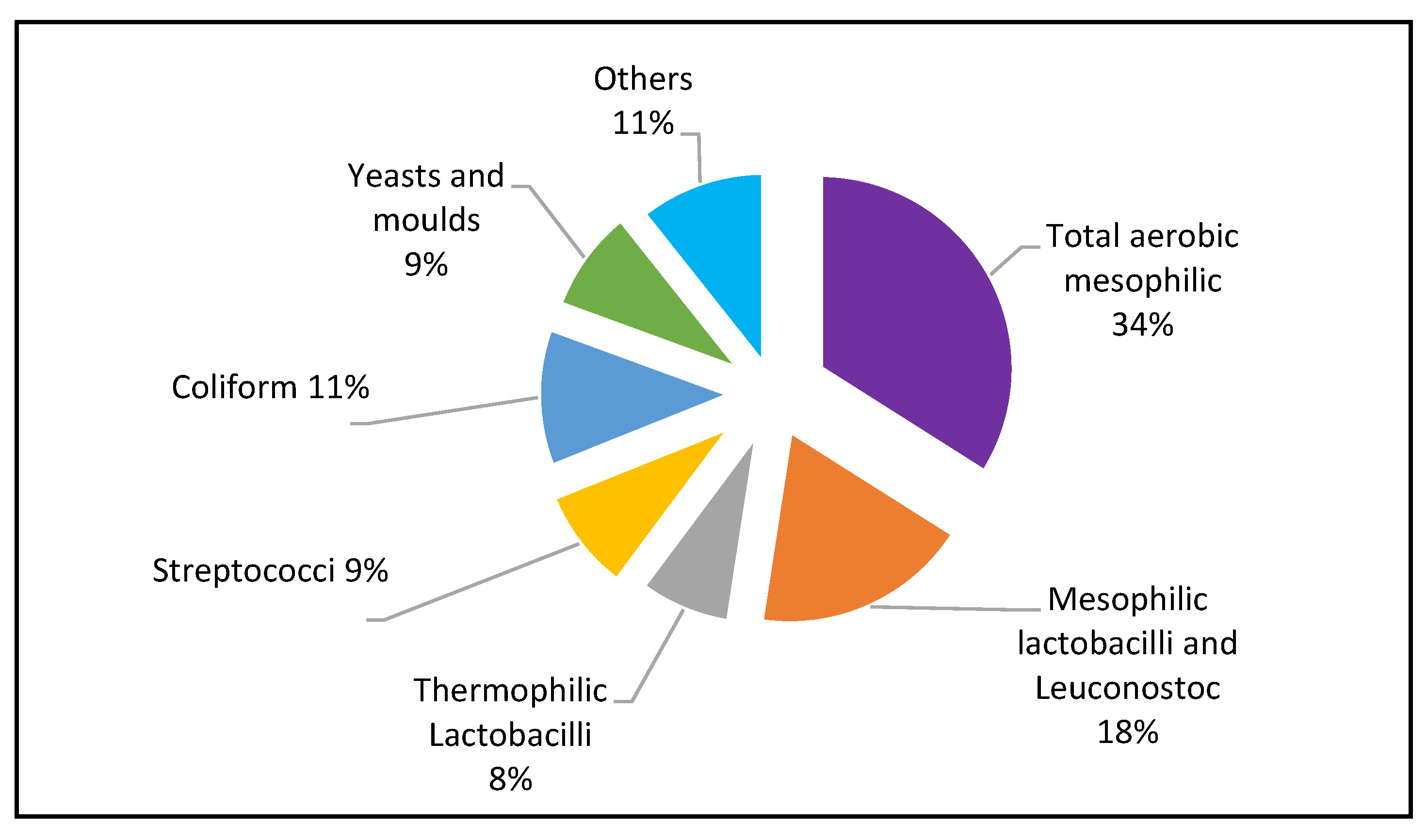1. Introduction
Dairy products especially yoghurt, are important food among cattle keepers in Kenya and Uganda. The traditional African herders ferment raw milk as a means of preservation of the milk due to a lack of refrigeration facilities but also because yoghurt or acidified milk is a quenching drink, especially for the nomads. Milk is a highly nutritious and highly perishable food (1). In the traditional sector of Africa, milking is carried out by hand, in the open air or generally under poor conditions (2). Rarely the udder is washed before milking, if done, the water is of variable sources other than tap water, contributing to the poor quality of milk and milk products (3). Contamination during milking is one of the sources of microorganisms in raw milk (4). Effective hygiene practices to the hands of the milker, washing of the udder and the milking equipment and the general surrounding environment are inadequate (5). Besides, cooling and storage facilities are absent. The traditional farmer mitigates the impact of poor handling by fermenting the milk. The microbiological aim of fermentation is to achieve a pH fall that prevents or reduces the growth of pathogens. However, milk is safe to consume.
Yoghurt is one of the most popular fermented milk products. The milk is fermented spontaneously using raw milk acidified by indigenous microflora in the milk (6). Some of these microorganisms are components of the starter cultures (7) whilst others are spoilage and/or pathogenic microorganisms (8). Although data regarding milk quality and the incidence of pathogens in milk from large commercial dairy farms is well documented (9), there is limited or absence of data in the literature regarding the microbiological quality and pathogen prevalence in Northern Uganda. Understanding microorganisms present in traditional fermented milk is necessary since their presence is directly connected to the quality of the product and the health of the consumers. The microbiological quality assessment of yoghurt is mainly concerned with the protection of the consumers against exposure to any health hazard and ensuring that the material is not suffering microbiological deterioration during its anticipated shelf life (10).
2. Materials and Methods
2.1. Collection of and Transportation of Samples
Six samples of full cream typical 8 h old of typical indigenous traditional African fermented milk were collected in duplicates from a Kalenjin farm in (Kenya) labelled as KE and two farms in Uganda; the Karamojong, labelled (UG 1) and Acholi in Gulu (UG 2). The samples were collected during the rainy season (July - September) in sterile plastic milk bottles. The samples were immediately put on ice in an ice box and transported to the laboratory. On arrival in the laboratory, pH, titratable acidity, and microbiological analyses of the samples were taken within four hours then after 24, 48 and 72 h to check the microbiological growth during storage. Broth dilution and pour plate methods were used for the microbial analyses (11). The remaining samples were then stored in a fridge (4oC). The yoghurt samples were prepared according to the Official Methods of Analysis Chemist (AOAC )(12).
2.2. pH Measurement
The pH of the samples was measured with a Mettler Toledo Delta 320 pH meter, at room temperature (20oC ± 2). The pH electrode was firstly calibrated at pH 4 and 7 with standard buffer solutions. The calibrated pH electrode was inserted into a 10 ml sample. The readings were recorded accordingly. All measurements were carried out in triplicate.
2.3. Titratable Acidity of Fermented Milk Sample
20 g of well-shaken yoghurt or un-fermented milk was weighed accurately into a 250-mL Erlenmeyer flask, 40 mL of boiled and cooled distilled water was added to it. With a sterile pipette, 2-3 drops of the indicator (phenolphthalein) were added to the milk as an indicator of the endpoint. The content of the flask was titrated against 0.1N sodium hydroxide (NaOH) until the sample changed colour to persistent light pink. The initial and final readings on the meniscus burette were recorded, prior to starting the titration and at the endpoint, respectively. The amount (mL) of 0.1N NaOH titrated was calculated by subtracting the initial volume from the final volume to give the amount of NaOH used to reach the endpoint. This was performed at least three times per sample. The per cent lactic acid was then calculated using the equation Eq [
1] below:
where:
Vt= Volume of titrant (ml NaOH)
N = Normality of titrant
90 = Equivalent weight for lactic acid
Vs = Volume of sample used (ml yoghurt/milk)
2.4. Sample Preparation for Analysis
10 millilitres (ml) of each sample were aseptically weighed and homogenised with 90 ml of sterile quarter-strength Ringer’s Solution (pH 7.2) using a Stomacher lab-blender (Seward Medical, London, UK) for 2 minutes. Serial dilutions (10−1 to 10−8) were prepared in the same diluent and duplicate counting plates were prepared. For pour plating, one millilitre of the sample was taken from the chosen dilution to obtain an expected count of 30 to 300 for Aerobic Mesophilic Bacterial Count, 15 to 150 for Coliform count, and 10 to 200 for Yeast and Mould count (13). The media and sample dilutions were gently mixed and allowed to set. All counts were made in duplicate plates. For surface plating, 0.1 ml of the dilutions were spread on the surface of dried media plates.
All media were prepared according to the manufacturers’ instructions. Sterile quarter-strength Ringer’s Solution (BR 0052, Thermo Fischer Scientific, Loughborough, UK) was used as an isotonic diluent for the microorganisms. The quarter Ringer solution was sterilized by autoclaving at 121˚C for 15 minutes. All media were prepared with deionized water. Glassware such as Petri dishes, test tubes, pipettes and flasks were sterilized in a hot oven at 160˚ C for one hour.
2.5. Microbial Analysis
The yoghurt samples were examined for Total Aerobic Mesophilic Bacterial Count. This estimates the number of viable aerobic bacteria per gram or millilitre of the product measured in colony-forming unit per ml (cfu/ml) according to the procedures of Abebe et al., (14). Samples were prepared as above (section 2.4). Aerobic mesophilic bacteria were counted on pour plates of Plate Count Agar (PCA), (Oxoid M325, Basingstoke, Hampshire, UK) incubated in an inverted position at 30oC for 48±1h (15).
Lactobacilli were enumerated on pour plates of de Man Rogosa and Sharpe agar (MRS, LAB098) at pH 5.5 (16) incubated in an inverted position incubated anaerobically in an anaerobic jar at 42±1°C for 48±2 h. A further analysis was carried out on MRS agar + Vancomycin for the enumeration of leuconostocs incubated anaerobically at 32oC for 48±2 h in Anaerobic jars (Biolab and Oxoid) with gas generating kits (Oxoid BR 38B). Streptococci were enumerated on M17 Agar (LAB092) and M17 broth (CM0817, pH 6.5), incubated aerobically for 48 ±2 h at 37±1°C (17).
For Salmonella identification, 25 ml of the sample was pre-enriched with 225 ml of Buffered Peptone Water (BPW) and incubated for 24 h at 37oC. A portion (0.1 ml) of the pre-enriched culture was transferred to 9.9 ml of Rappaport-Vassiliadis (RV) broth and incubated at 42oC for 24 h. A loopful of the enrichment broth was then transferred to Xylose Lysine Deoxycholate (XLD) agar and incubated at 37oC for 24 h. Characteristic Salmonella colonies having a slightly transparent zone of reddish colour and black centre were sub-cultured on nutrient agar and confirmed biochemically using Triple Sugar Iron (TSI) and Simon citrate agar according to the procedures of Gebeheyu et al. (18) with some modification.
Escherichia coli and coliform bacteria were enumerated on Violet Red Bile Agar (VRBA, Oxoid CM 107B Ltd Basingstoke, Hans UK and Violet red bile agar (Oxoid CM 107 with added MUG supplement BRO 71 E), Thermo Fischer Scientific, Loughborough, UK) (19) incubated aerobically for 24±2h at 37±1°C. The supplement containing 4-methylumbelliferyl-B-d-glucuronide (MUG) allowed the separate enumeration of E. coli which contain glucuronidase activity. The presence of E. coli was further tested using indole production in tryptone water (Oxoid, UK) with Kovac’s reagent (Biolife), as previously reported by Moushumi and Prabir (20).
For the general enumeration of Salmonella and Shigella spp., the sample (25 ml) was pre-enriched with 225 ml of Buffered Peptone Water (BPW) and incubated for 24h at 37oC. A portion (0.1 ml) of the pre-enriched culture was transferred to 10 ml Rappaport-Vassiliadis (RV) broth and incubated at 42oC for 24h. A loopful of the enrichment broth culture was then transferred to Xylose Lysine Deoxycholate (XLD) agar and incubated at 37oC for 24h. Characteristic Salmonella colonies having a slightly transparent zone of reddish colour and black centre were sub-cultured on nutrient agar and confirmed biochemically using Triple Sugar Iron (TSI) and Simon citrate agar (21). Red colonies only, were regarded to be Shigella.
Most Probable Number technique was used for the enumeration of Bacillus cereus using selective media mannitol yolk Polymyxin (MYP) B agar and polymyxin pyruvate egg mannitol bromothymol blue agar (PEMBA).(22)
For S. aureus counts were enumerated on Baird–Parker’s medium (Oxoid CM 0275 + SR054C) Staphylococcus aureus was detected using the reference method of the International Dairy Federation (23).
Listeria monocytogenes was enumerated in a well-mixed sample (25 ml), homogenized in 225 ml of Listeria Enrichment Broth A and B then incubated for 24h at 37oC (24) and on Listeria selective medium (Oxford formulation CM856, Oxoid UK) adjunct with Oxoid™ Listeria selective supplement (SR0140, Oxoid, UK). The latter was then incubated for 48 h at 30 °C. A loop full of the enrichment culture broth was streaked in duplicate onto Polymyxin-Acriflavine-Lithium Chloride-Ceftazidime-Aesculin-Mannitol (PALCAM) selective agar (Oxoid, CM877) and incubated for 48h at 37oC. Suspected Listeria monocytogenes colonies were further characterized using Gram staining and catalase test. The color of Listeria spp. colonies typically ranged from greyish green to brownish green with black zones of 1–3 mm diameter of aesculin hydrolysis. Five presumptive Listeria monocytogenes colonies were selected from each Petri dish of selective agar and cultivated on trypticase soy agar medium (CM0131, Oxoid, UK) supplemented with 0.6% yeast extract and subsequently placed into an incubator for 24 h at 30oC to perform further analyses, including examination of non-spore Gram-positive coccobacilli strains for catalase, umbrella growth in motility, nitrate reduction, MR/VP, β-hemolysis production biochemical tests (acid formation from glucose, rhamnose, xylose, and mannitol fermentation) and a further characterised using Gram stain and catalase test were carried out (24).
Yeast and mould counts were enumerated on Malt Extract Agar (MEA) (1.5% Agar No 2) (Oxoid) and Potato Dextrose Agar (+0.005 g/L chloramphenicol). The plates were incubated at 20 and 25 ± 1°C for 5 days. Yeast and mould colonies were counted separately (25).
Analytical Profile Index (API) Biochemical Test
The analytical profile index or API is a biological classification of bacteria based on biological tests, allowing fast identification. This system is developed for quick identification of clinically relevant bacteria and because of this, only known bacteria could be identified. The Biochemical and Physiological tests were carried out with the appropriate API strips to identify the presumptive bacteria.
Table 1.
Summary of culture and media used for the isolation of microorganisms in traditional African fermented milk (cfu/ml).
Table 1.
Summary of culture and media used for the isolation of microorganisms in traditional African fermented milk (cfu/ml).
| Medium for growth |
Microorganisms |
Time (Hours) |
Growth condition and incubation Temperature |
Growth condition and incubation Temperature |
| Plate Count Agar (Oxoid M325) |
Total aerobic mesophilic aerobic bacteria |
48±2h |
aerobic 30±1oC |
aerobic 30±1oC |
| MRS agar, LAB098) |
Mesophilic Lactobacilli
|
48±2h |
aerobic 35±1oC |
aerobic 35±1oC |
| MRS agar (LAB 098 + Vancomycin) |
Leuconostoc |
48±2h |
anaerobic 30±1oC |
anaerobic 30±1oC |
| MRS agar (pH 5.5) LAB098 |
Thermophilic Lactobacilli
|
48±2h |
anaerobic 42±1oC |
anaerobic 42±1oC |
| MRS agar (pH 6.) LAB098 |
Thermophilic Lactococci |
48±2h |
anaerobic 42±1oC |
anaerobic 42±1oC |
| M17 agar (LAB 092) |
Mesophilic Streptococci
|
48±2h |
anaerobic 30±1oC |
aerobic 35±1oC |
| Violet Red Bile Lactose agar with MUG supplement BRO 71 E), |
Non-Sorbitol E. coli
|
24 ±2h |
aerobic 37±1oC |
aerobic 37±1oC |
| Violet Red Bile Agar (VRBA) |
Total coliform |
24 ±2h |
aerobic 30±1oC |
aerobic 30±1oC |
| XLD |
Salmonella and Shigella spp. |
24 ±2h |
aerobic 37±1oC |
aerobic 37±1oC |
| Baird–Parker’s medium (Oxoid CM 0275 + SR054C) |
Staphylococcus aureus |
24 ±2h |
aerobic 37±1oC |
aerobic 37±1oC |
| Listeria Enrichment Broth A and B |
Listeria. Monocytogenes |
24 ±2h |
aerobic 30±1oC |
aerobic 30±1oC |
|
B. cereus agar |
B. cereus |
|
aerobic 30±1oC |
aerobic 30±1oC |
| 1.5% Malt Extract and Agar No. 2 |
Yeast and mould |
|
aerobic 25±1oC |
aerobic 25±1oC |
| PDA + chloramphenicol |
Mould |
|
aerobic 30±1oC |
aerobic 30±1oC |
3. Results
3.1. The Physiochemical Properties
Table 2 shows the mean pH and titratable values of the yoghurt samples.
The mean titratable acidity of the samples was 1.26 ± 0.1% in UG 1 sample, 0.92 ± 0.1% in UG 2 and TA 0.7± 0.1 % in the KE samples.
3.2. The Microbial Counts
The Total Aerobic Mesophilic Bacterial count is an indicator of the sanitary conditions of handling of raw milk and good-quality milk products (26). Table 3 shows the summaries of the microbial counts obtained from the tested traditional fermented milk samples. The results show unhygienic quality. The mean Total Aerobic Mesophilic bacteria counts in the samples were 5.14 x 109 cfu/ml.
The mean counts of mesophilic Lactobacilli on MRS (35±1oC) were 1.74 x 108 in UG 1, 2.12 x 106 in UG 2 and 5.9 x 107 in KE respectively (Table 3). The mean counts of mesophilic Lactococci on M17 agar (30±1oC) were 2.43 x 108 in UG 1 and UG 2 and 6.2 x 107 in KE. The mean counts of thermophilic lactobacilli on MRS (42oC) were 2.87 x 107, 1.25 x 109 cfu/ml and 1.48 x 106 cfu/ml in the UG 1, UG 2 and KE samples respectively. The mean Streptococci was higher in UG 1 (108 cfu/ml) followed by UG 2 (107 cfu/ml) and 106 cfu/ml KE. Table 3 shows the mean coliform counts were high in UG 1 (2.12 x 105), but 2 logs cfu/ml was lower in UG 2 and KE samples (2.12 x 103 cfu/ml). An important finding was the presence of E. coli (mean counts x 103 cfu/ml) and Salmonella (mean counts x 102 cfu/ml). The mean S. aureus counts were x 103 cfu/ml in UG 1 and e samples but higher (x 105 cfu/ml) in UG 2 (Table 3). The mean L. monocytogenes counts were 1.7 x 102 cfu/ml in UG 1 and 1.2 x 103 cfu/ml in KE but not detected in UG 2. Yeasts and mould counts were between x 107 -1011 cfu/ml.
Table 3.
Mean microbial counts in traditional African fermented milk (cfu/ml).
Table 3.
Mean microbial counts in traditional African fermented milk (cfu/ml).
| Medium |
UG 1 |
UG 2 |
KE |
Growth condition |
Plate Count Agar (PCA)
(for Total aerobic mesophilic bacteria) |
9.7 x 109
|
3.3 x 109
|
2.53 x 109
|
aerobic 30±1oC |
| (MRS agar) (for Mesophilic Lactobacilli) |
1.74 x 108
|
2.12x 106
|
5.9 x 107
|
aerobic 35±1oC |
MRS agar + Vacomycine
(for Leuconostoc) |
1.55 x 106
|
1.61x 105
|
6.2 x 107
|
anaerobic 30±1oC |
| M17 agar (for Mesophilic lactococci) |
1.17 x 108
|
3.7 x 108
|
6.2 x 107 |
anaerobic 30±1oC |
| MRS agar (Thermophilic Lactobacilli) |
2.87 x 107
|
1.54x 109
|
8.0 x 106
|
anaerobic 42±1oC |
|
MRS agar (for Thermophilic Lactococci |
2.63 x 106
|
1.25x 109
|
1.48 x 106
|
anaerobic 42±1oC |
| M17 agar (for Streptococci) |
1.74 x 108
|
6.2 x 107
|
3.7 x 106
|
aerobic 37±1oC |
| Violet Red Bile Lactose agar (for Non-Sorbitol E. coli) |
2.92 x 103
|
1.61x 103
|
4.1 x 103
|
aerobic 37±1oC |
Violet Red Bile Agar (VRBA),
(coliforms counts) |
2.12 x 105
|
4.2 x 103
|
1.56 x 103
|
aerobic 30±1oC |
| XLD (for Salmonella spp.) |
1.7 x 102
|
1.4 x 103
|
1.5 x 102
|
aerobic 37±1oC |
| XLD (for Shigella spp.) |
ND |
ND |
ND |
aerobic 37±1oC |
| Baird–Parker’s medium (for S. aureus) |
1.18 x 103
|
1.4 x 105
|
1.11 x 103
|
aerobic 37±1oC |
Listeria Enrichment Broth A and B
(for L. monocytogenes) |
1.7 x 102
|
ND |
1.2 x 103
|
aerobic 37±1oC |
| Bacillus cereus |
2.1 x 102
|
2.12x 103
|
1.35 x 103
|
aerobic 30±1oC |
| Yeasts and moulds (on 1.5% Malt Extract and Agar No. 2)) |
2.07 x 107
|
5.4 x 109
|
3.9 x 1011
|
aerobic 25±1oC |
| PDA + chloramphenicol (for Mould) |
4.0 x 108
|
6.7 x 107
|
2.16x 1010
|
aerobic 30±1oC |
3.3. The Microbial Analysis
22 different types of microorganisms were grouped according to their colony phenotypes and Gram stains. The prevalence of each group of microorganisms is presented in a pie chart (
Figure 1) expressed as percentage of the total number of the isolates (n) obtained from the samples. Aerobic mesophilic bacteria were the largest group of microorganisms comprising 34% of the total count. (
Figure 1). Mesophilic lactobacilli and
Leuconostoc group comprised 18%, while 8% of the isolates were thermophilic lactobacilli spp. and 9% were Streptococci species. The coliforms encompassed 11% of the counts. Yeasts and moulds (9%) and others (unidentified) microorganisms were 11% of the total count.
Identification of the Isolates with API Biochemical Analysis
After the Gram stain, the isolates were subjected to API biochemical analysis. Table 3 shows the predominant presumptuous microorganisms identified by API biochemical analysis.
Mesophilic aerobes grown on M17 and MRS agars at 35°C, dominated the samples. Bacillus cereus and S. aureus had the highest number of microorganisms in the group.
Lactic acid bacteria were the dominant groups of bacteria in all the samples identified by their phenotypic and microscopic appearance. They were grouped under Lactobacillus, Streptococcus, and Lactococcus spp. The phenotypic characteristics of the presumptuous Lactobacillus bulgaricus and Streptococcus thermophilus were comparable to the laboratory collection of Lactobacillus bulgaricus NCIMB 11778 and Streptococcus thermophilus NCIMB 10378, when Gram-stained and in API biochemical analysis. Ten isolates were grouped under Streptococci spp., and another 10 under the Lactobacilli of which, three isolates were presumptuously identified as Beta-bacterium (heterofermentative lactobacillus). Others included Leuconostoc and Enterococcus spp.
From the UG 1 samples, twelve different colonies were isolated and grouped according to their Gram stain reactions. From the Gram stain, three of the ten isolates; appeared rod-shaped and according to API 50 CH biochemical test, it was grouped as Lactobacillus spp. Ten of the twelve isolates were Gram-positive and coccoid in shape. They were grouped under the Streptococcus spp. Of the mesophilic lactobacilli, API test showed presumptuous L. cremoris, L. mesenteroides, Lactococcus spp. L. lactis spp.
Table 3.
Phenotypic and morphological characteristics of yeasts and moulds isolated from the samples.
Table 3.
Phenotypic and morphological characteristics of yeasts and moulds isolated from the samples.
| Isolate |
Macro-colony morphology (margin, colour, elevation, cell appearance |
UG1 |
UG2 |
KE |
| 1 |
Cream, smooth, oval shape entire and ellipsoidal cell |
√ |
√ |
√ |
| 2 |
Undulating, white top with green base, slightly convex, spheroidal to short ellipsoidal. (Blue colony on Kluveymyces Differential Medium) |
√ |
√ |
√ |
| 3 |
Yellow-green, powdery and pale yellowish on reverse Aspergillus flavus |
√ |
ND |
√ |
| 4 |
Dirty white with yellow spores at the centre, base orange, slightly radially furrowed (Microsporum spp.) |
√ |
√ |
√ |
| 5 |
Cream-yellow, powdery, and pale yellowish on reverse, capsulate margin, slightly raised centre, filamentous cells, > 85 mm colony diameter. |
√ |
√ |
√ |
| 6 |
White to cream, yellowish, wrinkled, nearly flat elevation, oval cells & Ellipsoidal |
√ |
√ |
ND |
| 7 |
White to cream coloured, flat with aerial mycelium (Aspergillus spp) |
√ |
√ |
√ |
| 8 |
Green with a red base |
√ |
√ |
√ |
| 9 |
White at the base and black spores at the top |
√ |
√ |
√ |
| 10 |
White pin head, clear zones around the colony |
√ |
√ |
√ |
| 11 |
Black, yellow to pale cream in the centre (Aspergillus) |
ND |
ND |
ND |
| 12 |
White measuring 1-4 mm, opaque and flat. Ropy to the touch |
√ |
√ |
√ |
| 13 |
Straw cream at the centre, base orange, slightly radially furrowed |
√ |
√ |
ND |
| 14 |
Well-formed white colonies (grew well on M17 too) (Aspergillus spp)) |
√ |
√ |
√ |
| 15 |
Green and pale yellow on reverse (Penicillium) |
ND |
ND |
ND |
| 16 |
White base with black conidiophores |
√ |
|
√ |
| 17 |
Greenish black, white mycelia at the margin, white in the centre (Rhizopus sp.) |
ND |
ND |
ND |
| 18 |
Greenish with surrounded by creamy-white ring at the margin (Penicillium) |
ND |
√ |
ND |
| 19 |
White to cream, smooth, glaucous dark green on obverse and pale yellow on reverse, |
√ |
√ |
√ |
| 20 |
Cotton white to cream on the obverse and yellow to orange on the reverse with dark brown exudate |
√ |
√ |
√ |
| 21 |
White colony, opaque and flat |
√ |
√ |
√ |
| 22 |
Bright red colonies |
ND |
ND |
ND |
Many of the Gram-positive mesophilic groups were presumptuously identified as belonging to the Bacillus, Staphylococcus and Enterococcus spp. which are common in the cattle environment. S. caprae which colonizes healthy human skin, nails, and nasal mucosa was identified in UG 2 sample. The Gram-positive diplococci or pairs or short chained isolates were grouped under the Enterococcus. The coliforms were dominated by E. coli. With API 20E, presumptuous E. faecalis, E. agglumeritus, E. durans were preliminarily identified. From the KE sample, seventeen different isolates were grouped. API Analyses showed that presumptively, four of the isolates were of Lactobacilli spp. Others were less distinct but grouped as Staphylococci species. Table 3 shows the phenotypic and morphological characteristics of some of the yeasts isolated from the traditional fermented milk. The results showed the diversity of yeasts and moulds isolated from the various samples (25). It was not easy to identify the isolates to the species level.
4. Discussion
In this study, the physicochemical, and microbiological attributes of typical traditional African yoghurt from Northern Uganda and western Kenya, were assessed to establish the status of microbial risks associated with the traditional fermented milk. Sour milk is processed at the household level by leaving the fresh raw milk to ferment naturally for 1 -3 days at ambient temperature. Fermentations are carried out spontaneously in gourds or earthenware pots. Sometimes sour milk from previous batches is added to speed up the fermentation process (26).
In the three days from production to analysis), the pH of the tested traditional fermented milk was low (2.9 -3.6). Makut et al. (27), Mathara et al. (28), Ifeanyi et al. (29) and Digbabul et al. (31) reported pH results ranging from pH 3.5- - 5.11 for traditionally fermented yoghurt. The low pH in this study was reflected in the titratable acidity which was 1.26 ± 0.1, 0.71 ± 0.1; 0.92 ± 0.1% for UG 1, UG 2 and KE respectively.
The Aerobic Mesophilic Bacterial count (AMBC) in fermented milk indicates the sanitary conditions during the production and handling of raw milk or post-fermentation contamination (32). The average AMBC obtained in the current study was very high (x 109 cfu/ml). This number failed to comply with the Health Protection Agency guidelines (33) for acceptable microbial limit (x 106 cfu/ml) in fermented milk products. In regards to the microbial quality of the tested samples, the AMBC was not significantly different (p>0.05) from each other.
The mean counts for mesophilic lactobacilli were highest in UG 1 (x 108) followed by KE (107), and lower in UG 2 (106 cfu/ml). However, the thermophilic lactobacilli were 107 cfu/ml in UG 1 but higher in UG 2 samples (109 cfu/ml) although lower (106 cfu/ml) and 3 logs lower in KE samples and UG 1 respectively. A high level of thermophilic lactobacilli was recovered in UG 2 sample with counts of 109 cfu/ml. The high AMBC (106 - 109 cfu/mL) could come from the already high numbers of bacteria in raw milk as observed by other researchers in raw milk taken from different areas of Africa (5, 8, 10, 12, 13, 16). Hot weather at the production areas also enhances the growth of microorganisms in the milk if contaminated before or during processing (33). Besides having high counts of AMBC, the yoghurt samples had a rich diversity of microorganisms, predominated by lactic acid bacteria and yeasts.
In Africa, fermentation is spontaneous with back slopping using the previously fermented milk as starters rather than specific starter cultures as elsewhere in the world. Thus it comes as no surprise that this typical African fermented milk harboured such a rich and diverse type of microbes, especially lactic acid bacteria. The level of the bacteria recovered in the samples is in agreement with those reported for Zambia by Yambayamba and Zulu, (5). Similarly, high bacterial counts (5.6 -7.5 log cfu/ml) were reported by Abdalla and Abdel Nabi (34) in zabadi (x 108 cfu/ml) of Sudan and Egypt; (34); in the traditional fermented milk of Zimbabwe (x 108 cfu/ml) (35); in the traditional fermented milk of Morocco (36). In South Africa, a high number of microorganisms (x 108 cfu/ml) was reported too (37, 38). This high number of mesophilic bacteria could be due to the warm ambient temperature (28-35oC) of the natural fermentation of the milk at the time. The presence of microorganisms in traditional fermented milk depends on the nature of the fermented milk and the temperature of the regions where they were obtained from (39). It also follows the level of contamination at the production site. Contamination can occur during milking, especially where hygiene practices such as pre-milking udder washings are poor (40). It is therefore important to remove both visible dirt and bacteria from the outer surface of the udder which are likely to contribute to the contamination of the raw milk. Most of the traditional herders in the region of study do not practice pre-udder washing (14).
Furthermore, other workers (6, 41, 42) noted that mesophilic bacteria such as Leuconostoc spp. are observed in traditional fermented milk products in regions with cold climates. Whereas, in warm regions, thermophilic bacteria such as Lactobacillus and Streptococcus dominate (43). This could explain the high numbers of mesophilic bacteria in these samples because they were fermented and collected during the rainy season and cooler months (25-35°C) in both Kenya and Uganda.
Lactic acid bacteria were in the range of 108 log cfu/ml. The counts of thermophilic lactobacilli and lactococcal were 2.87 x 107 in UG 1 and 1.54 x 109 in UG 2 and 1.74 x 108 in KE samples. Obadai and Dodd (44) reported counts of LAB in the range of x 108 to x 1010 in nyarmie, the traditional fermented milk of Ghana. This agrees with those reported by Owusu-Kwarteng et al. (45) and in nunu, of Ghana’s traditional fermented milk product and by Mathara et al., (46) of kule naoto in Kenyan traditional milk. In this report, the most dominant streptococci were S. thermophilus. The abundance of Lactobacillaceae and Streptococcaceae over other families suggested the dominance of LAB during the fermentation process, and this was equally reported in other studies (43, 46, 47). In addition, this high number of lactic acid bacteria could be due to the natural selection and/or temperature of fermentation. Fewer leuconostocs suggest that this group are unable to compete with other lactic acid bacteria in mixed cultures (48). This gives them a selective disadvantage over other lactic acid bacteria (48) and a selective advantage over thermophilic bacteria. Lactic-acid bacteria are generally recognized as safe (GRAS) as well as being part of the natural microbiota of various foods and are often used as starter cultures. Many LAB such as Lactococcus, Leuconostoc, Pediococcus, and Lactobacillus species demonstrate success in inhibiting microorganisms and other pathogens in yoghurt (49, 50).
Coliforms were high in UG 1 sample (x 105 cfu/ml) but lower in UG 2 and KE (x 103 cfu/ml) samples in this study. Counts of coliform in UG 1 samples suggested poor handling and processing conditions of the milk (51). Other pathogens such as E. coli, Salmonella species, Bacillus cereus and S. aureus were also recovered with counts between 103 and 104 cfu/ml. Hamama (52) reported similar results in ‘Lben’ and ‘Jben’ the Moroccan traditional fermented dairy products. Salmonella species are known pathogens that can cause food poisoning if contaminated milk or milk products are consumed. In the present study, Salmonella species were recovered in UG 1 samples irrespective of the low pH (pH 2.9). Salmonella, as enteric pathogens, encounter a low pH value in the environment, especially during its transit in the host. According to Foster (53), Salmonella species such as Salmonella typhimurium periodically confront acid environments during its life. In an experiment, Liyuwork et al. (54) observed antimicrobial resistance in Salmonella species isolates from dairy products in Addis Ababa. Chatti et al., (56) reported acid-resistant Salmonella isolated from food and waste water in Tunisia. Although Salmonella is supposed to be destroyed or inactivated during fermentation of highly acidic products such as yoghurt in which the pH value is less than 4.55, this is not the case in this study where the acidity is low, yet the pathogen was still detected in some of the samples. This could be due to the fact that Salmonella can survive in various environmental niches for long periods of time (53).
Many diseases are transmissible via milk products and pathogenic and acid-tolerant bacteria in acidic foods have recently been a cause of public health concern. Unpasteurised milk has been a major vehicle for the transmission of pathogens such as E. coli, L. monocytogenes and Salmonella (57). It can be assumed that other sources of contamination by microorganisms are unclean teats, milkers’ hands and the use of the same milking and fermentation vessels (58). The presence of coliforms has long been thought to indicate faecal contamination (57, 58), however, recent reports regarding this diverse group of bacteria indicate that only a fraction are faecal in origin, while the majority are environmental contaminants (59). Low counts of coliforms might be due to the high acidity of the products. However, coliforms were still recovered even in such high acidity product
Yeast and mould can build up on equipment surfaces and under the surface of the package lid which often contaminate the fermenting milk (59). The presence of yeast and mould in milk and its product is undesirable as they can cause changes in the product with reduced shelf life rendering it unacceptable for consumption (60). In this study, yeasts and moulds formed a high number of the components of the microbial population. The high number of yeasts and fungus in the products suggests a high presence of yeasts in the environment where the milk was fermented.
In addition, it indicates that yeasts are a significant part of the microflora of these naturally fermented milk products in these areas. Yeasts could be a common part of the flora of the milking parlour (25) and milk containers or fermentation vessels and could impact the overall quality of these products. Yeasts and mould can produce toxic metabolites which are not destroyed during fermentation. This finding agrees with the reports of Akabanda et al. (61) and Savova and Nikolova (62). In this study, several yeasts and mould species were also recovered in the traditional yoghurt similar to the report of Savova and Nikolova (62). Growth of yeasts is mostly undesirable in milk and dairy products because these microorganisms harbour a high risk of spoilage. However, yeasts play an important role in foodstuffs, as they are able to grow in a broad range of pH environments and usually adapt to coexistence with LAB in acidic environments (63). Saccharomyces cerevisiae, a lactose fermenting yeasts present in the yoghurt might have contributed to lowering the acidity of the products. Yeasts also contribute to the flavour of the product (25).
Getachew et al. (64) commented that the variety of microorganisms present in naturally fermented milk products creates rich and full flavours that are hard to imitate. However, the use of appropriate traditional equipment is crucial to pathogen control. Additionally, the equipment must be easy to clean and sanitize, to prevent the formation of niches where microorganisms can grow and settle, forming biofilms (65). Furthermore, lack of pasteurisation, inadequate storage and maturation conditions, the temperature of water used for cow udder washing, the practice of mixing milk lots, the type of milk container, use of refrigeration, and milk filtration are some of the major risk-enhancing factors in traditional milk fermentation (56).
To minimize contamination during milking, effective hygienic practices need to be applied to the hands of the milkers and udder of the animals, and the general environment such as reducing faecal sources of contamination, (66) as well as the milking equipment (67). Washing hands without detergent may not improve the hygienic conditions of milk and milk products (68). Poor drying practices following hand washing and the use of old and unclean clothes for other farm activities is a risk factor for milk contamination (69). Traditional knowledge plays a role in awareness creation in the community to manage their day-to-day activities in livestock management (70). The main advantage of spontaneous fermentation processes is that they are appropriate to rural situations, since they were, in fact, created by it.
Several reports on the microbiological quality of fermented milk of Africa from different countries give knowledge of the various microorganisms in yoghurt and other traditional fermented milk (70). However, there are still gaps that need to be filled regarding pathogen control in traditional milk fermentation environments as microorganisms in traditional dairy products continue to be identified. Although many countries have milk safety regulations and surveillance systems for monitoring foodborne pathogens to ensure food safety, such surveillance of milk and milk products is not conducted on a routine basis in most African countries. Consistency in the day-to-day implementation of milking procedures is an important part of good dairy farming practices for milking. The need to use the guide developed by Food and Agriculture Organisation (71) would help to improve the standard of milk quality at traditional farms and farming practices.
5. Conclusions
This study of the traditional fermented milk of Northern Uganda and Kalinjin showed that although the products had low pH the products, the yoghurt still harboured a high and variable load of bacteria thus, the products could pose health risks to the consumers. The presence of microorganisms such as E. coli, Salmonella spp. Bacillus spp. and Staphylococci spp indicate the need for improvement of hygiene in traditional fermented milk production among small traditional farmers. The cross-contamination of milk products with microorganisms is an ongoing risk throughout traditional milk production.
To improve the quality of the yoghurt, training and awareness raising on hygiene practices on the farm including cleaning and sanitizing hands before and after milking, udder washing, drying the udder with clean dry cloths, proper washing of milk equipment, and how to avoid cross-product contamination from the environment and equipment should be stepped up. This should include everyone in the household who is involved in milking and production, especially women. Additionally, a clear message on the dangers of consuming dairy products made from raw milk must be emphasised. Besides, the frequency of inspection of the dairy facilities cannot be overlooked.
Author Contributions
Conceptualization, BAO, DJH; Formal analysis, BAO, DJH, HG; Data Collection, BAO; Data Analysis, BAO, DJH, HG, EOG; Writing-original draft, BAO, DJH, HG, EOG; Writing-review BAO, DJH, HG, EOG; Methodology BAO, DJH HG, EOG; Supervision; HG, EOG; Editing, HG, DJH, EOG. The author(s) read and approved the final manuscript.
Funding
This research received no external funding but the support of the Government of the Republic of South Sudan through the salary payment of Betty Ogwaro during the study period.
Data Availability Statement
The data presented in this study are openly available.
Acknowledgments
We sincerely appreciate the support and collaboration of Dr. Lensio Onek Angole from the University of Juba in facilitating the sample collection.
Conflicts of Interest
The author declares no conflict of interest.
References
- Ahmad T., Butt M.Z., Aadil R.M., Abdallah Inam-ur-Raheem A., Bekhit M., El Din A. Guimaraes J.T., Balthaza, C.F., Rocha, R.S., Esmerino, E.A., Freitas, M.Q, Silva, M.C., Sameen, A., Cruz, A.G. Impact of nonthermal processing on different milk enzymes. Int J of Dairy Tech 2019, 481–495. [Google Scholar]
- Leone C, Thippareddi H, Ndiaye C., Niang I., Diallo Y, Singh M. Safety and Quality of Milk and Milk Products in Senegal-A Review. Foods. 2022, 2;11(21):3479. PMID: 36360092; PMCID: PMC9656659. [CrossRef]
- Moatsou G., Moschopoulou E.. Microbiology of raw milk. In: Özer BH, Akdemir-Evrendilek G, editors. Dairy microbiology and biochemistry: recent developments: Taylor & Francis Group, LLC; 2015:1–38.
- Aliyo A, Teklemariam Z. Assessment of Milk Contamination, Associated Risk Factors, and Drug Sensitivity Patterns among Isolated Bacteria from Raw Milk of Borena Zone. Ethiopia. J Trop Med. 2022. PMID: 35769792; PMCID: PMC9236756. [CrossRef]
- Yambayamba K., E. & Zulu M. P. Influence of the milking environment on the microbial quality of raw milk produced by smallholder farmers in Magoye. Uni of Zambia J of Sci and Tech. 2011, 15, 37–43. [Google Scholar]
- Leone C, Thippareddi H, Ndiaye C., Niang I, Diallo Y, Singh M. Safety and Quality of Milk and Milk Products in Senegal-A Review. Foods. 2022. 2:11(21):3479. [CrossRef]
- Landis EA., Oliverio A.M., McKenney E.A., Nichols L.M., Kfoury N., Biango-Daniels M., Shell L.K., Madden A.A., Shapiro L., Sakunala S., Drake K., Robbat A., Booker M., Dunn, RR., Fiere N., Wolfe BE. The diversity and function of sourdough starter microbiomes. Ecology Micro and Inf Dis. 2021. [CrossRef]
- Deddefo A., mamo G., Asfaw M., and Amenu, K. Factors affecting the microbiological quality and contamination of farm bulk milk by Staphylococcus aureus in dairy farms in Asella, Ethiopia. BMC Micro. 2023. 2365. [CrossRef]
- Washaya, S. , Jakata C., Tagwira M., Mupofu T. Bacterial Milk Quality along the Value Chain in Smallholder Dairy Production. Scientific World Journal. 2022, 21, 7967569. [Google Scholar] [CrossRef]
- El-Ansary, M.A. Assessment of Microbiological Quality of Yoghurt Sold in El-Behera Governorate. Alexandria J of Vet Sci. 2014, 43, 52–57. [Google Scholar] [CrossRef]
- Mukisa, I.M., Kyoshabire. Microbiological, physicochemical, and sensorial quality of small-scale produced stirred yoghurt on the market in Kampala city, Uganda. Nutr. Food Sci 2010, 40, 409–418. [Google Scholar] [CrossRef]
- AOAC Official Methods of Analysis Chemist (18th ed.) 2006.
- Duncan S. E. , Yaun , and Sumner S. S., and Bruhn J., Tech. Comm. , “Chapter 09 Microbiological Methods for Dairy Products”, Standard Methods for the Examination of Dairy Products. American Pub Health Ass. Washington, D.C., USA. 2012. [CrossRef]
- Abebe B., Yilma Z., and Nurfeta. Hygienic and microbial quality of raw whole cow’s milk produced in Ezha district of Gurage Zone, Southern Ethiopia. Wudpecker J of Agric Res. 2012, 1, 178–187. [Google Scholar]
- IOS (International Organization for Standardization). Milk and Milk Products-General Guidance for the Preparation of Test Samples, Initial Suspensions and Decimal Dilutions for Microbiological Examination: Lait Et Produits Laitiers-Lignes Directrices Générales Pour la Préparaton Des Échantillons Pour Essai, de la Suspension Mère Et Des Dilutions Décimales en Vue de L'examen Microbiologique. 2001. ISO.
- De Man, J.C. , Rogosa, M., Sharpe E.M. A medium for the cultivation of Lactobacilli. J App Bact. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Terzaghi, B.E. , Sandine W.E. Improved medium for lactic Streptococci and their bacteriophages. Appl Microb. 1975, 29, 807–813. [Google Scholar] [CrossRef]
- Gebeyehu, A. , Taye M. & Abebe R. Isolation, molecular detection, and antimicrobial susceptibility profile of Salmonella from raw cow milk collected from dairy farms and households in southern Ethiopia. BMC Microbiol 2022, 22, 84. [Google Scholar] [CrossRef]
- Feng P., Weagant F.S., Grant M.A., Burhardt W. Enumeration of Escherichia coli and the Coliform Bacteria. Bacteriological Analytical Manual (BAM). 2020. Chapter 4.
- Moushumi, B. and Prabir K.S. Microbiological quality of some retail spices in India. Food Res. Int. 2003, 469–474. [Google Scholar]
- IOS (International Organization for Standardization). Milk and Milk Products-General Guidance for the Preparation of Test Samples, Initial Suspensions and Decimal Dilutions for Microbiological Examination: Lait Et Produits Laitiers-Lignes Directrices Générales Pour la Préparaton Des Échantillons Pour Essai, de la Suspension Mère Et Des Dilutions Décimales en Vue de L'examen Microbiologique. 2001. ISO.
- Elbassiony, T.A. , Abd EL Mgeed A.S.M.; Ewida R.M. Prevalence of Some Spore Forming Food Poisoning Bacteria in Milk and Some Milk Products. J. Adv. Vet. Res. 2021, 11, 243–246. [Google Scholar]
- International Dairy Federation (IDF). International Dairy Federation. Milk and Milk-based Products — Enumeration of Staphylococcus aureus (IDF Standard 1990. 145: IDF, Brussels.
- Nero L.A, de Mattos M.R., Barros Mde A., Ortolani M.B., Beloti V., Franco B.D. Listeria monocytogenes and Salmonella spp. in raw milk produced in Brazil: Occurrence and interference of indigenous microbiota in their isolation and development. Zoonoses Public Health. 2008, 55, 299–305. [CrossRef]
- Gadaga, T.H. The occurrence and diversity of yeasts in Zimbabwean traditional fermented milk and their potential for use as starter cultures. PhD Thesis, Agricultural University of Norway. A°S Norway, ISBN 9-82-575-0444-0. 2000.
- Kunda, B. , Pandey G.S., and Muma J.B. Compositional and sanitary quality of raw milk produced by smallholder dairy farmers in Lusaka Province of Zambia. Livestock Research for Rural Development. 2015, 27. [Google Scholar]
- Makut, D. , Ogbonna A.I., Dalami H. An Assessment of the Bacteriological Quality of Different Brands of Yoghurt Sold in Keffi, Nasarawa State. Nigeria. J of Nat Sci and Res. 2014, 4, 19–22. [Google Scholar]
- Nduko, J.M. , Matofari J.W., Nandi Z.O., and Sichangi M.B. Spontaneously fermented Kenyan milk products. A review of the current state and future perspectives. African J of Food Sci. 2017, 11, 1–11. [Google Scholar]
- Ifeanyi, V.O. , Ihesiaba E.O., Muomaife O.M., & Ikenga C. Assessment of microbiological quality of yoghurt sold by street vendors in Onitsha metropolis, Anambra State, Nigeria. British Microbiology Research Journal. 2013, 3, 198. [Google Scholar]
- Digbabul B, Shember J, Amove J. Physicochemical, microbiological, and sensory evaluation of yoghurt sold in Makurdi metropolis. African Journal of Food Science and Technology 2014, 5, 129–135. [Google Scholar]
- Sulaiman I.M, Hsieh Y.H. Dairy in Human Health and Disease Across the Lifespan. Berlin, Germany: Springer. Foodborne pathogens in milk and dairy products: Genetic characterization and rapid diagnostic approach for food safety of public health importance. 2017: pp. 127–143.
- Health Protection Agency (HPA). Guidelines for Assessing the Microbiological Safety of Ready-to-Eat Foods London: Health Protection Agency. 2009.
- Majoie G.Ã, Mousse W., Haziz S.I.N.., Farid B.A.D., Ahouissou O.R., Adjanohoun A., Lamine B.M. Microbial quality of artisanal yoghurt and Degue products collected in schools of Cotonou and Abomey-Calavi (Benin). Afr. J. Food Sci. 2020, 14, 112–118. [CrossRef]
- Abdalla M.O.M and Abdel Nabi, S.Z. Evaluation of microbiological quality of Sudanese fermented dairy product ‘mish’ during storage. Adv J of Food Sci and Tech 2010, 2, 155–158. [Google Scholar]
- Moonga, H. B. , Schoustra, S. E., Linnemann, A. R., Kuntashula, E., Shindano, J., & Smid, E. J. The art of mabisi production: A traditional fermented milk. PLOS ONE. 2019, 14, e0213541. [Google Scholar] [CrossRef]
- Benkirane G., Ananou S., Dumas E., Ghnimi S., and Gharsallaoui A. Moroccan Traditional Fermented Dairy Products: Current processing Practices and Physiochemical and Microbiological Properties. A Review. J of Micro, Biotech and Food Sci. 2022. [CrossRef]
- Maleke M. S., Adefisoye, M. A., Doorsamy, W., & Adebo, O. A. Processing, nutritional composition and microbiology of amasi: A Southern African fermented milk product. Sci Afri, 2021. 12. [CrossRef]
- Taye, Y. , Degu T., Fesseha H., and Mathewos M. Isolation and Identification of lactic acid bacteria from cow Milk and Milk products. The Sci World Jl. 2021. [CrossRef]
- Jans, C. , Meile L., Kaindi, D. W. M., Kogi-Makau, W., Lamuka, P., Renault, P.,... & Bonfoh, B. African fermented dairy products–overview of predominant technologically important microorganisms focusing on African Streptococcus infantarius variants and potential future applications for enhanced food safety and security. Int J of Food Micro. 2017, 250, 27–36. [Google Scholar]
- Gleeson, D. , O’Brien B., Flynn J., O’Callaghan E., Galli F. Effect of pre-milking teat preparation procedures on the microbial count on teats prior to cluster application. Ir Vet J. 2009, 62, 461. [Google Scholar] [CrossRef]
- Haile, W. , Yilma Z., and Teklegiorgis Y. Incidence of pathogenic and indicator bacteria in raw and pasteurized milk in Hawassa city, rift valley of Southern Ethiopia. Afr J of Food Sci, 2012, 7. [Google Scholar]
- Benkerroom, N. Traditional Fermented Foods of North African Countries: Technology and Food Safety Challenges with Regard to Microbiological Risks. Comp Rev in Food Sci and Food Saf. 2013, 12, 54–89. [Google Scholar] [CrossRef]
- Ismail Ahmed, A. Isolation and Identification of LAB from Sudanese Traditional Fermented Camel (Camelus dromedarius) Milk Gariss. Open J Nutr Food Sci. 2022, 4, 1022. [Google Scholar]
- Obodai M., Dodd, C.E.R. Characterization of dominant microbiota of a Ghanaian fermented milk product, nyarmie, by culture- and nonculture-based methods. J of App Micr. 2006, 100, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Owusu-Kwarteng J. Tano-debrah K. Glover R.L.K., Akabanda F. Process characteristics and microbiology of fura produced in Ghana. Nat and Sci. 2010, 8, 41–51. [Google Scholar]
- Mathara, J.M. , Schillingera U., Kutima P.M., Mbugua S.K., Holzapfel W.H. Isolation, Identification, and characterisation of the dominant microorganisms of kule naoto: the Maasai traditional fermented milk in Kenya. Int J of Food Micro. 2004, 94, 269–27. [Google Scholar] [CrossRef] [PubMed]
- Teuber M., The genus Lactococcus. IN: Wood BJB., Holzapfel WH (ed). The Genera of Lactic acid Bacteria. The Lactic Acid Bacteria. 1995. Vol 2. Springer, Boston, MA.
- Togo, C.A. , Feresu, S.B., Mutukumira, A.N. Identification of Lactic Acid Bacteria isolated from Opaque beer (Chibuku) for potential use as a starter culture. The J of Food Tech in Afr. 2002, 7, 93–97. [Google Scholar]
- Webb, L.; Ma, L.; Lu, X. Impact of Lactic Acid Bacteria on the Control of Listeria monocytogenes in Ready-to-Eat Foods. Food Qual. Saf. 2022. [Google Scholar] [CrossRef]
- Nyambane, B. , Thari, W.M., Wangoh J., and. Njage P.M.K. Lactic acid bacteria and yeasts involved in the fermentation of amabere amaruranu, a Kenyan fermented milk. Food Sci & Nutri. 2014, 2, 692–699. [Google Scholar]
- Pyz-Lukasik, R. , Paszkiewicz W., Brodzki, P., Belkot, Z. Microbiological quality of milk sold directly from producers to consumers. J of Dairy Sci. 2015, 98, 4294–4301. [Google Scholar] [CrossRef]
- Hamama A. Moroccan traditional fermented dairy products. In: Ruskin, F.R. (Ed.). Applications of biotechnology to traditional fermented foods. National Academy Press, Washington DC. 1992. p: 75-79.
- Foster, J.W. Low pH Adaptation and the Acid ~Tolerance Response of Salmonella typhimurium. Cri Reviews in Micro. 2008, 21, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Liyuwork, T. , Biruhalem T., Sefinew A., Hailleleul N. Prevalence and antimicrobial resistance profile of Salmonella isolates from dairy products in Addis Ababa, Ethiopia. Afr. J of Micro Res 2013, 7943, 5045–5050. [Google Scholar]
- Bedassa, A. , Nahusenay H., Asefa Z., Sisay T; Girmay G., Kovac J., Vipham J.L., and Zewdu A. Prevalence and associated risk factors for Salmonella enterica contamination of cow milk and cottage cheese in Ethiopia. Int J Food Cont. 2023, 10, 2. [Google Scholar]
- Chatti A., Daghfous D., Landoulsi A. Acid resistance of Salmonella isolated from animals, food and wastewater in Tunisia. Ann Saudi Med. 2007, 27, 195–198. [CrossRef]
- Halkman HBD., Halkman, A.K. Indicator Organisms. Encyclopaedia of Food Microbiology (Second Edition). 2014.
- D’Amico and Donelly, C.W. Microbiological quality of raw milk used for small-scale artisan cheese production in Vermont: Effect of farm characteristics and practices. J of Dairy Sci. 2010, 9, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Somaratne, N.; Hallas, G. Review of Risk Status of Groundwater Supply Wells by Tracing the Source of Coliform Contamination. Water 2015, 7, 3878–3905. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeasts in dairy products. A Review. J. of App Bact. 1990, 68, 199–211. [Google Scholar] [CrossRef]
- Akabanda, F. , Owusu-Kwarteng, J., Glover, R.L.K., Tano-Debrah, K. Microbiological characteristics of Ghanaian traditional fermented milk product. Nunu. Nature Sci. 2010, 8, 178–187. [Google Scholar]
- Savova, L. and Nikolova, M. Isolation and Taxonomic study of yeast strain from Bulgarian Dairy products. J of Culture Coll. 2000, 3, 59–65. [Google Scholar]
- Bùchi NR., and Seller H. Yeast and moulds: yeasts in milk and dairy products. 2011. On ResearchGate. [CrossRef]
- Getachew, A., Tadie, A., Chercos, D.H., and Guadu, T. Level of Faecal Coliform Contamination of Drinking Water Sources and Its Associated Risk Factors in Rural Settings of North Gondar Zone, Ethiopia: A Cross-Sectional Community Based Study. Ethiop J Health Sci. 2018, 28, 227–223. [CrossRef]
- Carpentier, B.; Cerf, O. Review-Persistence of Listeria monocytogenes in Food Industry Equipment and Premises. Int. J. Food Micro. 2011, 145, 1–8. [Google Scholar] [CrossRef] [PubMed]
- ZelalemY. Quality factors that affect Ethiopian milk business: Experiences from selected dairy potential areas. Netherlands Development Organization, Addis Ababa, Ethiopia. 2010.
- Jans, C. , Meile L., Wambua D., Kaindi M., Kogi-Makau W., Lamuka P., Renault, P., Kreikemeyer B., Lacroix C., Hattendorf J., Zinsstag J., Schelling E.,m Fokou g., Bonfoh B. African fermented dairy products – Overview of predominant technologically important microorganisms focusing on African Streptococcus infantarius variants and potential future applications for enhanced food safety and security. Int J of Food Micro. 2017, 250, 27–36. [Google Scholar]
- Bereda A, Yesuf Kurtu M, Yilma Z. Handling, processing and utilization of Milk and Milk products in Ethiopia: a review. World J Dairy Food Sci. 2014, 9, 105–112. [Google Scholar]
- Mekonnen, Z. , Kidemu M. Abebe H., Semere M., Gebreyesus M., Workuy A., Tesfaye M., Chernet A. Traditional knowledge and institutions for sustainable climate change adaptation in Ethiopia. Current Res in Env Sust. 2021, 3, 100080. [Google Scholar]
- Okello AL, Bardosh K, Smith J, Welburn SC. One Health: Past successes and future challenges in three African contexts. PLoS Negl Trop Dis. 2014, 8, e2884. [Google Scholar]
- Food and Agriculture Organisation of the United Nations (FAO). Guide to good dairy farming practice: Milking Hygiene. 2023.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).