Submitted:
08 June 2023
Posted:
09 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Model
2.1. Coarse-Grained Molecular Dynamics Simulation Details
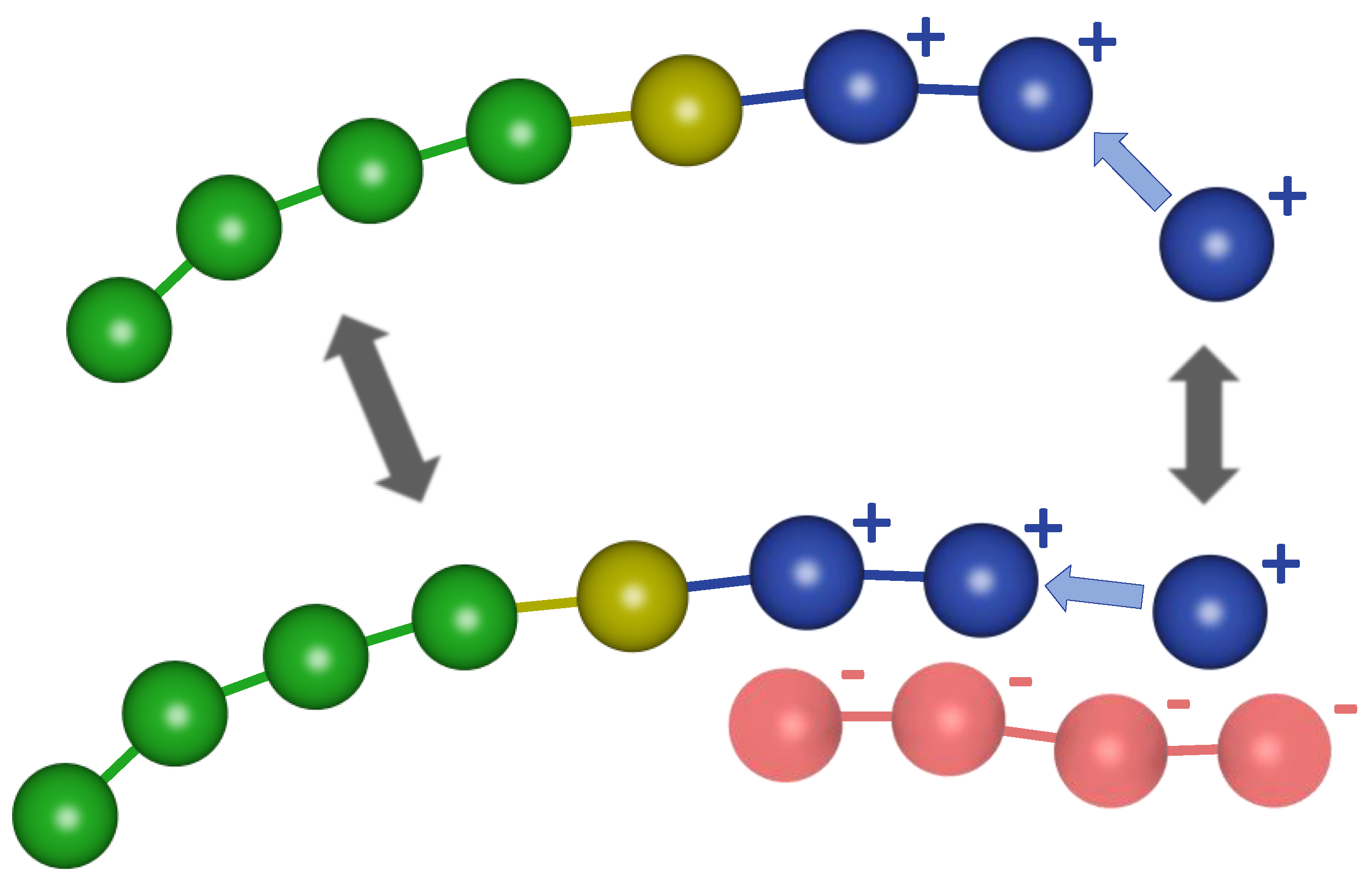
2.2. Modeling the Polymerization
2.3. Clustering Analysis
3. Results and Discussion
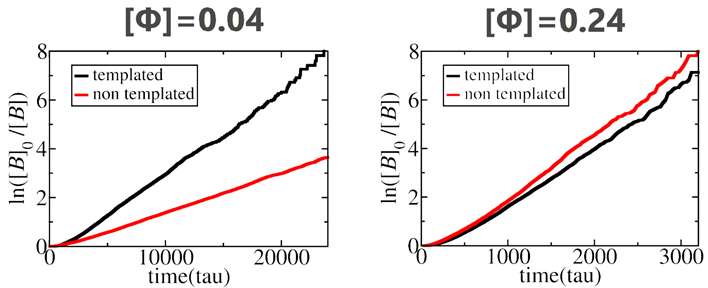
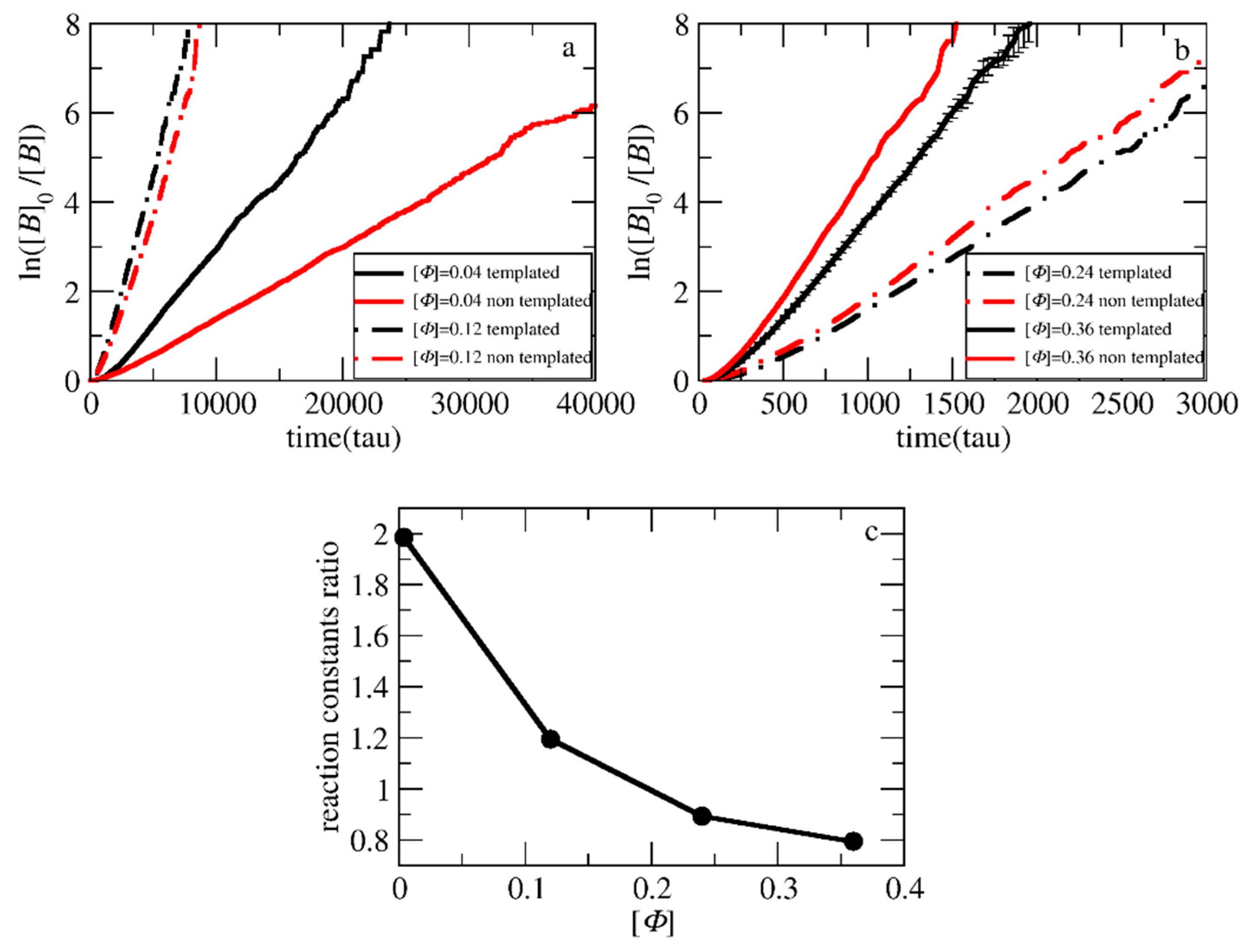
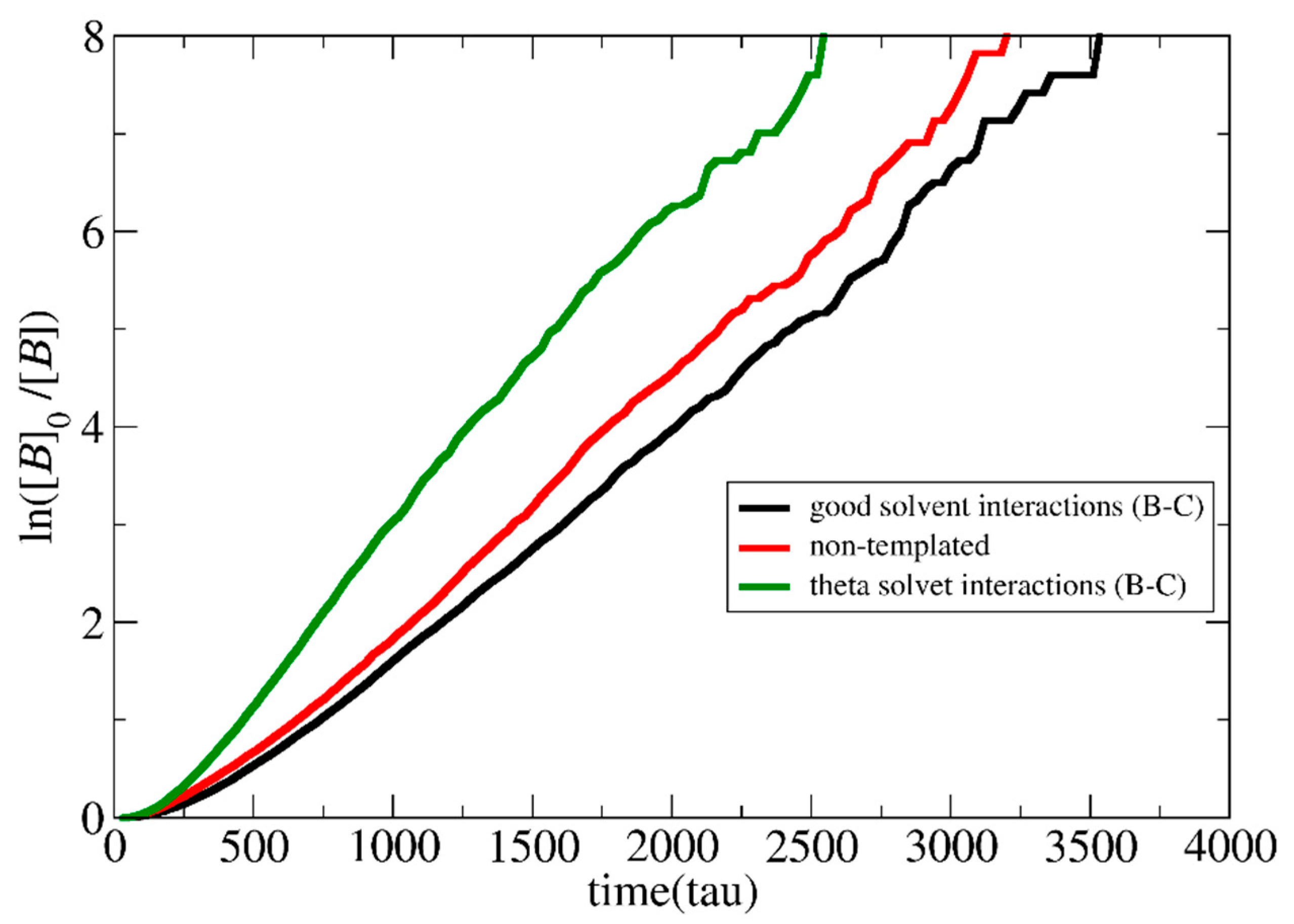
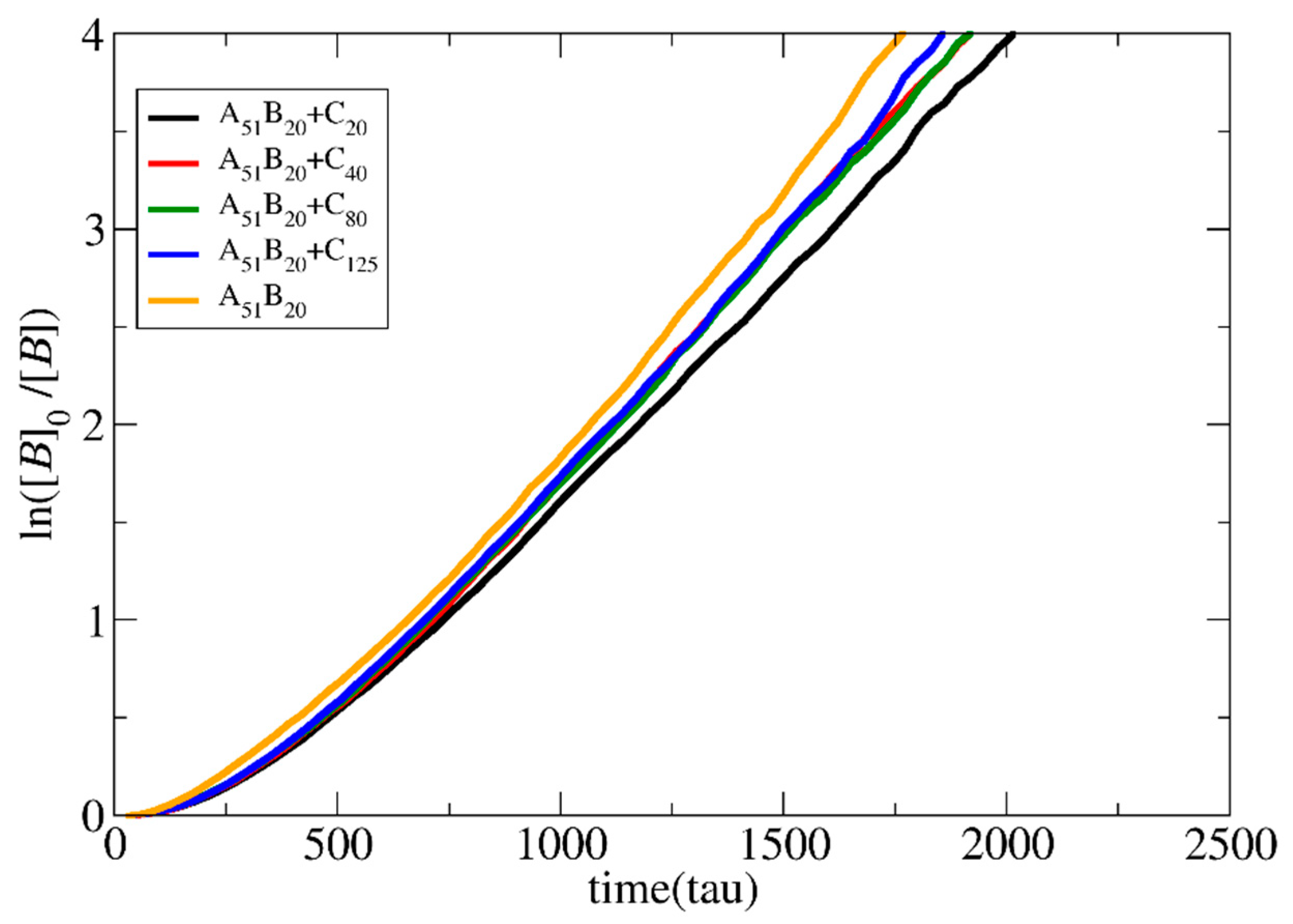
3.1. Polymerization Rate
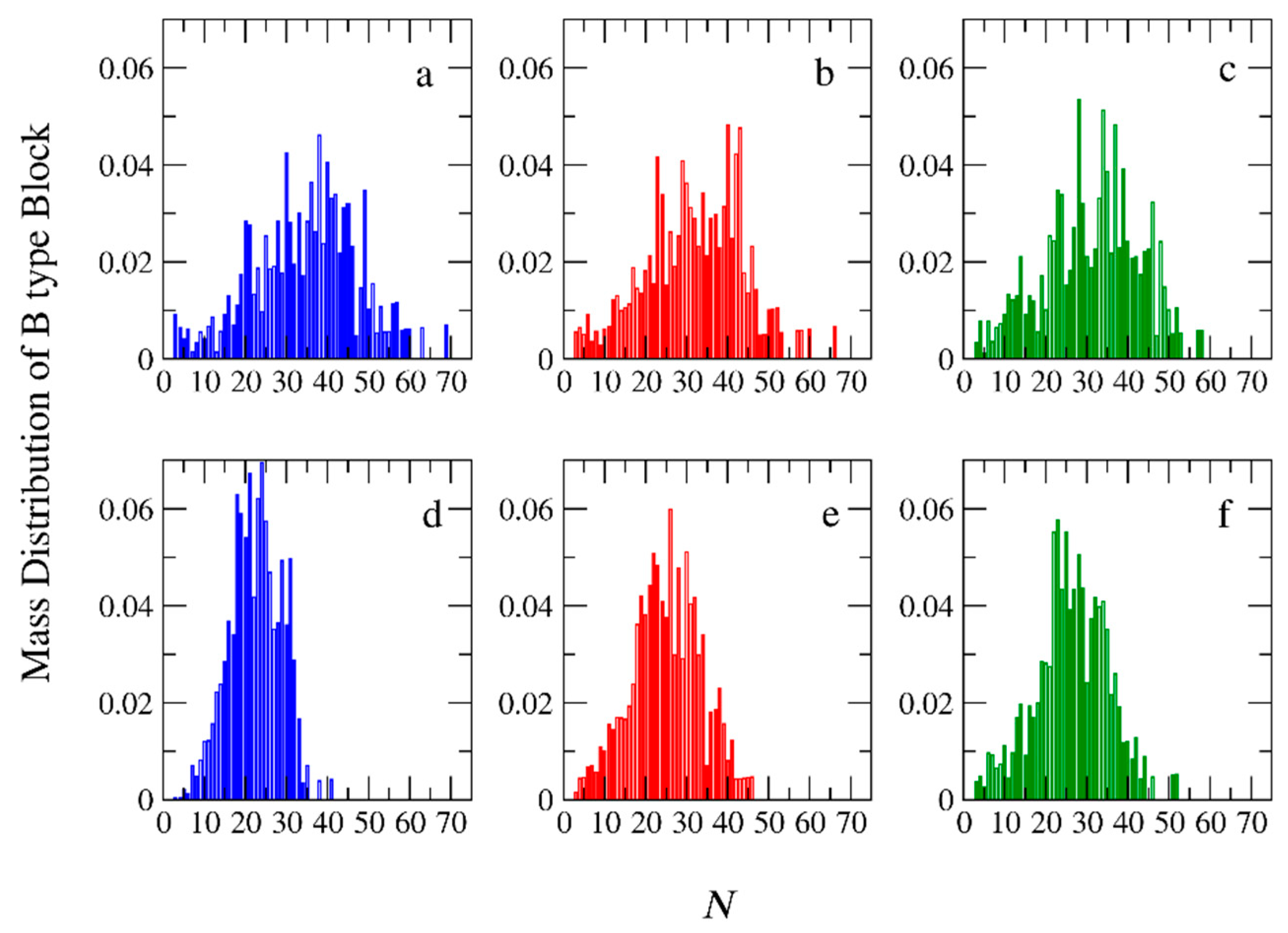
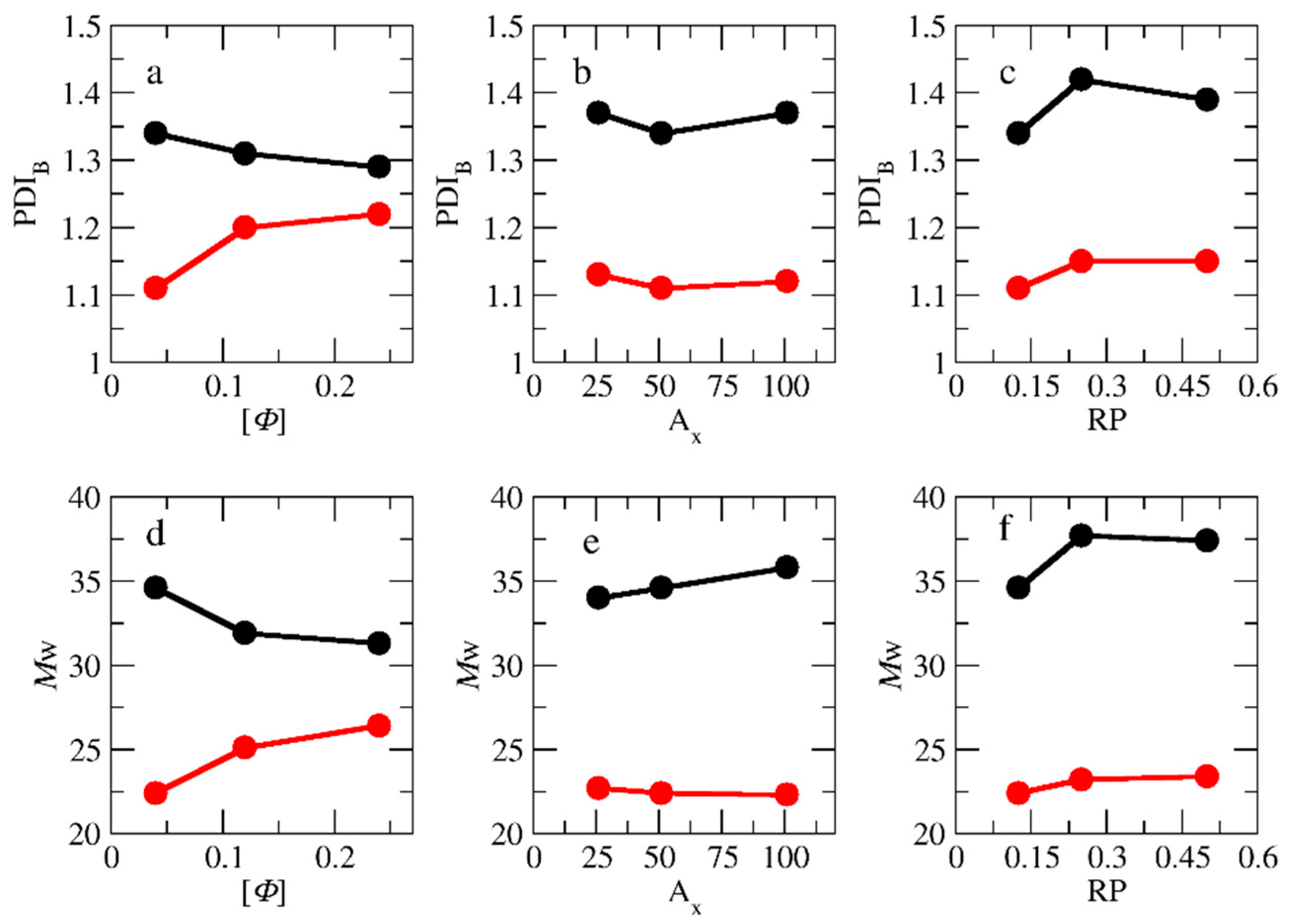
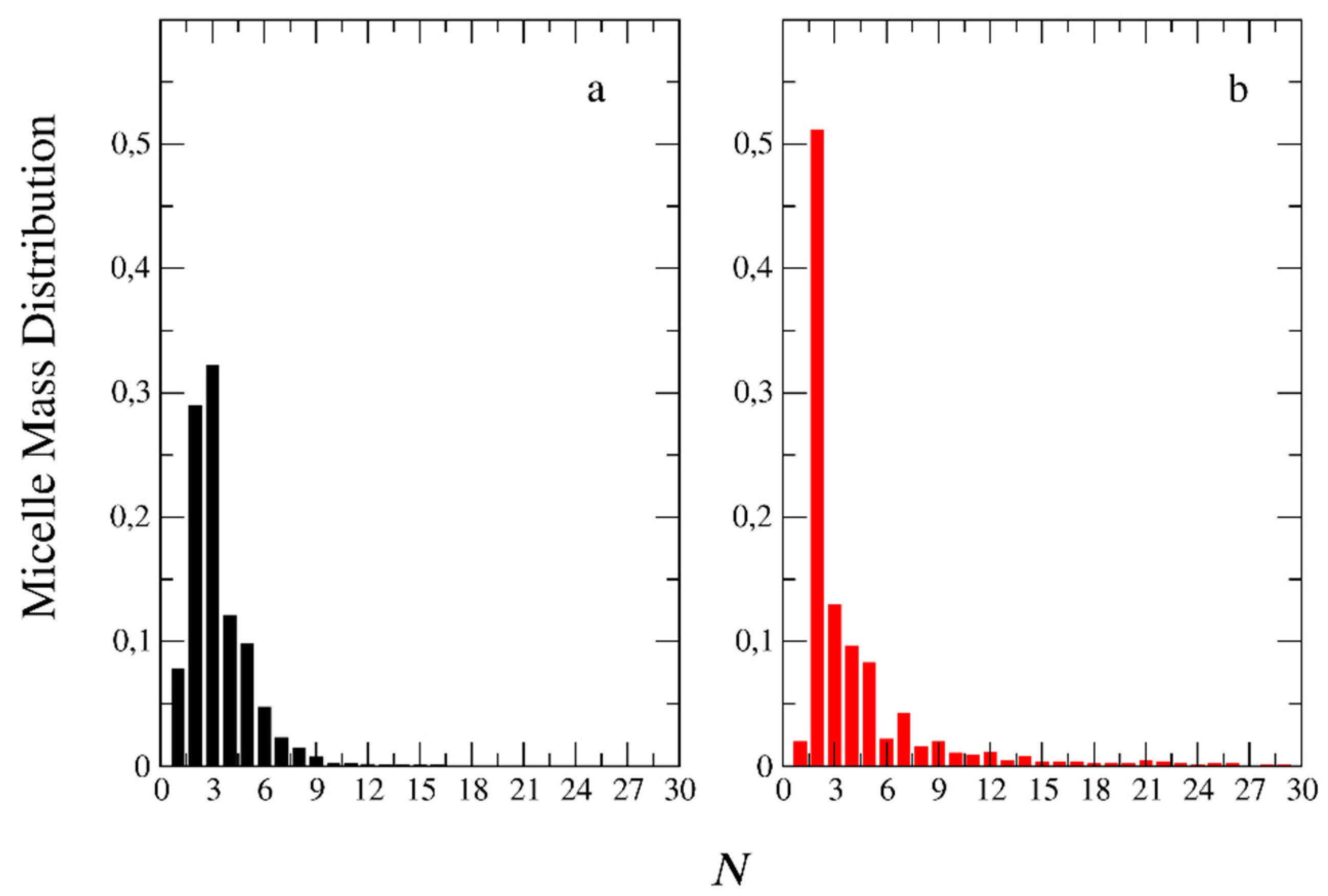
3.2. Molecular Weights and Polydispersity
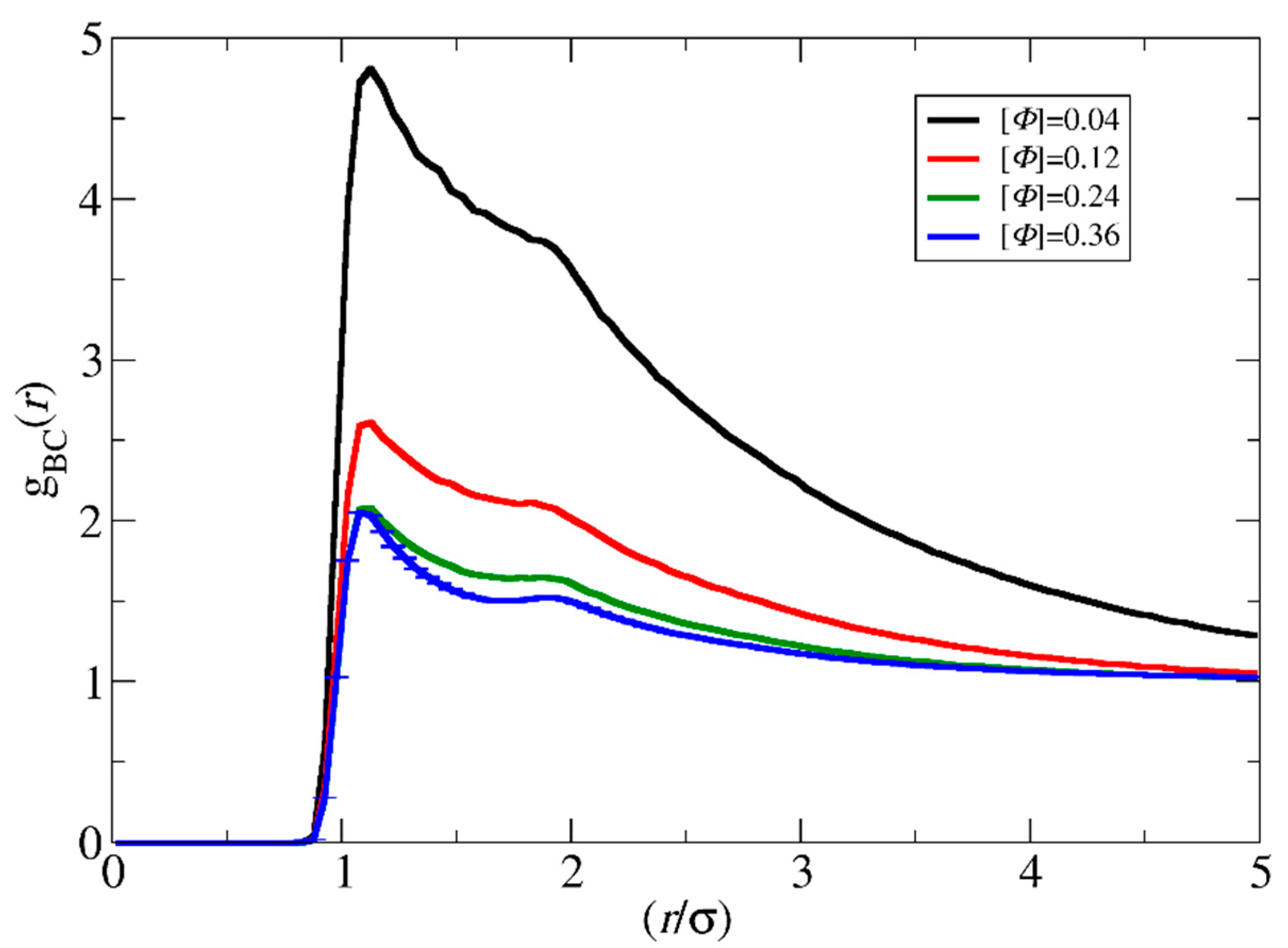
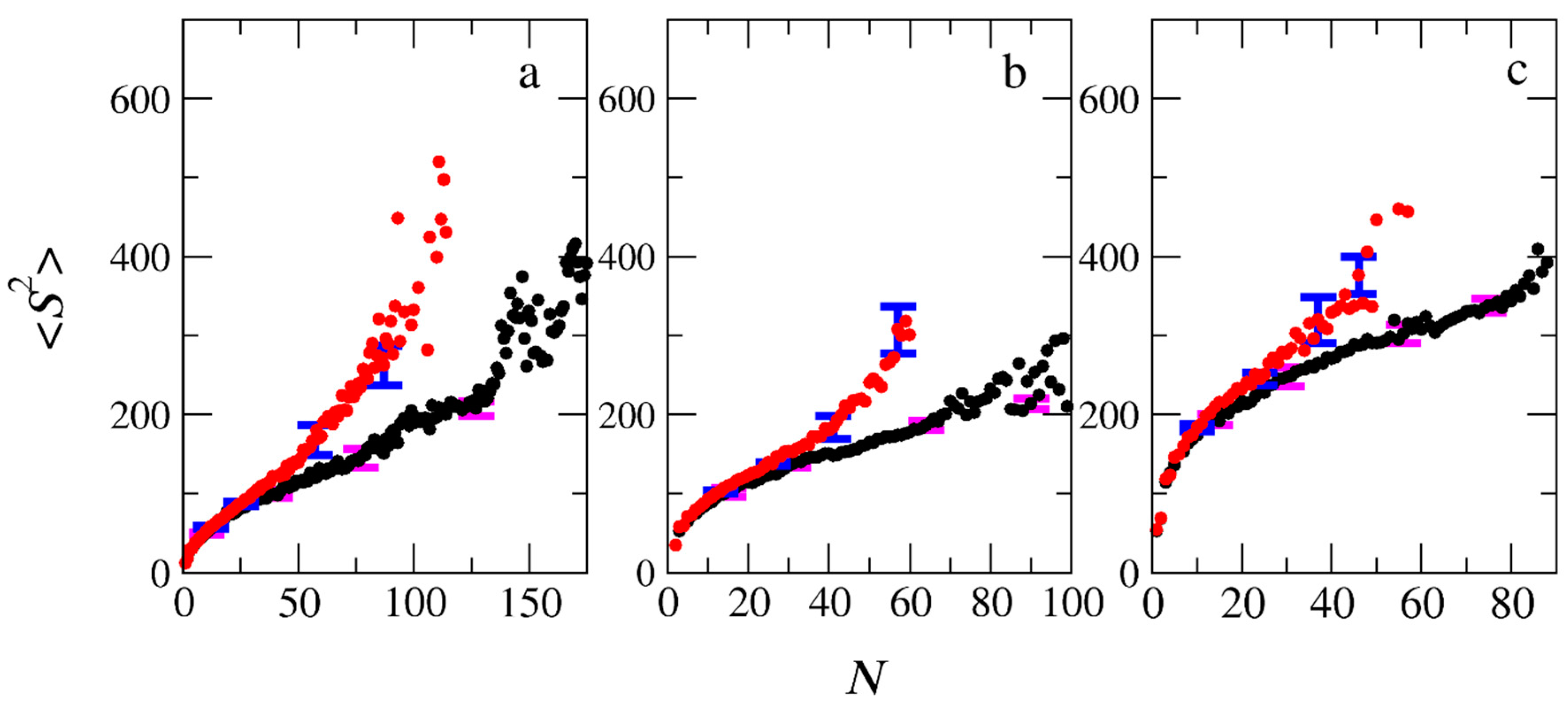
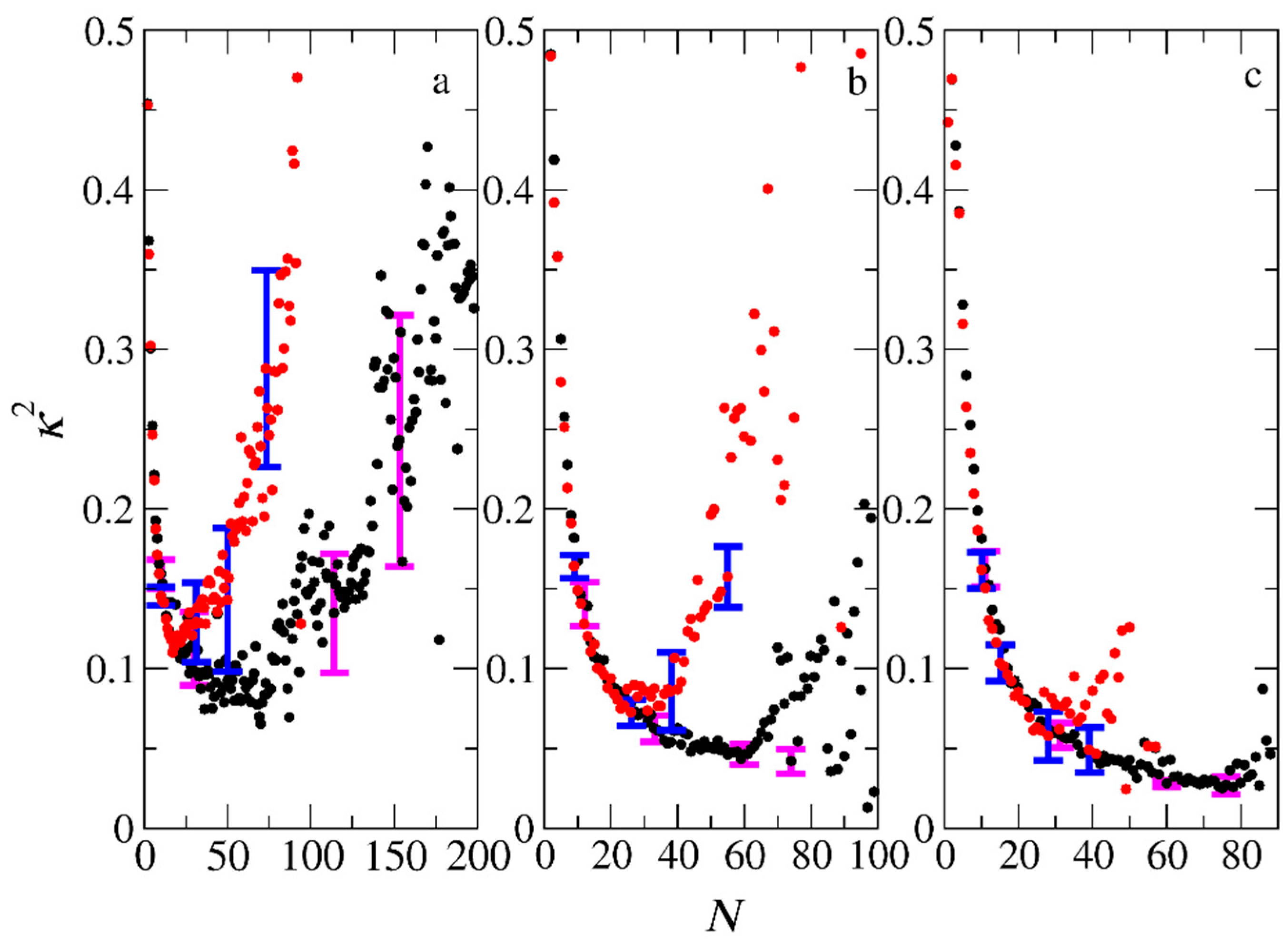
3.3. Micelle Size and Shape
4. Conclusions
Acknowledgments
References
- Magana, J. R.; Sproncken, C. C. M.; Voets, I. K. On Complex Coacervate Core Micelles: Structure-Function Perspectives. Polymers. 2020. https://doi.org/10.3390/POLYM12091953. [CrossRef]
- Harada, A.; Kataoka, K. Polyion Complex Micelle Formation from Double-Hydrophilic Block Copolymers Composed of Charged and Non-Charged Segments in Aqueous Media. Polymer Journal. 2018. https://doi.org/10.1038/pj.2017.67. [CrossRef]
- van der Kooij, H. M.; Spruijt, E.; Voets, I. K.; Fokkink, R.; Cohen Stuart, M. A.; van der Gucht, J. On the Stability and Morphology of Complex Coacervate Core Micelles: From Spherical to Wormlike Micelles. Langmuir 2012, 28 (40), 14180–14191. https://doi.org/10.1021/la303211b. [CrossRef]
- Shah, S.; Leon, L. Structural Dynamics, Phase Behavior, and Applications of Polyelectrolyte Complex Micelles. Current Opinion in Colloid and Interface Science. 2021. https://doi.org/10.1016/j.cocis.2021.101424. [CrossRef]
- Advances in the Structural Design of Polyelectrolyte Complex Micelles. Journal of Physical Chemistry B. 2021. https://doi.org/10.1021/acs.jpcb.1c01258. [CrossRef]
- Wu, H.; Ting, J. M.; Yu, B.; Jackson, N. E.; Meng, S.; de Pablo, J. J.; Tirrell, M. v. Spatiotemporal Formation and Growth Kinetics of Polyelectrolyte Complex Micelles with Millisecond Resolution. ACS Macro Lett 2020, 9 (11). https://doi.org/10.1021/acsmacrolett.0c00543. [CrossRef]
- Anraku, Y.; Kishimura, A.; Kamiya, M.; Tanaka, S.; Nomoto, T.; Toh, K.; Matsumoto, Y.; Fukushima, S.; Sueyoshi, D.; Kano, M. R.; Urano, Y.; Nishiyama, N.; Kataoka, K. Systemically Injectable Enzyme-Loaded Polyion Complex Vesicles as in Vivo Nanoreactors Functioning in Tumors. Angewandte Chemie - International Edition 2016, 55 (2). https://doi.org/10.1002/anie.201508339. [CrossRef]
- Yoon, H.; Dell, E. J.; Freyer, J. L.; Campos, L. M.; Jang, W. D. Polymeric Supramolecular Assemblies Based on Multivalent Ionic Interactions for Biomedical Applications. Polymer. 2014. https://doi.org/10.1016/j.polymer.2013.12.038. [CrossRef]
- Gioldasis, C.; Gergidis, L. N.; Vlahos, C. Micellization through Complexation of Oppositely Charged Diblock Copolymers: Effects of Composition, Polymer Architecture, Salt of Different Valency, and Thermoresponsive Block. Journal of Polymer Science 2021, 59 (2), 191–204. https://doi.org/10.1002/pol.20200754. [CrossRef]
- Gucht, J. van der; Spruijt, E.; Lemmers, M.; Cohen Stuart, M. A. Polyelectrolyte Complexes: Bulk Phases and Colloidal Systems. J Colloid Interface Sci 2011, 361 (2), 407–422. https://doi.org/10.1016/j.jcis.2011.05.080. [CrossRef]
- Li, C.; Magana, J. R.; Sobotta, F.; Wang, J.; Stuart, M. A. C.; van Ravensteijn, B. G. P.; Voets, I. K. Switchable Electrostatically Templated Polymerization. Angewandte Chemie - International Edition 2022, 61 (39). https://doi.org/10.1002/anie.202206780. [CrossRef]
- Ni, J.; Wan, Y.; Cai, Y.; Ding, P.; Cohen Stuart, M. A.; Wang, J. Synthesis of Anionic Nanogels for Selective and Efficient Enzyme Encapsulation. Langmuir 2022, 38 (10). https://doi.org/10.1021/acs.langmuir.1c03325. [CrossRef]
- Bos, I.; Terenzi, C.; Sprakel, J. Chemical Feedback in Templated Reaction-Assembly Networks. Macromolecules 2020, 53 (23). https://doi.org/10.1021/acs.macromol.0c01915. [CrossRef]
- Ding, Y.; Zhao, Q.; Wang, L.; Huang, L.; Liu, Q.; Lu, X.; Cai, Y. Polymerization-Induced Self-Assembly Promoted by Liquid-Liquid Phase Separation. ACS Macro Lett 2019, 8 (8). https://doi.org/10.1021/acsmacrolett.9b00435. [CrossRef]
- Gavrilov, A. A.; Chertovich, A. V. Simulation of the RAFT Polymerization in 3D: Steric Restrictions and Incompatibility between Species. Polym Chem 2022, 13 (15). https://doi.org/10.1039/d1py01624e. [CrossRef]
- Genzer, J. In Silico Polymerization: Computer Simulation of Controlled Radical Polymerization in Bulk and on Flat Surfaces. Macromolecules 2006, 39 (20). https://doi.org/10.1021/ma061155f. [CrossRef]
- Turgman-Cohen, S.; Genzer, J. Computer Simulation of Concurrent Bulk- and Surface-Initiated Living Polymerization. Macromolecules 2012, 45 (4). https://doi.org/10.1021/ma202679r. [CrossRef]
- Cheng, L.; Cao, D. Effect of Tail Architecture on Self-Assembly of Amphiphiles for Polymeric Micelles. Langmuir 2009, 25 (5), 2749–2756. https://doi.org/10.1021/la803839t. [CrossRef]
- Suek, N. W.; Lamm, M. H. Computer Simulation of Architectural and Molecular Weight Effects on the Assembly of Amphiphilic Linear-Dendritic Block Copolymers in Solution. Langmuir 2008, 24 (7), 3030–3036. https://doi.org/10.1021/la703006w. [CrossRef]
- Miliou, K.; Gergidis, L. N.; Vlahos, C. Polyelectrolyte Micelles in Salt-Free Solutions: Micelle Size and Electrostatic Potential. J Polym Sci B Polym Phys 2018, 56 (12), 924–934. https://doi.org/10.1002/polb.24608. [CrossRef]
- Plimpton, S. Fast Parallel Algorithms for Short-Range Molecular Dynamics. J Comput Phys 1995, 117 (1), 1–19. https://doi.org/10.1006/jcph.1995.1039. [CrossRef]
- Murat, M.; Grest, G. S. Molecular Dynamics Study of Dendrimer Molecules in Solvents of Varying Quality. Macromolecules 1996, 29 (4), 1278–1285. https://doi.org/10.1021/ma951219e. [CrossRef]
- Georgiadis, C.; Moultos, O.; Gergidis, L. N.; Vlahos, C. Brownian Dynamics Simulations on the Self-Assembly Behavior of AB Hybrid Dendritic-Star Copolymers. Langmuir 2011, 27 (2), 835–842. https://doi.org/10.1021/la104188q. [CrossRef]
- Moultos, O.; Gergidis, L. N.; Vlahos, C. Self-Assembly Behavior of Thermoresponsive Bis-Solvophilic Linear Block Terpolymers: A Simulation Study. Macromolecules 2012, 45 (5), 2570–2579. https://doi.org/10.1021/ma2025573. [CrossRef]
- Kalogirou, A.; Gergidis, L. N.; Moultos, O.; Vlahos, C. Entropic Effects, Shape, and Size of Mixed Micelles Formed by Copolymers with Complex Architectures. Phys Rev E Stat Nonlin Soft Matter Phys 2015, 92 (5). https://doi.org/10.1103/PhysRevE.92.052601. [CrossRef]
- Ni, R.; Cao, D.; Wang, W.; Jusufi, A. Conformation of a Spherical Polyelectrolyte Brush in the Presence of Oppositely Charged Linear Polyelectrolytes. Macromolecules 2008, 41 (14), 5477–5484. https://doi.org/10.1021/ma800827x. [CrossRef]
- Kalogirou, A.; Moultos, O. A.; Gergidis, L. N.; Vlahos, C. Micellization Properties of Θ-Shaped, Figure-Eight-Shaped and Linked Rings Copolymers. Macromolecules 2014, 47 (16), 5851–5859. https://doi.org/10.1021/ma501053d. [CrossRef]
- Kalogirou, A.; Gergidis, L. N.; Miliou, K.; Vlahos, C. Complexation of Polyelectrolyte Micelles with Oppositely Charged Linear Chains. Journal of Physical Chemistry B 2017, 121 (8), 1982–1991. https://doi.org/10.1021/acs.jpcb.6b12709. [CrossRef]
- Stillinger, F. H. Rigorous Basis of the Frenkel-Band Theory of Association Equilibrium. J Chem Phys 1963, 38 (7). https://doi.org/10.1063/1.1776907. [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; Vanderplas, J.; Passos, A.; Cournapeau, D.; Brucher, M.; Perrot, M.; Duchesnay, É. Scikit-Learn: Machine Learning in Python. Journal of Machine Learning Research 2011, 12.
- Virtanen, P.; Gommers, R.; Oliphant, T. E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat Methods 2020, 17 (3). https://doi.org/10.1038/s41592-019-0686-2. [CrossRef]
- Hagberg, A. A.; Schult, D. A.; Swart, P. J. Exploring Network Structure, Dynamics, and Function Using NetworkX. In 7th Python in Science Conference (SciPy 2008); 2008.
- Asad Ayoubi, M.; Zhu, K.; Nyström, B.; Olsson, U.; Almdal, K.; Khokhlov, A. R.; Piculell, L. Morphological Investigation of Polydisperse Asymmetric Block Copolymer Systems of Poly(Styrene) and Poly(Methacrylic Acid) in the Strong Segregation Regime. J Polym Sci B Polym Phys 2013, 51 (23). https://doi.org/10.1002/polb.23389. [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




