1. Introduction
Winged bean [
Psophocarpus tetragonolobus (L.) DC] (2n = 18), a leguminous crop being cultivated for its green pods, tuberous roots, mature seeds, and leaves belongs to the
Fabaceae family. Winged bean contributes to the enhancement of food and nutrition security in the tropical region especially the sub-Saharan Africa (SSA) [
1]. Most part of winged bean, including pods, seeds, leaves, flowers, and tubers are edible and are rich in protein and other nutrients [
2]. Winged bean can be grown as a grain legume, green vegetable, tuber-crop, forage and as a cover crop [
2]. In comparison to major legumes, winged bean has a higher percentage of crude protein (30-38%) in the seeds compared to cowpea (23%), pigeon pea (22%), and lima beans (23%) [
3], [
4] and the tuberous root contains about 20% protein and 25–30% carbohydrates [
5]. The fresh young bean pod contains Vitamin C, thiamin pyridoxine (Vitamin B6), niacin, riboflavin and other minerals such as iron, copper, manganese, and calcium [
6]. Roasted and boiled tuberous roots of winged bean have been reported to improve the nutritional needs in Papua New Guinea, Indonesia, Malaysia, Myanmar, Ghana, and Nigeria [
5]. Aside nutritional benefits, winged bean has a high ability to fix atmospheric nitrogen to soil thus, making it a good option in cover cropping [
7].
Despite the enormous attributes of winged bean as a crop with food security potential in SSA, the production of winged bean is limited by multiple factors among which include the lack of genetic improvement with respect to the agronomic performance, nutritional, and biochemical components, lack of value chain demand, and lengthy life cycle. The lack of research focus on winged bean to facilitate the development of improved variety as well as the expansion of knowledge on the utilization of the crop has affected the economic potential of the crop in the society and has resulted in winged bean being categorized among the underutilized / orphan crops.
Assessing the genetic diversity of the available winged bean germplasm is a starting point to bring winged bean into the limelight of leguminous crops of economic importance. This is essential as it will provide an understanding of the extent and the level of genetic variability in the winged bean genetic resources and assist in identifying the genes controlling the expression of essential biological functions that could be exploited for winged bean improvement. The knowledge of genetic diversity helps crop improvement experts to identify and select progenitors with good characteristics for progeny development from where selection exercise can be carried out towards targeted breeding objectives. These characters include high yield potential, biotic and abiotic stress tolerance/resistance, and food quality attributes, directed towards combating food security and facilitating sustainable agriculture.
Genetic diversity in crops can be assessed using classical and molecular approaches. Of these two approaches, molecular assessment has proven to be more accurate and effective owing to the shortcomings attributable to classical or morphological approach [
32]. Though, the classical approach which is based on the differentiation in the morphology of the crop has been helpful, the high influence of the environment on the expression of the traits has been the major setback in the use of this approach. An attempt to overcome the limitation of classical approach led to the development of molecular approach. Molecular approach, which involves the use of molecular markers and good understanding of agro-morphological variation existing in germplasm of most legume crop has greatly facilitated the development of improved genotypes with desirable agronomic attributes [
8]. Of the molecular markers that have been used in profiling winged bean, microsatellite markers have been the most preferred due to the absence of single nucleotide polymorphisms (SNPs) markers [
6], [
7], [
9]. Microsatellites are co-dominant, abundant in genomes and highly polymorphic and are preferred for crops where SNPs are yet to be discovered [
7].
The aims of this study were to: (i) estimate the variance components and heritability for the traits of economic importance in winged bean and identify accession(s) with superior performance for these traits, (ii) assess the level of genetic diversity among 15winged bean accession using phenotypic and microsatellite (SSR) markers. This study will contribute to the development of the protocol to evaluate genetic diversity and choice of parental selection for future winged bean improvement.
2. Materials and Methods
2.1. Germplasm and Experimental site
Seeds of 15winged bean accessions were obtained from the Genetic Resources Centre (GRC) of the International Institute of Tropical Agriculture (IITA), Ibadan, Oyo State, Nigeria. Details of the accessions are presented in
Table 1. The experiments were carried out at the Research Field and Bioscience Center of IITA, Ibadan, located at the transition forest savanna agro-ecology of Nigeria (latitude 70° 30′ N, longitude 30° 54′ E, altitude 227.2 m above sea level, an alfisol soil of the Egbeda series and an average annual rainfall of 1,308 mm and monthly rainfall ranging between 0.05 and 86.5 mm; and the minimum and maximum temperatures ranging between 20 and 27 °C.
Seed viability assessment
The experiment to assess viability of the seeds of 15winged bean accessions was conducted at the GRC germination laboratory. Ten (10) seeds of each accession were scarified mechanically by cutting through the seed coat opposite the micropyle with a scalpel blade so as to allow water imbibition to break external dormancy and were treated with fungicide (mancozeb) to prevent seed infection and death. Seedburo K-22 germination paper sheets (also known as Kimpak or crepe paper) were cut and arranged in the transparent polyethylene boxes (7 to 14 mm layers per box) and autoclaved. Seeds were sown in polyethylene boxes according to standard operating laboratory procedure. Emergence count for each accession was done at 10 and 15 days after sowing. Each box was examined for number of germinated seeds, percentage dead seeds, normal seedling vigor and percentage of germination (Plate 1A and 1B).
2.2. Agro-morphological characterization
Scarified seeds of the 15 winged bean accessions were planted at IITA Ibadan Research Station in May 2017 and 2018. Entries were arranged using randomized complete block design (RCBD) with three replicates. Plot size was 5 x 5 m with spacing of 1 m and 1 m between plants within a row. No fertilizer was applied during the evaluation process and weeding was done as at when needed to keep the plot weed free. Plants were staked at eight weeks after planting and were protected from insect attacks with 0.5% karate and Cyperdeforce (lambda- cyhalothrin) from the period of flower bud initiation to pod maturity. Ten plants were tagged in each plot for data collection.
Figure 1.
Plate 1A and 1B: Winged bean germination test (1A) Plate from incubation at 10days after sowing, (1B) Plant at 15days after sowing in the germination room.
Figure 1.
Plate 1A and 1B: Winged bean germination test (1A) Plate from incubation at 10days after sowing, (1B) Plant at 15days after sowing in the germination room.
2.3. Data collection
Data were collected on vegetative and reproductive characters of winged bean. A total of 13 quantitative traits were scored. The quantitative traits were measured using seed counter, metric rulers, and Vennier caliper and weighed using weighing balance. The descriptions of the traits can be found in
Table 2
2.4. Statistical analyses
Analysis of variance (ANOVA) was conducted with a mixed linear model using the lmer Test package in R Software [
10] following the alpha lattice model below;
where Y
ijk is the phenotypic performance of accession for trait under consideration, µ is the average accession performance, Gen
i is the effect of accession i, Rep
j is the effect of replication j, Year
l is the effect of the planting year, Gen x Year
(il) is the effect of the interaction of genotype by year, and Error
ijl is the residual effect.
Replication was considered as a random effect whereas accessions were considered as fixed effect. Error (δ
2e), genotypic (δ
2g) and phenotypes (δ
2p) variances were calculated from expected mean squares (EMS) of ANOVA following Kresovich [
11]
Genotypic by environment interaction variance;
Where MSg = mean square of genotype; MSgl = mean square due to accession by year interaction; MSe = error mean square (mean square of error); l = number of year; r = number of replications
Broad-sense heritability (H
2), phenotypic coefficient of variance (PCV), and genotypic coefficient of variance (GCV) were calculated using the values derived from respective variance components. Broad-sense heritability (H
2) was classified as low (<30%), medium (30–60%), and high (>60%), according to Johnson et al. [
12]. Following Deshmukh et al. [
13], phenotypic and genotypic coefficients of variation greater than 20% were rated as high, between 10 and 20% were rated medium and lower than 10% were regarded as low.
where; δ
2p = phenotypic variance, δ
2g = genotypic variance, δ
2gl = genotype by year interaction variance; δ
2e: residual variance, r = number of replication; l = number of year; µ: grand mean of the trait
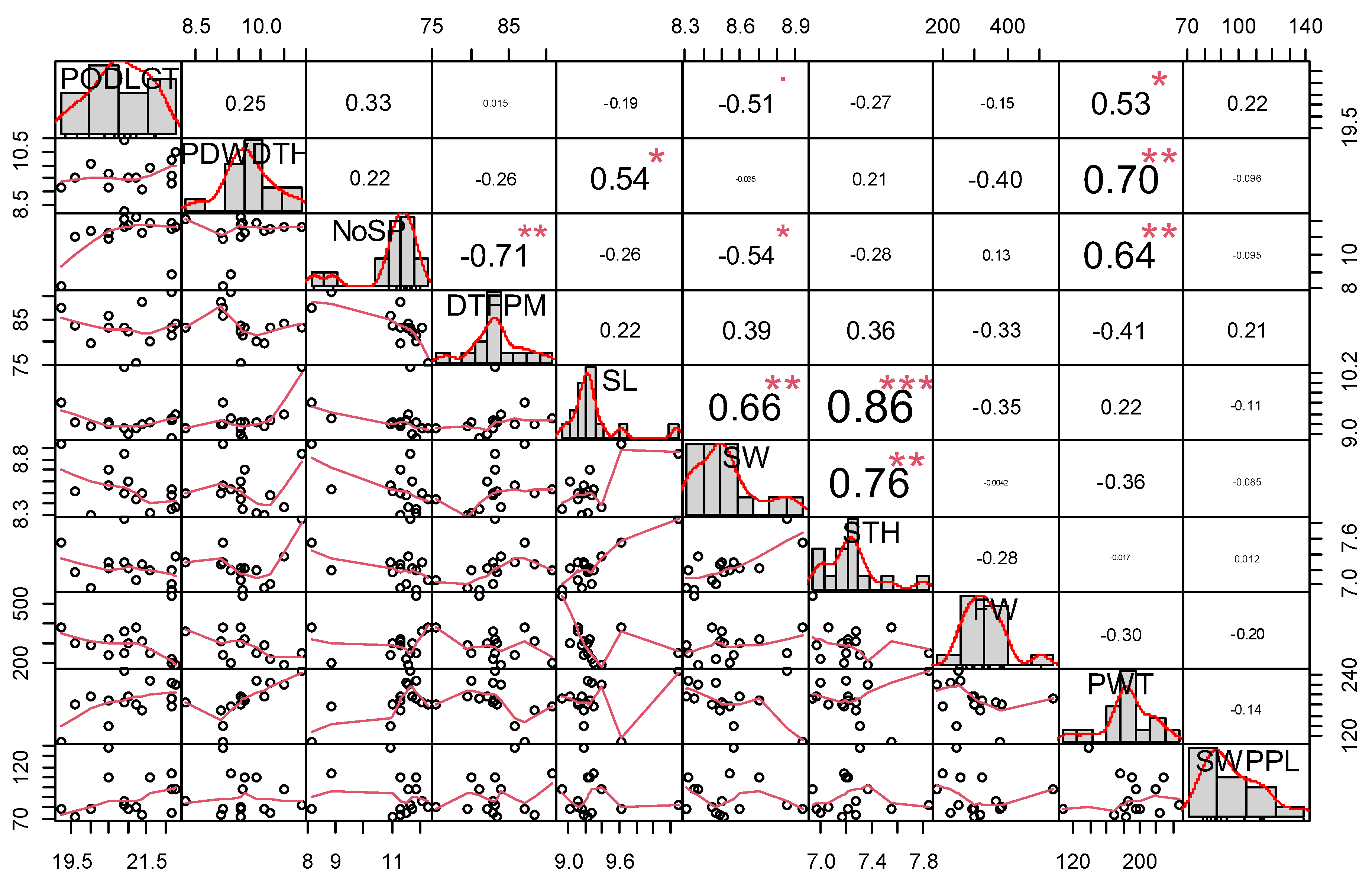
The Degree of relationships or associations among the assessed traits were determined using the Pearson’s correlation coefficients and visualized using ggpairs function in ggplot2 package [
14]. Principal component analysis (PCA) was done using the PRCOMP function implemented in R [
15] to identify the important traits that contributed to the observed genotypic variation. Hierarchical cluster analysis was done based on Ward.D2 method using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA). The final hierarchical cluster was built and viewed using the Dendextend package [
16] and circlize package [
17] in R. The optimum number of clusters was identified using the NbClust package [
18]. Path coefficient analysis was based on structural equation modeling and implemented using the Lavaan package [
19]. In this model, seed weight per plant and number of seeds per plant were considered as response variables against the agronomic traits as predictor variables. The path diagram from the Lavaan outputs was constructed using the semPlot package [
20] to depict the direct effect of these traits on seed weight per plant and number of seeds per plant for suitability for indirect selection.
2.5. Molecular characterization
Young leaf samples of three-week old winged bean plants were taken for genomic DNA (gDNA) extraction using a modified sodium dodecyl sulfate (SDS) extraction protocol [
21]. The leaf samples were collected randomly from three replicates in tens for the 15 accessions. The resultant genomic DNA was checked for degradation and quality using agarose gel electrophoresis method by running the extracted genomic DNA samples on 1% agarose gel and visualized under UV fluorescence using gel documentation system (ENDURO
TM GDS). DNA quantity and purity were checked using a Nanodrop spectrophotometer (ND- 1000) (Thermo Fisher Scientific, USA).
The SSR sequences were based on the primer sequence reported by Vatanparast et al. [
22]. The length of nucleotides were nine for tri-nucleotide and one for tetra-nucleotide repeat motifs for both forward and reverse primer as shown in (
Table 3). The microsatellite regions were amplified using EconoTaq PLUS 2X Master Mix Catalog No. 30035 (Lucigen). PCR reaction volume of 20 µL containing 40 ng of g DNA 1 µL, 10 mM of each primer 1µL, 10 µL EconoTaq PLUS 2X Master Mix and 7 µL nuclease free water (Catalog No. E476 (AMRESCO LLC, OH 44139, USA). The PCR amplification was performed in a thermocyler for each reaction with an initial denaturation at 95
0C for 5 min followed by 35 cycles consisting of 95
0C at 30 s (denaturation), 50
0C for 30 s (annealing), and 72
0C at 30 s (extension) a final extension at 72
0C for 10 min. The amplified product were visualized by agarose gel electrophoresis (Sunrise 96, Biometra, Germany), at 100 volts for two hours. The PCR products from ten SSRs markers were co-loaded for fragment analysis, was performed using a 1:10 dilution of the fluorescently labelled PCR amplicons, LIZ500 sizing standard and Hi-DiTM Formamide Catalog No. 4311320 (Thermo Fisher Scientific, Carlsbad, USA) mixture and denatured at 95°C for five minutes. After denaturation, the fragments analysis was performed using a 50 cm capillary array, POP-7TMon an ABI PRISM™ 3500xl. Genetic Analyser (Applied Biosystems, Thermo Fisher Scientific, Carsbad, USA), and peak and alleles sizes were scored using GeneScan™ Software and interpreted using GeneMapper
®v5.0. (Applied Biosystems, Thermo Fisher Scientific, Carlsbad, USA).
Data files of the molecular data were assembled in a database (Genemapper v 5.0) [
23] and allele sizes was checked for congruency and adjusted according to the allelic references provided in gene mapper manual for microsatellites markers. Data obtained were exported to an excel sheet. The presence or absence of fragment size was transformed into a binary character matrix (1 for presence and 0 for absence). Descriptors of genetic diversity, such as allele number per marker, allele frequency, gene diversity and polymorphic information content (PIC) were calculated using Power Marker v 3.25 software [
24]. The polymorphic information content (PIC) of each SSR marker was calculated using the Power Marker v 3.25 software. PIC gives an indication of the discriminatory power of an SSR locus by considering not only the number of alleles that are expressed but also the relative frequencies of those alleles. Phylogenetic relations were determined by cluster analysis using unweighted pair group method with arithmetic averages (UPGMA). An analysis of molecular variance (AMOVA) was performed to study the differences between the UPGMA clusters. AMOVA pairwise comparisons between groups and estimation of molecular diversity (expected heterozygosity (He) within groups were conducted using GenAlex 6.5 [
25]
3. Results
3.1. Variability in agronomic traits of 15 winged bean accessions
Results of the analysis of variance (ANOVA) showing the statistical differences for accessions, year, and accessions x year interaction is presented in
Table 4. Combined ANOVA revealed accessions x year interaction effects at
p < 0.05 for all the assessed traits indicating the year influence on the observed phenotypic expression of the accessions for these traits. The interaction effects was significant for days to plant maturity and seed weight, suggesting that accession performance was year dependent. Accession effect was significant at
p < 0.001 for all the traits evaluated, indicating significant differences in the observed phenotypic performance of the accessions. Significant variation at
p < 0.05 was observed for accessions effects for all measured traits, indicating that the accession performance differs for all the traits evaluated. Year effect was significant for pod width at
p < 0.001 and days to plant maturity at
p < 0.05, indicating the existence of year differences with respect to these traits. Both accessions and year effects were significant at
p < 0.001 for all the studied traits in both years.
3.2. Genetic variances and broad-sense heritability of agronomic and yield traits in winged bean accessions
Genotypic and phenotypic variance components, genotypic and phenotypic coefficients of variation, and broad-sense heritability of agronomic traits in winged bean accessions are presented in
Table 4. Genotypic coefficients of variation (GCV) varied from a moderate classification of 15.62% for pod weight and 17.67% for seed weight to high classification of 23.24% for fodder weight. A similar result was recorded for phenotypic coefficients of variation (PCV) that varied from a moderate classification of 10.31% for number of seeds per pod 18.32% for pod weight, and 21.26% for seed weight to a high classification of 29.71% for fodder weight. Broad-sense heritability (H
2) ranged between 18.92% (moderate) and 72.67% (high). High H
2 (>40%) was observed in all the estimated parameters except for days to first plant maturity, seed length, seed thickness and pod width.
3.3. Dimension reduction analysis of yield and agronomic traits
The results of the PCA showed that the first three principal components (PCs) contributed largely to the observed phenotypic variance (
Figure 2A). These PCs had eigenvalues greater than one and accounted for 76.89% of the total genetic variation in the study (
Table 5). The first PC accounted for 34.36% of the variation, with major contributions from number of seeds per pod, seed width, and seed thickness (
Table 5 and
Figure 2B). The second PC accounted for 26.42%, with major contributions from pod width, seed length, fodder weight and pod weight (
Table 5 and
Figure 2B). The third PC accounted for 16.12%, with major contributions pod length and seed weight (
Table 5). The genotype by traits biplot revealed that accessions TPt-6A and TPt-6 had good performance for pod weight. Accession TPt-48 had good performance for pod length. Accession TPt-43 had good performance for fodder weight, and accessions TPt-32 and TPt-153 had good performance for days to first pod maturity (
Figure 2C).
3.4. Relationships among agronomic and yield traits in 15 winged bean accession
The relationship among evaluated winged bean traits is presented in
Figure 3. A significant positive relationship was observed between pod weight and pod length (r = 0.53, p< 0.05), pod width and seed length (r = 0.54, p< 0.05) and pod weight (r = 0.70, p< 0.01). Number of seeds per pod and pod weight (0.64, p< 0.01) seed thickness and seed length (r = 0.86,
p < 0.001) seed width (r = 0.76), number of seed per pod (0.64). Seed length and seed weight (r = 0.66, p< 0.01), seed thickness and seed length (r = 0.86, p< 0.001) and seed weight (r = 0.76, p< 0.01). A negative significant relationship was observed for days to first plant maturity and number of seeds per pod (r =
-0.71,
p < 0.01). A significant positive relationship indicates similar direction in trait performance, while a significant negative relationship indicates opposite direction in traits expression.
3.5. Diversity among 15 winged bean accession based on Gower’s distance derived from morphological characteristics
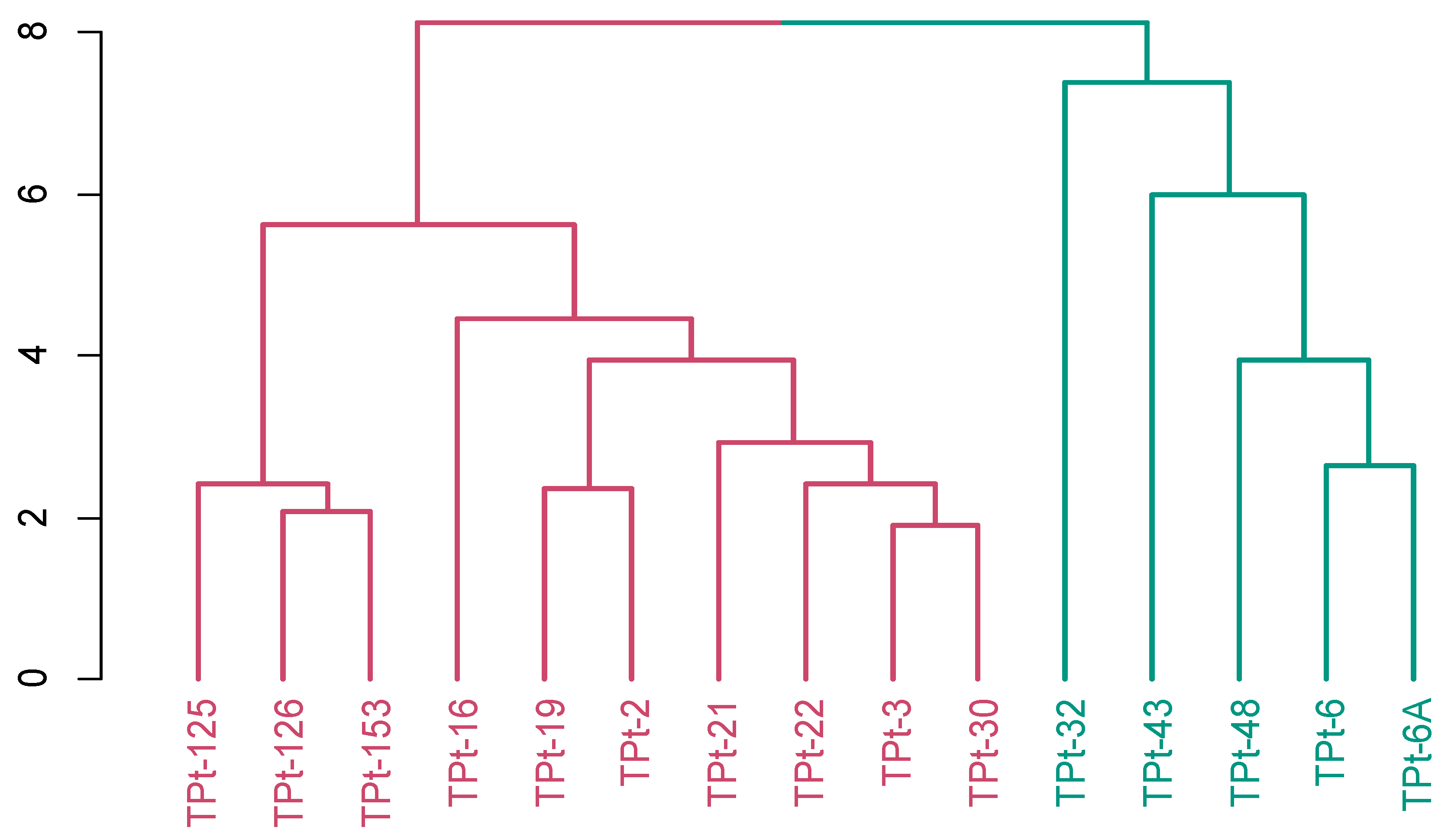
Hierarchical clustering employed for the grouping of the 15winged bean accessions based on Gower’s distance using the evaluated agronomic and yield traits produced two clusters (
Figure 4). Cluster one consisted of ten accession (TPt-125, TPt-126, TPt-153, TPt-16, TPt-19,TPt-2, TPt-21, TPt-22, TPt-3 and TPt-30) characterized by seed length, seed width and seed thickness while cluster two had five accessions as, (TPt-32,TPt-43, TPt-48, TPt-6 and TPt-6A ) characterized by number of seeds per plot and days to first plant maturity
Table 6.
Cluster Descriptions of the two hierarchical clusters formed on the basis of ten measured traits of the 15 winged bean accessions.
Table 6.
Cluster Descriptions of the two hierarchical clusters formed on the basis of ten measured traits of the 15 winged bean accessions.
| Traits |
Cluster one – Red (10) |
Cluster two – Green (5) |
|
| |
Min |
Max |
Mean |
Min |
Max |
Mean |
F-value |
| PODLGT |
19.20 |
22.20 |
20.66a |
19.60 |
22.30 |
21.24a |
1.20ns |
| PDWDTH |
9.15 |
10.92 |
9.64a |
8.30 |
10.51 |
9.63a |
0.00ns |
| NoSP |
8.13 |
11.63 |
10.18b |
11.00 |
12.25 |
11.65a |
8.30* |
| DTFPM |
83.20 |
90.80 |
86.04a |
75.50 |
88.50 |
82.08b |
4.78* |
| SL |
9.19 |
10.29 |
9.53a |
8.92 |
9.39 |
9.16b |
5.82* |
| SW |
8.54 |
8.94 |
8.72a |
8.31 |
8.60 |
8.44b |
16.65** |
| STH |
7.18 |
7.85 |
7.42a |
6.93 |
7.70 |
7.14b |
6.44* |
| FW |
203.00 |
380.00 |
280.40a |
185.00 |
546.00 |
316.50a |
0.51ns |
| PWT |
108.00 |
247.00 |
172.20a |
170.00 |
228.00 |
198.00a |
1.98ns |
| SWPPL |
78.50 |
138.90 |
104.64a |
70.40 |
109.40 |
84.77a |
4.31ns |
3.6. Path analysis among assessed traits of 15 winged bean accession
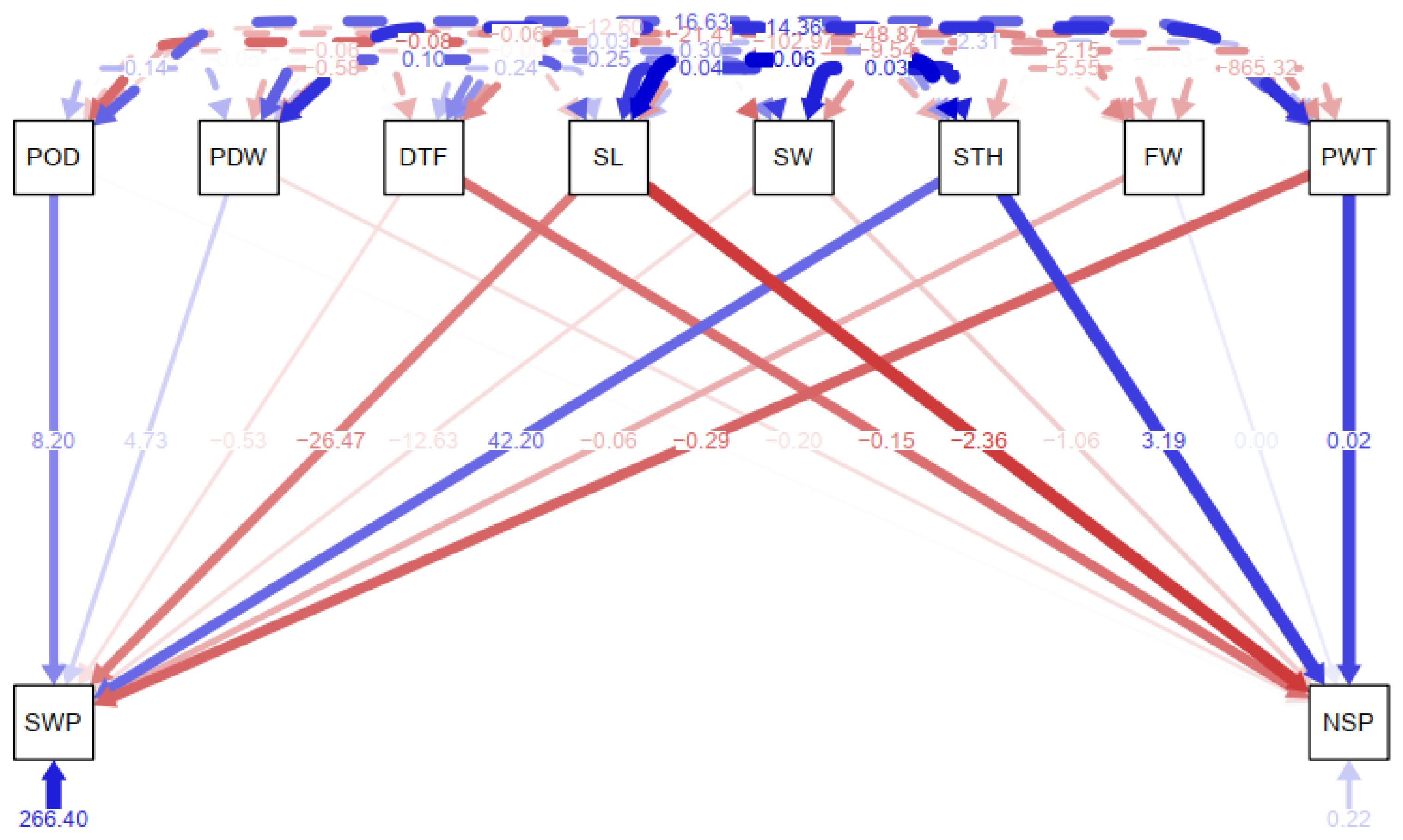
The path analysis conducted to depict the direct effects of agronomic traits on yield traits for suitability for indirect selection is presented in
Figure 5. The path analysis began with structural equation modeling where seed weight and number of seeds per pod were considered response variables against other correlated agronomic and yield traits. The chi-square test of the model fit was moderately significant (χ2 (4) = 2.455,
p = 0.653). Overall, fit indices were in good range (RMSEA = 0.00 [0.00, 0.09],
p = 0.81; CFI = 1.00; SRMR = 0.01). Pod weight significantly predicts the number of seed per pod (b = 0.02, SE = 0.007, p = 0.003) such that a one-unit increase in pod weight will bring about a 0.02-unit increase in number of seeds per pod. Seed thickness significantly predicts the number of seeds per pod (b = 3.19, SE = 1.38, p = 0.021) such that a one-unit increase in seed thickness was associated with a 3.19-unit increase in number of seeds per pod. Days to maturity significantly predicts the number of seeds per pod (b = -0.15, SE = 0.054, p = 0.006) such that a one-unit increase in days to maturity was associated with a 0.15-unit decrease in the number of seeds per pod.
3.7. Molecular diversity among 15 winged bean accessions
3.7.1. Polymorphism as detected by Simple Sequence repeats (SSRs)
The microsatellite markers, allele frequency, gene diversity, number of alleles amplified in each locus, and PIC values are presented in
Table 7. The ten microsatellite loci amplified the winged bean DNA. The ten microsatellite loci detected a total of 42 polymorphic alleles. The number of alleles for each SSR loci ranged between 3 and 6, with an average value of 4.2 alleles per locus. The PIC values for the SSRs varied from 0.0888 to 0.4606 with a mean of 0.2178. SSRs 879, and 3111, both tetra nucleotide primers detected the highest number of fragments (six) and, therefore, were the most informative primers while each of SSRs 24,704, 747, and 854 gave the least fragments (three).
3.7.2. Analysis of molecular variance (AMOVA)
From the 150 individual plants that constituted in the 15 winged bean accessions, AMOVA showed between population variation of 5% and within population variation of 95%. This suggests the presence of low genetic differentiation among the accessions as a whole germplasm. However, high intra accession diversity exist among the individuals of each accession (
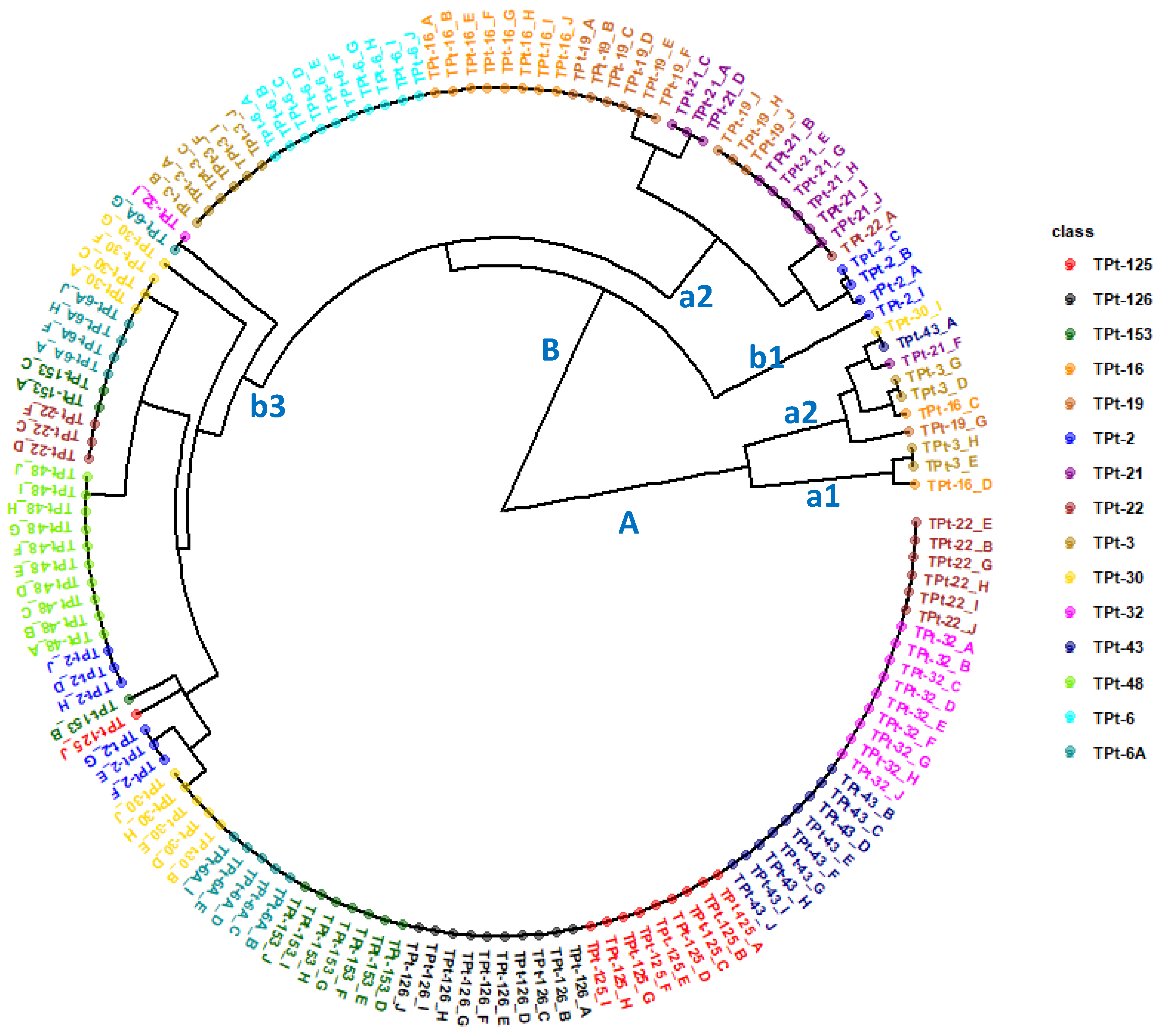
Table 8). The phylogeny diagam from the Jaccard dissimilarity matrix revealed the presence of two genetic groups or clusters A and B (
Figure 6). Cluster A can be subdivided into two subgroups (a1 and b1) comprising individuals from different winged bean accessions while cluster B can be partitioned into three subgroups (b1, b2, and b3). Though the 150 individuals were from 15 accessions (10 plants from each accessions), the phylogeny results did not show a complete segregation of the ten plants from each accession into the same subgroup for many accessions of the winged bean. Only three of the 15 winged bean accessions (TPt-6, TPt-126, and TPt-48) had all the ten individuals grouped together showing no intra accession variation. Aside these, nine accessions (TPt-32, TPt-43, TPt-125, TPt-16, TPt-153, TPt-19, TPt-21, TPt-22 and TPt-3) had over 50% of their individuals grouped together. The rest of the accessions had less than 50% of their individuals clustered together.
4. Discussion
4.1. Variability in agronomic and yield traits of winged bean as identifiers of gene reservoirs for winged bean improvement
Winged bean production is challenged by numerous constraints, including, but not limited to, low yield, long life cycle, photoperiodic sensitivity and indeterminate growth habit and flowering, which require the attention of legume breeders in countries where the crop exists [
26], [
27]. The genetic variability for yield and agronomic traits existing among the winged bean accessions considered in this study suggests the presence of gene reservoir for genetic improvement of winged bean when the focus is on improved yield potential and other important agronomic traits. Breeding for improved yield and other desirable agronomic traits can be challenging for breeders especially with crops with minimal information on the indices that determines response to selection [
28]. The medium to high heritability estimates observed for seed weight per plant, pod weight, fodder weight, seed width, number of seeds per pod, and pod length are good indices that suggest the ease of these traits to genetic improvement in winged bean breeding. These traits had major contributions to the observed genetic variability among the studied winged bean accessions as detected through moderate to high genetic correlation coefficient estimates. Previous studies have also reported high heritability estimates for seed weight, pod weight, fodder weight, seed width, and number of seeds per pod [
5]. Genetic variability is particularly useful as it facilitates the selection of good progenitors for progeny development from where selection can be applied towards the development of winged bean ideotypes [
29]. In our study, hierarchical clustering, which explains genetic similarities among accessions that were grouped in the same cluster revealed a cluster with 10 members characterized by longer, wider, and thicker seeds, and another cluster with five members characterized by higher number of seeds per pod and earliness.
4.2. Relationships among assessed traits for indirect selection in winged bean improvement
In breeding winged bean for improved yield performance, positive and strong relationship between pod weight, pod length, pod width, and number of seeds per pod, as well as between seed length and seed width and thickness could provide a useful guide especially when consideration is given to indirect selection. Adegboyega et al. [
5], Tanzi et al. [
26], and Schinavito et al. [
27] similarly observed positive relationships between seed yield and other traits (pod width, seed length, fodder weight, and pod weight) in a panel of winged bean accessions studied for genetic diversity and impact of staking on winged bean production. One of the challenges in winged bean improvement is lengthy life cycle, photoperiodic sensitivity and indeterminate growth habit and flowering of the genotypes [
34]. Thus, any means to select for improved yield and good seed characteristics through indirect selection for other correlated agronomic trait will be of advantage, this study revealed that number of seeds per pod could be predicted by pod weight, days to maturity, seed length, and seed thickness.
4.3. Microsatellite markers reveal intra-accession variation within the winged bean germplasm
For effective conservation and utilization of germplasm in breeding programs, it is necessary to assess the genetic diversity using molecular tools [
30]. In our study, 10 SSR primers were used to assess the genetic diversity in 15 winged bean accessions to ascertain the presence or absence of intra-accession variability. This was targeted at providing useful information that will facilitate proper conservation and utilization of assembled winged bean germplasm.
The amplification of winged bean SSRs recorded an average PIC value of 0.22 which is comparable to recent studies in pigeon pea [
31], munged bean [
32], and common bean [
33]. Though, this value is lower than that reported for cowpea [
34] and Bambara groundnut [
35]. The observed low PIC values in this study could arise from the small population size used for genotyping as well as the limited number of SSR primers used for profiling. In previous studies, Chandra et al. [
7] successfully used RAPDs and ISSRs to capture considerable genetic diversity among 24 winged bean accessions in which a PIC of 0.17 and 0.213 were reported for RAPDs and ISSR primers, respectively. Wong et al. [
6] equally reported a PIC of 0.16 for 18 primers and suggested that it might be because of the low validation rate of polymorphic markers that were screened which was in turn due to the limited number of accessions that were screened. The PIC values are expected to increase with increased number of accessions covering a broader range of geographical origins.
In this study, the analysis of molecular variance revealed within group variability of 95% as compared to between groups of 5% which suggests the presence of large intra-accession variability compared to inter-accession variability. This is not surprising as the accessions used in this study have been collected from different locations/origins/countries mostly from local farmers and conserved in IITA- gene bank. The phylogeny analysis further revealed the nature of the intra-accession similarities and dissimilarities that exist among studied winged bean germplasm. It should be noted that the clustering of the genotypes that constitute the accessions in the study was not based on geographical origins as a co-factor.
The results of this aspect of the study may not necessarily indicate high level of genetic diversity as diversity is not conditioned only by the allelic divergence at many loci but also by complementary alleles with dominance or epistatic genetic effects [
36], [
37]. Our study, therefore, validated the existence of a high level of intra-accession diversity in winged bean germplasm. The results of the molecular analysis in combination with that of the phenotypic characterization presented in this report may form the basis for further studies aimed at exploiting existing variation in winged bean germplasm for its improvement.
Author Contributions
Conceptualization, S.T.E., A.A.M., A.M.T.; Methodology, S.T.E, A.A.M., O.O.; Supervision A.A.M. and A.M.T.; Writing original draft, S.T.E.; Manuscript review and editing, S.T.E., A.A.M, B.A.A., A.I.I., O.O.A., F.B., A.M.T.
Funding
This study is funded by the Crop Trust through the Genetic Resource Center, IITA, Ibadan, Nigeria.
Data Availability Statement
Data can be obtained upon request from the corresponding author.
Acknowledgments
The authors express gratitude to members of staff of the seed bank of the Genetic Resources Center, IITA, Ibadan, members of staff Bioscience Centre of IITA, and Inqaba biotech South Africa
Conflicts of Interest
The authors declare that the research was conducted in the absence of any potential conflict of interest.
References
- Tanzito, G.; Ibanda, P.A.; Ocan, D.; Lejoly, J. Use of charcoal (biochar) to enhance tropical soil fertility: A case of Masako in Democratic Republic of Congo. J. Soil Sci. Environ. Manag. 2020, 11, 17–29. [Google Scholar] [CrossRef]
- Sriwichai, S.; Monkham, T.; Sanitchon, J.; Jogloy, S.; Chankaew, S. Dual-Purpose of the Winged Bean (Psophocarpus tetragonolobus (L.) DC.), the Neglected Tropical Legume, Based on Pod and Tuber Yields. Plants 2021, 10, 1746. [Google Scholar] [CrossRef]
- Aletor, V.A.; Aladetimi, O.O. Compositional evaluation of some cowpea varieties and some under-utilized edible legumes in Nigeria. Food/Nahr. 1989, 33, 999–1007. [Google Scholar] [CrossRef]
- Koshy, E.P.; John, P.; Scaria, S. Winged bean: The wings that carry away malnutrition. Acad. Rev. 1999, 3, 77–80. [Google Scholar]
- Adegboyega, T.T.; Abberton, M.T.; AbdelGadir, A.H.; Dianda, M.; Maziya-Dixon, B.; Oyatomi, O.A.; Ofodile, S.; Babalola, O.O. Evaluation of Nutritional and Antinutritional Properties of African Yam Bean (Sphenostylis stenocarpa (Hochst ex. A. Rich.) Harms.) Seeds. J. Food Qual. 2020, 2020, 1–11. [Google Scholar] [CrossRef]
- Wong, Q.N.; Tanzi, A.S.; Ho, W.K.; Malla, S.; Blythe, M.; Karunaratne, A.; Massawe, F.; Mayes, S. Development of Gene-Based SSR Markers in Winged Bean (Psophocarpus tetragonolobus (L.) DC.) for Diversity Assessment. Genes 2017, 8, 100. [Google Scholar] [CrossRef]
- Mohanty, C.S.; Verma, S.; Singh, V.; Khan, S.; Gaur, P.; Gupta, P.; Nizar, M.A.; Dikshit, N.; Pattanayak, R.; Shukla, A.; et al. Characterization of winged bean (Psophocarpus tetragonolobus (L.) DC.) based on molecular, chemical and physiological parameters. Am. J. Mol. Biol. 2013, 03, 187–197. [Google Scholar] [CrossRef]
- Egan, A.N.; Schlueter, J.; Spooner, D.M. Applications of next-generation sequencing in plant biology. Am. J. Bot. 2012, 99, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Ojuederie, O.B.; Nkang, N.A.; Odesola, K.A.; Igwe, D.O. Genetic diversity assessment of winged bean (Psophorcarpus tetragonolobus) accessions revealed by Inter-Simple Sequence Repeat (ISSR) markers. J Plant Biol Crop Res 2020, 3, p1014. [Google Scholar]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest Package: Tests in linear mixed effects models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef]
- Kresovich, S. Quantitative genetics in maize breeding: by A.R. Hallauer and J.B. Miranda; Iowa State University Press, Ames, Iowa, U.S.A. 50010; 1989 (Second edition), xii + 468 pp.; hardcover US$44.95; ISBN 0-8138-1522-3. Field Crop. Res. 1990, 23, 78–79. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Genotypic and Phenotypic Correlations in Soybeans and Their Implications in Selection1. Agron. J. 1955, 47, 477–483. [Google Scholar] [CrossRef]
- Deshmukh, S.N.; Basu, M.S.; Reddy, P.S. Genetic variability, character association and path coefficients of quantitative traits in Virginia bunch varieties of groundnut. Indian J. Agric. Sci. 1986, 56. [Google Scholar]
- AKassambara, F. Mundt, “Package ‘ factoextra,’” CRAN- R Packag., 2020.
- RC Team, “A language and environment for statistical computing. R Foundation for Statistical Computing.” Vienna, Austria, 2017.
- Galili, T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlizeimplements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- MCharrad, N. Ghazzali, V. Boiteau, and A. N. Maintainer, “Determining the Best Number of Clusters in a Data Set,” Cran, 2015.
- Rosseel, Y. lavaan: AnRPackage for Structural Equation Modeling. J. Stat. Softw. 2012, 48, 1–36. [Google Scholar] [CrossRef]
- Epskamp, S. semPlot: Unified Visualizations of Structural Equation Models. Struct. Equ. Model. A Multidiscip. J. 2015, 22, 474–483. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: version II. Plant Mol. Biol. Report. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Vatanparast, M.; Shetty, P.; Chopra, R.; Doyle, J.J.; Sathyanarayana, N.; Egan, A.N. Transcriptome sequencing and marker development in winged bean (Psophocarpus tetragonolobus; Leguminosae). Sci. Rep. 2016, 6, 29070. [Google Scholar] [CrossRef]
- Chatterji, S.; Pachter, L. Reference based annotation with GeneMapper. Genome Biol. 2006, 7, R29–R29. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Peakall, R.O.D. and Smouse, P.E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Tanzi, A.S.; Ho, W.K.; Massawe, F.; Mayes, S. Development and interaction between plant architecture and yield-related traits in winged bean (Psophocarpus tetragonolobus (L.) DC.). Euphytica 2019, 215, 36. [Google Scholar] [CrossRef]
- Schiavinato, M.A.; Válio, I.F. Influence of photoperiod and temperature on the development of winged bean plants. Rev. Bras. De Fisiol. Vegetal. 1996, 8, 105–110. [Google Scholar]
- Adejumobi, I.I.; Agre, P.A.; Onautshu, D.O.; Adheka, J.G.; Cipriano, I.M.; Monzenga, J.C.L.; Komoy, J.L. Assessment of the yam landraces (Dioscorea spp.) of DR Congo for reactions to pathological diseases, yield potential, and tuber quality characteristics. Agriculture 2022, 12, p599. [Google Scholar] [CrossRef]
- Amoo, I.A.; Adebayo, O.T.; Oyeleye, A.O. Chemical evaluation of winged beans (Psophocarpus tetragonolobus), Pitanga cherries (Eugenia uniflora) and orchid fruit (Orchid fruit myristica). Afr. J. Food Agric. Nutr. Dev. 2006, 6. [Google Scholar]
- Li, G.; Ra, W.-H.; Park, J.-W.; Kwon, S.-W.; Lee, J.-H.; Park, C.-B.; Park, Y.-J. Developing EST-SSR markers to study molecular diversity in Liriope and Ophiopogon. Biochem. Syst. Ecol. 2011, 39, 241–252. [Google Scholar] [CrossRef]
- Dutta, S.; Kumawat, G.; Singh, B.P.; Gupta, D.K.; Singh, S.; Dogra, V.; Gaikwad, K.; Sharma, T.R.; Raje, R.S.; Bandhopadhya, T.K.; et al. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol. 2011, 11, 17–17. [Google Scholar] [CrossRef]
- Gupta, S.K.; Bansal, R.; Gopalakrishna, T. Development and characterization of genic SSR markers for mungbean (Vigna radiata (L.) Wilczek). Euphytica 2013, 195, 245–258. [Google Scholar] [CrossRef]
- Blair, M.W.; Hurtado, N.; Chavarro, C.M.; Muñoz-Torres, M.C.; Giraldo, M.C.; Pedraza, F.; Tomkins, J.; Wing, R. Gene-based SSR markers for common bean (Phaseolus vulgaris L.) derived from root and leaf tissue ESTs: an integration of the BMc series. BMC Plant Biol. 2011, 11, 50–10. [Google Scholar] [CrossRef]
- Gupta, S.K.; Gopalakrishna, T. Development of unigene-derived SSR markers in cowpea (Vigna unguiculata) and their transferability to otherVignaspecies. Genome 2010, 53, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Molosiwa, O.O.; Aliyu, S.; Stadler, F.; Mayes, K.; Massawe, F.; Kilian, A.; Mayes, S. SSR marker development, genetic diversity and population structure analysis of Bambara groundnut [Vigna subterranea (L.) Verdc.] landraces. Genet. Resour. Crop. Evol. 2015, 62, 1225–1243. [Google Scholar] [CrossRef]
- Adebayo, M.A.; Kolawole, A.O.; Adebayo, T.A. Assessment of new generation of drought-tolerant maize (Zea mays L.) hybrids for agronomic potential and adaptation in the derived savanna agro-ecology of Nigeria. Int J Agron Agric Res 2015, 7, 45–54. [Google Scholar]
- Adebayo, M.A.; Menkir, A.; Gedil, M.; Blay, E.; Gracen, V.; Danquah, E.; Funmilayo, L. Diversity assessment of drought tolerant exotic and adapted maize (Zea mays L.) inbred lines with microsatellite markers. J. Crop. Sci. Biotechnol. 2015, 18, 147–154. [Google Scholar] [CrossRef]
Figure 2.
PCA screen plot (A), variable contribution (B) and accession biplot (C) accounting for the total variability observed in PC1 and PC2. PODLGT= Pod length; PDWDTH=Pod Width; NoSP=Number of Seeds per Pod; DTFPM= Days to first plant maturity; SL= Seed Length; SW= Seed Width; STH= Seed Thickness; FW= Fodder Weight; PW= Pod weight; SWPPL= Seed Weight.
Figure 2.
PCA screen plot (A), variable contribution (B) and accession biplot (C) accounting for the total variability observed in PC1 and PC2. PODLGT= Pod length; PDWDTH=Pod Width; NoSP=Number of Seeds per Pod; DTFPM= Days to first plant maturity; SL= Seed Length; SW= Seed Width; STH= Seed Thickness; FW= Fodder Weight; PW= Pod weight; SWPPL= Seed Weight.
Figure 3.
Correlation coefficients among agronomic and yield traits. PODLGT= Pod length; PDWDTH=Pod Width; NoSP=Number of Seed per Pod; DTFPM= Days to first plant maturity; SL= Seed Length; SW= Seed Width; STH= Seed Thickness; FW= Fodder Weight; PW= Pod weight; SWT= Seed Weight; *, **, *** = significant at p < 0.05, 0.01, and 0.001 respectively.
Figure 3.
Correlation coefficients among agronomic and yield traits. PODLGT= Pod length; PDWDTH=Pod Width; NoSP=Number of Seed per Pod; DTFPM= Days to first plant maturity; SL= Seed Length; SW= Seed Width; STH= Seed Thickness; FW= Fodder Weight; PW= Pod weight; SWT= Seed Weight; *, **, *** = significant at p < 0.05, 0.01, and 0.001 respectively.
Figure 4.
Hierarchical clustering showing the grouping of 15 winged bean accessions into two clusters using ten agronomic and yield traits based on the Gower’s dissimilarity matrix. Cluster one (Red) and cluster two (Green).
Figure 4.
Hierarchical clustering showing the grouping of 15 winged bean accessions into two clusters using ten agronomic and yield traits based on the Gower’s dissimilarity matrix. Cluster one (Red) and cluster two (Green).
Figure 5.
Path coefficient analysis between response and independent winged bean variables. PODLGT= Pod length; PDWDTH=Pod Width; NoSP=Number of Seeds per Pod; DTFPM= Days to first plant maturity; SL= Seed Length; SW= Seed Width; STH= Seed Thickness; FW= Fodder Weight; PW= Pod weight; SWT= Seed Weight. Red indicates direct negative impact while Blue indicate direct positive impact.
Figure 5.
Path coefficient analysis between response and independent winged bean variables. PODLGT= Pod length; PDWDTH=Pod Width; NoSP=Number of Seeds per Pod; DTFPM= Days to first plant maturity; SL= Seed Length; SW= Seed Width; STH= Seed Thickness; FW= Fodder Weight; PW= Pod weight; SWT= Seed Weight. Red indicates direct negative impact while Blue indicate direct positive impact.
Figure 6.
Phylogenetic relationships of 150 individuals from 15 winged bean accession from Jaccard’s dissimilarity matrix.
Figure 6.
Phylogenetic relationships of 150 individuals from 15 winged bean accession from Jaccard’s dissimilarity matrix.
Table 1.
Accession numbers, sources and qualitative morphological characters of the studied 15 winged bean accessions.
Table 1.
Accession numbers, sources and qualitative morphological characters of the studied 15 winged bean accessions.
| S/N |
Accession no |
Source |
SC |
SS |
FLC |
STEMCLR |
PPS |
PS |
LSS |
| 1 |
TPt-2 |
Nigeria |
Brownish orange |
Oval |
Pastel violet |
Green |
absent |
Flat on suture |
Deltoid-large |
| 2 |
TPt-3 |
Nigeria |
Yellowish brown |
Oval |
Light violet |
purple |
present |
Flat on suture |
Ovate Lanceolate-medium |
| 3 |
TPt-6 |
Nigeria |
Yellowish brown |
Oval |
Pastel violet |
Green |
present |
Flat on side |
Ovate-large |
| 4 |
TPt-16 |
Indonesia |
Brownish orange |
Round |
Light violet |
Green |
absent |
Flat on side |
Ovate Lanceolate-large |
| 5 |
TPt-19 |
Nigeria |
Yellowish brown |
Oval |
Pale blue |
Green |
absent |
Flat on suture |
Deltoid-large |
| 6 |
TPt-21 |
Papua New Guinea |
violet brown |
Round |
Light violet |
Green |
absent |
Flat on sides |
Deltoid-medium |
| 7 |
TPt-22 |
Papua New Guinea |
Brownish Yellow |
Round |
Pastel violet |
purple |
absent |
Flat on sides |
Deltoid-large |
| 8 |
TPt-32 |
Unknown |
Yellowish brown |
Oval |
Pale violet |
Green |
absent |
Flat on suture |
Deltoid-large |
| 9 |
TPt-43 |
Unknown |
Tan |
Oval |
Light violet |
Greenish purple |
present |
Flat on sides |
Deltoid-large |
| 10 |
TPt-48 |
Unknown |
Yellowish brown |
Oval |
Pale violet |
Green |
present |
Flat on suture |
Deltoid-large |
| 11 |
TPt-125 |
Unknown |
Tan |
Oval |
Pastel violet |
Green |
absent |
Flat on suture |
Deltoid-large |
| 12 |
TPt-126 |
Unknown |
Yellowish brown |
Oval |
Light violet |
Green |
absent |
Flat on sides |
Deltoid-large |
| 13 |
TPt-153 |
Unknown |
Light brown |
Oval |
Light violet |
Greenish purple |
absent |
Flat on sides |
Deltoid-large |
| 14 |
TPt-6A |
Nigeria |
Brownish orange |
Oval |
Light violet |
Green |
absent |
Flat on suture |
Deltoid-large |
| 15 |
TPt-30 |
Unknown |
Brownish orange |
Round |
Pastel violet |
Green |
absent |
Flat on sides |
Deltoid-large |
Table 2.
Descriptions of the 13 quantitative traits measured on the studied 15 winged bean accessions in 2017 and 2018 seasons.
Table 2.
Descriptions of the 13 quantitative traits measured on the studied 15 winged bean accessions in 2017 and 2018 seasons.
| S/N |
Traits |
Description of measurement |
Collection Period |
| 1 |
Days to First Flower (DTFF) |
number of days from planting to when a plant in a plot emerged first flower |
6 WAP |
| 2 |
Days to First Pod (DTFP), |
number of days from planting to when a plant in a plot emerged first plant |
8WAP |
| 3 |
Days to 50% Flower (DT5F) |
number of days from planting to when 50% of the plants in a plot emerged flower |
6-8WAP |
| 4 |
Vine length (VL7WAP) |
measured as the distance between the stem and the last leaf at the top node |
6-7 WAP |
| 5 |
Number of pods per peduncle (NPPP) |
counting the number of pods for tagged plant on a plot |
8-12 WAP |
| 6 |
Pod length (PODLGTH) |
measured from the point of attachment to the tip of the pod |
At Maturity |
| 7 |
Pod width (PODWDTH) |
measured from the edge of one wing to that of the opposite wing at the middle of the pod |
At Maturity |
| 8 |
Number of seeds per pods (NSP) |
Counted and averaged over ten tagged plants in a plot. |
At Harvest |
| 9 |
Seed weight (SW) |
measured using a sensitive digital scale as mean weight of ten dry seeds |
At Harvest |
| 10 |
Seed thickness (STH) |
measured using a vennier caliper as mean thickness of ten dry seeds |
At Harvest |
| 11 |
Seed length (SL) |
measured using a vennier caliper as mean length of ten dry seeds |
At Harvest |
| 12 |
Seed width (SDTHW) |
measured using a vennier caliper as mean width of ten dry seeds |
At Harvest |
| 13 |
Fodder weight (FW) |
measured as the weight of leaf mass or abundance of leaf mass at maturity |
At Harvest |
Table 3.
Names and sequences of the 10 SSR primers used for winged bean PCR amplification.
Table 3.
Names and sequences of the 10 SSR primers used for winged bean PCR amplification.
| SSR Primer name |
dyes |
5′ Forward sequence 3′ |
5′ Reverse sequence 3′ |
| 24 |
6-Fam |
ACC TCA TAG AGG AAT ACG AC |
CAA TAT GTG GAG GAA GTA GA |
| 704 |
Atto-532 |
GAT TGT TGT GAG ATT GAA GT |
ATG CAA ATA GCT TAC AAA AG |
| 747 |
6-Fam |
ACT TTG TGA AAA TGA AGG TA |
AAT TTA ATA TGG CTG CTA AA |
| 854 |
Atto-532 |
CTC TAA AAT TCT CAC ACT CG |
CGA ATT TCT TTC AAT TCT TA |
| 860 |
Atto-532 |
TGA GGA AAA TAA AAA GAA AA |
CGA GTG TGA GAA TTT TAG AG |
| 879 |
Atto-565 |
GCA ACA CTT TAG CTC ATT AT |
GAA CTT CAA CAC TAT TCC AA |
| 1104 |
Atto-565 |
CTT CAA CTG CTT GTT CTA CT |
TAA AGA AGA AAG AGG AAA GG |
| 3111 |
6-Fam |
AGT TGG AAA GTA GCA GAG TT |
GGT GTG AGA AGC ATA ATA AA |
| 5819 |
Atto-550 |
AAT AAT GTC AAT TAC GCA GT |
GAA CTG AAG CCA TGT AGT AG |
| 11100 |
Atto-550 |
AAT AGA AGG CTT GGT GTC |
CTT CCT CTT CTC TTC GTC T |
Table 4.
Traits mean squares, genetic variance, coefficient of variation, and broad-sense heritability for agronomic and yield traits in winged bean.
Table 4.
Traits mean squares, genetic variance, coefficient of variation, and broad-sense heritability for agronomic and yield traits in winged bean.
| Source |
DF |
PODLGT |
PDWDTH |
NoSP |
DTFPM |
SL |
SW |
STH |
FW |
PWT |
SWPPL |
| Accessions |
14 |
5.45* |
2.42** |
3.12** |
83.78* |
0.62* |
0.20** |
0.33* |
49038* |
7227.3*** |
2265.16*** |
| Year |
1 |
2.99 |
1736.4*** |
2.46 |
551.19* |
0.337 |
0.024 |
0.10 |
36 |
788 |
46.06 |
| Accessions*Year |
14 |
3.21 |
1.71 |
1.74 |
75.14* |
0.515 |
0.122 |
0.27 |
10927 |
2033.2 |
705.95* |
| Residual |
|
1.54 |
0.99 |
1.59 |
6.16 |
0.574 |
0.283 |
0.41 |
145.06 |
41.546 |
18.75 |
| CV |
|
7.36 |
10.52 |
14.63 |
7.39 |
6.16 |
3.29 |
5.76 |
48.04 |
21.95 |
20.85 |
| Mean |
|
21.04 |
9.63 |
11.16 |
83.39 |
9.28 |
8.53 |
7.24 |
304.32 |
189.39 |
91.39 |
| δ2g |
|
0.37 |
0.12 |
0.59 |
1.44 |
0.02 |
0.01 |
0.01 |
5003.26 |
875.00 |
260.87 |
| δ2p |
|
0.91 |
0.40 |
1.32 |
13.96 |
0.10 |
0.03 |
0.06 |
8173.12 |
1204.00 |
377.53 |
| GCV (%) |
|
2.90 |
3.57 |
6.86 |
1.44 |
1.51 |
1.40 |
1.42 |
23.24 |
15.62 |
17.67 |
| PCV (%) |
|
4.53 |
6.60 |
10.31 |
4.48 |
3.47 |
2.15 |
3.26 |
29.71 |
18.32 |
21.26 |
| H2 (%) |
|
40.99 |
29.26 |
44.31 |
10.31 |
18.92 |
42.75 |
18.83 |
61.22 |
72.67 |
69.10 |
Table 5.
Principal component analysis and contributions of agronomic and yield traits to the genetic variability of 15 winged bean accession.
Table 5.
Principal component analysis and contributions of agronomic and yield traits to the genetic variability of 15 winged bean accession.
| Variable |
Dim.1 |
Dim.2 |
Dim.3 |
| PODLGT |
-0.529 |
0.301 |
0.559 |
| PDWDTH |
-0.118 |
0.877 |
-0.046 |
| NoSP |
-0.775 |
0.295 |
-0.262 |
| DTFPM |
0.667 |
-0.147 |
0.574 |
| SL |
0.648 |
0.703 |
-0.168 |
| SW |
0.881 |
0.088 |
-0.271 |
| STH |
0.767 |
0.472 |
-0.115 |
| W |
-0.202 |
-0.550 |
-0.550 |
| PWT |
-0.519 |
0.799 |
-0.010 |
| SWPPL |
0.035 |
-0.093 |
0.694 |
| Eigen value |
3.436 |
2.642 |
1.612 |
| Percentage of variance (%) |
34.357 |
26.424 |
16.117 |
| Cumulative of variance (%) |
34.357 |
60.781 |
76.898 |
Table 7.
Locus number, allele frequency, number of alleles per locus, gene diversity and PIC of ten SSR markers used for profiling 15 winged bean accessions.
Table 7.
Locus number, allele frequency, number of alleles per locus, gene diversity and PIC of ten SSR markers used for profiling 15 winged bean accessions.
| Locus no |
Allele frequency |
No of Alleles |
Gene Diversity |
PIC†
|
| SSR-24 |
0.9400 |
3.0000 |
0.1144 |
0.1109 |
| SSR-704 |
0.9333 |
3.0000 |
0.1263 |
0.1218 |
| SSR-747 |
0.9400 |
3.0000 |
0.1144 |
0.1109 |
| SSR-854 |
0.9333 |
3.0000 |
0.1263 |
0.1218 |
| SSR-860 |
0.9200 |
4.0000 |
0.1508 |
0.1462 |
| SSR-879 |
0.6267 |
6.0000 |
0.5041 |
0.4229 |
| SSR-1104 |
0.9267 |
4.0000 |
0.1387 |
0.1342 |
| SSR-3111 |
0.5800 |
6.0000 |
0.5419 |
0.4596 |
| SSR-5819 |
0.9533 |
5.0000 |
0.0903 |
0.0888 |
| SSR-11100 |
0.5400 |
5.0000 |
0.5522 |
0.4606 |
| Mean |
0.8293 |
4.2000 |
0.2459 |
0.2178 |
Table 8.
Analysis of Molecular Variance showing among and within population variance for 150 individual of 15 winged bean accession.
Table 8.
Analysis of Molecular Variance showing among and within population variance for 150 individual of 15 winged bean accession.
| Variation |
df |
SS |
MS |
Est. Var. |
% |
| Among Pops |
2 |
1.733 |
0.867 |
0.013 |
5% |
| Within Pops |
147 |
33.180 |
0.226 |
0.226 |
95% |
| Total |
149 |
34.913 |
|
0.239 |
100% |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).