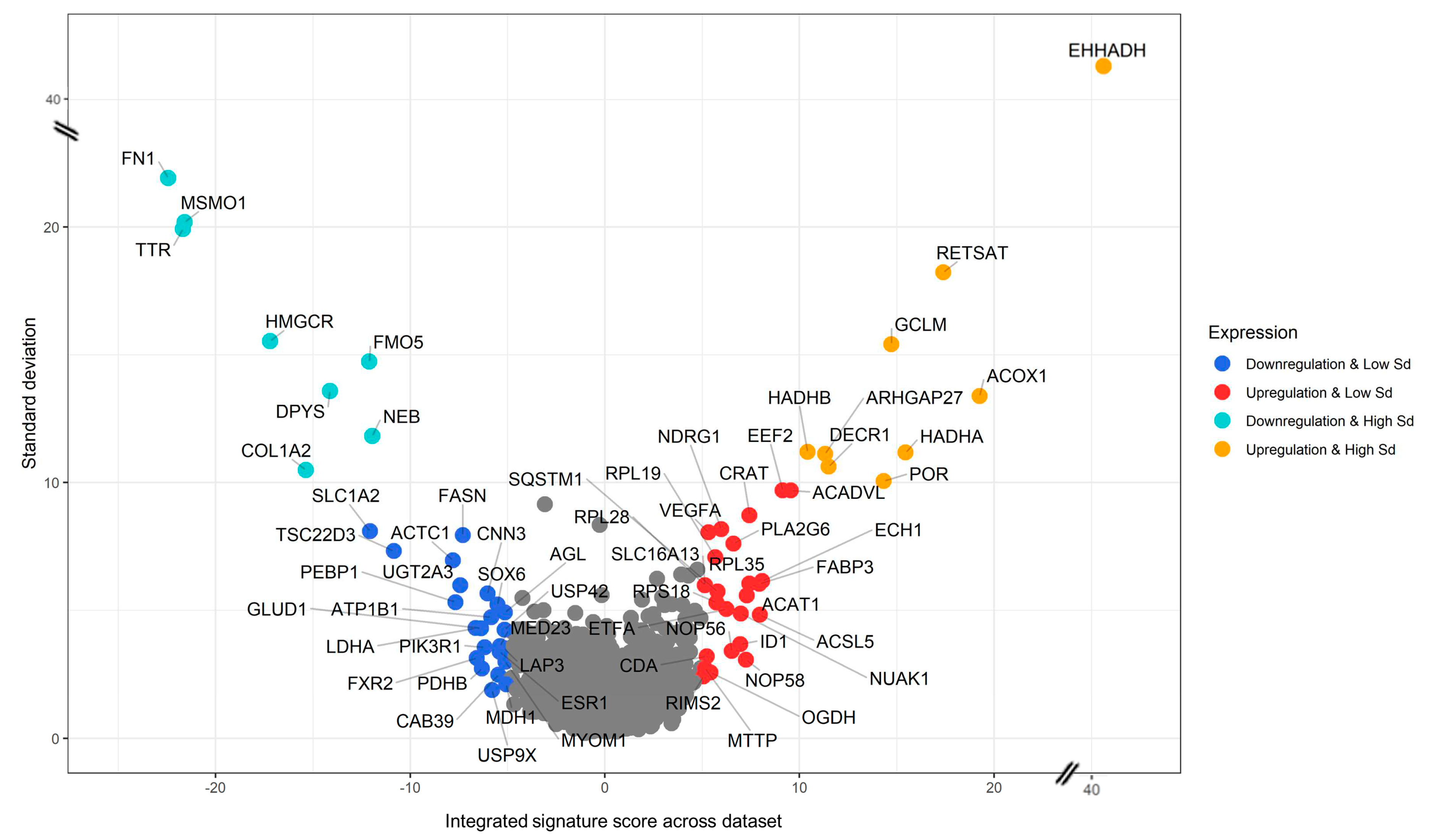
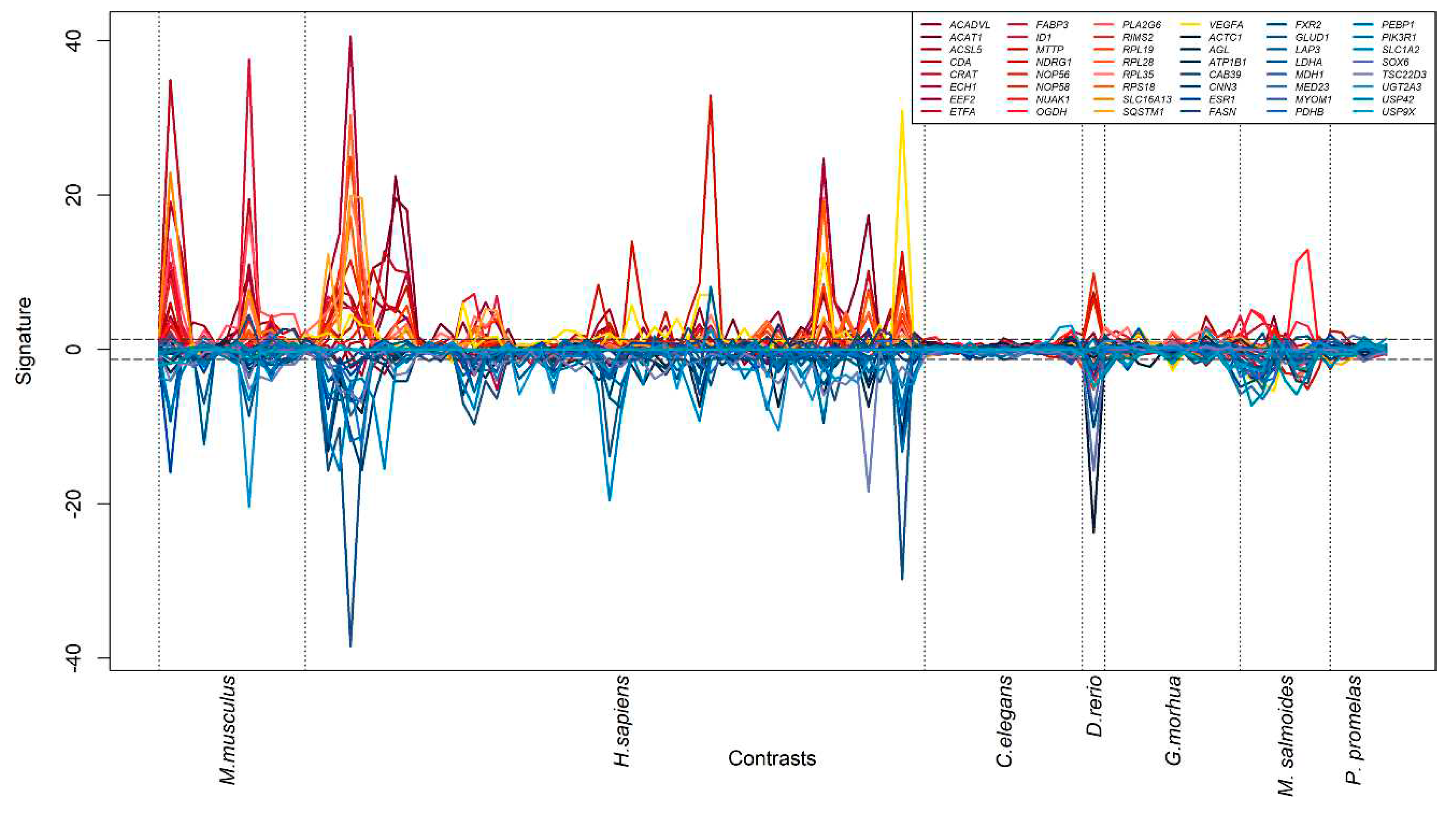
3.3. Generation of a cross-species PFAS responses
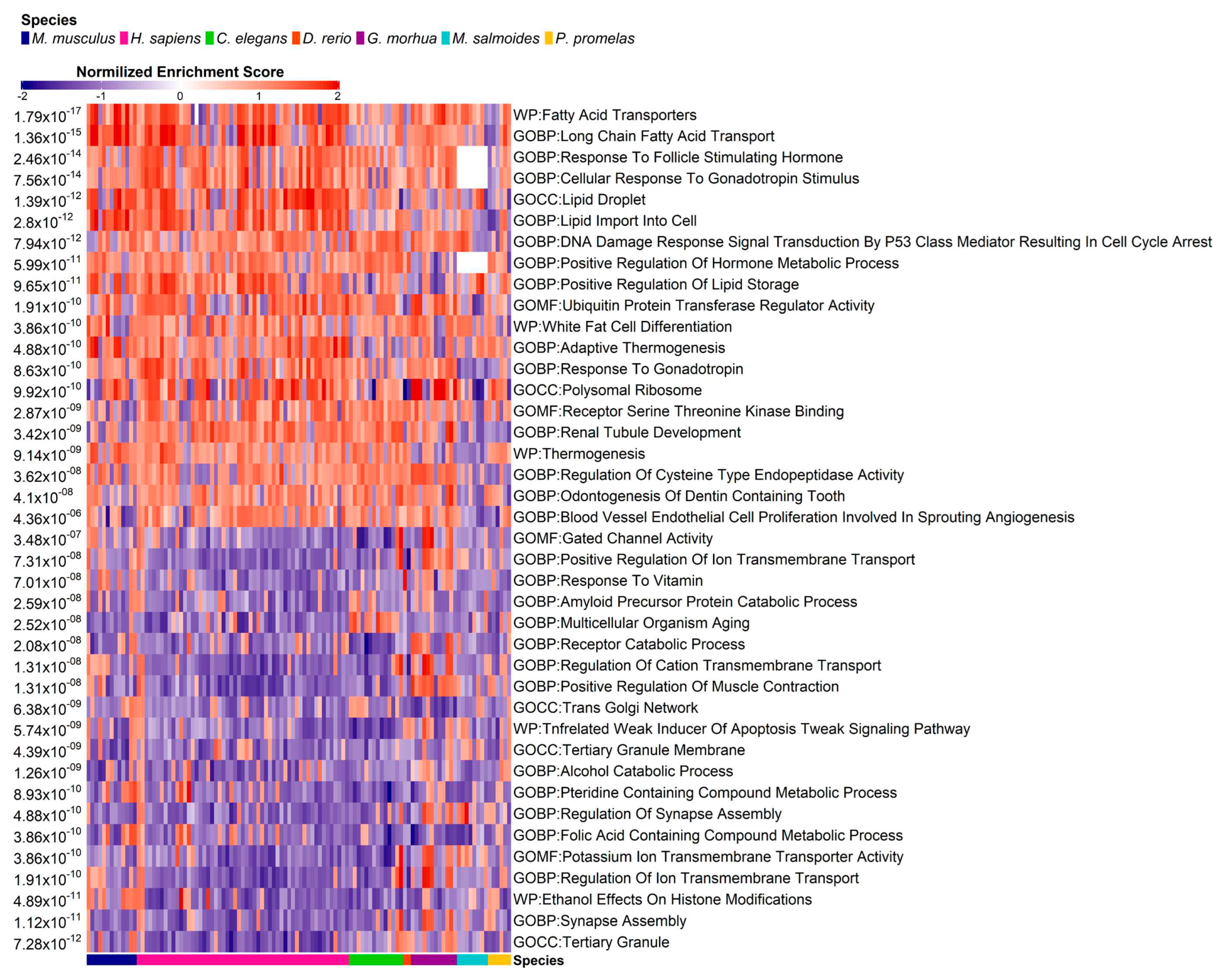
3.4. Pathway Enrichment Analysis
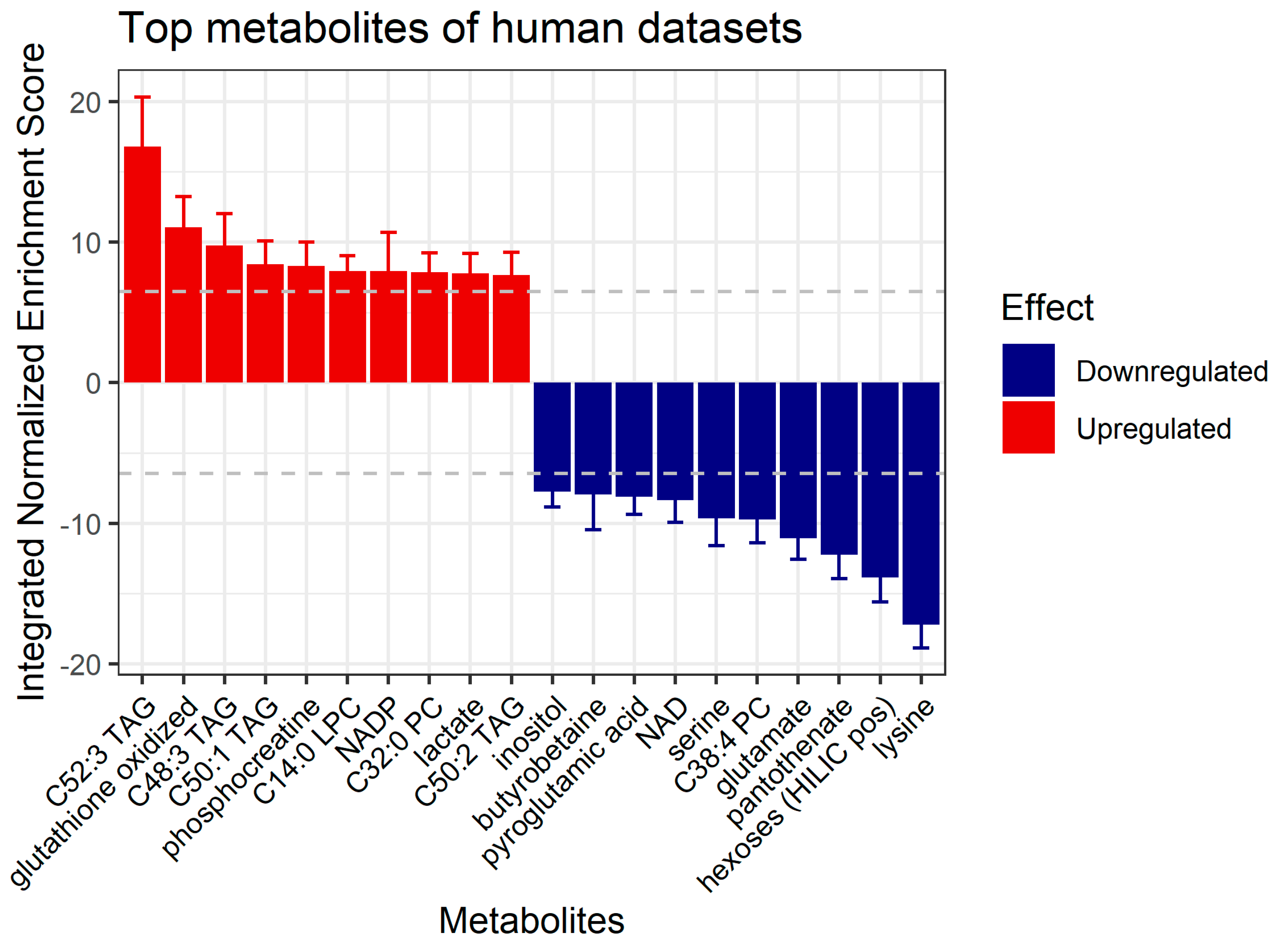
3.5. Prediction of Affected Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buck, R.C.; Franklin, J.; Berger, U.; Conder, J.M.; Cousins, I.T.; de Voogt, P.; Jensen, A.A.; Kannan, K.; Mabury, S.A.; van Leeuwen, S.P.J. Perfluoroalkyl and Polyfluoroalkyl Substances in the Environment: Terminology, Classification, and Origins. Integr. Environ. Assess. Manag. 2011, 7, 513–541. [Google Scholar] [CrossRef]
- Panieri, E.; Baralic, K.; Djukic-Cosic, D.; Buha Djordjevic, A.; Saso, L. PFAS Molecules: A Major Concern for the Human Health and the Environment. Toxics 2022, 10, 44. [Google Scholar] [CrossRef]
- OECD Reconciling Terminology of the Universe of Per-and Polyfluoroalkyl Substances: Recommendations and Practical Guidance; OECD Paris, France, 2021.
- PubChem Classification Browser. Available online: https://pubchem.ncbi.nlm.nih.gov/classification/#hid=120 (accessed on 3 March 2023).
- Gaines, L.G.T.; Sinclair, G.; Williams, A.J. A Proposed Approach to Defining Per- and Polyfluoroalkyl Substances (PFAS) Based on Molecular Structure and Formula. Integr. Environ. Assess. Manag. 2023. [Google Scholar] [CrossRef]
- CompTox Chemicals Dashboard Available online:. Available online: https://comptox.epa.gov/dashboard/chemical-lists/PFASSTRUCTV5 (accessed on 2 March 2023).
- Organisation for Economic Co-operation and Development Toward a New Comprehensive Global Database of Per- and Polyfluoroalkyl Substances (PFASs): Summary Report on Updating the OECD 2007 List of per- and Polyfluoroalkyl Substances (PFASs) Available online: https://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=ENV-JM-MONO(2018)7&doclanguage=en (accessed on 14 June 2023).
- Gaines, L.G.T. Historical and Current Usage of Per- and Polyfluoroalkyl Substances (PFAS): A Literature Review. Am. J. Ind. Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- Podder, A.; Sadmani, A.H.M.A.; Reinhart, D.; Chang, N.-B.; Goel, R. Per and Poly-Fluoroalkyl Substances (PFAS) as a Contaminant of Emerging Concern in Surface Water: A Transboundary Review of Their Occurrences and Toxicity Effects. J. Hazard. Mater. 2021, 419, 126361. [Google Scholar] [CrossRef]
- Piva, E.; Ioime, P.; Dall’Ara, S.; Fais, P.; Pascali, J.P. Per- and Polyfluoroalkyl Substances (PFAS) Determination in Shellfish by Liquid Chromatography Coupled to Accurate Mass Spectrometry. Drug Test. Anal. 2022, 14, 1652–1659. [Google Scholar] [CrossRef]
- Brase, R.A.; Mullin, E.J.; Spink, D.C. Legacy and Emerging Per- and Polyfluoroalkyl Substances: Analytical Techniques, Environmental Fate, and Health Effects. Int. J. Mol. Sci. 2021, 22, 995. [Google Scholar] [CrossRef]
- Beale, D.J.; Sinclair, G.M.; Shah, R.; Paten, A.M.; Kumar, A.; Long, S.M.; Vardy, S.; Jones, O.A.H. A Review of Omics-Based PFAS Exposure Studies Reveals Common Biochemical Response Pathways. Sci. Total Environ. 2022, 845, 157255. [Google Scholar] [CrossRef]
- Fan, L.; Tang, J.; Zhang, D.; Ma, M.; Wang, Y.; Han, Y. Investigations on the Phytotoxicity of Perfluorooctanoic Acid in Arabidopsis Thaliana. Environ. Sci. Pollut. Res. Int. 2020, 27, 1131–1143. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hu, W.; Li, X.; Hu, F.; Liu, Y.; Xie, T.; Liu, B.; Xi, Y.; Su, Z.; Zhang, C. Effects of Perfluoroalkyl Substances on Root and Rhizosphere Bacteria: Phytotoxicity, Phyto-Microbial Remediation, Risk Assessment. Chemosphere 2022, 289, 133137. [Google Scholar] [CrossRef] [PubMed]
- Lucas, K.; Gaines, L.G.T.; Paris-Davila, T.; Nylander-French, L.A. Occupational Exposure and Serum Levels of Per- and Polyfluoroalkyl Substances (PFAS): A Review. Am. J. Ind. Med. 2023, 66, 379–392. [Google Scholar] [CrossRef]
- Babayev, M.; Capozzi, S.L.; Miller, P.; McLaughlin, K.R.; Medina, S.S.; Byrne, S.; Zheng, G.; Salamova, A. PFAS in Drinking Water and Serum of the People of a Southeast Alaska Community: A Pilot Study. Environ. Pollut. Barking Essex 1987 2022, 305, 119246. [Google Scholar] [CrossRef] [PubMed]
- LaKind, J.S.; Naiman, J.; Verner, M.-A.; Lévêque, L.; Fenton, S. Per- and Polyfluoroalkyl Substances (PFAS) in Breast Milk and Infant Formula: A Global Issue. Environ. Res. 2023, 219, 115042. [Google Scholar] [CrossRef] [PubMed]
- LaKind, J.S.; Verner, M.-A.; Rogers, R.D.; Goeden, H.; Naiman, D.Q.; Marchitti, S.A.; Lehmann, G.M.; Hines, E.P.; Fenton, S.E. Current Breast Milk PFAS Levels in the United States and Canada: After All This Time, Why Don’t We Know More? Environ. Health Perspect. 2022, 130, 25002. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Meng, L.; Ma, D.; Cao, H.; Liang, Y.; Liu, H.; Wang, Y.; Jiang, G. The Occurrence of PFAS in Human Placenta and Their Binding Abilities to Human Serum Albumin and Organic Anion Transporter 4. Environ. Pollut. Barking Essex 1987 2021, 273, 116460. [Google Scholar] [CrossRef]
- Pascali, J.P.; Piva, E.; Bonasoni, M.P.; Migliavacca, C.; Seidenari, A.; Fais, P. Analysis and Distribution of Per- and Polyfluoroalkyl Substances in Decidua and Villi Placenta Explants. Environ. Res. 2023, 229, 115955. [Google Scholar] [CrossRef] [PubMed]
- Piva, E.; Giorgetti, A.; Ioime, P.; Morini, L.; Freni, F.; Faro, F.L.; Pirani, F.; Montisci, M.; Fais, P.; Pascali, J.P. Hair Determination of Per- and Polyfluoroalkyl Substances (PFAS) in the Italian Population. Toxicology 2021, 458, 152849. [Google Scholar] [CrossRef]
- Cui, Q.; Pan, Y.; Wang, J.; Liu, H.; Yao, B.; Dai, J. Exposure to Per- and Polyfluoroalkyl Substances (PFASs) in Serum versus Semen and Their Association with Male Reproductive Hormones. Environ. Pollut. Barking Essex 1987 2020, 266, 115330. [Google Scholar] [CrossRef]
- Sunderland, E.M.; Hu, X.C.; Dassuncao, C.; Tokranov, A.K.; Wagner, C.C.; Allen, J.G. A Review of the Pathways of Human Exposure to Poly- and Perfluoroalkyl Substances (PFASs) and Present Understanding of Health Effects. J. Expo. Sci. Environ. Epidemiol. 2019, 29, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Fenton, S.E.; Ducatman, A.; Boobis, A.; DeWitt, J.C.; Lau, C.; Ng, C.; Smith, J.S.; Roberts, S.M. Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research. Environ. Toxicol. Chem. 2021, 40, 606–630. [Google Scholar] [CrossRef]
- Ehrlich, V.; Bil, W.; Vandebriel, R.; Granum, B.; Luijten, M.; Lindeman, B.; Grandjean, P.; Kaiser, A.-M.; Hauzenberger, I.; Hartmann, C.; et al. Consideration of Pathways for Immunotoxicity of Per- and Polyfluoroalkyl Substances (PFAS). Environ. Health Glob. Access Sci. Source 2023, 22, 19. [Google Scholar] [CrossRef]
- Ding, N.; Harlow, S.D.; Randolph, J.F.; Loch-Caruso, R.; Park, S.K. Perfluoroalkyl and Polyfluoroalkyl Substances (PFAS) and Their Effects on the Ovary. Hum. Reprod. Update 2020, 26, 724–752. [Google Scholar] [CrossRef]
- Green, M.P.; Harvey, A.J.; Finger, B.J.; Tarulli, G.A. Endocrine Disrupting Chemicals: Impacts on Human Fertility and Fecundity during the Peri-Conception Period. Environ. Res. 2021, 194, 110694. [Google Scholar] [CrossRef] [PubMed]
- Rickard, B.P.; Rizvi, I.; Fenton, S.E. Per- and Poly-Fluoroalkyl Substances (PFAS) and Female Reproductive Outcomes: PFAS Elimination, Endocrine-Mediated Effects, and Disease. Toxicology 2022, 465, 153031. [Google Scholar] [CrossRef]
- Wang, W.; Hong, X.; Zhao, F.; Wu, J.; Wang, B. The Effects of Perfluoroalkyl and Polyfluoroalkyl Substances on Female Fertility: A Systematic Review and Meta-Analysis. Environ. Res. 2023, 216, 114718. [Google Scholar] [CrossRef]
- Steenland, K.; Winquist, A. PFAS and Cancer, a Scoping Review of the Epidemiologic Evidence. Environ. Res. 2021, 194, 110690. [Google Scholar] [CrossRef]
- Girardi, P.; Merler, E. A Mortality Study on Male Subjects Exposed to Polyfluoroalkyl Acids with High Internal Dose of Perfluorooctanoic Acid. Environ. Res. 2019, 179, 108743. [Google Scholar] [CrossRef] [PubMed]
- Dunder, L.; Lind, P.M.; Salihovic, S.; Stubleski, J.; Kärrman, A.; Lind, L. Changes in Plasma Levels of Per- and Polyfluoroalkyl Substances (PFAS) Are Associated with Changes in Plasma Lipids - A Longitudinal Study over 10 Years. Environ. Res. 2022, 211, 112903. [Google Scholar] [CrossRef] [PubMed]
- Canova, C.; Barbieri, G.; Zare Jeddi, M.; Gion, M.; Fabricio, A.; Daprà, F.; Russo, F.; Fletcher, T.; Pitter, G. Associations between Perfluoroalkyl Substances and Lipid Profile in a Highly Exposed Young Adult Population in the Veneto Region. Environ. Int. 2020, 145, 106117. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Barregard, L.; Xu, Y.; Scott, K.; Pineda, D.; Lindh, C.H.; Jakobsson, K.; Fletcher, T. Associations between Perfluoroalkyl Substances and Serum Lipids in a Swedish Adult Population with Contaminated Drinking Water. Environ. Health Glob. Access Sci. Source 2020, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Batzella, E.; Zare Jeddi, M.; Pitter, G.; Russo, F.; Fletcher, T.; Canova, C. Associations between Mixture of Perfluoroalkyl Substances and Lipid Profile in a Highly Exposed Adult Community in the Veneto Region. Int. J. Environ. Res. Public. Health 2022, 19, 12421. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.M.; Kotlarz, N.; Knappe, D.R.U.; Lea, C.S.; Collier, D.N.; Richardson, D.B.; Hoppin, J.A. Drinking Water-Associated PFAS and Fluoroethers and Lipid Outcomes in the GenX Exposure Study. Environ. Health Perspect. 2022, 130, 97002. [Google Scholar] [CrossRef] [PubMed]
- Roth, K.; Yang, Z.; Agarwal, M.; Liu, W.; Peng, Z.; Long, Z.; Birbeck, J.; Westrick, J.; Liu, W.; Petriello, M.C. Exposure to a Mixture of Legacy, Alternative, and Replacement per- and Polyfluoroalkyl Substances (PFAS) Results in Sex-Dependent Modulation of Cholesterol Metabolism and Liver Injury. Environ. Int. 2021, 157, 106843. [Google Scholar] [CrossRef]
- Rowan-Carroll, A.; Reardon, A.; Leingartner, K.; Gagné, R.; Williams, A.; Meier, M.J.; Kuo, B.; Bourdon-Lacombe, J.; Moffat, I.; Carrier, R.; et al. High-Throughput Transcriptomic Analysis of Human Primary Hepatocyte Spheroids Exposed to Per- and Polyfluoroalkyl Substances as a Platform for Relative Potency Characterization. Toxicol. Sci. Off. J. Soc. Toxicol. 2021, 181, 199–214. [Google Scholar] [CrossRef]
- Attema, B.; Janssen, A.W.F.; Rijkers, D.; van Schothorst, E.M.; Hooiveld, G.J.E.J.; Kersten, S. Exposure to Low-Dose Perfluorooctanoic Acid Promotes Hepatic Steatosis and Disrupts the Hepatic Transcriptome in Mice. Mol. Metab. 2022, 66, 101602. [Google Scholar] [CrossRef]
- Feng, Z.; McLamb, F.; Vu, J.P.; Gong, S.; Gersberg, R.M.; Bozinovic, G. Physiological and Transcriptomic Effects of Hexafluoropropylene Oxide Dimer Acid in Caenorhabditis Elegans during Development. Ecotoxicol. Environ. Saf. 2022, 244, 114047. [Google Scholar] [CrossRef]
- Dasgupta, S.; Reddam, A.; Liu, Z.; Liu, J.; Volz, D.C. High-Content Screening in Zebrafish Identifies Perfluorooctanesulfonamide as a Potent Developmental Toxicant. Environ. Pollut. Barking Essex 1987 2020, 256, 113550. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.J.; Schmid, J.E.; Lau, C.; Abbott, B.D. Activation of Mouse and Human Peroxisome Proliferator-Activated Receptor-Alpha (PPARα) by Perfluoroalkyl Acids (PFAAs): Further Investigation of C4-C12 Compounds. Reprod. Toxicol. Elmsford N 2012, 33, 546–551. [Google Scholar] [CrossRef]
- Khan, E.A.; Zhang, X.; Hanna, E.M.; Yadetie, F.; Jonassen, I.; Goksøyr, A.; Arukwe, A. Application of Quantitative Transcriptomics in Evaluating the Ex Vivo Effects of Per- and Polyfluoroalkyl Substances on Atlantic Cod (Gadus Morhua) Ovarian Physiology. Sci. Total Environ. 2021, 755, 142904. [Google Scholar] [CrossRef]
- Reardon, A.J.F.; Rowan-Carroll, A.; Ferguson, S.S.; Leingartner, K.; Gagne, R.; Kuo, B.; Williams, A.; Lorusso, L.; Bourdon-Lacombe, J.A.; Carrier, R.; et al. Potency Ranking of Per- and Polyfluoroalkyl Substances Using High-Throughput Transcriptomic Analysis of Human Liver Spheroids. Toxicol. Sci. Off. J. Soc. Toxicol. 2021, 184, 154–169. [Google Scholar] [CrossRef]
- Imir, O.B.; Kaminsky, A.Z.; Zuo, Q.-Y.; Liu, Y.-J.; Singh, R.; Spinella, M.J.; Irudayaraj, J.; Hu, W.-Y.; Prins, G.S.; Madak Erdogan, Z. Per- and Polyfluoroalkyl Substance Exposure Combined with High-Fat Diet Supports Prostate Cancer Progression. Nutrients 2021, 13, 3902. [Google Scholar] [CrossRef] [PubMed]
- Heintz, M.M.; Chappell, G.A.; Thompson, C.M.; Haws, L.C. Evaluation of Transcriptomic Responses in Livers of Mice Exposed to the Short-Chain PFAS Compound HFPO-DA. Front. Toxicol. 2022, 4, 937168. [Google Scholar] [CrossRef]
- Pfohl, M.; Marques, E.; Auclair, A.; Barlock, B.; Jamwal, R.; Goedken, M.; Akhlaghi, F.; Slitt, A.L. An ’Omics Approach to Unraveling the Paradoxical Effect of Diet on Perfluorooctanesulfonic Acid (PFOS) and Perfluorononanoic Acid (PFNA)-Induced Hepatic Steatosis. Toxicol. Sci. Off. J. Soc. Toxicol. 2021, 180, 277–294. [Google Scholar] [CrossRef]
- Collí-Dulá, R.C.; Martyniuk, C.J.; Streets, S.; Denslow, N.D.; Lehr, R. Molecular Impacts of Perfluorinated Chemicals (PFASs) in the Liver and Testis of Male Largemouth Bass (Micropterus Salmoides) in Minnesota Lakes. Comp. Biochem. Physiol. Part D Genomics Proteomics 2016, 19, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Jorquera, I.A.; Colli-Dula, R.C.; Kroll, K.; Jayasinghe, B.S.; Parachu Marco, M.V.; Silva-Sanchez, C.; Toor, G.S.; Denslow, N.D. Blood Transcriptomics Analysis of Fish Exposed to Perfluoro Alkyls Substances: Assessment of a Non-Lethal Sampling Technique for Advancing Aquatic Toxicology Research. Environ. Sci. Technol. 2019, 53, 1441–1452. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets--Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Farrell, C.M.; Feldgarden, M.; Fine, A.M.; Funk, K.; et al. Database Resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. 2023, 51, D29–D38. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinforma. Oxf. Engl. 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2; Use R!; Springer International Publishing: Cham, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Mercatelli, D.; Lopez-Garcia, G.; Giorgi, F.M. Corto: A Lightweight R Package for Gene Network Inference and Master Regulator Analysis. Bioinforma. Oxf. Engl. 2020, 36, 3916–3917. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinforma. Oxf. Engl. 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.S.; Irizarry, R.A. A Framework for Oligonucleotide Microarray Preprocessing. Bioinforma. Oxf. Engl. 2010, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A Bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinforma. Oxf. Engl. 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Alvarez, M.J.; Shen, Y.; Giorgi, F.M.; Lachmann, A.; Ding, B.B.; Ye, B.H.; Califano, A. Functional Characterization of Somatic Mutations in Cancer Using Network-Based Inference of Protein Activity. Nat. Genet. 2016, 48, 838–847. [Google Scholar] [CrossRef]
- Hu, Y.; Flockhart, I.; Vinayagam, A.; Bergwitz, C.; Berger, B.; Perrimon, N.; Mohr, S.E. An Integrative Approach to Ortholog Prediction for Disease-Focused and Other Functional Studies. BMC Bioinformatics 2011, 12, 357. [Google Scholar] [CrossRef]
- Schilder, B. Orthogene: An R Package for Easy Mapping of Orthologous Genes across Hundreds of Species. 2023.
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinforma. Oxf. Engl. 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Sergushichev, A. Fast Gene Set Enrichment Analysis 2019, 060012.
- Cavicchioli, M.V.; Santorsola, M.; Balboni, N.; Mercatelli, D.; Giorgi, F.M. Prediction of Metabolic Profiles from Transcriptomics Data in Human Cancer Cell Lines. Int. J. Mol. Sci. 2022, 23, 3867. [Google Scholar] [CrossRef]
- Li, H.; Ning, S.; Ghandi, M.; Kryukov, G.V.; Gopal, S.; Deik, A.; Souza, A.; Pierce, K.; Keskula, P.; Hernandez, D.; et al. The Landscape of Cancer Cell Line Metabolism. Nat. Med. 2019, 25, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Powell, P.K.; Wolf, I.; Jin, R.; Lasker, J.M. Metabolism of Arachidonic Acid to 20-Hydroxy-5,8,11, 14-Eicosatetraenoic Acid by P450 Enzymes in Human Liver: Involvement of CYP4F2 and CYP4A11. J. Pharmacol. Exp. Ther. 1998, 285, 1327–1336. [Google Scholar] [PubMed]
- Ni, K.-D.; Liu, J.-Y. The Functions of Cytochrome P450 ω-Hydroxylases and the Associated Eicosanoids in Inflammation-Related Diseases. Front. Pharmacol. 2021, 12, 716801. [Google Scholar] [CrossRef]
- Gao, H.; Cao, Y.; Xia, H.; Zhu, X.; Jin, Y. CYP4A11 Is Involved in the Development of Nonalcoholic Fatty Liver Disease via ROS-induced Lipid Peroxidation and Inflammation. Int. J. Mol. Med. 2020, 45, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Ouyang, T.; Liu, H.; Cao, L.; Chen, W. Perfluoroalkyl Substance (PFAS) Exposure and Risk of Nonalcoholic Fatty Liver Disease in the Elderly: Results from NHANES 2003-2014. Environ. Sci. Pollut. Res. Int. 2023. [Google Scholar] [CrossRef]
- Glinos, D.A.; Garborcauskas, G.; Hoffman, P.; Ehsan, N.; Jiang, L.; Gokden, A.; Dai, X.; Aguet, F.; Brown, K.L.; Garimella, K.; et al. Transcriptome Variation in Human Tissues Revealed by Long-Read Sequencing. Nature 2022, 608, 353–359. [Google Scholar] [CrossRef]
- Chhibber, A.; French, C.E.; Yee, S.W.; Gamazon, E.R.; Theusch, E.; Qin, X.; Webb, A.; Papp, A.C.; Wang, A.; Simmons, C.Q.; et al. Transcriptomic Variation of Pharmacogenes in Multiple Human Tissues and Lymphoblastoid Cell Lines. Pharmacogenomics J. 2017, 17, 137–145. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kim, Y.-S.; Yoo, K.-J.; Lee, H.-J.; Lee, D.-R.; Yeo, C.Y.; Baek, K.-H. The Expression of Usp42 during Embryogenesis and Spermatogenesis in Mouse. Gene Expr. Patterns GEP 2007, 7, 143–148. [Google Scholar] [CrossRef]
- Calvert, L.; Green, M.P.; De Iuliis, G.N.; Dun, M.D.; Turner, B.D.; Clarke, B.O.; Eamens, A.L.; Roman, S.D.; Nixon, B. Assessment of the Emerging Threat Posed by Perfluoroalkyl and Polyfluoroalkyl Substances to Male Reproduction in Humans. Front. Endocrinol. 2021, 12, 799043. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Qadri, S.; Luukkonen, P.K.; Ragnarsdottir, O.; McGlinchey, A.; Jäntti, S.; Juuti, A.; Arola, J.; Schlezinger, J.J.; Webster, T.F.; et al. Exposure to Environmental Contaminants Is Associated with Altered Hepatic Lipid Metabolism in Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2022, 76, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Geiger, S.D.; Xiao, J.; Ducatman, A.; Frisbee, S.; Innes, K.; Shankar, A. The Association between PFOA, PFOS and Serum Lipid Levels in Adolescents. Chemosphere 2014, 98, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; McConnell, R.; Catherine, C.; Xu, S.; Walker, D.I.; Stratakis, N.; Jones, D.P.; Miller, G.W.; Peng, C.; Conti, D.V.; et al. Perfluoroalkyl Substances and Severity of Nonalcoholic Fatty Liver in Children: An Untargeted Metabolomics Approach. Environ. Int. 2020, 134, 105220. [Google Scholar] [CrossRef]
- Yang, Z.; Fu, L.; Cao, M.; Li, F.; Li, J.; Chen, Z.; Guo, A.; Zhong, H.; Li, W.; Liang, Y.; et al. PFAS-Induced Lipidomic Dysregulations and Their Associations with Developmental Toxicity in Zebrafish Embryos. Sci. Total Environ. 2023, 861, 160691. [Google Scholar] [CrossRef]
- Filatov, M.; Khramova, Y.; Parshina, E.; Bagaeva, T.; Semenova, M. Influence of Gonadotropins on Ovarian Follicle Growth and Development in Vivo and in Vitro. Zygote Camb. Engl. 2017, 25, 235–243. [Google Scholar] [CrossRef]
- Roepke, T.A.; Sadlier, N.C. REPRODUCTIVE TOXICOLOGY: Impact of Endocrine Disruptors on Neurons Expressing GnRH or Kisspeptin and Pituitary Gonadotropins. Reprod. Camb. Engl. 2021, 162, F131–F145. [Google Scholar] [CrossRef]
- Cowland, J.B.; Borregaard, N. Granulopoiesis and Granules of Human Neutrophils. Immunol. Rev. 2016, 273, 11–28. [Google Scholar] [CrossRef]
- Tang, Z.-R.; Zhang, R.; Lian, Z.-X.; Deng, S.-L.; Yu, K. Estrogen-Receptor Expression and Function in Female Reproductive Disease. Cells 2019, 8, 1123. [Google Scholar] [CrossRef]
- Stratakis, N.; V Conti, D.; Jin, R.; Margetaki, K.; Valvi, D.; Siskos, A.P.; Maitre, L.; Garcia, E.; Varo, N.; Zhao, Y.; et al. Prenatal Exposure to Perfluoroalkyl Substances Associated With Increased Susceptibility to Liver Injury in Children. Hepatol. Baltim. Md 2020, 72, 1758–1770. [Google Scholar] [CrossRef]
- Szilagyi, J.T.; Avula, V.; Fry, R.C. Perfluoroalkyl Substances (PFAS) and Their Effects on the Placenta, Pregnancy, and Child Development: A Potential Mechanistic Role for Placental Peroxisome Proliferator-Activated Receptors (PPARs). Curr. Environ. Health Rep. 2020, 7, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, E.; Stratakis, N.; Basagaña, X.; Brantsæter, A.L.; Casas, M.; Fossati, S.; Gražulevičienė, R.; Småstuen Haug, L.; Heude, B.; Maitre, L.; et al. Prenatal and Postnatal Exposure to PFAS and Cardiometabolic Factors and Inflammation Status in Children from Six European Cohorts. Environ. Int. 2021, 157, 106853. [Google Scholar] [CrossRef]
- Gardener, H.; Sun, Q.; Grandjean, P. PFAS Concentration during Pregnancy in Relation to Cardiometabolic Health and Birth Outcomes. Environ. Res. 2021, 192, 110287. [Google Scholar] [CrossRef]
- Bloom, M.S.; Commodore, S.; Ferguson, P.L.; Neelon, B.; Pearce, J.L.; Baumer, A.; Newman, R.B.; Grobman, W.; Tita, A.; Roberts, J.; et al. Association between Gestational PFAS Exposure and Children’s Adiposity in a Diverse Population. Environ. Res. 2022, 203, 111820. [Google Scholar] [CrossRef]
- Hamilton, K.J.; Hewitt, S.C.; Arao, Y.; Korach, K.S. Estrogen Hormone Biology. Curr. Top. Dev. Biol. 2017, 125, 109–146. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Krust, A.; Gansmuller, A.; Dierich, A.; Chambon, P.; Mark, M. Effect of Single and Compound Knockouts of Estrogen Receptors Alpha (ERalpha) and Beta (ERbeta) on Mouse Reproductive Phenotypes. Dev. Camb. Engl. 2000, 127, 4277–4291. [Google Scholar] [CrossRef]
- Rønnekleiv, O.K.; Qiu, J.; Kelly, M.J. Arcuate Kisspeptin Neurons Coordinate Reproductive Activities with Metabolism. Semin. Reprod. Med. 2019, 37, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Gieske, M.C.; Kim, H.J.; Legan, S.J.; Koo, Y.; Krust, A.; Chambon, P.; Ko, C. Pituitary Gonadotroph Estrogen Receptor-Alpha Is Necessary for Fertility in Females. Endocrinology 2008, 149, 20–27. [Google Scholar] [CrossRef]
- Couse, J.F.; Bunch, D.O.; Lindzey, J.; Schomberg, D.W.; Korach, K.S. Prevention of the Polycystic Ovarian Phenotype and Characterization of Ovulatory Capacity in the Estrogen Receptor-Alpha Knockout Mouse. Endocrinology 1999, 140, 5855–5865. [Google Scholar] [CrossRef]
- Houck, K.A.; Patlewicz, G.; Richard, A.M.; Williams, A.J.; Shobair, M.A.; Smeltz, M.; Clifton, M.S.; Wetmore, B.; Medvedev, A.; Makarov, S. Bioactivity Profiling of Per- and Polyfluoroalkyl Substances (PFAS) Identifies Potential Toxicity Pathways Related to Molecular Structure. Toxicology 2021, 457, 152789. [Google Scholar] [CrossRef]
- Zhao, Z.; Bo, Z.; Gong, W.; Guo, Y. Inhibitor of Differentiation 1 (Id1) in Cancer and Cancer Therapy. Int. J. Med. Sci. 2020, 17, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Hu, J.; Chen, F.; Lecomte, N.; Basnet, H.; David, C.J.; Witkin, M.D.; Allen, P.J.; Leach, S.D.; Hollmann, T.J.; et al. ID1 Mediates Escape from TGFβ Tumor Suppression in Pancreatic Cancer. Cancer Discov. 2020, 10, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Phelps, D.W.; Palekar, A.I.; Conley, H.E.; Ferrero, G.; Driggers, J.H.; Linder, K.E.; Kullman, S.W.; Reif, D.M.; Sheats, M.K.; DeWitt, J.C.; et al. Legacy and Emerging Per- and Polyfluoroalkyl Substances Suppress the Neutrophil Respiratory Burst. J. Immunotoxicol. 2023, 20, 2176953. [Google Scholar] [CrossRef] [PubMed]
- Grandjean, P.; Timmermann, C.A.G.; Kruse, M.; Nielsen, F.; Vinholt, P.J.; Boding, L.; Heilmann, C.; Mølbak, K. Severity of COVID-19 at Elevated Exposure to Perfluorinated Alkylates. PloS One 2020, 15, e0244815. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.; Jöud, A. Susceptibility to COVID-19 after High Exposure to Perfluoroalkyl Substances from Contaminated Drinking Water: An Ecological Study from Ronneby, Sweden. Int. J. Environ. Res. Public. Health 2021, 18, 10702. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, D.; Holding, A.N.; Giorgi, F.M. Web Tools to Fight Pandemics: The COVID-19 Experience. Brief. Bioinform. 2021, 22, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Omoike, O.E.; Pack, R.P.; Mamudu, H.M.; Liu, Y.; Strasser, S.; Zheng, S.; Okoro, J.; Wang, L. Association between per and Polyfluoroalkyl Substances and Markers of Inflammation and Oxidative Stress. Environ. Res. 2021, 196, 110361. [Google Scholar] [CrossRef]
- Barton, K.E.; Zell-Baran, L.M.; DeWitt, J.C.; Brindley, S.; McDonough, C.A.; Higgins, C.P.; Adgate, J.L.; Starling, A.P. Cross-Sectional Associations between Serum PFASs and Inflammatory Biomarkers in a Population Exposed to AFFF-Contaminated Drinking Water. Int. J. Hyg. Environ. Health 2022, 240, 113905. [Google Scholar] [CrossRef]
- Meneguzzi, A.; Fava, C.; Castelli, M.; Minuz, P. Exposure to Perfluoroalkyl Chemicals and Cardiovascular Disease: Experimental and Epidemiological Evidence. Front. Endocrinol. 2021, 12, 706352. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, T.; Walker, D.I.; Thomas, D.C.; Qiu, C.; Chatzi, L.; Alderete, T.L.; Kim, J.S.; Conti, D.V.; Breton, C.V.; et al. Dysregulated Lipid and Fatty Acid Metabolism Link Perfluoroalkyl Substances Exposure and Impaired Glucose Metabolism in Young Adults. Environ. Int. 2020, 145, 106091. [Google Scholar] [CrossRef]
- Fragki, S.; Dirven, H.; Fletcher, T.; Grasl-Kraupp, B.; Bjerve Gützkow, K.; Hoogenboom, R.; Kersten, S.; Lindeman, B.; Louisse, J.; Peijnenburg, A.; et al. Systemic PFOS and PFOA Exposure and Disturbed Lipid Homeostasis in Humans: What Do We Know and What Not? Crit. Rev. Toxicol. 2021, 51, 141–164. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of Its Protective Roles, Measurement, and Biosynthesis. Mol. Aspects Med. 2009, 30, 1–12. [Google Scholar] [CrossRef]
- Dale, K.; Yadetie, F.; Müller, M.B.; Pampanin, D.M.; Gilabert, A.; Zhang, X.; Tairova, Z.; Haarr, A.; Lille-Langøy, R.; Lyche, J.L.; et al. Proteomics and Lipidomics Analyses Reveal Modulation of Lipid Metabolism by Perfluoroalkyl Substances in Liver of Atlantic Cod (Gadus Morhua). Aquat. Toxicol. Amst. Neth. 2020, 227, 105590. [Google Scholar] [CrossRef]
- Ojo, A.F.; Xia, Q.; Peng, C.; Ng, J.C. Evaluation of the Individual and Combined Toxicity of Perfluoroalkyl Substances to Human Liver Cells Using Biomarkers of Oxidative Stress. Chemosphere 2021, 281, 130808. [Google Scholar] [CrossRef]
- He, L.; Liu, Y.; Liu, D.; Feng, Y.; Yin, J.; Zhou, X. Exogenous and Endogenous Serine Deficiency Exacerbates Hepatic Lipid Accumulation. Oxid. Med. Cell. Longev. 2021, 2021, 4232704. [Google Scholar] [CrossRef]
- Payne, M.; Stephens, T.; Lim, K.; Ball, R.O.; Pencharz, P.B.; Elango, R. Lysine Requirements of Healthy Pregnant Women Are Higher During Late Stages of Gestation Compared to Early Gestation. J. Nutr. 2018, 148, 94–99. [Google Scholar] [CrossRef]
- Van Winkle, L.J.; Galat, V.; Iannaccone, P.M. Lysine Deprivation during Maternal Consumption of Low-Protein Diets Could Adversely Affect Early Embryo Development and Health in Adulthood. Int. J. Environ. Res. Public. Health 2020, 17, 5462. [Google Scholar] [CrossRef] [PubMed]
- Schlett, K. Glutamate as a Modulator of Embryonic and Adult Neurogenesis. Curr. Top. Med. Chem. 2006, 6, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Depeint, F.; Bruce, W.R.; Shangari, N.; Mehta, R.; O’Brien, P.J. Mitochondrial Function and Toxicity: Role of the B Vitamin Family on Mitochondrial Energy Metabolism. Chem. Biol. Interact. 2006, 163, 94–112. [Google Scholar] [CrossRef] [PubMed]
- Wielsøe, M.; Long, M.; Ghisari, M.; Bonefeld-Jørgensen, E.C. Perfluoroalkylated Substances (PFAS) Affect Oxidative Stress Biomarkers in Vitro. Chemosphere 2015, 129, 239–245. [Google Scholar] [CrossRef]
- Labine, L.M.; Oliveira Pereira, E.A.; Kleywegt, S.; Jobst, K.J.; Simpson, A.J.; Simpson, M.J. Comparison of Sub-Lethal Metabolic Perturbations of Select Legacy and Novel Perfluorinated Alkyl Substances (PFAS) in Daphnia Magna. Environ. Res. 2022, 212, 113582. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





