1. Introduction
The fusion of machine learning and biomedical and bioengineering research has brought a paradigm shift in the way we understand, diagnose, and treat an array of health conditions. With rapid advancements in technology and an influx of high-dimensional data, the role of Machine Learning (ML) and especailly the AutoML has become central to the process of knowledge discovery in these fields [
1]. However, to provide a machine learning solution create a burden to bimedical and bioengineering researchers who have to search additional support to develop or test different tools for each steps of their studies. The burgeoning complexity of biomedical and bioengineering research underscores the need for the automatic generation and optimization of machine learning models that can keep pace with this data-driven evolution [
2].
AutoML frameworks provide a robust solution to this rising demand. They offer end-to-end pipelines that encompass all necessary steps from data pre-processing to hyperparameter tuning and model evaluation, automating labor-intensive and error-prone manual tasks [
3]. By significantly reducing the time taken for the model development process, they allow researchers to focus on interpreting and applying results, thus accelerating the pace of discovery in the biomedical and bioengineering field.
Despite the undeniable potential of AutoML, several gaps and challenges persist in its implementation in biomedical and bioengineering research. Firstly, the unique characteristics of biomedical data, including high dimensionality, heterogeneity, and inherent noise, require specialized pre-processing and analytical approaches not adequately addressed by current general-purpose AutoML frameworks. Secondly, these frameworks often lack interpretability, a crucial requirement in the medical field, where understanding the decision-making process of a model is as essential as its predictive accuracy [
4]. Finally, integrating domain knowledge into the AutoML process remains an open challenge, although it could greatly improve the quality of models generated and the applicability of their predictions [
5]. Therefore, to address these challenges requires a new framework Paradise to enable machine itself to learn domain specific knowledge and use the knowledge to make the automotive decisions on AutoML process.
This paper introduces an Automatic Semantic Machine Learning Microservice Framework designed to bridge these gaps. Each microservice is shorted as AIMS that should implemented based on domain specific knowledge. It is tailored to the specific needs of biomedical and bioengineering research and places emphasis on enhancing model interpretability, incorporating domain knowledge, and handling the intricacies of biomedical data. Our proposed framework aims to streamline the research process, augment the quality of scientific exploration, and provide a foundation for significant self-learning AutoML in biomedical and bioengineering research. In additional, three case studies are tested and discussed at the end.
2. Related Work, Limitations and Technology Background
2.1. Related Work and Current Limitations
There is a multitude of AI tools currently available to aid biomedical research and various automation frameworks have emerged to streamline the process. For instance, machine learning platforms like Google’s TensorFlow [
6] and Scikit-learn [
7] have been widely utilized in biomedical research for tasks such as image analysis, genomics, and drug discovery.
Google’s AutoML [
8], TPOT [
9] and H2O’s AutoML [
10] are some of the popular AutoML tools used for automating the machine learning pipeline. These platforms optimize the process by automating tasks like data pre-processing, feature selection, model selection, and hyperparameter tuning, which are traditionally labor-intensive and error-prone.
Other than these general-purpose tools, there are also specialized AI platforms tailored for biomedical research. DeepChem [
11], for instance, is a machine learning library specifically designed for drug discovery and toxicology, offering specialized features not available in general-purpose libraries.
However, while these tools have made significant strides in advancing biomedical research, there are several limitations associated with their use.
General-purpose ML and AutoML tools, such as TensorFlow and Google’s AutoML, are not specifically designed for handling the unique characteristics of biomedical data such as high dimensionality, heterogeneity, and inherent noise. This often necessitates significant manual pre-processing before data can be fed into these tools [
12].
Furthermore, these tools often lack interpretability, an essential requirement in biomedical research where understanding the decision-making process of a model is as important as its predictive accuracy [
4].
While specialized tools like DeepChem offer features tailored for biomedical applications, they do not cover the entire spectrum of biomedical research and are limited in their scope. Additionally, the automatic integration of domain knowledge into the machine learning process is an ongoing challenge and is not well-addressed by current tools [
4].
Therefore, there are many recent discussions on multiple-biomedical task handling with self-learning, self-optimisations and self-configuration processes such as [
13] focuses on data science processing automation with optimisation and [
3] focuses on feature selection and model training.
2.2. Multiple-Task AI System Research
Industrial AI leading research groups such as Google AI and Meta AI understood that data-driven AI technologies have issues with performing complex tasks. For example, creating human conversations with contextual understanding, or detecting early signs of disease from images. In addition, data-driven AI is resource intensive and suffers from algorithm bias [
14]. Thus, the multiple-task enabled AI systems with a knowledge-driven approach present a pathway toward a solution to these problems. Why is it thought that a knowledge-driven approach is necessary and crucial for the multiple-task system? There are two reasons:
The information acquired from different tasks may present value that can be used as the basis to build new ML models for new tasks without requiring the high-cost processing to re-capture the same feature characteristics.
Updating knowledge through validation is a relatively consistent process that will be less prone to bias from noisy data.
On completion of this research, two new ideas from Google and Meta have been published.
Google present an experimental process based on knowledge-mutation [
15,
16]. Here the knowledge refers to base neural network transformers. To begin with, the experimental environment contains transformers which can work on different tasks (different image datasets for the classification problems). Then, when a new task arrives, the most related transformer will be triggered to do a mutation process. The mutation process can edit the base model by inserting a new layer, removing a layer, or doing both according to the performance optimisation. In the end, a new mutated adapter is created to enable dealing with a similar task next time. Whenever a new task with a new dataset arrives, the mutation process is executed based on the latest mutated model.
Meta research group presents a world model approach to acquiring knowledge very much in the spirit of actor-critic reinforcement learning [
17]. The system architecture is a combination of smaller modules - configurator, perception, world model, cost, short-term memory, and actor - that feed into each other. The world model module is responsible for maintaining a model of the world that can then be used to both; estimate missing information about the world, and predict plausible future states of the world. The perception module will receive signals to estimate the current state of the world and for a given task the configurator module will have trained the perception module to extrapolate the relevant signal information. Then in combination, the perception, world model, cost, short-term memory, and actor modules feed into the configurator module which configures the other modules to fulfil the goals of the task. Finally, the actor module is handed the optimal action to perform as an action. This has an effect on the real world which the perception module can then capture which in turn triggers the process to repeat. That is, each action will produce a piece of state-changing knowledge feedback to the world model for continuous learning.
Both Google and Meta’s visions derive from the previous hyperparameter optimisation-based AutoML processes [
18], For example, AutoKeras [
19], a neural architecture auto-search framework is proposed to perform network morphism guided by Bayesian optimisation and utilising a tree-structured acquisition function optimisation algorithm. The searching framework selects the most promising Keras implemented NN for a given dataset.
The above experimental results show improvements in tackling complex AI tasks and possible pathways toward human-level AI systems. However, there are two main limitations:
The knowledge definition is too narrow and only uses the generated neural network as the knowledge limits the capability of recording all valuable outcomes through the learning experience.
There is no unified knowledge representation structure for knowledge inference (machine thinking).
Do we already have a knowledge representation framework from our AI research in the past 70 years? The answer is yes.
2.3. Knowledge Representation and Reasoning
Knowledge representation and reasoning (KRR) are always the core research areas in AI systems [
20]. The knowledge-enhanced machine learning approach attracts less attention that the data-driven approaches. However, KRR is still key in developing the future generation of AI systems development [
21], even Deep Neural Networks (DNN) can create KRR but just in a different form [
22]. In our vision, the KRR should not only extract knowledge from data but also learn knowledge from system actions that can support the reasoning process. Knowledge reasoning can be seen as the fundamental building block that allows machines to simulate humankind’s thinking and decision-making [
23]. With generations of development on KRR, the current most promising approach is the knowledge graph (KG) [
24] derived from the semantic web [
25] community. A knowledge graph has two layers of representation structure: 1. pre-defined ontology and vocabularies and 2. instances of triple statements (e.g. dog isA Animal, where the dog is an instance, isA is a predict while Animal is a concept vocabulary defined in the ontology). The reasoning part is to apply the logical side of the ontology, such as description logic (e.g. is the dog an animal? reasoning result is ’Yes’) [
26]. There are many complex types of ontologies developed in the last decade to solve different KRR problems and applications. The most important development of ontology-driven reasoning is to encode dynamic uncertainty [
27], probability [
28] and causality [
29]. Therefore, the KG-based KRR framework can be applied to implement our proposed vision.
2.4. Services and Machine Learning Ontologies
The web services community has researched auto-configuration or service composition for many years by applying a variety of dynamic integration methods. There are two trends in service composition research:
Directly extracts the services description file (e.g. WSDL) and Quality of Services (QoS) into a mathematical model with a logical framework for composing services such as a linear logic approach [
30] and genetic algorithms [
31,
32]. The major limitation is that there are no formal specifications for modelling and reasoning. Therefore, the processes are mostly hard-coded to match the logic framework.
The other trend is to apply Semantic Web standards for semantically encoding services description and their QoS properties (Semantic Web Services SWS)[
33,
34]. The main benefit is that semantic annotation has an embedded logical reasoning framework to deal with composition tasks.
On the one hand, the semantic web services (SWS) trend has greater strength for integrating the KRR approach with the same semantic infrastructure and reasoning logic. Currently, there are three standards of OWL-S (composition-oriented ontology), WSMO (task-goal matching oriented ontology), and WSDL-S (invocation-oriented ontology). On the other hand, there are two differences between our vision’s microservice to normal SWS. The first one is that AIMSs have simpler input and output requirements to perform an efficient composition process. The other is that the purpose of each microservice is to deal with data analytic or machine learning tasks. Therefore, the AIMS ontology needs to be defined by modifying current machine learning ontology standards. Researchers have realised that there is a need to have a machine learning ontology and some recent proposals in this domain are: the Machine Learning Schema and Ontologies (MLSO) introduces twenty-two top-layer concepts and four categories of lower-layer vocabularies (the detailed ontology design is in [
35]); the Machine Learning Ontology (MLO) proposes to describe machine learning algorithms with seven top layer concepts of Algorithm, Application, Dependencies, Dictionary, Frameworks, Involved, and MLTypes [
36].
2.5. The Gaps
By reviewing the current state of the art, we found that there are remaining research gaps to achieve our goal.
Self-supervised knowledge generation during the machine learning process and solution creation. In the past, knowledge generation system mainly refers to expert systems that acquire knowledge from human expertise or systems that transform existing knowledge from one presentation to the other. [
29] presents an automatic process of disease causality knowledge generation from HTML-text documents. However, it still doesn’t fully address the problem of how to automatically learn valuable knowledge from the whole task-solution-evaluation machine learning life cycle. Considering human-level intelligence, we always learn either directly from problem-solving or indirectly through other human expertise (e.g. reading a book or watching a video) or a combination of bother (e.g. reflecting on the opinions of others).
Provisioning knowledge-guided auto-ML solution. In contrast to the first gap, there are no significant research works on using the knowledge to assist in providing an AI solution. Again, compared to human-level intelligence, we always try to apply acquired knowledge or knowledge-based reasoning to solve the problem. We can consider that the transformer process [
37] is a step forward in this direction. We can treat well-trained AI models as a type of knowledge to apply to different tasks in a similar problem domain. However, there is still no defined framework that can specify what knowledge is required and how to use the knowledge to find a solution to new tasks [
15].
3. The Framework Architecture
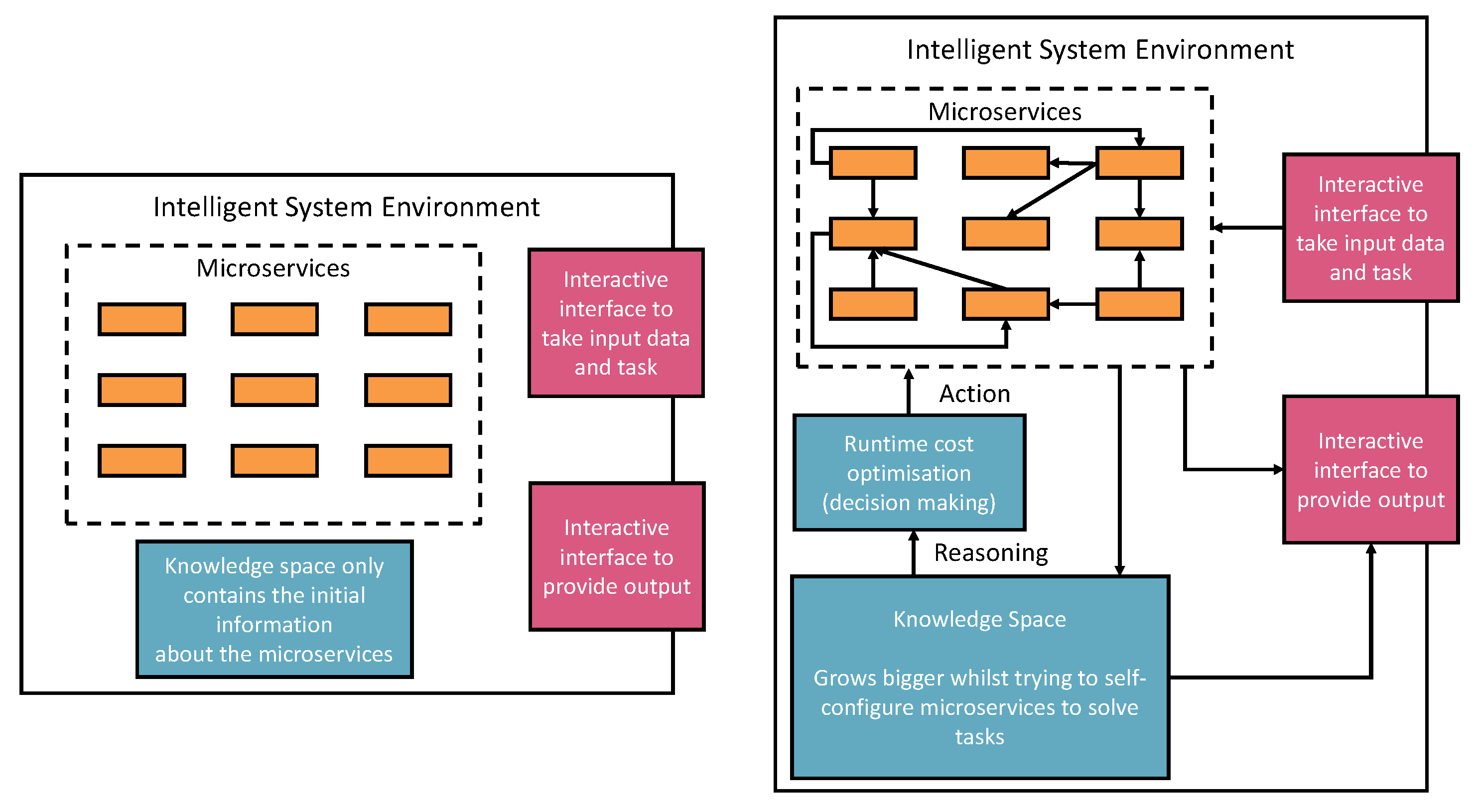
Figure 1 represents our vision of Self-supervised Knowledge Learning with the AIMSs Engineering approach. The left part of the figure presents the initial settings of the intelligent environment. The initial environment only contains default AIMSs information such as purposes, I/O requirements, and invokable URI (detailed AIMS metadata ontology will be introduced in the next section). However, the initial settings are ready for doing four things:
Registering new AIMSs from outside the environment. The registration process is through the interactive interface according to the defined microservice ontology. Therefore, human involvement in Machine Leanring microservice engineering is a core part of this vision, which defines humans as an educator to teach basic skills and capabilities to deal with different tasks. Then, the environment will reuse these skills and capabilities to acquire knowledge. The knowledge will provide powerful reasoning sources to independently deal with complex tasks, decision-making, and creating new pipelines.
Taking tasks with a variety of inputs, such as CSV data files, images, text and audio data. The environment auto-configures on the default AIMSs and provides solutions to the tasks. The success or failure outcome will be recorded as knowledge. The microservice human engineering process will start if there are no suitable AIMSs to deal with the task.
The environment can compose multiple AIMSs to complete a task if one single microservice cannot achieve it.
The environment can start learning, representing, and storing knowledge in the knowledge space as knowledge graph data. The knowledge is derived from processing input data, the auto-configuration process, and task outcomes. The knowledge size will increase and thus provide better optimisations, auto-configuration, and feedback to the system user.
To realise the vision presented in
Figure 1, we will discuss the related existing technologies and their research outcomes that can be adapted into our research next.
4. Self-Supervised Knowledge Learning for Solution Generation
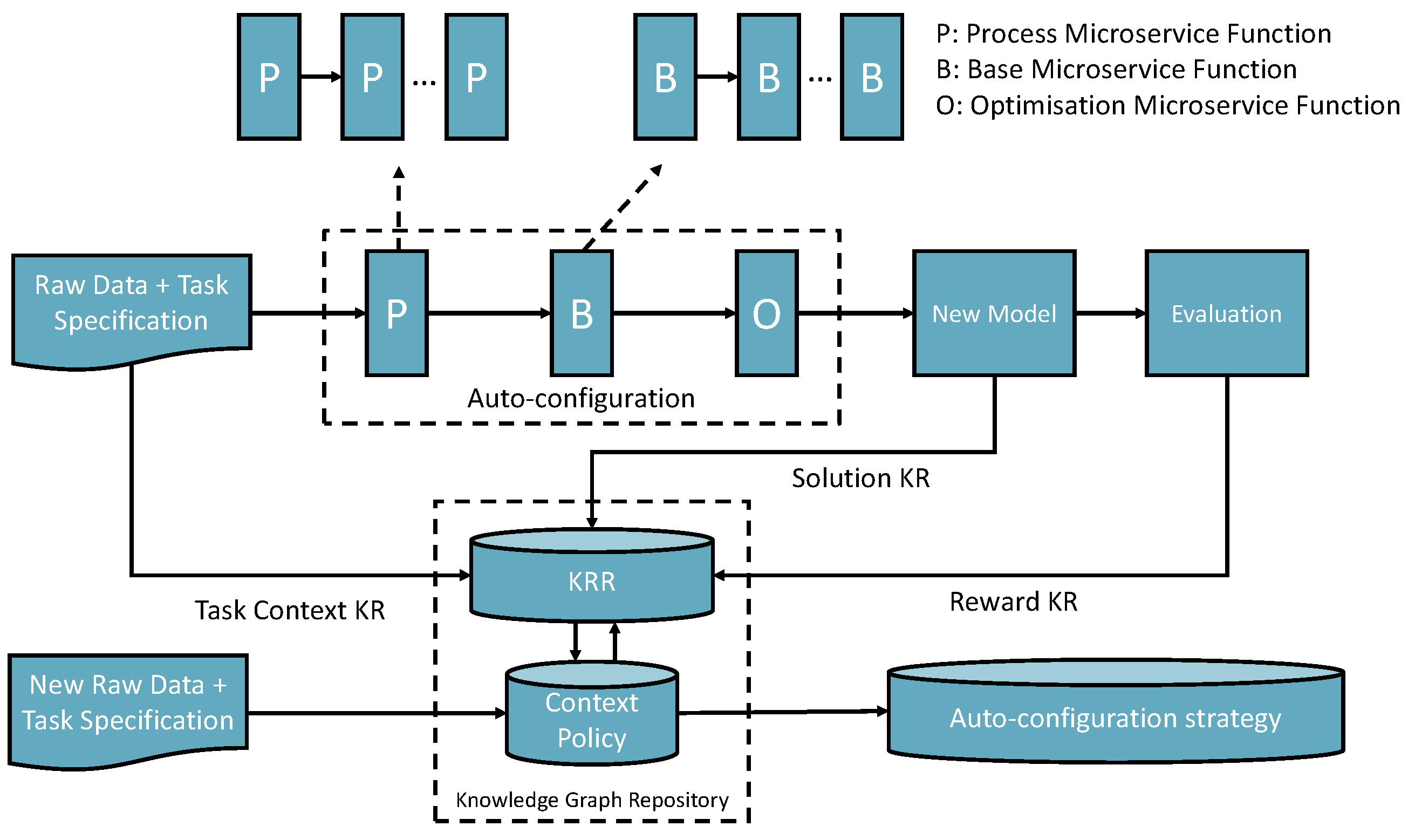
The self-supervised knowledge learning approach involves three types of auto-configuration transfer learning methods.
Figure 2 presents the overall learning framework.
The first method is knowledge space searching and transferring: A task with a dataset (referred to as a task-context) arrives, and there is no previous knowledge related to the task-context. Therefore, the knowledge space will be searched to try to find a possible microservice that can match the context to complete the task or search for a pipeline (workflow) that contains multiple I/O compatible AIMSs together towards the best and successful completion which can be optimised. The task-context and the optimized solution are recorded as task input and output knowledge. The evaluation will generate rewards for the policy knowledge space. In addition, the knowledge learnt from the process will be recorded to update the world knowledge space.
The second one is the mutation of a previously generated context-matched solution. If the new task-context matches with a previously recorded task-context in the knowledge space, then the previous solution will be loaded to adapt to the new datasets and the optimisation process. Finally, a new mutated solution is created and recorded as new knowledge with the new evaluation rewards and world knowledge of the KRR environment.
The third one is the continuous learning mutation method based on the reinforcement learning approach. With the growth of the KRR statements, the auto-mutation will take place using world knowledge to re-train the solutions according to the rewards. The third learning method takes place offline only but continues doing an update when KRR is updated.
5. Experimental Implementation
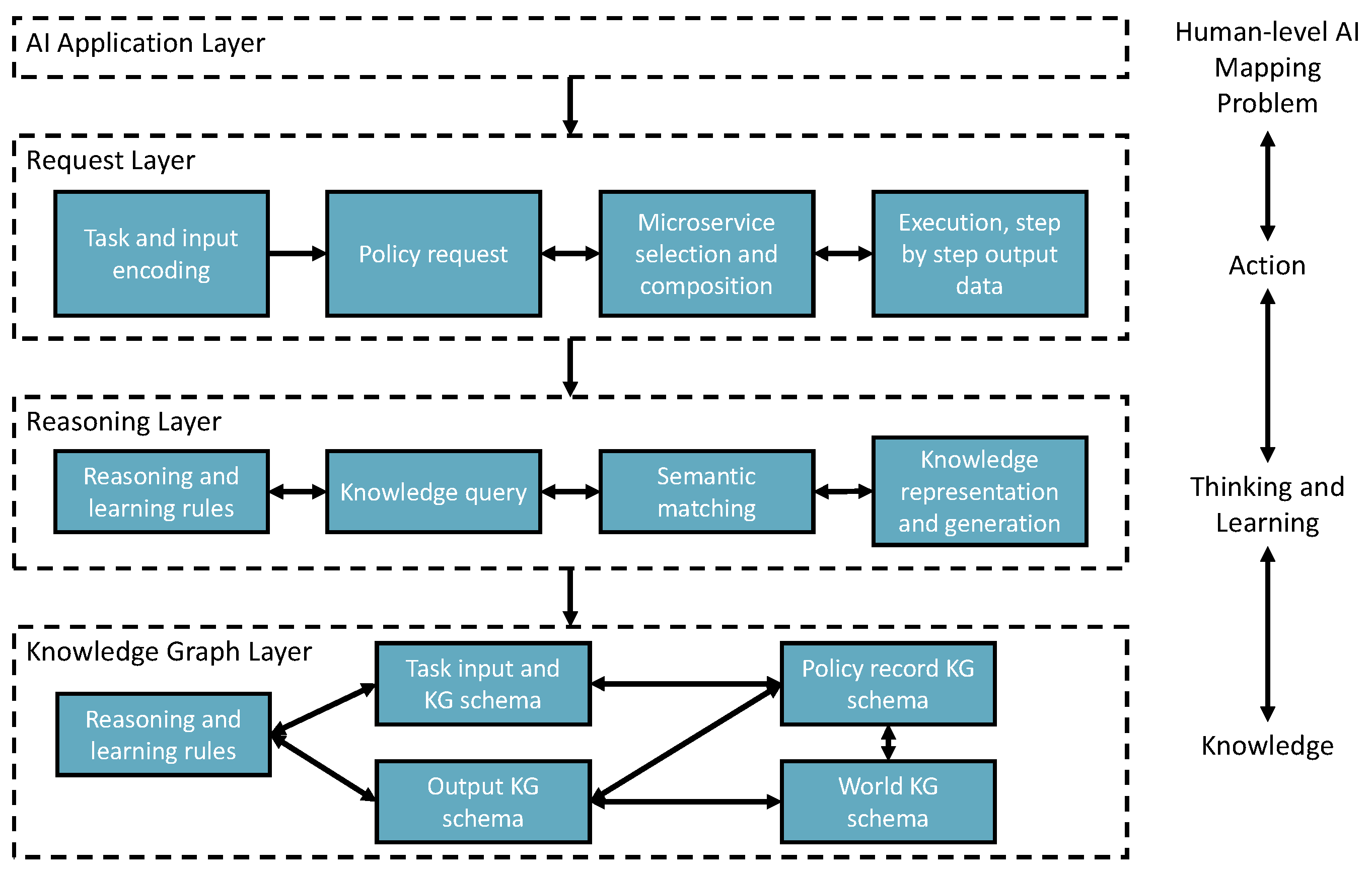
5.1. Overview of the Implementation Structure
Figure 3 shows a three-layer implementation of the vision. It also shows how these layers map to the automotive solution provisioning process.
Request layer takes tasks and inputs from AI applications to trigger the solution searching and self-learning processes. Task-context is semantically encoded to enable starting the policy knowledge to explore the environment for learning, creating, or finding solutions.
Reasoning layer takes the request layer’s semantic reasoning tasks for semantic matching, reasoning and doing reinforcement learning mechanism. Finally, the policy will be recorded in the knowledge graph layer. In addition, the newly added AIMSs are registered to the environment with semantic annotations through knowledge registration and generation components.
Knowledge Graph layer remembers the knowledge data in the knowledge graph triple store based on different types of knowledge schema.
5.2. Knowledge Ontology Implementation
5.2.1. AI Microservice Knowledge Modelling
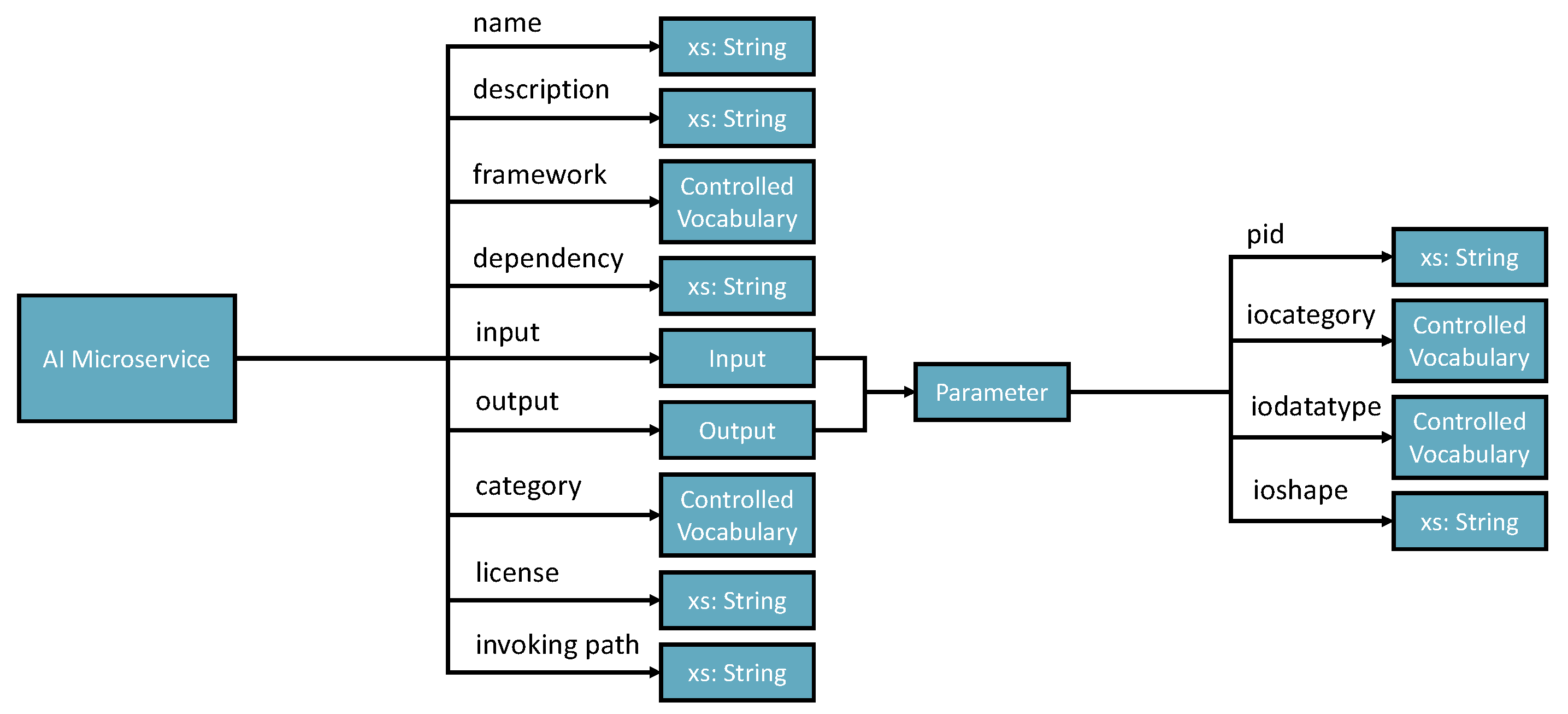
AIMS registration ontology defines 9 parameters (see
Figure 4).
name - must be no duplication in the system, and the registration process will check the name’s legibility.
description - a short presentation of the AIMS for human understanding.
framework - indicates the programming framework used to develop the AIMS. Normally, it should be just one framework as AIMS is designed to be decoupled and ideally single responsibility.
dependency - describes the required programming libraries that need to be pre-configured to enable the AIMS to work.
input and output - specifies the parameters that should be in the input and output messages.
category - tells what AI-related domain the ms works on, such as supervised classification, unsupervised clustering, image classification base model, and more.
license - identifies the use conditions and copyright of the AIMS.
invoke path - contains the portal for accessing the AIMS. The path can be a local path or URI of a restful API.
5.2.2. Task-Context Knowledge Modelling
Each given task triggers a context knowledge creation that collects the knowledge of:
the type of input data - a controlled variable that majorly includes normal dataset (e.g. CSV file), image, and text.
task domain - free text to record the specific application domain
desire output type - records the output required to complete the task successfully.
the features - the dataset or data presents the initial characters of the input data. For example, the number of columns and column names will be remembered as part of the context knowledge of the task.
5.2.3. Policy Knowledge Modeling
The output of the performed task can be categorised into two types failure and success. Both failure and success need to update the policy knowledge link to the task-input context. Failure has no solution registered to the knowledge but records which AIMSs have been successfully invoked (can be an empty list) until the step that cannot continue going further. So the failure experience will tell the system administrators (people) what AIMS(s) are required to create a solution. The success registers the solution location and changes the policy with the reward value. If the solution contains a workflow of composed AIMSs, then the workflow will also be registered as knowledge with the normalized rewards for each of the AIMS.
The ontology was designed as:
policy context - links to a task-context
policy state - 1 is success and 0 is failure
solution iloc - the location where the solution can be loaded and executed.
workflow - presents a pipeline solution that composes multiple AIMSs.
solution reward - the reward value stored for the policy that can be the recommended guidance for supporting the creation of a new task solution.
5.2.4. World Knowledge Modeling
The world knowledge presents the facts learned from the task solution creation process and outputs. There are three types of world knowledge recorded in the current environment:
feature optimisation outcomes - the features selected in the optimisation are valuable, and these features will be reused to create a classification model if the new dataset features are the same.
answers for a certain text topic - a generated text answer for a question. The answer quality will be reported as a reward value feedback from humans back to the policy knowledge.
image RBG vectors - map to a classification label. The reward process is the same as the answers.
More world knowledge can be expanded in the environment. By having these commonsense and policy records, reinforcement can be performed to improve the solution accuracy incrementally.
5.3. Environment Initialization
The experiment environment is developed by Python in a local single-computer environment. We simplified the AIMS as a .py module in the environment to be invoked and registered. The full implementation can be found in GitHub repository
(https://github.com/semanticmachinelearning/AISMK). We initialised the environment with three types of AIMS
1. Data Processing AIMSs that include CSV file to a Panda service, Data training and split service, data quality control services, data normalisation service, Image process Service, data quality control service.
2. ML AIMSs that include clustering services, classification services, GPT-neo-1.3B text generation services [
38,
39], ViT image classification transformers [
40] and Seanborn visualisation services.
3. RFECV optimisation services.
6. Scenario Evaluation and Lesson Learned
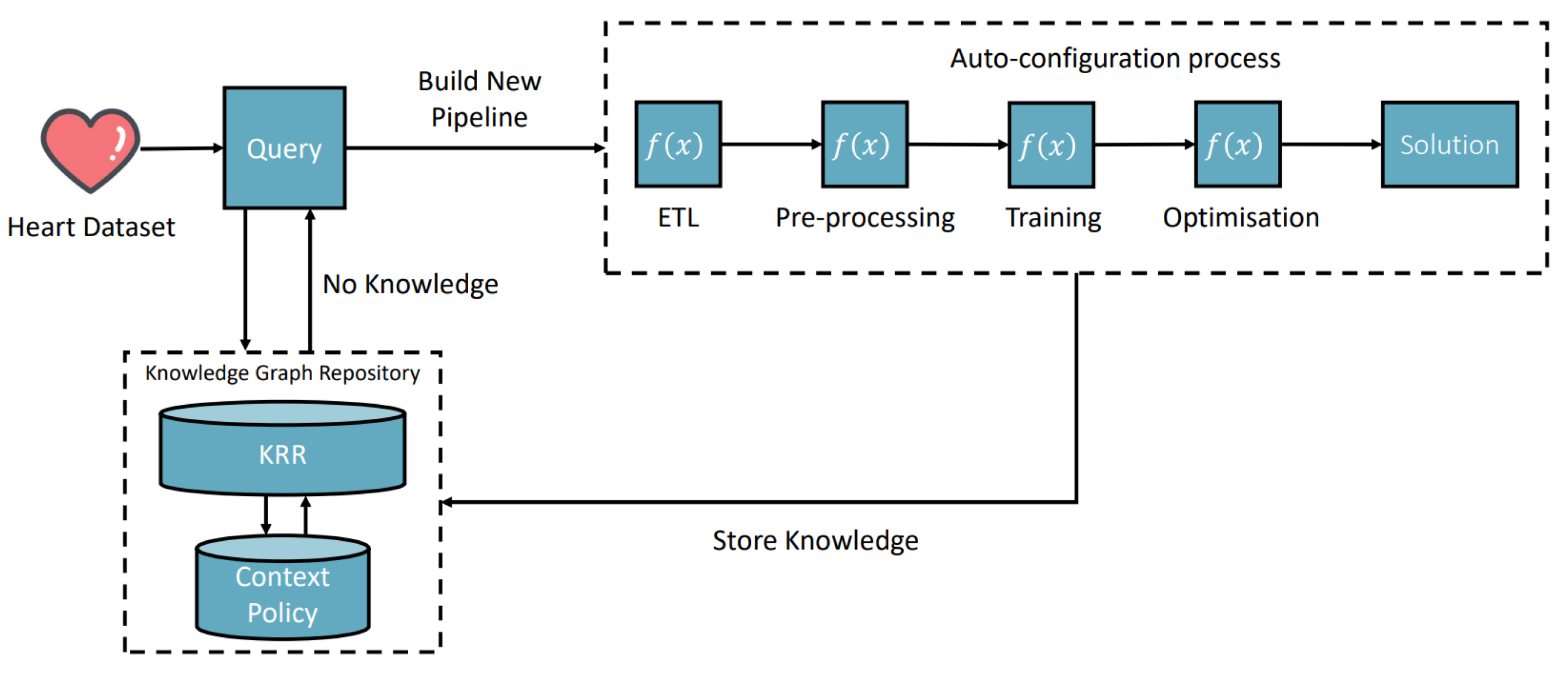
6.1. Heart Disease Classification Scenario
Figure 5 presents one of our use case scenarios in the medical domain. The task context is:
The application domain is medical and the desired output is an optimised classification pipeline model.
Figure 5 illustrates how the process starts by searching existing knowledge (no knowledge found) and creates a workflow containing four AI microservices for loading the CSV data, splitting the data, classification pipeline creation, and optimisation to get the accuracy of 96.8% model. Different pieces of knowledge are learned and recorded in the system environment.
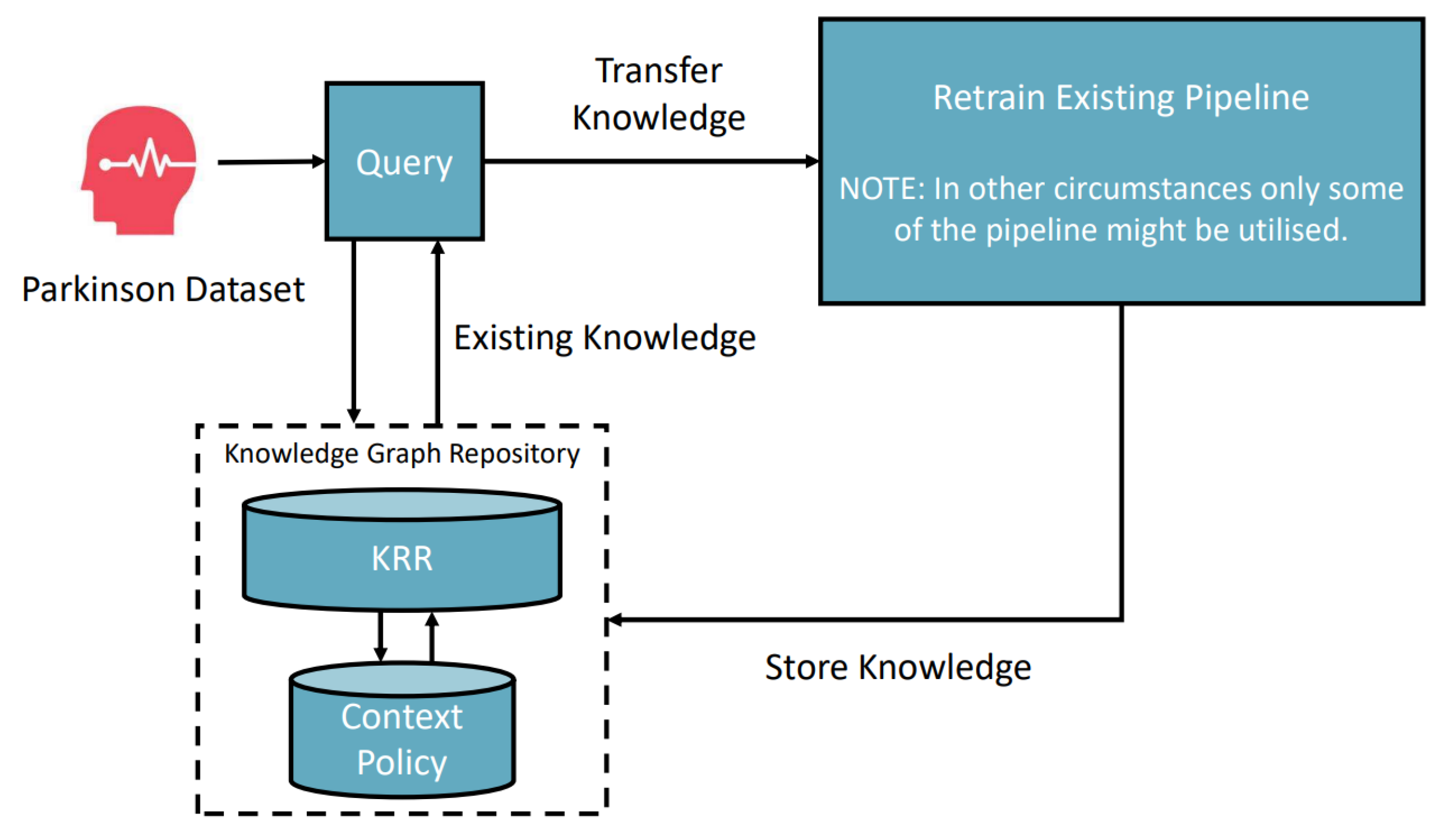
6.2. Parkinson Disease Classification Scenario
The second task-context is:
Figure 6 depicts the scenario in which a similar task of classifying Parkinson’s disease is fed in, the framework starts searching for a solution. As the system environment has pre-knowledge, gained through the previous heart disease classification, and since the only difference is the dataset, the framework can use the classification pipeline and retrain it to be optimised for the new dataset. We can call this process a composition transfer learning process. The novelty is that the system environment can solve different tasks by applying contextual knowledge of the problem. Thus, the framework can automatically deal with all types of data if the required models are semantically registered in the framework. Through this composition transfer learning, the whole automatic pipeline can produce a 94.6% accurate model.
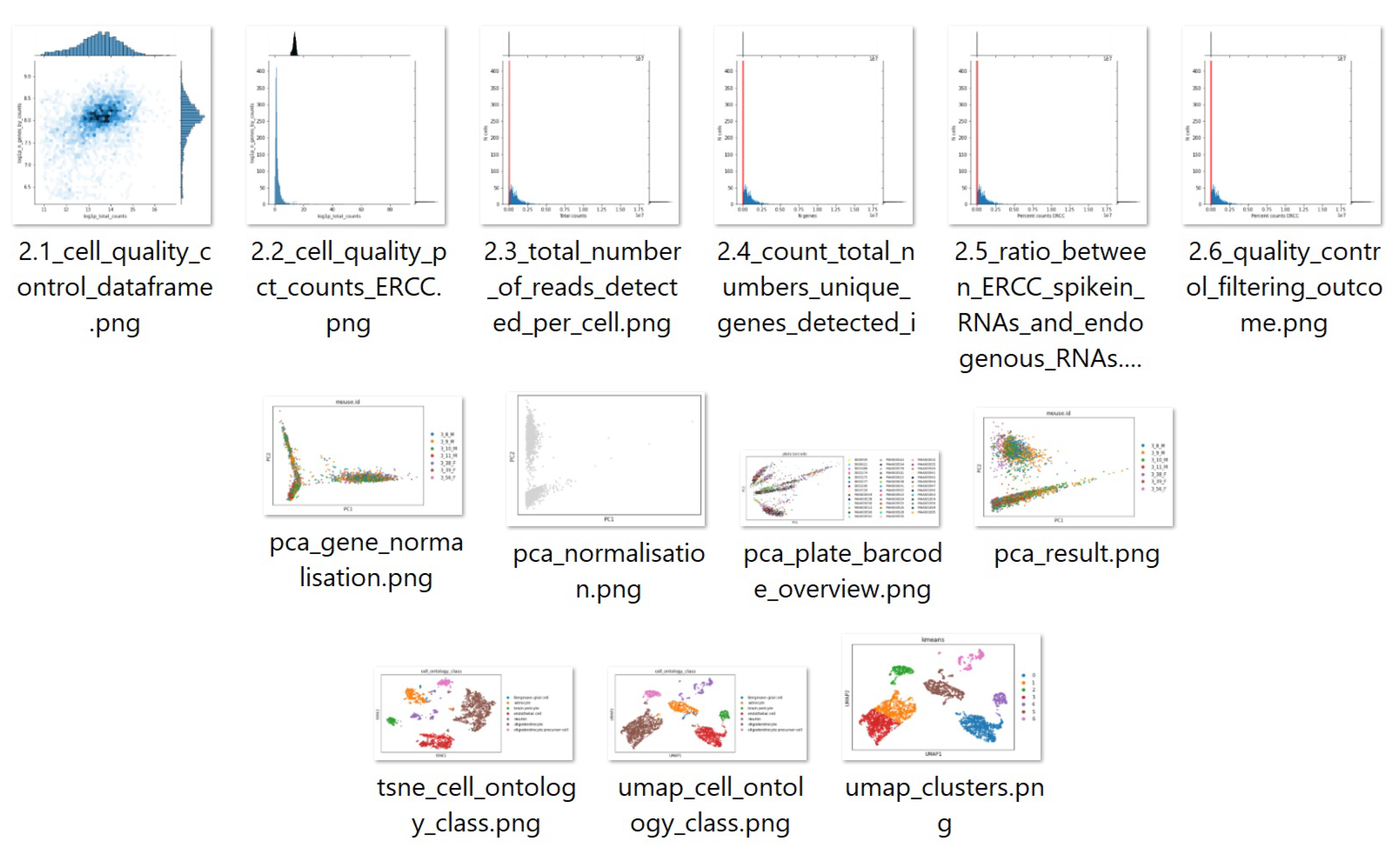
6.3. A Complex Scenario: Mouse Brain Single-Cell RNASeq Downstream Analysis
In this section, we use a clustering analysis case study to highlight how the proposed framework can solve a real-world downstream single-cell data analysis task. The clustering analysis task works on a mouse brain single-cell RNASeq dataset. The dataset is publicly available through a workshop tutorial at [
41]. There are five sequential processing and analysis steps:
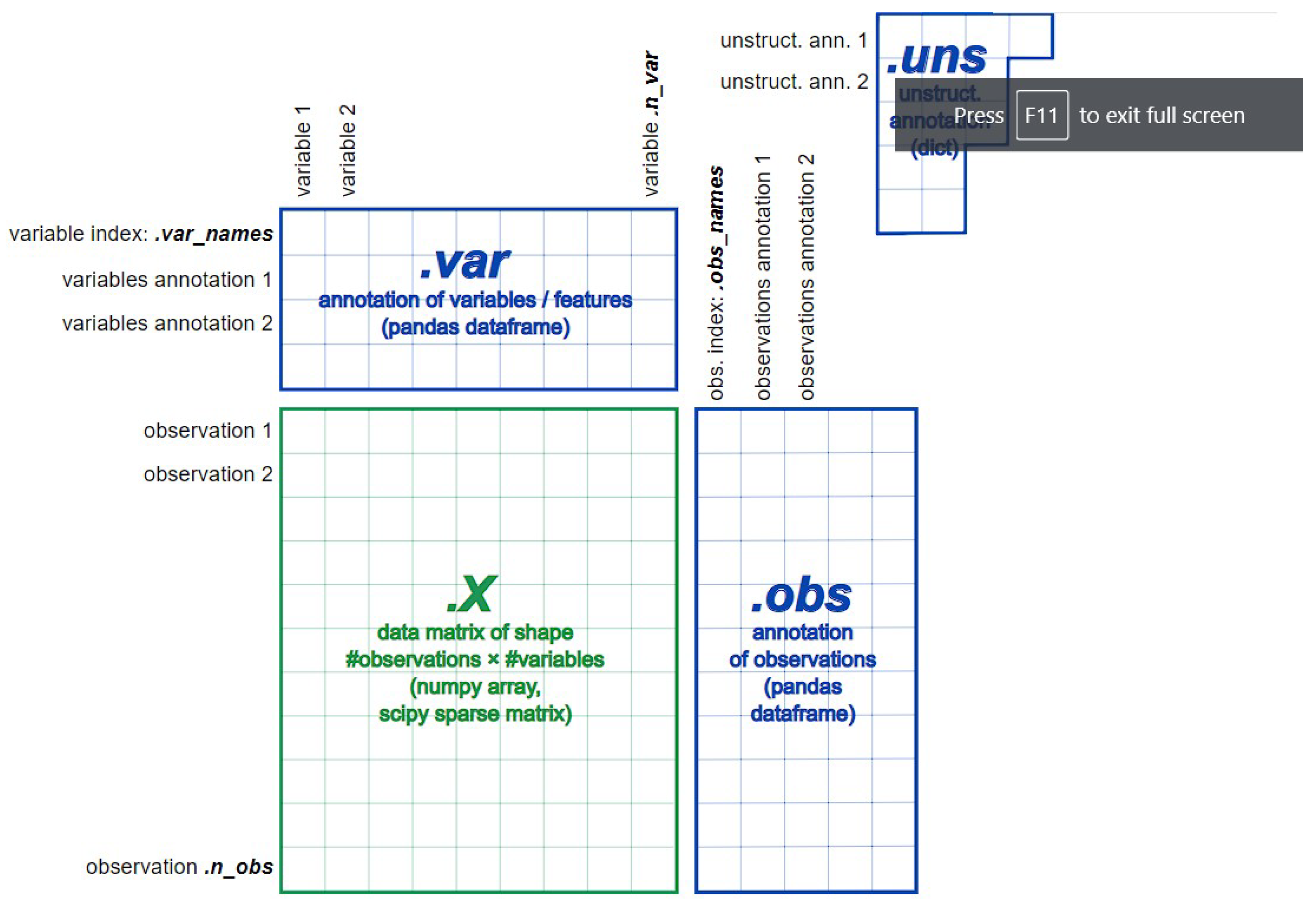
Data semantic transforming and loading: For instance, applying AnnData structure [
42], where AnnData stores observations (samples) of variables/features in the rows of a matrix (see
Figure 7).
Data quality control: This aims to find and remove the poor quality cell observation data which were not detected in the previous processing of the raw data. The low-quality cell data may potentially introduce analysis noise and obscure the biological signals of interest in the downstream analysis.
Data normalisation: Dimensionality reduction and scaling of the data. Biologically, dimensional reduction is valuable and appropriate since cells respond to their environment by turning on regulatory programs that result in the expression of modules of genes. As a result, gene expression displays structured co-expression, and dimensionality reduction by the algorithm such as principle component analysis can group those co-varying genes into principle components, ordered by how much variation they explained.
Data feature embedding: Further dimensionality reduction using advanced algorithms, such as t-SNE and UMAP. They are powerful tools for visualising and understanding big and high-dimensional datasets.
Clustering analysis: Group cells into different clusters based on the embedded features.
Based on the above five steps, we developed extra four microservices which include AnnData loading, two feature embedding services (t-SNE and UMAP), and clustering services (Louvain graphical clustering algorithms). The other existing microservices can be involved should be different types of normalisation (PCA or CPM algorithm), K-mean clustering algrithm.
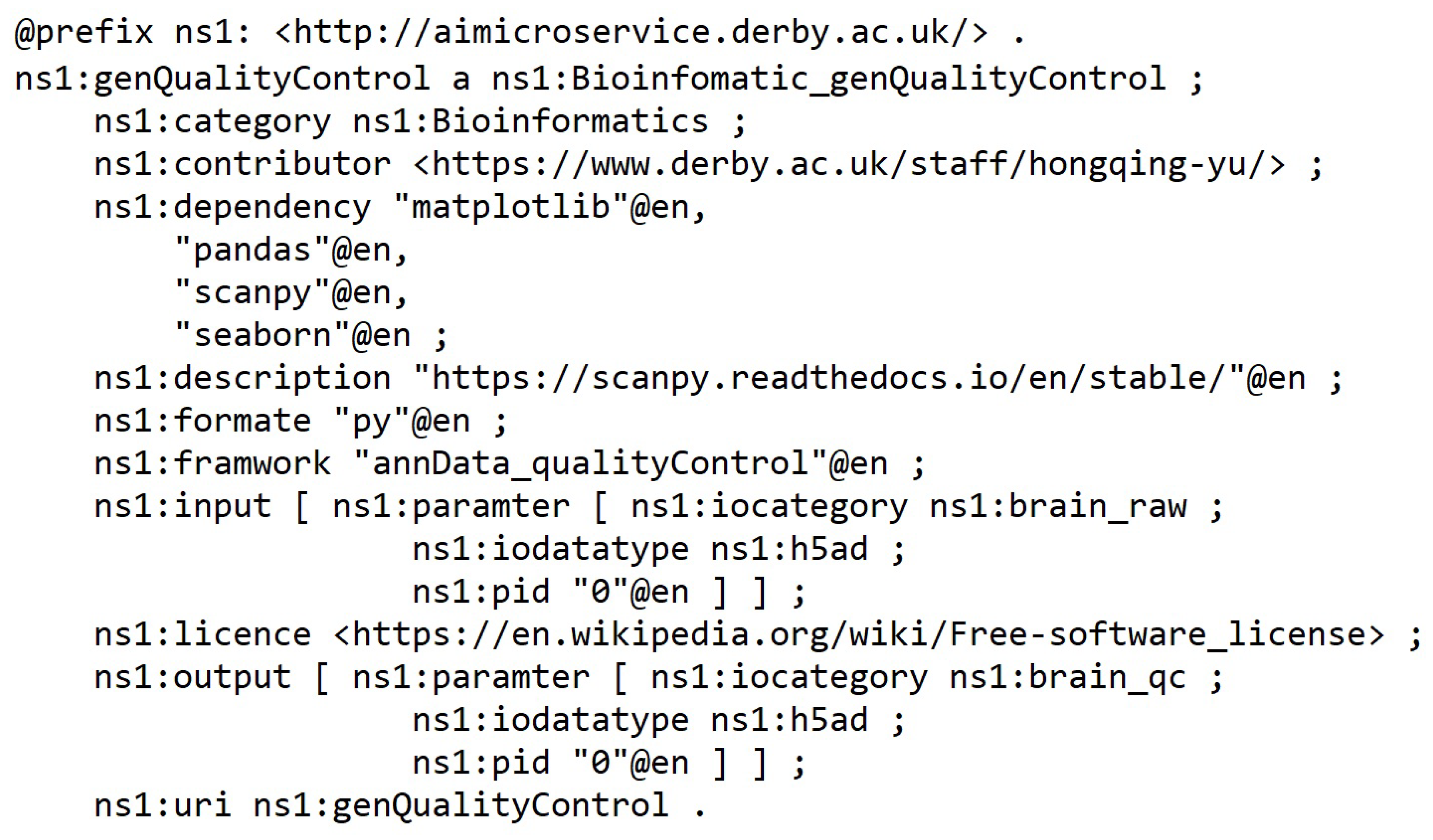
The microservices were semantically registered into the framework through the interface.
Figure 8 depicts an example of quality control microservice semantic description in the knowledge graph repository.
With all the microservices registered, researchers can start expressing the analysis task to stop, interact and provide feedback at any stage during the process of automatically creating the solution. The researchers can also see visualisations of outputs produced by different steps (see
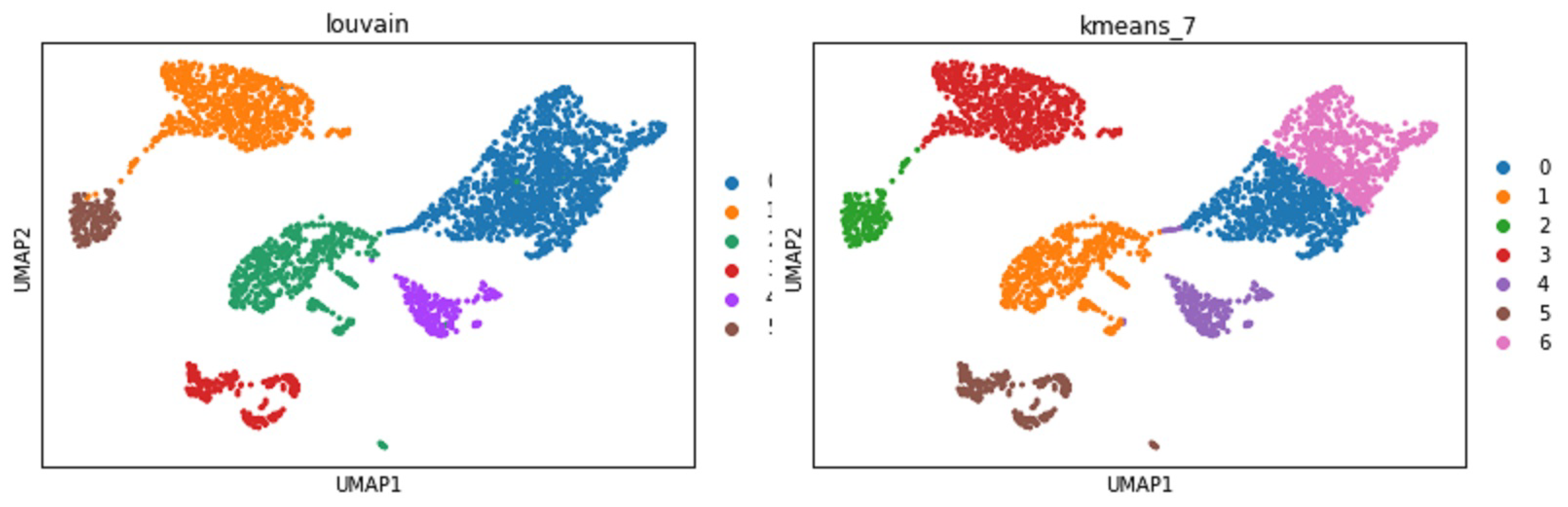
Figure 9). Therefore, researchers can provide preferences for selecting microservices if there are options.
A realistic example is that a researcher can specify a clustering task applied to the mouse brain single-cell RNASeq dataset. The framework will first try to see if a single microservice can complete this task. The answer is ’no’ because no semantic-matched microservice can take the RNASeq CSV input and provide the clustering output. At this juncture, the microservice that can take the RNASeq CSV will be invoked to process the data into the next step with the output of AnnData. If there are multiple choices in the composition sequence, all possibilities will be invoked to run unless the previous knowledge in the policies has a priority. The possibilities have multiple solutions at the end for researchers to analyse for giving professional feedback to the system. The feedback will help greatly with the knowledge graph policies. For example, suppose the researcher gives feedback to the system that UMAP is the better embedding method than t-SNE but has no priority on the clustering methods. In that case, the framework will produce two possible clustering results shown in
Figure 10.
7. Discussion
By evaluating the performance of the test scenarios, we believe the combination of KRR and automation of AIMSs is one possible approach to developing human-level AI systems. We implement the environment that has AIMSs that can take text, CSV files, and images as default settings with splitting data, classification, prediction, and optimisation AIMSs. It shows that system can generate and optimise solutions for different types of tasks by applying or creating knowledge. However, there are some limitations that need to be addressed in future work:
The benefit of applying a triple KG structure to encode KRR elements is the unification, standardization, and ease of adoption by different applications. However, if the KG grows too big, the efficiency of referencing based on the complex graph query is very slow. Especially, if the different types of knowledge are stored separately, then the union querying on the graph is very costly. The future work direction is to efficiently embed the Knowledge Graph into a more efficient vector space [
43].
The current implementation cannot take multi-modal inputs that belong to one task to generate a solution (Multi-modal Machine Learning). Humans can take images, data, sound, and smell together to complete the task, but it is still a challenge for the machine to merge different types of inputs together to solve the problem [
44,
45,
46].
When we started writing this paper, the Google research papers were published introducing the mutation of Neural Network (NN) to deal with multiple image classification tasks [
15,
16]. This latest paper provides some new ideas to us that not only data or services based mutation but also actions of inserting or removing NN hidden layers can be stored as knowledge in the future.
8. Conclusions
Our proposed Automatic Semantic Machine Learning Microservice Framework (AIMS) presents a novel approach to managing the complex demands of machine learning in biomedical and bioengineering research. The AIMS framework utilizes a self-supervised knowledge learning strategy to ensure automatic and dynamic adaptation of machine learning models, making it possible to keep pace with the evolving nature of biomedical research. By placing emphasis on model interpretability and the integration of domain knowledge, the framework facilitates an improved understanding of the decision-making process, enhancing the relevance and applicability of the generated models.
The three case studies presented underscore the framework’s effectiveness in various biomedical research scenarios, demonstrating its capacity to handle different types of data and research questions. As such, the AIMS framework not only offers a robust solution to current challenges in biomedical and bioengineering research but also sets a promising direction for future developments in automated, domain-specific machine learning. Further studies are required to evaluate the AIMS framework’s performance across a wider range of biomedical and bioengineering applications and to refine its capabilities for even more efficient and precise knowledge discovery.
Author Contributions
The contributions of the work are: Conceptualization, Hong Qing Yu; methodology, Hong Qing Yu, Sam O’Neill; Development and enhancement, Hong Qing Yu, Sam O’Neill; validation: Ali Kermanizadel, Hong Qing Yu; formal analysis, Sam O’Neill, Ali Kermanizadel, Hong Qing Yu; data curation, Hong Qing Yu.; writing original draft preparation, Hong Qing Yu; writing, review and editing, Sam O’Neill and Ali Kermanizadel; visualization, Hong Qing Yu, Sam O’Neill; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
References
- Obermeyer, Z.; Emanuel, E.J. Predicting the future — big data, machine learning, and clinical medicine. New Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Briefings Bioinform. 2017, 19, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Waring, J.; Lindvall, C.; Umeton, R. Automated machine learning: Review of the state-of-the-art and opportunities for healthcare. Artif. Intell. Med. 2020, 104, 101822. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Lin, H.; Liu, X.; others. A document level neural model integrated domain knowledge for chemical-induced disease relations. BMC Bioinform. 2018, 19. [Google Scholar] [CrossRef]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Goodfellow, I.; Harp, A.; Irving, G.; Isard, M.; Jia, Y.; Jozefowicz, R.; Kaiser, L.; Kudlur, M.; Levenberg, J.; Mané, D.; Monga, R.; Moore, S.; Murray, D.; Olah, C.; Schuster, M.; Shlens, J.; Steiner, B.; Sutskever, I.; Talwar, K.; Tucker, P.; Vanhoucke, V.; Vasudevan, V.; Viégas, F.; Vinyals, O.; Warden, P.; Wattenberg, M.; Wicke, M.; Yu, Y.; Zheng, X. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems, 2015. Software available from tensorflow.org.
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; Vanderplas, J.; Passos, A.; Cournapeau, D.; Brucher, M.; Perrot, M.; Duchesnay, E. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Inc., G. Google Cloud AutoML, 2021.
- Olson, R.S.; Moore, J.H. TPOT: A Python Automated Machine Learning Tool, 2020.
- H2O.ai. H2O AutoML, 2021.
- Ramsundar, B.; Eastman, P.; Walters, P.; Pande, V. Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More. O’Reilly Media 2019.
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef]
- Mustafa, A.; Rahimi Azghadi, M. Automated Machine Learning for Healthcare and Clinical Notes Analysis. Computers 2021, 10. [Google Scholar] [CrossRef]
- Ntoutsi, E.; Fafalios, P.; Gadiraju, U.; Iosifidis, V.; Nejdl, W.; Vidal, M.E.; Ruggieri, S.; Turini, F.; Papadopoulos, S.; Krasanakis, E.; Kompatsiaris, I.; Kinder-Kurlanda, K.; Wagner, C.; Karimi, F.; Fernandez, M.; Alani, H.; Berendt, B.; Kruegel, T.; Heinze, C.; Broelemann, K.; Kasneci, G.; Tiropanis, T.; Staab, S. Bias in data-driven artificial intelligence systems—An introductory survey. WIREs Data Min. Knowl. Discov. 2020, 10, e1356. [Google Scholar] [CrossRef]
- Gesmundo, A.; Dean, J. An Evolutionary Approach to Dynamic Introduction of Tasks in Large-scale Multitask Learning Systems, 2022. [CrossRef]
- Gesmundo, A.; Dean, J. muNet: Evolving Pretrained Deep Neural Networks into Scalable Auto-tuning Multitask Systems, 2022. [CrossRef]
- LeCun, Y. A Path Towards Autonomous Machine Intelligence, 2022.
- Yao, Q.; Wang, M.; Escalante, H.J.; Guyon, I.; Hu, Y.; Li, Y.; Tu, W.; Yang, Q.; Yu, Y. Taking Human out of Learning Applications: A Survey on Automated Machine Learning. CoRR 2018, abs/1810.13306, [1810.13306].
- Jin, H.; Song, Q.; Hu, X. Auto-Keras: An Efficient Neural Architecture Search System, 2018. [CrossRef]
- Sharma, L.; Garg, P.K. Knowledge representation in artificial intelligence: An overview. Artificial intelligence 2021, 19–28. [Google Scholar]
- Cozman, F.G.; Munhoz, H.N. Some thoughts on knowledge-enhanced machine learning. Int. J. Approx. Reason. 2021, 136, 308–324. [Google Scholar] [CrossRef]
- Hu, Z.; Yang, Z.; Salakhutdinov, R.; Xing, E. Deep neural networks with massive learned knowledge. Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, 2016, pp. 1670–1679.
- Chen, X.; Jia, S.; Xiang, Y. A review: Knowledge reasoning over knowledge graph. Expert Syst. Appl. 2020, 141, 112948. [Google Scholar] [CrossRef]
- Chen, X.; Jia, S.; Xiang, Y. A review: Knowledge reasoning over knowledge graph. Expert Syst. Appl. 2020, 141, 112948. [Google Scholar] [CrossRef]
- Berners-Lee, T.; Hendler, J.; Lassila, O. The semantic web. Sci. Am. 2001, 284, 34–43. [Google Scholar] [CrossRef]
- Baader, F.; Horrocks, I.; Lutz, C.; Sattler, U. Introduction to description logic; Cambridge University Press, 2017.
- Zhang, Q. Dynamic Uncertain Causality Graph for Knowledge Representation and Probabilistic Reasoning: Directed Cyclic Graph and Joint Probability Distribution. IEEE Trans. Neural Networks Learn. Syst. 2015, 26, 1503–1517. [Google Scholar] [CrossRef]
- BOTHA, L.; MEYER, T.; PEÑALOZA, R. The Probabilistic Description Logic. Theory Pract. Log. Program. 2021, 21, 404–427. [Google Scholar] [CrossRef]
- Yu, H.Q.; Reiff-Marganiec, S. Learning Disease Causality Knowledge From the Web of Health Data. Int. J. Semant. Web Inf. Syst. (IJSWIS) 2022, 18, 1–19. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, E.; Yu, H.Q.; Clapworthy, G.J. A Linear Logic Approach to the Composition of RESTful Web Services 2015. 10, 245–271. [CrossRef]
- Allameh Amiri, M.; Serajzadeh, H. QoS aware web service composition based on genetic algorithm. 2010 5th International Symposium on Telecommunications, 2010, pp. 502–507. [CrossRef]
- Qiang, B.; Liu, Z.; Wang, Y.; Xie, W.; Xina, S.; Zhao, Z. Service composition based on improved genetic algorithm and analytical hierarchy process. Int. J. Robot. Autom. 2018, 33. [Google Scholar] [CrossRef]
- Yu, H.Q.; Zhao, X.; Reiff-Marganiec, S.; Domingue, J. Linked Context: A Linked Data Approach to Personalised Service Provisioning. 2012 IEEE 19th International Conference on Web Services, Honolulu, HI, USA, June 24-29, 2012; Goble, C.A.; Chen, P.P.; Zhang, J., Eds. IEEE Computer Society, 2012, pp. 376–383. 24 June. [CrossRef]
- Dong, H.; Hussain, F.; Chang, E. Semantic Web Service matchmakers: State of the art and challenges. Concurr. Comput. Pract. Exp. 2013, 25. [Google Scholar] [CrossRef]
- Publio, G.C.; Esteves, D.; Lawrynowicz, A.; Panov, P.; Soldatova, L.N.; Soru, T.; Vanschoren, J.; Zafar, H. ML-Schema: Exposing the Semantics of Machine Learning with Schemas and Ontologies. ArXiv 2018, abs/1807.05351.
- Braga, J.; Dias, J.; Regateiro, F. A MACHINE LEARNING ONTOLOGY. Frenxiv Papers 2020. [Google Scholar] [CrossRef]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A Survey on Deep Transfer Learning. Artificial Neural Networks and Machine Learning – ICANN 2018; Kůrková, V.; Manolopoulos, Y.; Hammer, B.; Iliadis, L.; Maglogiannis, I., Eds. Springer International Publishing, 2018, pp. 270–279.
- Black, S.; Leo, G.; Wang, P.; Leahy, C.; Biderman, S. GPT-Neo: Large Scale Autoregressive Language Modeling with Mesh-Tensorflow 2021. If you use this software, please cite it using these metadata. [CrossRef]
- Gao, L.; Biderman, S.; Black, S.; Golding, L.; Hoppe, T.; Foster, C.; Phang, J.; He, H.; Thite, A.; Nabeshima, N. ; others. The Pile: An 800GB Dataset of Diverse Text for Language Modeling. arXiv 2020, arXiv:2101.00027 2020. [Google Scholar]
- Bhojanapalli, S.; Chakrabarti, A.; Glasner, D.; Li, D.; Unterthiner, T.; Veit, A. Understanding Robustness of Transformers for Image Classification. Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), 2021, pp. 10231–10241.
- Luecken, M.D.; Theis, F.J. Current best practices in single-cell RNA-seq analysis: A tutorial. J. Mol. Syst. Biol. 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- Cannoodt, R. anndata: ’anndata’ for R, 2022. https://github.com/dynverse/anndata.
- Le, T.; Huynh, N.; Le, B. Link Prediction on Knowledge Graph by Rotation Embedding on the Hyperplane in the Complex Vector Space. Artificial Neural Networks and Machine Learning – ICANN 2021; Farkaš, I., Masulli, P., Otte, S., Wermter, S., Eds.; Springer International Publishing: Cham, 2021; pp. 164–175. [Google Scholar]
- Ramachandram, D.; Taylor, G.W. Deep multimodal learning: A survey on recent advances and trends. IEEE Signal Process. Mag. 2017, 34, 96–108. [Google Scholar] [CrossRef]
- Baltrušaitis, T.; Ahuja, C.; Morency, L.P. Multimodal machine learning: A survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 41, 423–443. [Google Scholar] [CrossRef]
- Li, J.; Hong, D.; Gao, L.; Yao, J.; Zheng, K.; Zhang, B.; Chanussot, J. Deep learning in multimodal remote sensing data fusion: A comprehensive review. arXiv 2022, arXiv:2205.01380 2022. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).