1. Introduction
Since AlexNet[
1] first used deep learning to win the ImageNet[
2] competition in 2012, deep learning-based approaches have comprehensively outperformed well-designed traditional feature extraction algorithms such as SIFT[
3], HOG[
4], and so on. The use of convolutional neural networks(CNN) and other deep networks for image classification, detection, and segmentation has become a general solution. However, in agricultural applications, the problem of crop disease classification is a rather difficult problem. Crop disease detection is often characterized by both Fine-Grained Visual Classification[
5] and Few-Shot Learning[
6], with the former requiring a large number of images to learn common features, and the latter requiring making good judgments on scarce samples. Unlike distinguishing images such as birds[
7], dogs[
8], aircraft[
9], crop diseases have much smaller inter-class differences, and the number of individual disease samples is sparse, and in many cases it is even necessary to distinguish disease classes by some details.
Datasets are an extremely important factor for the performance of deep learning models, but in many cases we cannot fully guarantee that the samples are sufficient, especially when the image classification task is subdivided into a field such as crop disease classification, the dataset samples are often more scarce. Because of this, many existing methods in the field of crop disease classification[
10,
11,
12,
13,
14,
15] tend to use images with the same dataset partly as training set and partly as validation set, and then use cross-validation to evaluate the performance of the model.
However, datasets from the same distribution tend to overfit our models, and the models may learn features that are not related to the disease, such as the land or the healthy leaves around the disease, etc., thus achieving a fairly high validation accuracy. The complexity of the models dictates that it is possible that they are remembering all the samples and not really learning the features between the diseases. We want to be able to collect a test set from the real world, which differs significantly from the background of the training set we used and is representative of the scenarios the models see in the real world. This way we can directly use the test set in the validation phase to get a realistic view of the trained model’s perception of real-world crop diseases and thus give a more pertinent evaluation benchmark for the model. We call this real-world crop disease test set collected from the Internet Plant Real-World.
In addition to enriching the dataset, using suitable image augmentation methods can be equally effective in improving the model’s utilization of the dataset and improving the model performance from that. Various classical deep learning models and the image augmentation methods they use are shown in
Table 1. From the perspective of data distribution, healthy crops are abundant in the real world, while crop diseases are inherently rare, and this a priori knowledge needs to be leveraged. The long-tail distribution of the image data also tells us a second message, that there is a large amount of background information about the crops in the real world. We believe that the real-world background information in the sample is another piece of a priori information that cannot be ignored ,and should be given sufficient attention and utilized ,instead of eliminating it as an "impurity" that interferes with model performance in all problems. Unlike many previous unsupervised image augmentation techniques, we propose a supervised image augmentation method which we call Negative Contrast to improve the performance of the model. In our experiments, we found that the Negative Contrast can further improve the test accuracy based on Health Augmentation, and the improvement is more obvious in the case of sparse data sets.
In particular, our contributions include:
We elaborated a small and more realistic dataset Plant Real-World for 4 crops, with a training set and a test set.
Health Augmentation: a coding approach to encode health images as negative samples in a classification task is proposed, along with a simple adjustment of the network structure, with significant improvements in the average test accuracy across crops and across models.
Negative Contrast: An image augmentation method is presented to further improve the generalization performance of the model by supervised deduction of disease regions and adding the processed images to the training set, which improves significantly on samples with scarce disease data and improves the test accuracy by up to 30.8%.
2. Related Work
Crop Disease Dataset.There are a variety of publicly available crop disease data, the more common ones are PlantVillage[
23], which contains 54,303 images of plant disease leaves for image classification, and PlantDoc[
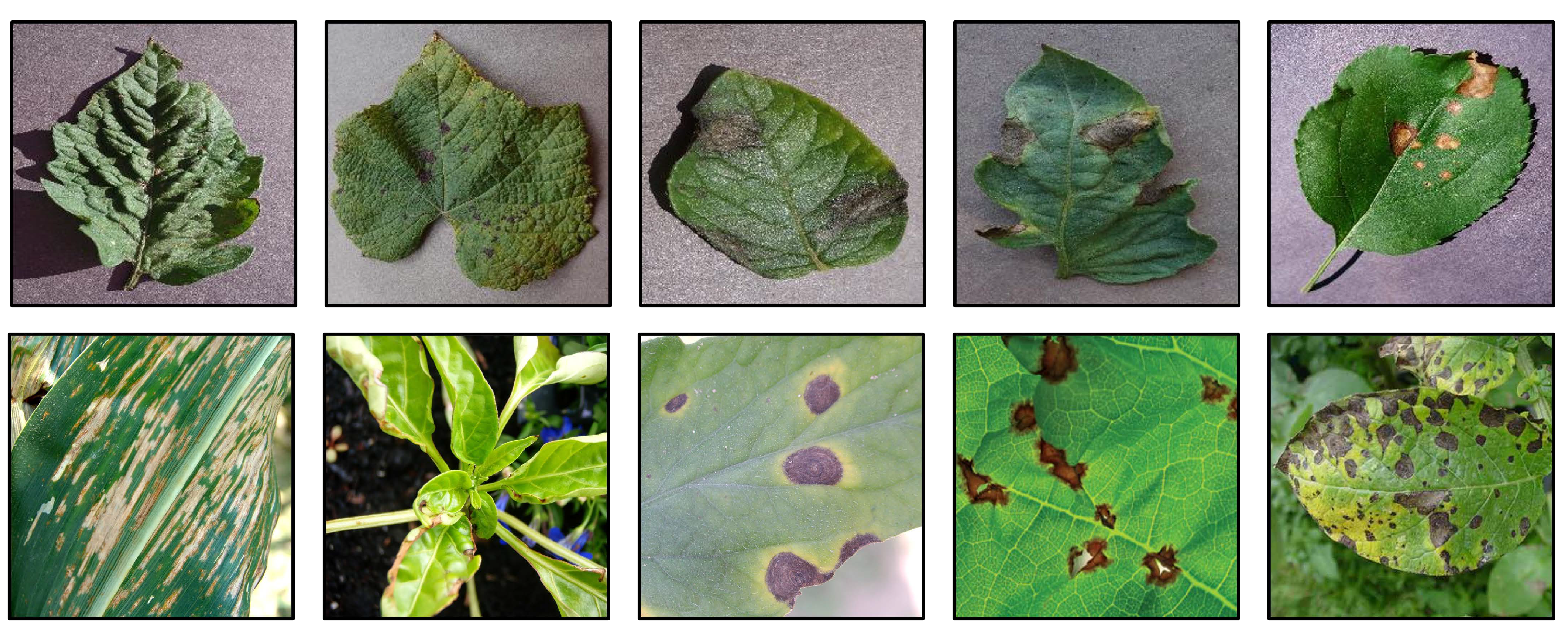
24], which covers 13 plant species and 17 types of diseases with 2598 data points for object detection. Some images of PlantDoc and PlantVillage are shown in
Figure 1. Currently, the most used image classification task for plant disease classification is Plant Village, which contains disease images corresponding to 14 different plants and healthy images of 12 plants, each of which is a 256*256 image containing three channels of R, G, and B. Given the diversity of plants and the sufficient number of images in this dataset, and to reflect the variability, some current works[
11,
13] have used a combination of the public dataset Plant Village and the private dataset for training to improve the performance of the model.
Crop Disease Classification.The rapid development of convolutional neural networks has provided a new direction for the crop disease classification problem. There has been a lot of work on this problem with a variety of studies. [
10] proposed the use of CNN models for disease classification on rice disease images and finally obtained 95.48% accuracy. [
25] used deep CNN models on four disease classification tasks for cucumber and obtained 93.4% accuracy on the test set for the first time. [
26] used DenseNet and Fine-tuning techniques when training the Plant Village test set, further improving the test progress to a rather impressive 99.75%. [
27] combined transfer learning and deep learning, where they first trained on ImageNet using VGG16 for training and then fine-tuned on a new crop disease dataset. [
13] embedded CBAM modules into ResNet and explored the effect of embedding location on classification accuracy, resulting in an accuracy of 97.59% on a common dataset consisting of PlantVillage and some actual images. [
11] proposed a new CNN structure that enabled their model to achieve a cross-validation accuracy of 99.58% with a more complex and comprehensive dataset of 27 diseases of 6 crops.
Image Augmentation.Image augmentation methods for deep learning can be classified into three categories, Model-free, Model-based, and Optimizing policy-based[
28].Model-free augmentation methods are model-independent and usually do not need to know the structure of the model in advance to be used. It can be divided into augmentation methods for single images and multiple images. In single images, simple geometric transformations such as rotation, flip, scale, elastic distortion, jittering, etc. are common. There are also complex transformations such as Hide-and-Seek[
29], which splits an image into patches that are randomly blocked, Cutout[
30], which applies axed-size masks to random positions in each image, Random Erasing[
31], which randomly selects a rectangular region and replaces it with random pixel values, and GridMask[
32] that applies a multi-scale grid mask to an image to simulate masking, etc. Among multiple images, the more popular ones are Mixup[
33], which linearly fuses images and labels, CutMix[
34], which spatially fuses two images and linearly fuses labels, Mosaic[
35], which spatially fuses four images and their corresponding labels to enrich the context information of each class, and Co-Mixup[
36] that maximizes the silent signal of the input images and diversity among the augmented images and so on.
Model-based augmentation methods are a series of augmentation methods with GAN[
37] as the core, including BDA[
38] that uses CGAN to generate images optimized by a Monte Carlo EM algorithm, MFC-GAN[
39] that uses multiple fake classes to obtain obtain fine-grained image for minority class, and GAN-MBD[
40] that translates an image from one class to another while maintaining semantics by multi-branching discriminators, etc.
In addition to the two image augmentation methods mentioned above, some methods based on Optimizing policy exist, such as AutoAugment[
41], which uses reinforcement learning to determine the best optimization strategy, FastAA[
42], which uses efficient density matching to achieve augmentation strategy search, and SPA[
43], which achieves image augmentation by suitable samples, etc.
3. Negative Contrast Augmentation
3.1. A New Crop Disease Dataset in Real-World
In many previous works, it has gradually become a consensus that the number and variability of dataset images play a crucial role in the performance of deep learning models. In the field of crop disease classification, a widely known dataset is PlantVillage. The conventional method of model testing during the training process is generally to use a portion of the training set as the test set during each training iteration, and observe the trend of accuracy change with the increase of the number of iterations to determine whether the model is learning effectively, and finally the highest accuracy rate in each iteration is obtained to indicate the best performance of the model.
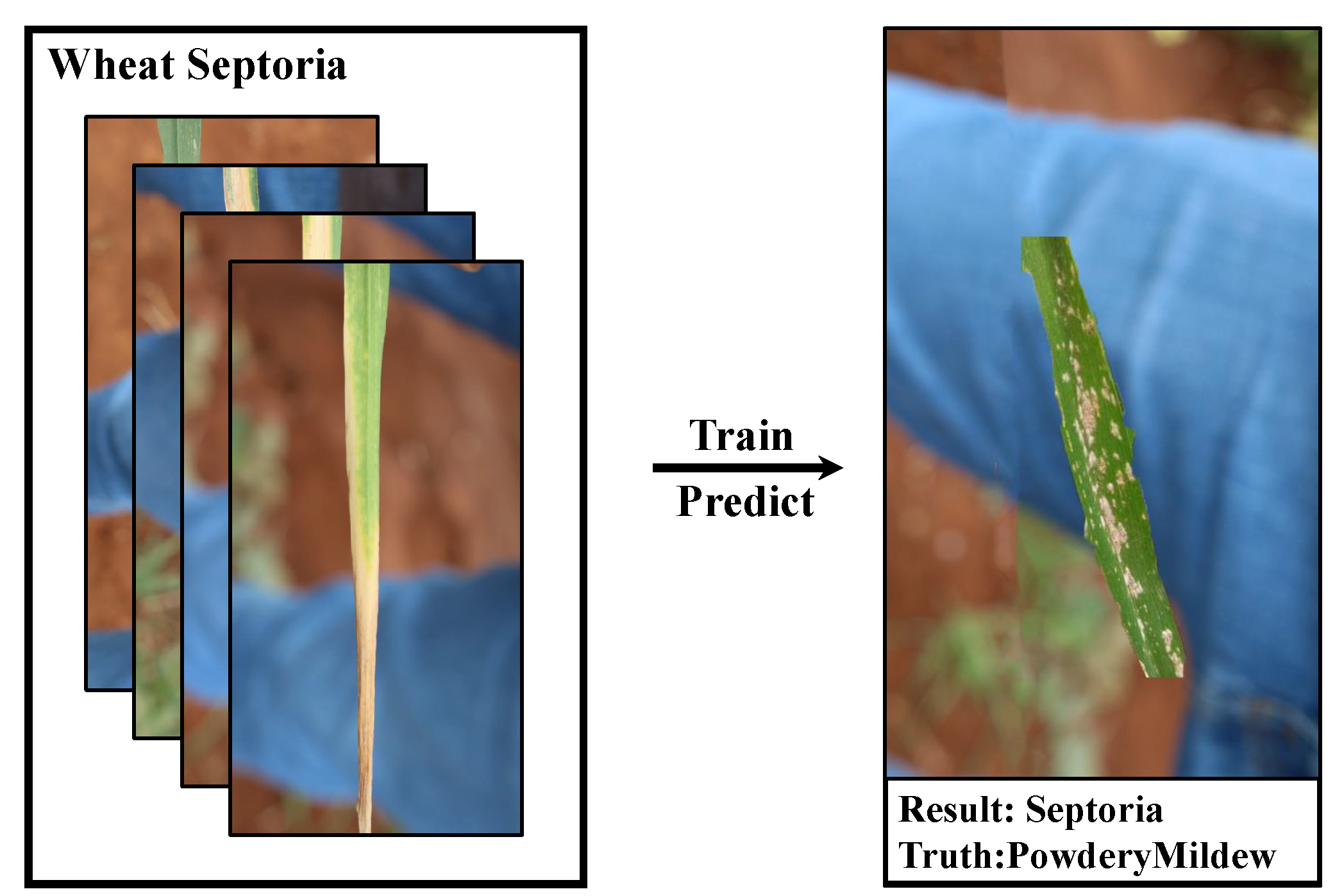
Our study also follows the same training setting. We found experimentally that using samples from the same large dataset that are often homogeneously distributed (i.e., come from the same background or have many similar discriminative features) leads to models that are apparently highly accurate (even approaching 100%) when trained, but dramatically less accurate when tested with real-world crop diseases. We found in our experiments that the trained model does have a high sensitivity to these backgrounds after we recombine the images with simple excisions, which is not good for the generalization performance of the model. As shown in
Figure 2.
Therefore, we believe that using a real-world test set in the iterative process can better reflect the performance of a model. We collected a variety of disease images from the Internet to form a real-world test set Plant Real-World. The statistics of each disease category in the Plant Real-World are shown in
Table 2, and the partial test images of Plant Real-World are shown in
Figure 3.
3.2. Health Augmentation
At present, convolutional neural networks have become the obvious choice for extracting image features and achieving image classification. In the training process of image classification tasks, background information is often treated as an irrelevant factor that interferes with the training. When the similarity between the trained classes of images is high, the background information can easily be used by the model as a basis for discriminating classes as well. One of the most straightforward approaches is to increase the number of samples so that the training set can contain a wide variety of backgrounds to improve generalization performance, but this is expensive and difficult to achieve in many cases. For many practical cases where it is sometimes not possible to increase the sample size, we can use healthy samples of crops, from the PlantVillage dataset or something else.
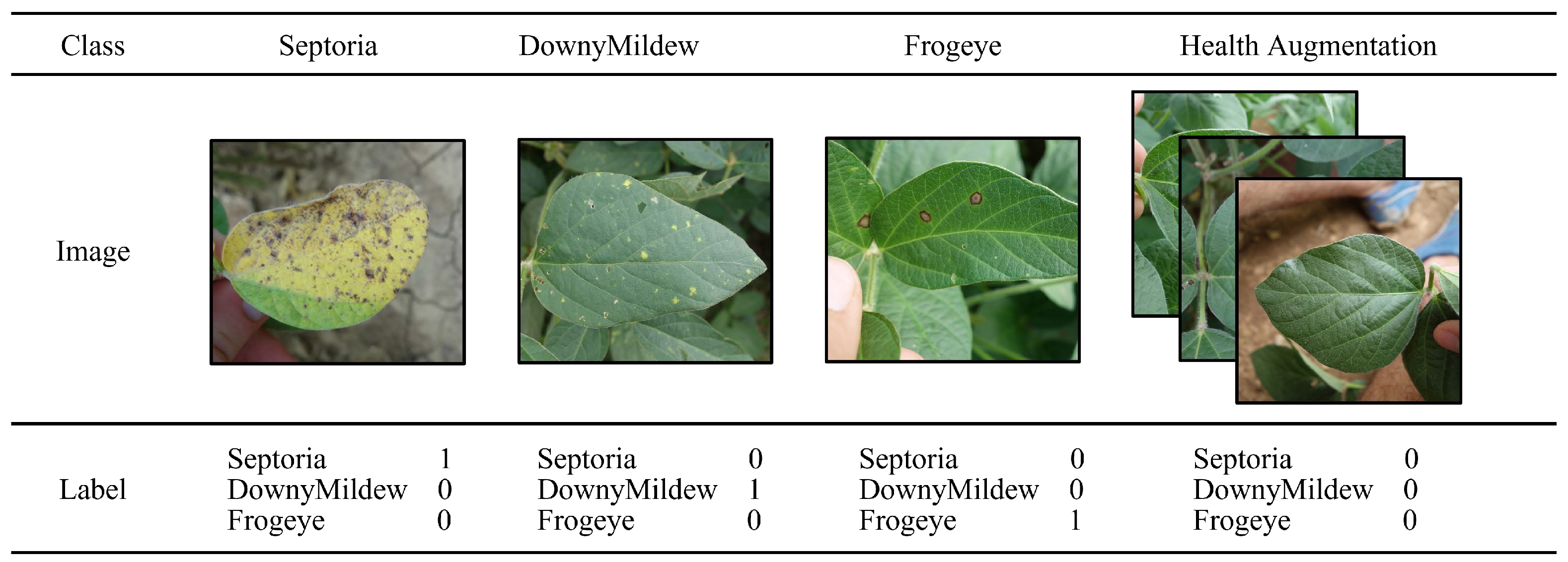
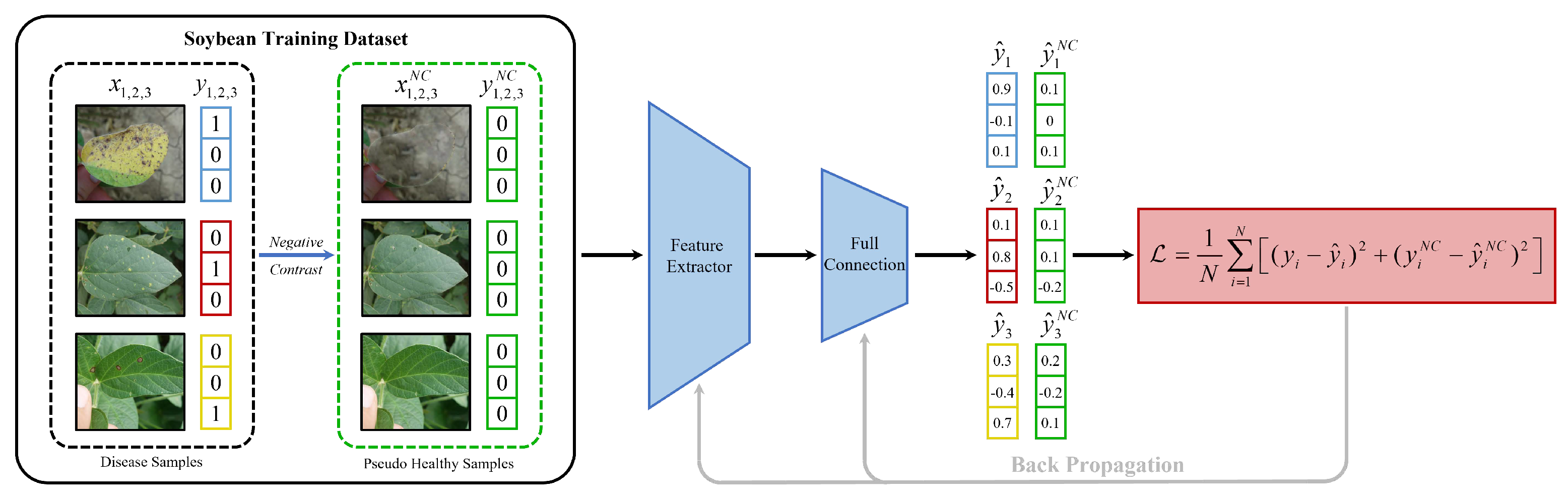
For some existing datasets, healthy samples are treated as a category, while for the task of disease classification, we found in our experiments that a healthy sample alone as a category can impair the performance of disease classification. A simple scenario is that when a healthy leaf has a few small spots on it, the model will prefer to identify the leaf with the spots as healthy as a category rather than as a disease, which is clearly inconsistent with the goal of the disease classification task, i.e., to correctly classify diseases. Therefore, the health category cannot be treated the same as the regular disease.In our approach, we demonstrate a new way of encoding healthy samples, as shown in
Figure 4.
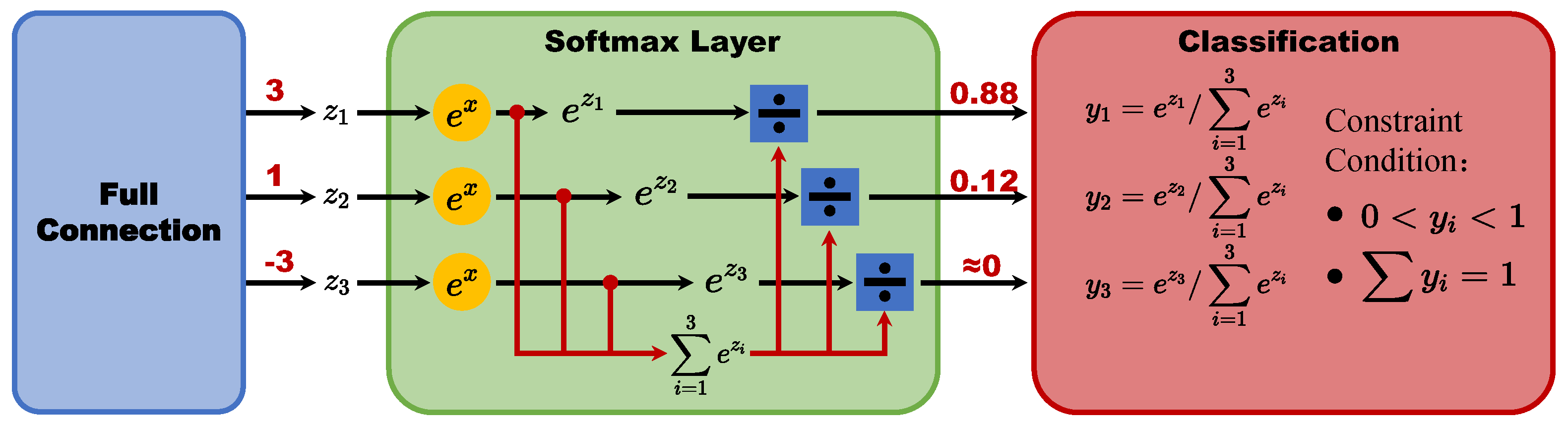
On the other hand, we can revisit the steps of deep learning image classification.For example, the ResNet50 image classification network uses a multilayer convolutional kernel as a feature extractor to extract features from images, then connects a fully connected layer for classification, and finally obtains the probability corresponding to each target class after probability normalization through the Softmax layer.
The structure of Softmax Layer is shown in
Figure 5. However, such a probability has two strict constraints, one is that the probability must be greater than 0 and less than 1, and the other is that the probability sum is 1. Such a definition does not allow the network to have an all-0 output, and requires the network to give a maximum probability result. we consider that such a structure denies the network to learn healthy samples, which limits the learning potential of the network and needs to be adjusted.
Given these two points, we propose our approach which we call Health Augmentation.Our adjustment is a total of two steps.
- (1)
Remove the softmax layer.
- (2)
Change the loss funciton from cross-entropy to mean squared error.
When we finish executing step 1, we need to adjust the loss function at the same time, which is step 2. The most common loss function used for multi-classification tasks is Cross-Entropy, which is expressed by the following formula:
Since the Softmax Layer layer is removed, the output value domain of each neuron in the last layer changes from (0,1) to
. This makes it easy for the loss function Cross-Entropy to fail to transmit the error forward. The reason for this is that the true number of the log function is not allowed to be less than or equal to 0 and thus the cross-entropy loss function cannot calculate the error of the back propagation. Therefore, we adjusted the expression of the loss function using mean squared error
so that the network without the Softmax Layer can also work properly.
We applied Health Augmentation on MoblieNetV2, ResNet50 and ShuffleNetV2 for training and testing based on the Plnat Real-World dataset, and the experimental results are shown in
Table 3.The experimental results show that Health Augmentation can make full use of healthy samples to further improve the accuracy of disease classification. Health Augmentation’s performance has remained consistent across models and crops.
3.3. Negative Contrast
In the crop disease classification task with sparse samples, [
27] has tried to improve the classification results by training on ImageNet and then performing transfer learning on the current task. However, the imbalance of the data and the scarcity of some samples may also be a result of the sample distribution itself. [
44] showed that the distribution of the number of object classes in an image follows Zipf’s law, i.e., the frequencies of certain events are inversely proportional to their rank r. In the field of crop disease classification, the same problem of highly unbalanced distribution of disease samples exists, for example, in the test set of Plant Real-World, the number of samples of Mildew for wheat disease is only 7, yet the number of samples of StripeRust sample size was 207. For some of the extremely scarce samples, the training results in very easy overfitting, which leads to poor performance in real-world applications. How to efficiently use the scarce data is also a question worth thinking about.
In this context, we propose a new image augmentation method: deducting diseased areas and using content-aware filling of PhotoShop to generate "pseudo healthy samples" for training, which we call
Negative Contrast(later referred to as NC). The most similar image augmentation method to ours is Cut, Paste and Learn [
45]. However, the biggest difference between our method and Cut, Paste and Learn is that our selected diseased areas are not directly fused with different backgrounds for training, but are generated as "pseudo healthy samples" for training. Based on Health Augmentation’s altered network structure, NC uses pseudo healthy samples in an expansion of the training set. The entire implementation pipeline of NC is shown in the
Figure 6.
We found that the "pseudo healthy samples" generated by this method have the same role as natural crop healthy samples, both of which can be used as healthy samples, participating in the training using the exact same settings as Health Augmentation. This can be used as a data augmentation method for scarce samples when health samples are also difficult to obtain in more adequate quantities, requiring only a small amount of manual processing to obtain a new training set with twice the amount of original data.We cut the dataset with different proportions of the training set, and the smallest training set size is only 5% of the original one, with only 5-7 images per category. With such a small sample size, compared to the model using Health Augmentation , NC can obtain further improvements in test accuracy. To ensure comparability of the experiments, we used the same number of health samples in NC and Health Augmentation.
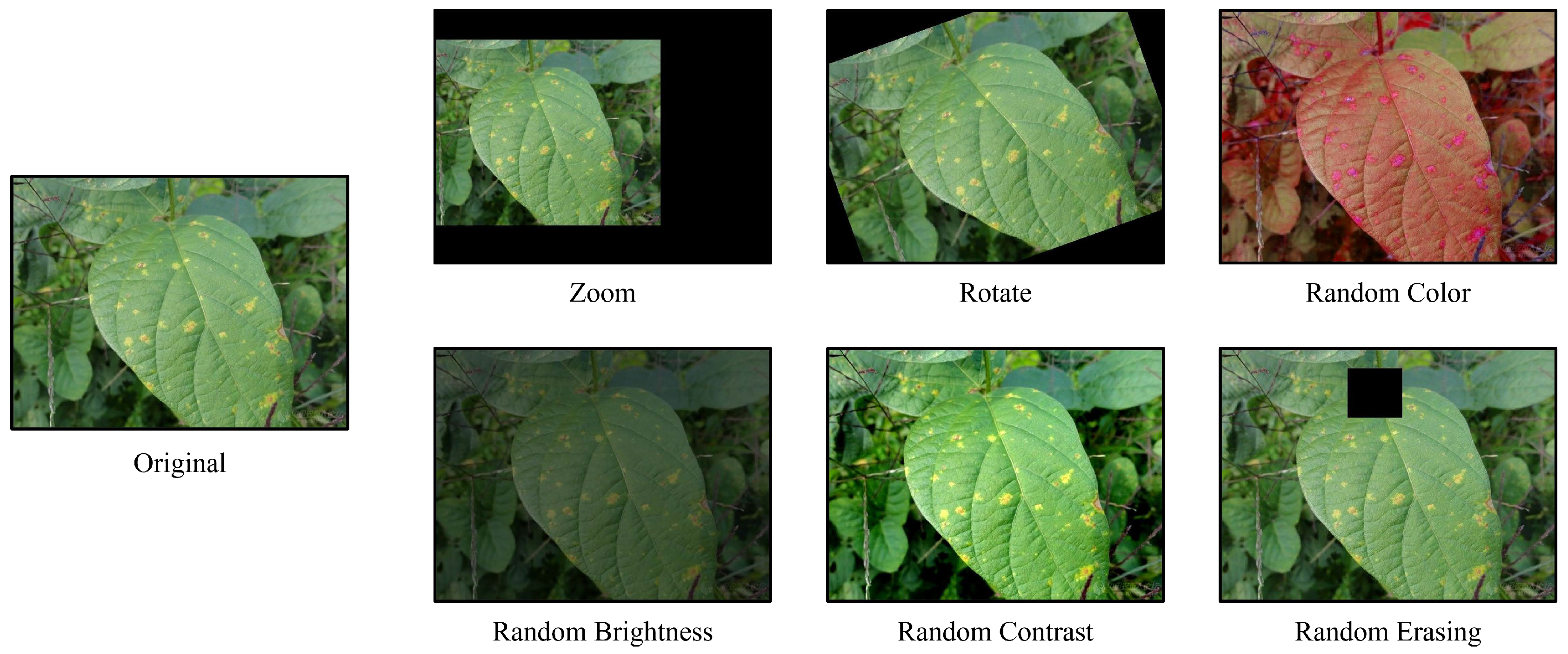
The experimental results of the sparse samples on Plant Real-World are shown in
Table 4.As an image augmentation method, we compare the performance of NC with the six commonly used image augmentation methods. After we performed image augmentation on the original images using each of these six methods separately and tested the models which is trained on these transformed-dataset on Plant Real-World, the illustration of different augmentation methods is shown in
Figure 7. Finally, the average accuracy of the three models (ResNet50, ShuffleNetV2, MobileNtV2) are listed in
Table 5 as the experimental results.
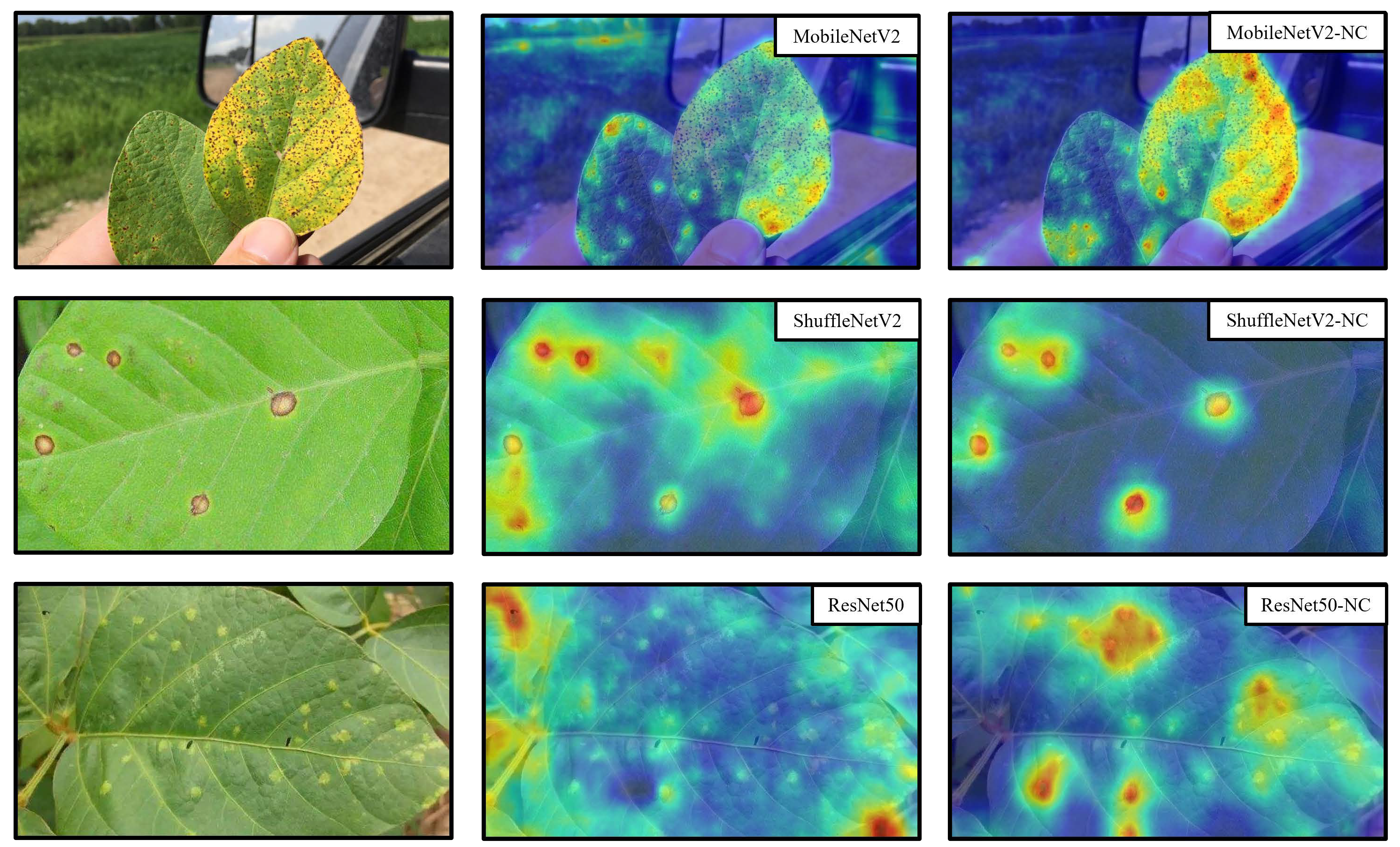
To further investigate whether Health Augmentation and Negative Contrast are accurate for the region of interest of the disease, we used the Grad-CAM[
46] tool to perform an attentional heat map analysis on these three types of models that have been trained on Plant Real-World. The implementation is as follows:
In order to obtain the class-discriminative attentional heat map Grad-CAM
in general architectures, the first thing is to compute the gradient of
with respect to feature maps
A of a convolutional layer,
. The critical weights that captures the ‘importance’ of feature map
k for a target class
c can be obtained with these gradients:
A linear combination of feature map
and weight
using the RELU activation function will give us our heat map:
As the heat map in
Figure 8 shows: compared to the models trained normally and using the Health Augmentation method, when the model is trained using NC, the model pays stronger attention to the area of the disease and weaker attention to the unrelated surroundings and the healthy leaves unrelated to the disease, which explains intuitively why the improved model tested by NC has higher accuracy.
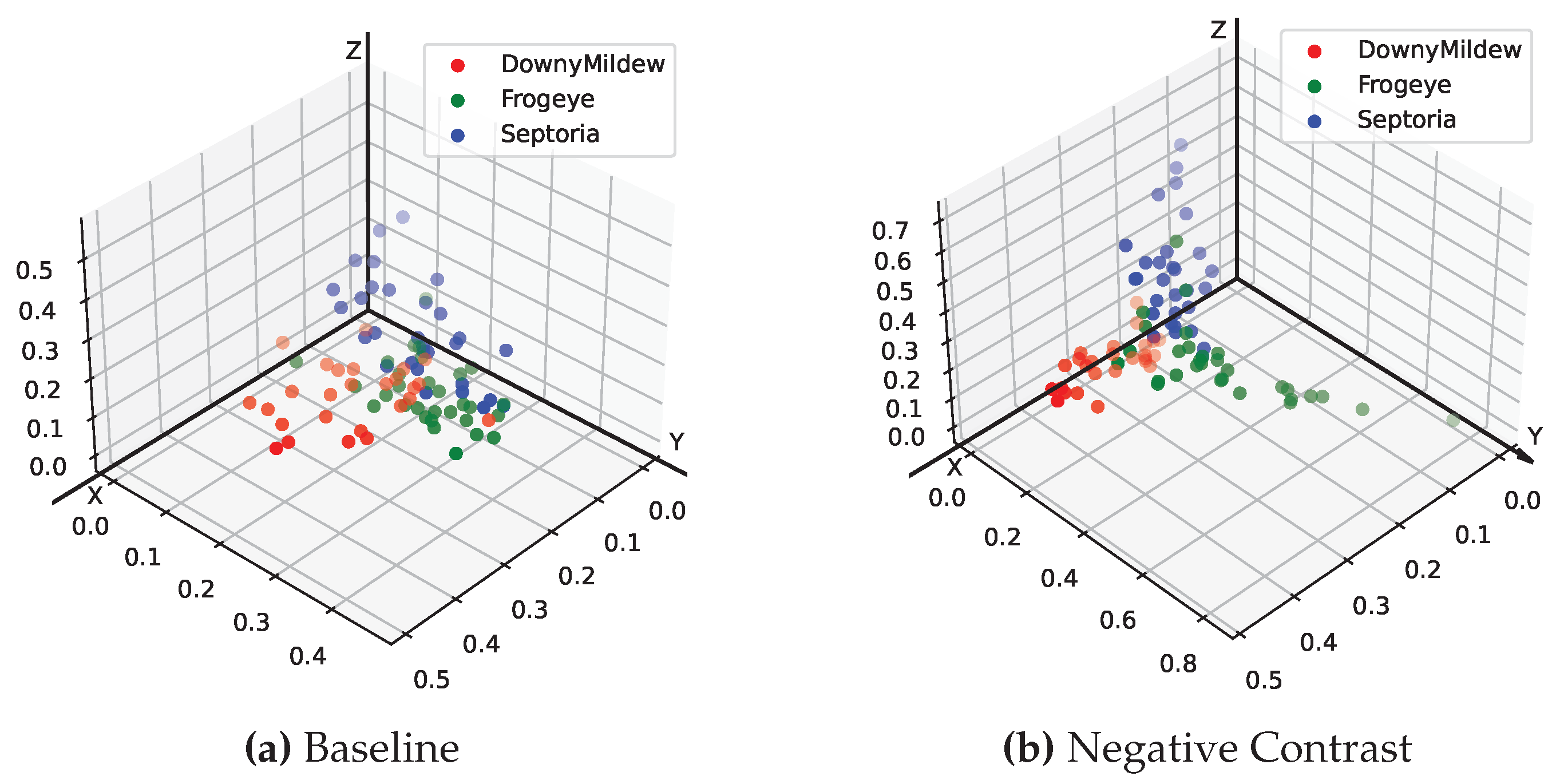
In addition to the heat map analysis, we wanted to explore the deeper reasons why the NC method worked. Inspired by the t-SNE[
47] method of dimensionality reduction visualization, we performed the visualization of the last layer of the fully connected layer feature space on the models trained by both normal and NC methods, respectively. For presentation purposes, we used a soybean dataset with only three disease categories for direct 3D spatial visualization of feature points. As can be seen in
Figure 9, the model trained using the NC method has a farther distance between different categories of diseases and a closer distance between the same categories on the feature space, reflecting a more superior classification performance.
4. Discussion
The main reason why people can accurately determine the disease of crops is based on the color, shape, etc. of the disease spot area, which is also a starting point of Negative Contrast. So we consider that it is more similar to Fine-Grained Visual Classification, and try to solve it. We reviewed many recent results on Fine-Grained Visual Classification[
48,
49,
50,
51,
52,
53] and found that these models are too huge. On some of the older mobile devices we actually tested, they had one-tenth the computational power of today’s state-of-the-art computer devices. Manipulating these latest models on a computer is extremely slow, not to mention the wide variety of handheld devices that are actually used, and the overly large models create difficulties for deployment in the field of crop disease classification. Therefore a more suitable technique for identifying crop diseases in lightweight models is urgently needed. This is why we have specifically tested two lightweight models MobileNetV2, ShuffleNetV2 in our experiments.
We also found through subsequent experiments that if the disease characteristics of the training set are more typical and each image is extremely representative, then it may not be necessary to have an excessively large number of samples to achieve a good model accuracy. We trained the training set with different proportions of cuts for each of the three typical diseases of soybean, and found that with the minimum required proportion of images (5% of the original training set), i.e., only a few images for each disease, we could still achieve a fairly good classification accuracy on Plant Real-World with Negetive Contrast. We believe this is a worthy phenomenon that demonstrates that with a representative training set, we can significantly reduce the amount of samples required for the training set, which is extremely important in crop disease tasks.
At the same time, we found that NC also has limitations in that it is somewhat dependent on the dataset for model enhancement performance, and more dependent on the dataset than on the model. In
Table 4, we can see that the various models have the smallest improvement on the corn dataset and the largest enhancement under the soybean dataset; however, the enhancement performance of each model is basically the same under the same dataset. At the same time, when there are many data samples, this method of supervised augmentation by hand will consume more human resources. Similarly, when the disease areas are irregular and intermingled with the background, we have to remove the entire disease area together with the background, so the accuracy improvement of this technique is not obvious for some crop diseases.
However, this does not prevent us from considering Negative Contrast as a general image augmentation technique. It achieves a fairly significant performance improvement for a wide range of models with a variety of data sets. It is simple, effective, and requires little change to the model structure, and can even be extended to other fields such as medical imaging and anomaly detection.
In addition, we believe that there are many more questions that deserve to be investigated. For example, how much can the number of samples needed be reduced by using Negative Contrast while maintaining the same accuracy; or give a theoretical proof of why such an approach works; or whether using such a technique in the field of target detection or segmentation will also improve the performance of the model, etc.
5. Conclusion
We provide a small real-world dataset of four crops, Plant Real-World, and secondly, we use Health Augmentation to improve model performance by leveraging health samples that are easily ignored by the public and suppressing the network’s learning of the background, thus significantly improving the network’s generalization performance on crop disease tasks. Finally we propose a generalized image augmentation method: Negative Contrast ,which can further improve the model’s testing accuracy in the real-world when samples are scarce.
References
- A. Krizhevsky, I. Sutskever, and G. E. Hinton, “Imagenet classification with deep convolutional neural networks,” Communications of the ACM, vol. 60, pp. 84 – 90, 2012.
- J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li, and L. Fei-Fei, “Imagenet: A large-scale hierarchical image database,” 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255, 2009.
- D. G. Lowe, “Object recognition from local scale-invariant features,” Proceedings of the Seventh IEEE International Conference on Computer Vision, vol. 2, pp. 1150–1157 vol.2, 1999.
- N. Dalal and B. Triggs, “Histograms of oriented gradients for human detection,” 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), vol. 1, pp. 886–893 vol. 1, 2005.
- T.-Y. Lin, A. RoyChowdhury, and S. Maji, “Bilinear cnn models for fine-grained visual recognition,” 2015 IEEE International Conference on Computer Vision (ICCV), pp. 1449–1457, 2015.
- J. Snell, K. Swersky, and R. S. Zemel, “Prototypical networks for few-shot learning,” ArXiv, vol. abs/1703.05175, 2017.
- C. Wah, S. Branson, P. Welinder, P. Perona, and S. J. Belongie, “The caltech-ucsd birds-200-2011 dataset,” 2011.
- A. Khosla, N. Jayadevaprakash, B. Yao, and L. Fei-Fei, “Novel dataset for fine-grained image categorization : Stanford dogs,” 2012.
- S. Maji, E. Rahtu, J. Kannala, M. B. Blaschko, and A. Vedaldi, “Fine-grained visual classification of aircraft,” ArXiv, vol. abs/1306.5151, 2013.
- Y. Lu, S. Yi, N. Zeng, Y. Liu, and Y. Zhang, “Identification of rice diseases using deep convolutional neural networks,” Neurocomputing, vol. 267, pp. 378–384, 2017. [CrossRef]
- V. Tiwari, R. C. Joshi, and M. K. Dutta, “Dense convolutional neural networks based multiclass plant disease detection and classification using leaf images,” Ecol. Informatics, vol. 63, p. 101289, 2021. [CrossRef]
- J. Chen, D. fu Zhang, Y. A. Nanehkaran, and D. Li, “Detection of rice plant diseases based on deep transfer learning.” Journal of the science of food and agriculture, 2020. [CrossRef]
- Y. Zhao, J. Chen, X. Xu, J. Lei, and W. Zhou, “Sev-net: Residual network embedded with attention mechanism for plant disease severity detection,” Concurrency and Computation: Practice and Experience, vol. 33, 2021. [CrossRef]
- A. Karlekar and A. Seal, “Soynet: Soybean leaf diseases classification,” Comput. Electron. Agric., vol. 172, p. 105342, 2020. [CrossRef]
- Ü. Atila, M. Uçar, K. Akyol, and E. Uçar, “Plant leaf disease classification using efficientnet deep learning model,” Ecol. Informatics, vol. 61, p. 101182, 2021. [CrossRef]
- K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778, 2016.
- G. Huang, Z. Liu, and K. Q. Weinberger, “Densely connected convolutional networks,” 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2261–2269, 2016.
- A. G. Howard, M. Zhu, B. Chen, D. Kalenichenko, W. Wang, T. Weyand, M. Andreetto, and H. Adam, “Mobilenets: Efficient convolutional neural networks for mobile vision applications,” ArXiv, vol. abs/1704.04861, 2017.
- B. Zoph, V. Vasudevan, J. Shlens, and Q. V. Le, “Learning transferable architectures for scalable image recognition,” 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 8697–8710, 2017. [CrossRef]
- H. Zhang, C. Wu, Z. Zhang, Y. Zhu, Z.-L. Zhang, H. Lin, Y. Sun, T. He, J. Mueller, R. Manmatha, M. Li, and A. Smola, “Resnest: Split-attention networks,” 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), pp. 2735–2745, 2020.
- H. Touvron, M. Cord, M. Douze, F. Massa, A. Sablayrolles, and H. J’egou, “Training data-efficient image transformers & distillation through attention,” in International Conference on Machine Learning, 2020.
- Z. Liu, Y. Lin, Y. Cao, H. Hu, Y. Wei, Z. Zhang, S. Lin, and B. Guo, “Swin transformer: Hierarchical vision transformer using shifted windows,” 2021 IEEE/CVF International Conference on Computer Vision (ICCV), pp. 9992–10 002, 2021.
- D. P. Hughes and M. Salathé, “An open access repository of images on plant health to enable the development of mobile disease diagnostics through machine learning and crowdsourcing,” ArXiv, vol. abs/1511.08060, 2015.
- D. P. Singh, N. Jain, P. Jain, P. Kayal, S. Kumawat, and N. Batra, “Plantdoc: A dataset for visual plant disease detection,” Proceedings of the 7th ACM IKDD CoDS and 25th COMAD, 2019.
- J. Ma, K. Du, F. Zheng, L. Zhang, Z. Gong, and Z. Sun, “A recognition method for cucumber diseases using leaf symptom images based on deep convolutional neural network,” Comput. Electron. Agric., vol. 154, pp. 18–24, 2018. [CrossRef]
- E. C. Too, L. Yujian, S. Njuki, and L. Yingchun, “A comparative study of fine-tuning deep learning models for plant disease identification,” Comput. Electron. Agric., vol. 161, pp. 272–279, 2019. [CrossRef]
- S. H. Lee, H. Goëau, P. Bonnet, and A. Joly, “New perspectives on plant disease characterization based on deep learning,” Comput. Electron. Agric., vol. 170, p. 105220, 2020. [CrossRef]
- M. Xu, S. Yoon, A. Fuentes, and D. S. Park, “A comprehensive survey of image augmentation techniques for deep learning,” ArXiv, vol. abs/2205.01491, 2022. [CrossRef]
- K. K. Singh and Y. J. Lee, “Hide-and-seek: Forcing a network to be meticulous for weakly-supervised object and action localization,” 2017 IEEE International Conference on Computer Vision (ICCV), pp. 3544–3553, 2017.
- T. Devries and G. W. Taylor, “Improved regularization of convolutional neural networks with cutout,” ArXiv, vol. abs/1708.04552, 2017.
- Z. Zhong, L. Zheng, G. Kang, S. Li, and Y. Yang, “Random erasing data augmentation,” in AAAI Conference on Artificial Intelligence, 2017.
- P. Chen, S. Liu, H. Zhao, and J. Jia, “Gridmask data augmentation,” ArXiv, vol. abs/2001.04086, 2020.
- H. Zhang, M. Cissé, Y. Dauphin, and D. Lopez-Paz, “mixup: Beyond empirical risk minimization,” ArXiv, vol. abs/1710.09412, 2017.
- S. Yun, D. Han, S. J. Oh, S. Chun, J. Choe, and Y. J. Yoo, “Cutmix: Regularization strategy to train strong classifiers with localizable features,” 2019 IEEE/CVF International Conference on Computer Vision (ICCV), pp. 6022–6031, 2019.
- A. Bochkovskiy, C.-Y. Wang, and H.-Y. M. Liao, “Yolov4: Optimal speed and accuracy of object detection,” ArXiv, vol. abs/2004.10934, 2020.
- J.-H. Kim, W. Choo, H. Jeong, and H. O. Song, “Co-mixup: Saliency guided joint mixup with supermodular diversity,” ArXiv, vol. abs/2102.03065, 2021.
- I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. C. Courville, and Y. Bengio, “Generative adversarial nets,” in NIPS, 2014.
- T. Tran, T. T. Pham, G. Carneiro, L. J. Palmer, and I. D. Reid, “A bayesian data augmentation approach for learning deep models,” ArXiv, vol. abs/1710.10564, 2017.
- A. Ali-Gombe and E. Elyan, “Mfc-gan: Class-imbalanced dataset classification using multiple fake class generative adversarial network,” Neurocomputing, vol. 361, pp. 212–221, 2019. [CrossRef]
- Z. Zheng, Z. Yu, Y. Wu, H. Zheng, B. Zheng, and M. Lee, “Generative adversarial network with multi-branch discriminator for imbalanced cross-species image-to-image translation,” Neural networks : the official journal of the International Neural Network Society, vol. 141, pp. 355–371, 2021. [CrossRef]
- E. D. Cubuk, B. Zoph, D. Mané, V. Vasudevan, and Q. V. Le, “Autoaugment: Learning augmentation strategies from data,” 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 113–123, 2019.
- R. Hataya, J. Zdenek, K. Yoshizoe, and H. Nakayama, “Faster autoaugment: Learning augmentation strategies using backpropagation,” in European Conference on Computer Vision, 2019.
- T. Takase, R. Karakida, and H. Asoh, “Self-paced data augmentation for training neural networks,” Neurocomputing, vol. 442, pp. 296–306, 2020. [CrossRef]
- X. Zhu, D. Anguelov, and D. Ramanan, “Capturing long-tail distributions of object subcategories,” 2014 IEEE Conference on Computer Vision and Pattern Recognition, pp. 915–922, 2014.
- D. Dwibedi, I. Misra, and M. Hebert, “Cut, paste and learn: Surprisingly easy synthesis for instance detection,” 2017 IEEE International Conference on Computer Vision (ICCV), pp. 1310–1319, 2017.
- R. R. Selvaraju, A. Das, R. Vedantam, M. Cogswell, D. Parikh, and D. Batra, “Grad-cam: Visual explanations from deep networks via gradient-based localization,” International Journal of Computer Vision, vol. 128, pp. 336–359, 2017.
- L. van der Maaten and G. E. Hinton, “Visualizing data using t-sne,” Journal of Machine Learning Research, vol. 9, pp. 2579–2605, 2008.
- R. Du, D. Chang, A. K. Bhunia, J. Xie, Y.-Z. Song, Z. Ma, and J. Guo, “Fine-grained visual classification via progressive multi-granularity training of jigsaw patches,” in ECCV, 2020.
- P. Zhuang, Y. Wang, and Y. Qiao, “Learning attentive pairwise interaction for fine-grained classification,” in AAAI, 2020. [CrossRef]
- S. Min, H. Yao, H. Xie, Z. Zha, and Y. Zhang, “Multi-objective matrix normalization for fine-grained visual recognition,” IEEE Transactions on Image Processing, vol. 29, pp. 4996–5009, 2020. [CrossRef]
- Z. Wang, S. Wang, S. Yang, H. Li, J. Li, and Z. Li, “Weakly supervised fine-grained image classification via guassian mixture model oriented discriminative learning,” 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 9746–9755, 2020.
- C. Liu, H. Xie, Z. Zha, L. Ma, L. Yu, and Y. Zhang, “Filtration and distillation: Enhancing region attention for fine-grained visual categorization,” in AAAI, 2020.
- R. Ji, L. Wen, L. Zhang, D. Du, Y. Wu, C. Zhao, X. Liu, and F. Huang, “Attention convolutional binary neural tree for fine-grained visual categorization,” 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 10 465–10 474, 2020.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).