1. Introduction
The ongoing global pandemic known as the 2019 coronavirus disease (COVID-19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). While the majority of patients with mild or moderate symptoms have a positive prognosis (1), there are cases where individuals progress to a severe or critical phase, characterized by severe respiratory failure requiring mechanical ventilation and intensive care unit hospitalization (2). Moreover, this infection can have an impact on various organ systems, such as the neurological, cardiovascular, gastrointestinal, renal, hematological, and immunological systems (3). Individuals with pre-existing cardiovascular diseases, high blood pressure, diabetes, and metabolic syndrome face an increased risk of mortality (1). Furthermore, a significant portion of patients experience persistent symptoms and comorbidities that are associated with organ damage. The long-term consequences of these complications remain uncertain.
Understanding the underlying reasons for the significant variation in individual responses to SARS-CoV-2 infection is essential for the development of effective therapies (4). However, this aspect remains poorly comprehended, posing a major challenge in therapeutic advancements. Recent investigations into the interactions between host cell membranes and the virus have provided valuable insights into the role of cellular lipids in the viral entry process (5). Specifically, sphingolipids (SL) have been identified as influential factors in the entry of bacteria and other viruses into cells (6,7). SL are essential components of eukaryotic cell membranes, serving as structural elements, signaling molecules, and modulators of enzyme activity (8). Apart from their structural functions, certain SL exhibit bioactivity and are associated with various pathological conditions, including inflammation-related disorders like atherosclerosis, rheumatoid arthritis, inflammatory bowel disease, type II diabetes, obesity, cancer, and neurological diseases (9–11). Furthermore, SL play a critical role in regulating viral replication and the innate immune response (6,12,13). Due to their versatility, these molecules have extensive involvement in both normal physiological processes and pathological states.
The metabolic processes involving SL are highly complex. Ceramide (Cer) holds significant importance in the SL metabolic pathway and can be generated through the breakdown of sphingomyelin (SM) or through De novo synthesis (14). The initial step involves serine palmitoyltransferase, which converts serine and palmitoyl-coenzymeA into 3-ketodihydrosphingosine. Subsequently, 3-ketodihydrosphingosine reductase converts it to sphinganine (14). Ceramide synthase then adds an acyl-fatty acid to sphinganine, forming dihydroceramide (dH-Cer). Dihydroceramide D4-saturase, located in the endoplasmic reticulum (ER), converts dH-Cer into Cer (14). Following its production, Cer needs to be transported from the ER to the Golgi organelle to facilitate the synthesis of sphingomyelin (SM) (15). Moreover, Cer can undergo transformation into various crucial SL (15). Ceramidase is an enzyme responsible for converting Cer into sphingosine (Sph) (2-amino-4-trans-octadecene-1,3-diol). Sphingosine kinases 1 (SphK1) and 2 (SphK2) can then phosphorylate Sph to generate the active lipid sphingosine-1-phosphate (S1P) (16). Sphingomyelin synthase converts Cer into SM, while sphingomyelinase (SMase) converts SM back into Cer. These conversion reactions occur in lysosomes (acid SMase) or on the cell surface (neutral SMase) (8,17).
Emerging evidence suggests that SL play a crucial role in modulating SARS-CoV-2 infection (5,18). Studies conducted on animal models have demonstrated that SARS-CoV-2 infection leads to an increase in SL levels, both within cells and in the serum (19). The significance of SL extends beyond their role as physical components of cell membranes and ligands. They also have an impact on the localization and activity of proteins involved in receptor-mediated signaling (8). In patients with COVID-19, plasma concentrations of Cer were found to be elevated (20,21), particularly in those with severe respiratory symptoms (22). Furthermore, reduced levels of serum Sph were strongly associated with symptomatic COVID-19 compared to asymptomatic cases (23).
In light of previous findings documenting changes in SL patterns among COVID-19 patients, our research takes a step further by integrating targeted mass spectrometry-based sphingolipidomics with the measurement of specific inflammatory mediators, clinical parameters, and the gene expression of SL-metabolizing enzymes. Notably, our study includes patients who have recovered from COVID-19, providing a comprehensive understanding of the disease trajectory. Through this multifaceted approach, our primary objective is to identify biomarkers that can serve as diagnostic and prognostic indicators, while also unraveling the intricate role of SL in the pathophysiological pathways of COVID-19. In line with this, our study emphasizes that SARS-CoV-2 infection induces a notable increase in SM levels in the plasma of patients with severe illness. The implications of these alterations warrant careful consideration, as they could potentially influence disease progression and outcomes.
2. Material and Methods
2.1. Study Design and Blood Collection
This prospective study was undertaken between June and November of 2020, prior to the introduction of COVID-19-vaccination. We used stringent and reasonable inclusion and exclusion criteria: adults who tested positive for COVID-19 (n=204), as determined by analyzing nasopharyngeal swabs using a genomic RNA assay with RT-PCR (Biomol OneStep Kit/COVID-19-Instituto de Molecular Biology of Paraná-IBMP Curitiba/PR, Brazil) or using serology-specific IgM and IgG antibodies tests (SARS-CoV-2 antibody test®, GuangzhouWondfo Biotech Co., Ltd., Guangzhou, China), and controls (healthy volunteers - n=55) who tested negative for SARS-CoV-2; and exclusion for children under 18 years of age, and pregnant or lactating women. Patients classified as asymptomatic to mild (n=36), moderate (n=60), severe (n=67), or critical (n=41) had blood samples collected. The criteria for the clinical classification of patients were set at the time of sample collection, based on a modified statement in the Novel Coronavirus Pneumonia Diagnosis and Treatment Guideline (7th edition) (24,25). Patients at two medical centers: Santa Casa de Misericórdia de Ribeirão Preto and Hospital São Paulo in Ribeirão Preto, São Paulo, Brazil, were given a venous puncture to gather peripheral blood samples upon their first admission and/or during their hospitalization. The blood samples of healthy controls and non-hospitalized participants were taken either at the Centre for Scientific and Technological Development "Supera Park" (Ribeirão Preto, São Paulo, Brazil) or at the residences of patients receiving at-home care. Convalescent participants were recruited (n=77) and included in the study based on the following inclusion criteria: men and women aged 30 to 69 years, approximately 30 days since the resolution of acute clinical signs of COVID-19 or medical discharge (in case of hospitalization). The exclusion criteria were as follows: acute or chronic clinical illnesses without medical supervision; anemia; use of immunosuppressive medicines; pregnant women; hormone replacement; smokers; and heavy alcohol or drug use. Additionally, a health status evaluation was conducted. Blood was obtained through venipuncture in EDTA tubes using a vacuum collection method (BD Vacutainer® EDTA K2 Franklin Lakes, New Jersey, United States) from all patients with COVID-19, healthy controls, and convalescent individuals. After centrifugation, plasma and buffy coat (middle layer containing leukocytes and thrombocytes, utilized for genomic RNA extraction) were separated and immediately frozen at -80°C either alone or with 0.5 mL of TRIzol reagent, respectively. All samples of blood were processed within four hours of their collection. For sphingolipidomics analysis, plasma was immediately kept in methanol (1:1 v/v) and analyzed using tandem-mass spectrometry (LC-MS/MS).
2.2. Ethical Considerations
In accordance with the international ethics guidelines, the participants informed consents were obtained as approved by the Conselho Nacional de Ética em Pesquisa (CONEP) and the Human Ethical Committees of the Faculty of Pharmaceutical Sciences of Ribeirão Preto and the School of Physical Education and Sport of Ribeirão Preto – University of São Paulo (USP). The research protocol received the certificate of Presentation and Ethical Appreciation for Immunocovid study (CAAE: 30525920.7.0000.5403), and AEROBICOVID study (CAAE: 33783620.6.0000.5659, and CAAE: 33783620.6.3001.5403). The sample size was decided by the convenience of sampling, the availability of participants at partner hospitals, their willingness to participate, and the local pandemic conditions.
2.3. Laboratory and Data Collection
Each patient's electronic medical records were checked thoroughly. Information collected for this study includes socio-demographic details, comorbidities, medical history, clinical symptoms, regular laboratory tests, immunological tests, clinical interventions, and outcomes. The information was documented on a standardized record form. Laboratory exams performed for the first time within 24 hours of admission were the primary endpoint for data collection. The secondary endpoint was clinical outcome (discharged or deceased). Blood exams were carried out in the clinical analysis laboratories of the patients' individual hospitals for those who were hospitalized. Blood analyses of healthy participants and non-hospitalized patients were performed at Serviço de Análises Clínicas (SAC), Departamento de Análises Clínicas, Toxicológicas e Bromatológicas of the Faculdade de Ciências Farmacêuticas de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, São Paulo, Brazil. Automated assays were performed to determine liver and kidney function, myocardial enzyme spectrum, coagulation factors, red blood cells, haemoglobin, platelets, total and differential leukocytes.
2.4. Cytokine Measurements
Plasma concentrations of interleukin (IL)-6, IL-8, IL-1β, IL-10, and tumor necrosis factor (TNF) were measured using a BD Cytometric Bead Array (CBA) Human Inflammatory Kit (BD Biosciences, San Jose, CA, USA) in accordance with the manufacturer's instructions. Briefly, following sample preparation, the cytokine beads were counted using a flow cytometer (FACS Canto TM II; BD 316 Biosciences, San Diego, CA, USA), and analyses were performed using FCAP Array (3.0) software (BD Biosciences, San Jose, CA, USA). The cytokine concentrations were represented as pg/mL.
2.5. Lipid Extraction and Sample Preparation for LC-MS/MS
All steps were executed at room temperature if not specified otherwise (26,27). Briefly, plasma samples (250 μL) were adjusted to 1 ml volume with PBS and transferred into a glass centrifuge vial. After adding 10 μl of the internal standard (10 μM Cer/Sph Mixture II, LM6005, Avanti® Polar Lipids), 300 μl of 18.5% HCl, 1 ml of MeOH, and 2 ml of CHCl3 were added before the content were vortexed for 30 min at 50 rpm in a tube rotator. Samples were centrifuged for 3 min at 2000 g, the lower organic phase was moved into a new glass tube and to the leftover aqueous phase was added another 2 ml of CHCl3, to repeat the lipid extraction before combining the two organic phases and vacuum-dry the solvent in a speed-vac for 45 min at 60°C. The sample was resuspended in 100 μl of MeOH:CHCl3 (4:1, v/v), and vortexed for 1 min before being stored at -80°C.
2.6. Sphingolipids Quantification by LC-MS/MS
SL were measured in plasma samples by investigators blinded for the experiments as described previously (26,28). In brief, liquid chromatography was performed using an Ascentis Express C18 column (Supelco, St. Louis, MO, USA) with 100 × 2.1 mm and a particle size of 2.7 μm, which was kept at 40°C for the whole procedure, in an ultra-high-performance liquid chromatography (UHPLC) system (Nexera X2; Shimadzu, Kyoto, Japan). The column was equilibrated for 20 min, followed by the injection of 10 μL of the sample onto the HPLC column. Elution was carried out under a binary gradient system consisting of Phase A, comprised of H
2O containing 1% formic acid, and Phase B, comprised of MeOH. Gradient elution was performed for 20 min at a flow rate of 0.5 mL/min, using the following setup: 0.0 - 1 min – 30% B, 1.1 - 2.5 min - 85% B, 2.5 - 5.0 min –100% B, 5.0 -15 min – hold 100% B, 15.1 - 20 min – re-equilibrated with 30% B. The HPLC system was directly connected to a TripleTOF 5600
+ mass spectrometer (SCIEX-Foster, CA, USA). An electrospray ionisation source (ESI) in positive ion mode was used for high-resolution multiple-reaction monitoring (MRM
HR) scanning. An atmospheric-pressure chemical ionization probe (APCI) was used for external calibrations of the calibrated delivery system (CDS). Automatic mass calibration (<2 ppm) was performed periodically after each of the five sample injections using APCI Positive Calibration Solution (Sciex-245 Foster, CA, USA) injected via direct infusion at a flow rate of 300 μL/min. Additional instrumental parameters were as follows: nebulizer gas (GS1), 50 psi; turbo gas (GS2), 50 psi; curtain gas (CUR), 25 psi; electrospray voltage (ISVF), +4.5 kV; temperature of the turbo ion spray source, 500°C. The dwell time was 10 ms, and a mass resolution of 35,000 was achieved at
m/z 400. Data acquisition was performed using Analyst
TM software (SCIEX- Foster, CA, USA). Qualitative identification of the lipid species was performed using PeakView
TM (SCIEX-Foster, CA, USA), and the mass transitions for all standards and analytes were reported in
Table S2, such as typical retention times. MultiQuant
TM (SCIEX-Foster, 252 CA, USA) was used for the quantitative analysis, which allows the normalization of the peak area of individual molecular ions using an internal standard for each family of SL (Cer/Sph Mixture II - Avanti Polar Lipids - LM6005). The raw area of the analytes was corrected monoisotopically (
Figure S4 – Equation I), in order to compensate the natural abundance of
13C and
14C isotopes that is not considered in our analytical method. For the quantification of SL species, the area ratio of each lipid against the corresponding internal standard, multiplied by the concentration of the IS (
Figure S4 – Equation II), provides the real analyte concentration (pmol/mL) in plasma. Any SL for which no standards are available is quantitated using the calibration curve of its closest counterpart. Additional methodology details are provided in the
Figures S5-S12.
2.7. RNA Extraction and Analysis
The buffy coat was processed via two stages in order to obtain total RNA. Initially, the frozen samples were thawed, and then 0.5 mL of TRIzol reagent was added prior to 5 min. incubation at room temperature. Then, 0.2 mL of chloroform was added and the mixture was incubated for 3 min at room temperature before being centrifuged at 12,000 g for 15 min at 4°C. The RNA-containing upper phase was transferred to a new tube containing an equal volume of ethanol (1:1 v/v). After this initial stage, 0.7 mL of each sample was transferred to a spin cartridge containing a clear silica membrane to continue the extraction with on-column DNase treatment (Life Technologies, PureLink DNase Set, Catalog No. 12185010) according to the manufacturer's specifications (Life Technologies, PureLink RNA Mini Kit, Catalog No. 12183018A). RNA concentration was determined using a Qubit Fluorometer (Life Technologies, Qubit RNA BR Assay Kit, Catalog No. Q10211), the purity was determined using the 260/280 nm and 260/230 nm absorbance measurement ratios in a Nanodrop spectrophotometer (Thermo Fisher Scientific), and the quality of purified total RNA was determined using the RNA Integrity Number (RIN) values on a Bioanalyzer instrument (Agilent Technologies, Agilent 2100 Bioanalyzer system with RNA 6000 Nano kit, Catalog No. 5067-1511).
2.8. Transcriptome Profiling
Using the Clariom S Human Assay (Applied Biosystems, Clariom S Assay human, Catalog No. 902927) for single-sample (cartridge array) processing on the GeneChip 3000 instrument system (Applied Biosystems, GeneChip WT Pico Reagent Kit, Catalog No. 902622) in a high-throughput facility (Thermo Fisher Scientific, Microarray Research Services Laboratory, Santa Clara, CA, USA), whole transcript expression arrays were generated from 66 samples distributed in healthy controls (n=12) and patients diagnosed with COVID-19 (n=54).
2.8.1. Bioinformatics Analysis of Transcriptome Data
Gene expression of enzymes involved in the SL pathway were obtained from whole blood leukocytes transcriptomic data available in the ArrayExpress database (
http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-11240 relative to the participants of this research consortium. This dataset contained pre-processed transcriptomic profiling from 66 samples distributed in healthy controls (n=12) and patients diagnosed with COVID-19 (n=54), subgrouped into the different clinical classification: mild (n=12), moderate (n=14), severe (n=14), and critical (n=14). The bioinformatic analyses were performed using R 4.1.2 libraries (29) in RStudio environment (30) and Bioconductor libraries (31). We used preprocessed expression data deposited at probe set level and obtained gene-based expression by collapsing the probes using the maximum mean method with collapseRows function of function of the WGCNA 1.71 package employing gene to probe annotation available at same dataset. Differential expression for whole transcriptome between previously described clinical groups were performed with
limma 3.50.1 using age, sex, body mass index (BMI), hypertension, diabetes, and outcome as co-variates. Differentially expressed genes (DEGs) were defined on Benjamini and Hockberg (32) adjusted
p-values < 0.05 in at least one pair of clinical groups. Graphical representations of generated data were constructed using
ggplot2 3.3.5.
2.8.2. Validation of Microarray Data by Reverse Transcription Quantitative Real-Time PCR
The RNA samples utilized for microarray-based gene expression profiling were confirmed using two-step RT-qPCR. Briefly, complementary DNA was generated from 300 ng of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied BiosystemsTM) according to the manufacturer's instructions. The qPCR reactions were prepared with 1X iTaqTM Universal SYBR® Green Supermix (BIO-RAD), 100 nM of each gene-specific primer combination, and 8 ng of cDNA, then amplified in an AriaMX Real-time PCR machine (Agilent, USA). qPCR cycling consisted of an initial denaturation for 30 seconds at 95 °C, followed by 40 cycles of 95 °C for 5 seconds and 60 °C for 20 seconds; melting curves were included to ensure the absence of nonspecific reactions. Using the Real-time PCR Miner algorithm, the efficiency of primer pairs was measured (33). Each sample was tested in duplicate, and gene expression analyses were performed according to the ΔCq model using the β-Actin as a reference gene (34).
2.9. Statistical Analysis
The data were evaluated for a normal distribution using the Shapiro-Wilk normality test and D'Agostino & Pearson test. The parametric data were analyzed using unpaired
t-tests or one-way ANOVA followed by Tukey’s multiple comparison tests. For non-parametric data, Mann-Whitney or Kruskal-Wallis test were used, followed by Dunn’s post-tests. The chi-square test was used to assess associations among categorical variables and Covid-19. The results were obtained using GraphPad Prism software (version 9.0) and the differences were considered statistically significant at
p < 0.05. The dependence on multiple variables were analyzed using significant Spearman’s correlations at
p < 0.05. Data were presented in the Correlation matrix using the R package qgraph (35). Partial Least Squares Discriminant Analysis (PLS-DA) was performed using the MetaboAnalyst 5.0 online software (
https://www.metaboanalyst.ca/). The relevance of sphingolipid metabolites was assessed using the variable importance in projection (VIP) score and this was subsequently used to identify the potential biomarkers (36). The accuracy of the predictor was determined by the area under the curve (AUC) of the receiver-operating characteristic (ROC). The AUCs, with 95% confidence intervals, were calculated to assess the diagnostic value; AUC > 0.70 was considered clinically relevant. Multivariate Binomial Logistic Regression Analysis was performed using the Jamovi software (Version 1.6 - 2021) to assess the association of sphingolipids with the prognosis and clinical outcome of severity and mortality, adjusting the model for the confounding variables age, sex, comorbidity, Body mass index (BMI), Hospitalization days, blood glucose and Neutrophil-Lymphocyte Ratio (NLR).
3. Results
3.1. Characterization of Study Participants
Patients' blood samples were collected during the height of the pandemic in the clinical emergency setting, ensuring a diverse representation without specific selection or matching. A total of 204 patients diagnosed with COVID-19 were included in this study, categorized based on the severity of their symptoms: mild (n=36), moderate (n=60), severe (n=67), and critical (n=41). These groups were compared to convalescent patients (n=77) and a healthy control group (n=55). Detailed demographic characteristics, clinical manifestations, and laboratory findings of the patients, convalescent individuals, and control group are provided in
Table 1. As expected, the groups differed significantly in terms of age, with advanced age being a well-established risk factor for hospitalization and severity of COVID-19 infection. There was a significant difference in the distribution of sex among the convalescent group. The most prevalent comorbidities observed in COVID-19 patients were hypertension (44.1%), diabetes (30.1%), cardiovascular disorders (21%), increased body mass index (BMI) (28.4 ± 5.9), history of smoking (19%), and neurological disorders (17%). The common initial symptoms reported by patients included cough, dyspnea, dysgeusia, and anosmia, followed by diarrhea, fever, myalgia, and hyperactive delirium.
Regarding hematological characteristics, severe and critical patients exhibited a significant decrease in hemoglobin, erythrocyte count, and lymphocyte count, along with a significant increase in total leukocyte count, neutrophil count, Neutrophil Lymphocyte Ratio (NLR), and blood glucose levels compared to patients with mild/moderate symptoms and the control group. Hematological parameters were assessed only at the time of admission. Oxygen saturation was significantly lower in COVID-19 patients compared to convalescent individuals and the control group. Approximately 65% of patients required oxygen administration, with the majority receiving oxygen therapy via a Venturi Mask (17.1%), High Flow Nasal oxygen support (31.9%), or invasive mechanical ventilation (16.2%). The average hospitalization duration for severe and critical patients was 9 ± 4 days. In terms of medical treatment, glucocorticoids were prescribed to 61% of patients, Azithromycin to 59.3%, Ceftriaxone to 45.6%, Oseltamivir to 29.4%, Hydroxychloroquine to 13.2%, and anticoagulants to 8.8% of the total patients, aligning with the standard care protocols in place during the patients' hospitalization period.
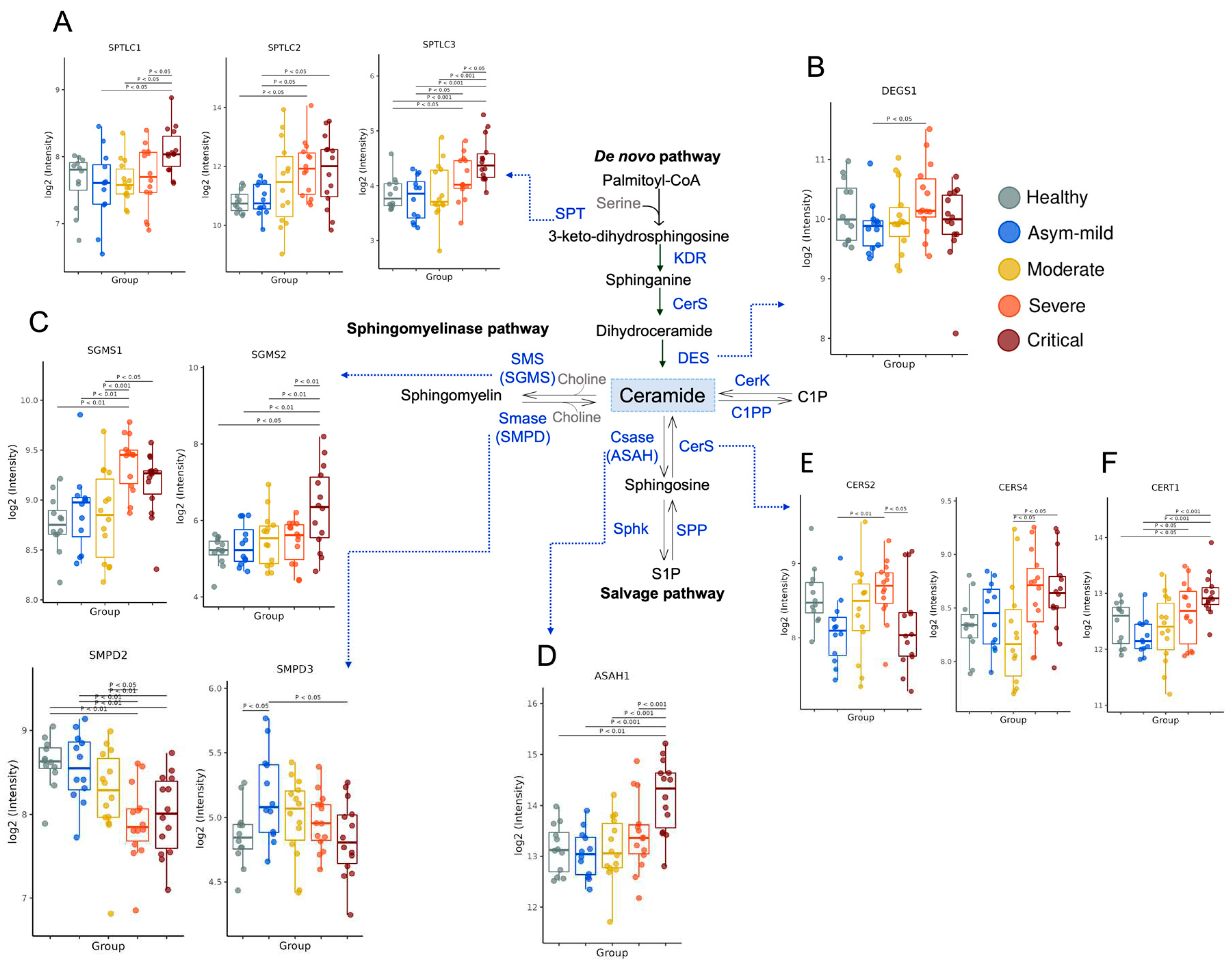
3.2. COVID-19 Severity Increased Gene Expression of Key Enzymes Involved in SM and Cer Synthesis
The metabolism of SL is highly intricate (8). We sought to investigate whether the gene expression of enzymes involved in their production was regulated in blood cells from the buffy coat. Cer serves as a crucial component in the metabolic flux of SL and can be generated through either
De novo synthesis or the breakdown of SM. The
De novo pathway begins with the action of serine-palmitoyltransferase (SPT).
SPTLC2 showed up-regulation in severe and critical patients, while
SPTLC1 and
SPTLC3 were primarily elevated in critical patients (
Figure 1A). Sequential reactions introduce variations to the core structure, leading to Cer formation. The enzyme sphingolipid-4-desaturase (
DEGS1) adds a double bond to the sphingoid base, while
DEGS2 incorporates a hydroxide group (OH). Notably,
DEGS1 expression was high in severe cases (
Figure 1B), and
DEGS2 displayed a tendency to decrease with severity (
Figure S1B). In the catabolic pathways of SL, SM, ceramide-1-phosphate (C1P), and glycosphingolipids are hydrolyzed, resulting in Cer formation. The expression levels of Cer-metabolizing enzymes derived from SM, such as sphingomyelinases (
SMPD2, and
SMPD3), were found to be down-regulated in COVID-19 severity (
Figure 1C). Furthermore, the others expression of enzymes associated with this pathway, such as ceramide synthase 2 (
CERS2) and ceramide synthase 4 (
CERS4) (
Figure 1E), as well as ceramide transfer protein (
CERT1) (
Figure 1F), was increased in severe COVID-19 patients. However, no significant regulation was observed in the gene expression of
CERS1,
CERS3,
CERS5, and
CERS6 within the same COVID-19 group (
Figure S1D). Cer can be converted into several important SL. The enzyme ceramidase generates sphingosine (Sph), and the expression of acid ceramidase 1 (
ASAH1) was elevated in critical patients (
Figure 1D), while
ASAH2 showed no influence (Supplementary
Figure 1E). Conversely, the expression levels of enzymes involved in the conversion of Cer to SM, such as sphingomyelin synthase 1 (
SGMS1), were significantly increased in COVID-19 patients in a severity-dependent manner (
Figure 1C). Intriguingly,
SGMS2 demonstrated an increase only in critically ill patients (
Figure 1C). Moreover, the majority of genes that promote Cer accumulation exhibited slight expression levels in severely affected patients, whereas genes involved in Cer-to-SM synthesis displayed significant up-regulation.
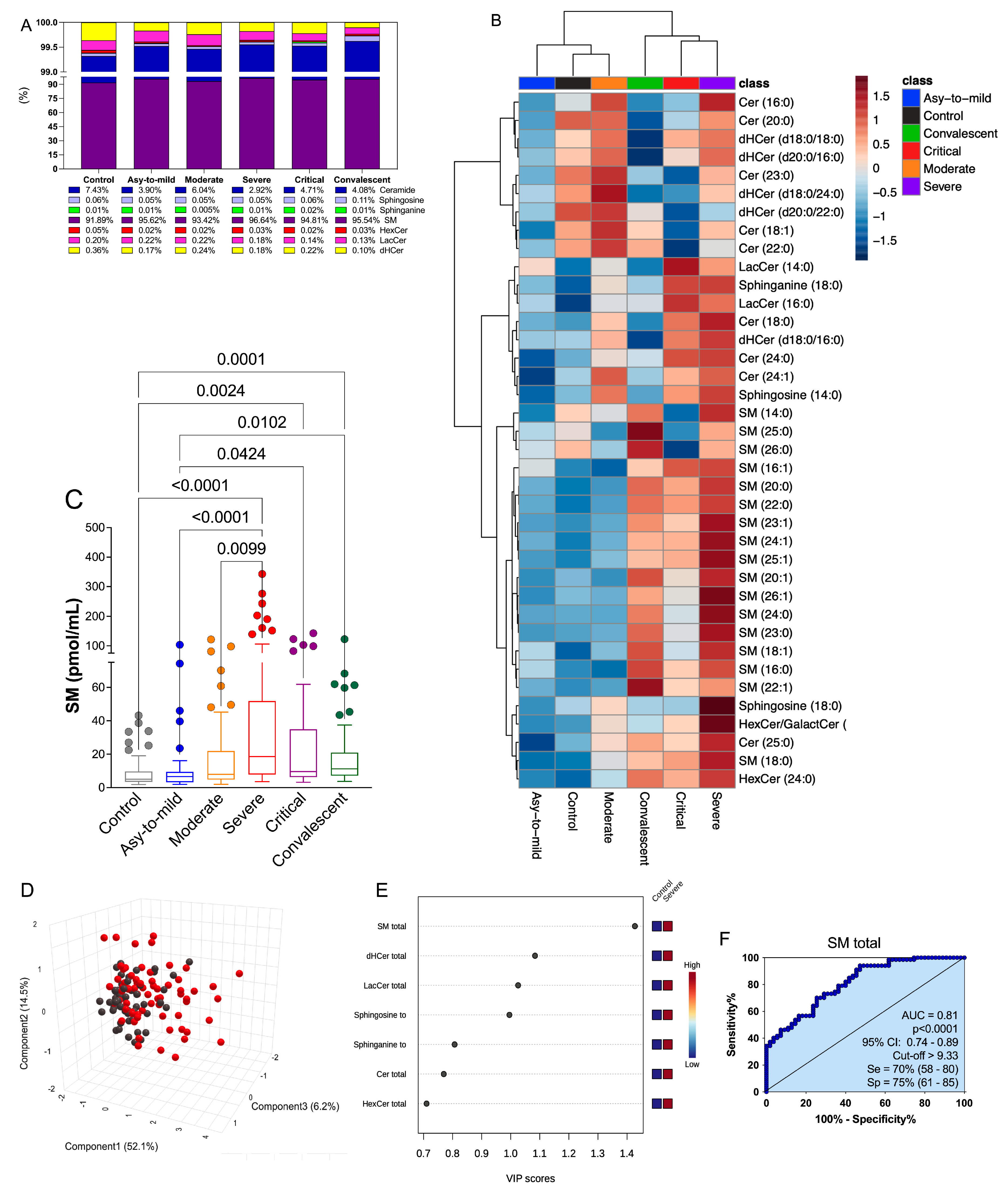
3.3. Plasma SM Profile Is Associated with COVID-19 and Can Be a Potential Biomarkers for Assessing Severity Disease
The plasma SL profile of the study participants was determined using LC-MS/MS targeted sphingolipidomics (
Figure 2). A total of thirty-eight SL species, classified into seven subclasses, were identified in the plasma of the volunteers.
Table S1 and
Figure S5 provide a comprehensive list of the measured lipids, including their annotations, retention time, and fragment panel. The data points were organized into distinct groups for analysis: i) COVID-19 patients categorized according to disease severity (ranging from asymptomatic to mild, moderate, severe, and critical); ii) healthy control subjects; and iii) convalescent individuals (30 days post COVID-19 recovery). Examining the overall SL plasma composition in the control group, SM was found to be the predominant species (91%), followed by Cer (7%), dihydroceramide (dHCer) (0.4%), lactosylceramide (LaCer) (0.2%), sphingosine (Sph) (0.06%), hexaglycosylceramide (HexCer) (0.05%), and sphinganine (0.01%) (
Figure 2A). Comparing the SL profiles of COVID-19 patients with those of the control group, we observed an increased percentage of SM production correlated with the severity of symptoms. The highest production was observed in the plasma of severe patients (96.64%), accompanied by slightly decreased total levels of other SL classes, specifically Cer, which exhibited reduced percentage levels in patients with more severe clinical manifestations. Interestingly, the total SL percentage profile of convalescent individuals closely resembled that of the control group, indicating a restoration of lipid homeostasis (
Figure 2A).
Quantifying the production of each SL class in plasma (measured in pmol/mL), we found a statistically significant increase in the total production of SM species in COVID-19 patients compared to both the control and convalescent groups (
Figure 2C). Moreover, we demonstrated that statistical differences existed between the different severity groups of COVID-19 patients, control subjects, and convalescent individuals for shinganine (
Figure S2A), dHCer (
Figure S2B), Cer (
Figure S2C), LacCer (
Figure S2D), HexCer (
Figure S2E), and Sph (
Figure S2F). These differences underscored the increased production of Cer and its derivative metabolites (dHCer, LacCer, and HexCer) in severe and critical COVID-19 patients. On this sense, several SL classes increased the production with the disease severity, but SM levels seem to be more prominent in the total percentage ratio. The analysis of the principal metabolic species associated with sphingolipid (SL) subclasses revealed the predominant identification of 38 lipids, represented in the heatmap plots based on COVID-19 severity (
Figure 2B). Average values of each SL species across all groups showcased a significant remodeling of plasma SL species in individuals with COVID-19, with certain species exhibiting prevalence by specific groups.
Given the extensive sphingolipidomic data, we conducted a comprehensive analysis to identify potential biomarkers and a signature associated with the progression of COVID-19. Initially, we focused on discerning the differences between severe COVID-19 patients and healthy participants. The 3D score plot in
Figure 2D, generated through partial least squares discriminant analysis (PLS-DA), clearly demonstrates the presence of a distinct sphingolipid (SL) profile associated with the disease. To identify the most predictive and differentiating features that could aid in sample classification, we utilized the "Variable importance in projection" (VIP) score, which is derived from PLS-DA and signifies the key factors contributing to group disparities (
Figure 2E). A VIP score greater than 1 indicates that a metabolite may serve as a potential biomarker (37). Notably, the subclass of SLs known as total SM exhibited the highest VIP score, suggesting its potential as a biomarker for assessing the severity of COVID-19 (
Figure 2E). Following the identification of prospective biomarkers using the VIP score plots, we further evaluated their effectiveness and predictive power in distinguishing severe COVID-19 groups from controls using receiver operating characteristic (ROC) curve analysis. In this regard, we specifically focused on total SM (
Figure 2F). The area under the curve (AUC) for total SM exceeded the threshold considered clinically significant (AUC=0.81, p<0.0001). These findings highlight the potential utility of SM as a biomarker for assessing the severity of COVID-19.
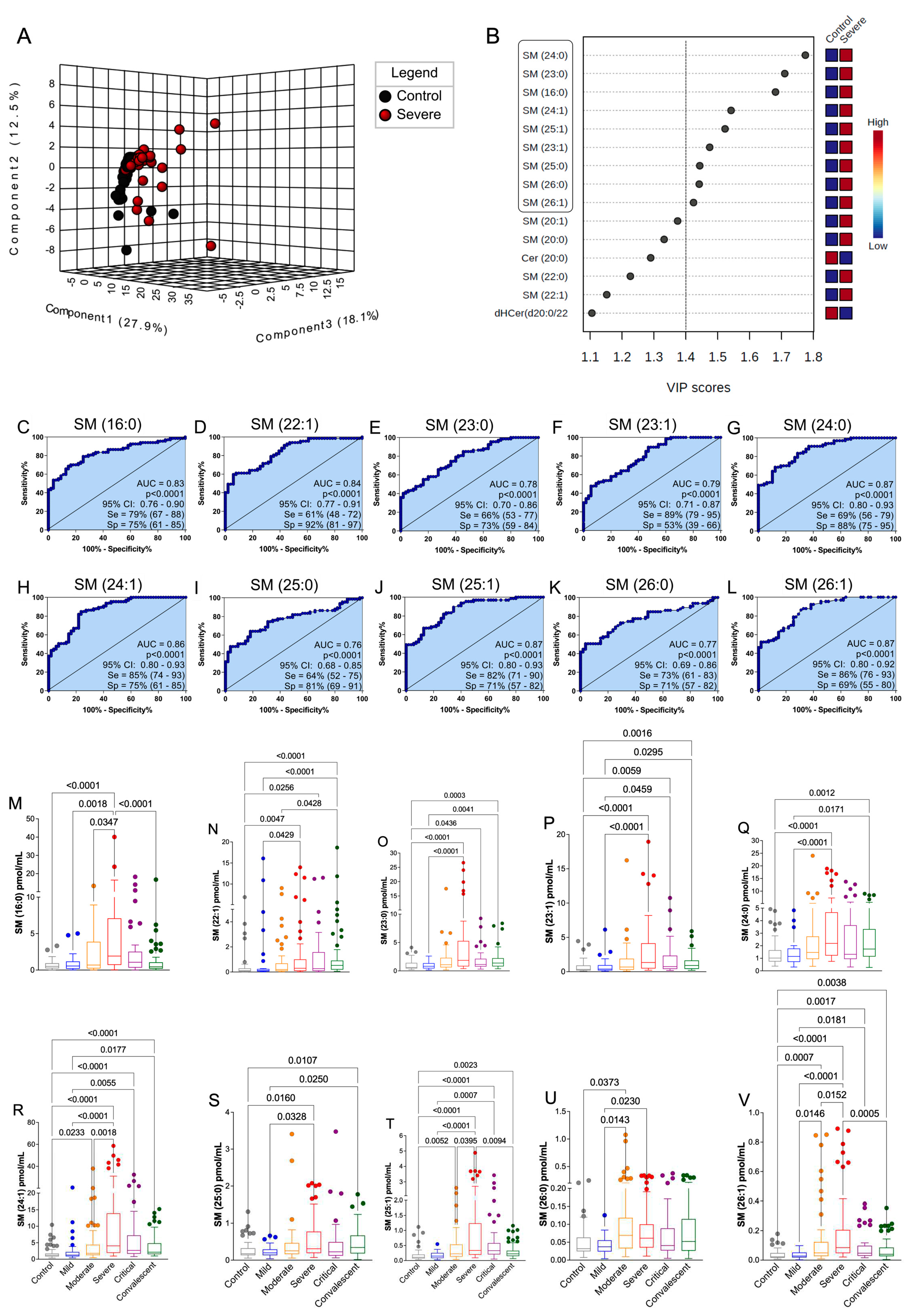
3.4. The Discovered Plasma SM-Species Panel Effectively Distinguished Severity COVID-19
After conducting PLS-DA and VIP score analyses for total SM, we proceeded to assess the significance of individual SM species using the same statistical method (
Figure 3A, 3B). Remarkably, we identified SM (d18:1)/24:0, /23:0, /16:0, /24:1, /25:1, /23:1, /25:0, /26:0, and /26:1 with VIP scores exceeding 1.4 in the severe patient group, indicating their potential as biomarkers for COVID-19 severity (
Figure 3B). To further investigate these altered SM compounds, we employed ROC curves (
Figure 3C-L) and boxplots graphics (
Figure 3M-V). All ten selected SM species exhibited excellent diagnostic performance (AUC > 0.70; p<0.0001), effectively distinguishing severe COVID-19 patients from healthy controls (
Figure 3C-L). Additionally, the median levels of these SM biomarkers exhibited significant increases across control, mild, moderate, severe, and critical COVID-19 patient groups (
Figure 3M-V). Notably, in plasma samples from convalescent individuals, the levels of SM (d18:1)/16:0, /24:1, /25:1, /26:0, and /26:1 showed a substantial elevation compared to severe and critical patients (
Figure 3M, 3R, 3T, 3U, 3V). Other SM species with moderate or low VIP scores (<1.4) were evaluated for biomarker potential using ROC curve analysis (
Figure S3). SM (d18:1)/16:1, 18:0, 20:0, 20:1, and 22:0 demonstrated diagnostic suitability (AUC > 0.70; p<0.0001), while SM (d18:1)/14:0 did not (
Figure S3A-G). Importantly, the production of these SM species increased with the severity of COVID-19 patients and subsequently decreased in convalescent individuals (
Figure S3H-N), thereby supporting the association between SM and disease severity. Nevertheless, further confirmation of these findings requires multivariate analysis to determine their impact on disease outcome.
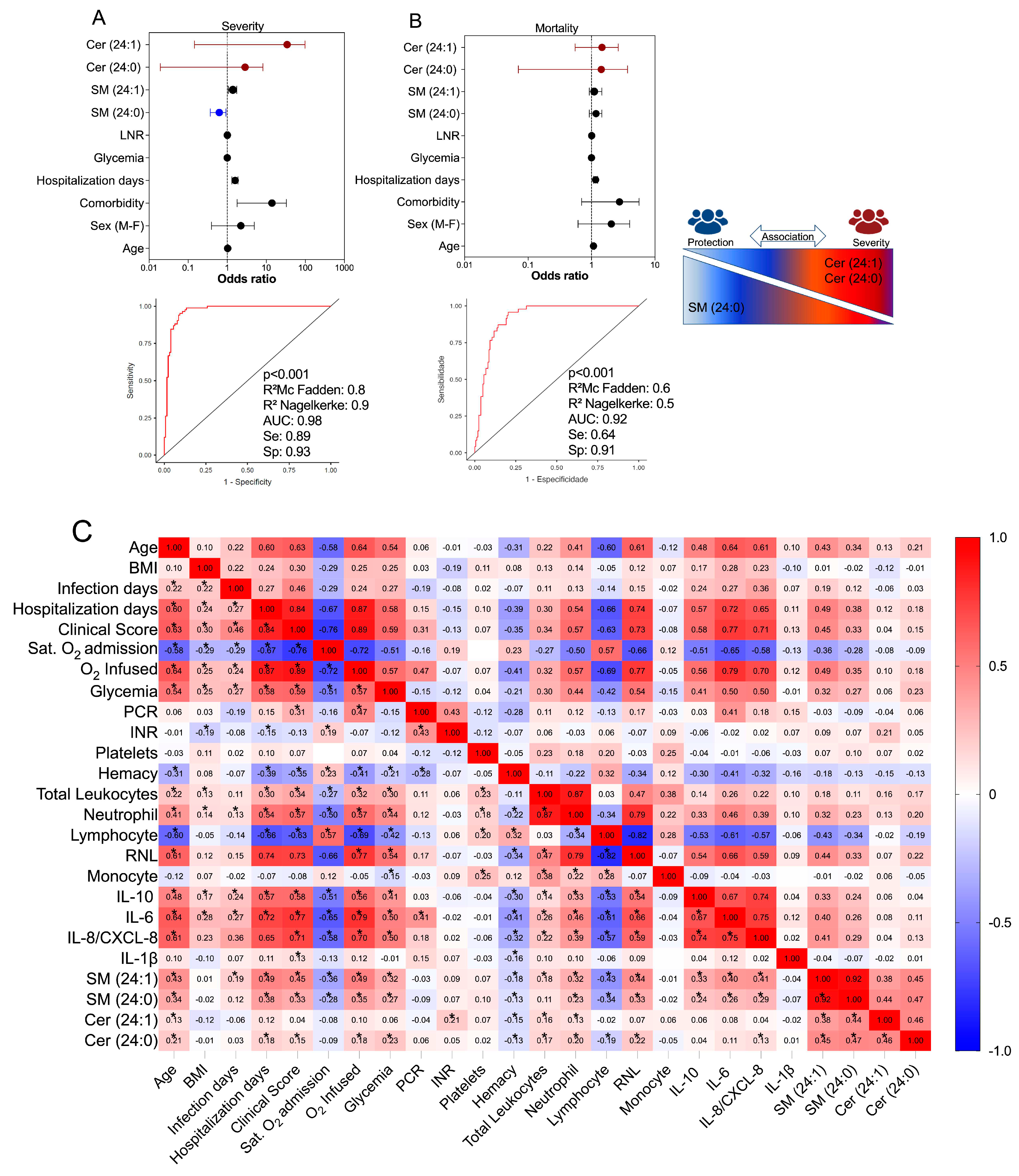
3.6. Multivariate Binomial Logistic Regression Determine the Association between Cer/SM Species and COVID-19 Clinical Severity and Mortality
The SM species previously identified as potential biomarkers for COVID-19 were further examined to determine their relationship with disease severity (
Figure 4A) and mortality outcomes (
Figure 4B) using multivariate binomial logistic regression. The statistical model was adjusted for established risk factors including age, sex, comorbidities, blood glucose, and NLR (Neutrophil-Lymphocyte Ratio). Similarly, the same statistical model was applied to assess the association of Cer species, which exhibited high production in the plasma of COVID-19 patients. Our findings indicated that the species Cer (d18:1/24:1) and Cer (d18:1/24:0) remained significant predictors of symptom intensity, with respective odds ratios of [3.82 (CI: 0.15-99), p=0.422] and [0.39 (CI: 0.01-8.20), p=0.552]. The regression model, which demonstrated statistical significance (p<0.001) and yielded high values for R
2MacFadden (0.8) and R
2Nagelkerke (0.9), along with a specificity of 0.93, sensitivity of 0.89, and an AUC of 0.98, suggested a possible protective effect of SM (d18:1/24:0) against disease severity [0.58 (CI: 0.36-0.92), p= 0.022] (
Figure 4A). Conversely, the levels of SM (d18:1/24:0) did not show a significant association with patient discharge from the hospital (
Figure 4B). The significant regression model (p<0.001), with R
2MacFadden = 0.6, R
2Nagelkerke = 0.5, specificity = 0.91, sensitivity = 0.64, and an AUC of 0.92, indicated that the Cer species (d18:1/24:1) and (d18:1/24:0) may be linked to mortality, with an odds ratio of [1.21 (CI: 0.55-2.63), p = 0.635] and [0.51 (CI: 0.07-3.69), p = 0.505] (
Figure 4B). However, no other SM species showed a substantial association with disease severity or death in COVID-19, as determined by the regression analyses.
3.5. Correlation of Value SM-Species with Immunological, Clinical, and Laboratory Markers in COVID-19
To investigate the relationship between plasma levels of SM species and clinical and laboratory features, as well as inflammatory mediator production, we employed Spearman's test. This analysis aimed to unravel the interplay between these lipid compounds and other parameters associated with the immunopathogenesis of COVID-19 (
Figure 4C). Following the identification and validation of SM (d18:1)/16:0, /24:1, /24:0, and Cer (d18:1)/24:1 and /24:0 as potential significant metabolites in relation to COVID-19, we proceeded to examine their associations. Our results revealed that both SM species and Cer species exhibited positive correlations with age, number of hospitalization days, clinical scores, glycemia, neutrophil count, NLR, and production of IL-10, IL-6, and IL-8. Conversely, they demonstrated negative correlations with oxygen saturation at the time of admission, hemacytometer count, and lymphocyte count. In terms of the strength of these associations, SM species displayed stronger and statistically significant connections to inflammatory parameters compared to Cer species. This finding suggests their involvement in the immunopathological processes of COVID-19. However, it is important to note that SM species may not necessarily contribute to disease severity. Instead, they might serve as counterpoint factors, influencing the balance of inflammation during the development of COVID-19. All the Speraman’s test statistical values were demonstrated in
Table S4.
4. Discussion
Given the intricate connection between lipids and the processes involved in the development of infectious diseases, it is plausible to explore the potential of analyzing alterations in the plasma lipid profile as a means to identify biomarkers. In this study, we employed our quantitative sphingolipidomics technology to evaluate clinical and inflammatory markers associated with plasma levels of SL and the expression of enzymes involved in the SL pathway in blood cells. Our sample consisted of a larger group of COVID-19 patients with varying degrees of severity, carefully characterized, and compared to convalescent individuals. While previous research on COVID-19 has predominantly focused on Cer, with only a limited number of studies investigating other SL metabolites, our findings present a novel discovery. We observed a progressive increase in the levels of SM class metabolites with the severity of the disease. Specifically, we identified elevated plasma levels of long-chain SM (d18:1) in severe COVID-19 patients when compared to those with mild symptoms and convalescent individuals. Interestingly, severe patients exhibited a higher proportion of SM in their overall SL output compared to patients with moderate symptoms and critical cases. This observation may indicate a pivotal point in the immunopathogenesis pathway of COVID-19, suggesting a bias towards SM synthesis in Cer metabolism.
To gain insight into the underlying mechanisms of SL procuction during COVID-19, we analyzed gene expression patterns of a comprehensive panel of enzymes involved in SL pathway in patients' peripheral blood cells, as the biochemical activity assays for these enzymes are yet to be developed. Overall, we identified altered expression of enzymes associated with De novo SL synthesis. Notably, serine-palmitoyltransferases (SPTLC1, SPTLC2, and SPTLC3) were upregulated in severe and critical forms of the disease, potentially contributing to increased production of shinganine and dHCer. Additionally, the expression of desaturase (DEGS1) was enhanced, leading to enhanced Cer production (11). The Cer molecules could then undergo modifications, such as the addition of phosphocholine by sphingomyelinase-pathway enzymes like SM synthase (SGMS1 and SGMS2) (38), which catalyze the conversion of Cer to SM. These enzymes were also upregulated in severe and critical COVID-19 patients. Alternatively, ceramidases (ASAH1), upregulated in critical illness, could deacylate Cer to form Sph (39). Notably, in severe forms of COVID-19, ceramide synthases (CERS2, CERS4, and CERT1) responsible for the synthesis of Cer from Sph (40) were upregulated. However, the expression of sphingomyelinases (SMPD2 and SMPD3), which catalyze the formation of Cer from SM (5), tended to decrease with increasing disease severity. Our data suggested an increased flux of metabolites towards Cer synthesis through multiple pathways, followed by elevated SM synthesis originating from this central pool of Cer, correlated with COVID-19 development.
SM is the primary SL present in cell membranes. Acid sphingomyelinase activity (aSMase), an enzyme found in lysosomes and in the cell membrane, is responsible for the conversion of SM to Cer. The aSMase/Cer system actively participates in host defense responses (7,41,42). SM, being a ubiquitous component of cells, is involved in various cellular activities, including cell division, proliferation, and autophagy (43). It also helps maintain a balance between pro-inflammatory and anti-inflammatory lipids, thus regulating the immune system in lung tissues (44). On the other hand, Cer predominantly participates in processes related to inflammation and damage (45). Cholesterol, SM, and phosphatidylcholines (PC) have implications for the immune system and play critical roles in macrophage activation (46), NK cell function (47), and the development and activity of T and B effector cells (48). Cer, specifically, are recognized as pro-inflammatory lipids in lung epithelial cells. In sepsis mouse models, inhibiting Cer synthesis by targeting aSMase through pharmacological means has shown potential in reducing organ damage caused by reactive oxygen species and inflammation (49,50). Additionally, Cer is involved in insulin signaling (51) and metabolic disorders like obesity in humans (52). This observation is intriguing since comorbidities such as obesity and type 2 diabetes are associated with worse outcomes in COVID-19. In this context, we observed a tendency to privilege SM synthesis than Cer in severe patients, but this event was not similarly observed in critical patients, suggesting that SM could be involved in a putative host mechanism to recovery the disease development from a worst prognostic.
Our findings also indicate a decrease in total SM counterparts and an increase in sphinganine, Sph, and Cer species in convalescent patients, consistent with reports of elevated levels of these SL species in symptomatic individuals compared to asymptomatic ones (23). This suggests a potential role involved in inflammation control. Moreover, we conducted ROC and correlation analyses to assess the relationship between these SL classes and the severity of COVID-19. We discovered that total plasma levels of SM with an AUC greater than 0.80 (p<0.001) have the potential to identify clinically severe symptoms. In fact, our findings demonstrated that SM (d18:1)/16:0, /22:1, /23:0, /23:1, /24:0, /24:1, /25:0, /25:1, /26:0, and /26:1 may serve as predictors of a poor prognosis (AUC > 0.7) for COVID-19. Additionally, the results demonstrated a moderate positive correlation between SM-total and inflammation markers in COVID-19, with a high disease clinical score. It is crucial to identify the mechanism by which SARS-CoV-2 increases SL levels. SM has the ability to bind with cholesterol and form lipid rafts, potentially facilitating virus entry into cellular surfaces (53–55). Conversely, depletion of host and viral SM has been observed to inhibit influenza virus infection (56). On the other hand, previous studies (20,22,23,57) have reported elevated levels of Cer and dHCer during COVID-19.
We have identified novel potential SM biomarkers that could predict the progression of COVID-19 towards severe symptoms. Specifically, SM species (d18:1/24:0) and (d18:1/24:1) were evaluated as potential severity biomarkers in our study. To account for confounding variables such as age, sex, comorbidities, body mass index (BMI), hospitalization days, glycemia, and the neutrophil-lymphocyte ratio (NLR), we used multivariate binomial logistic regression to assess their association with disease severity. Interestingly, our analysis revealed a tendency protective effect mediated by SM (24:0) against severe forms of COVID-19, as well as a risk factor mediated by Cer (24:1), based on a significant regression model (AUC=0.98; p<0.001). In fact, elevated levels of SM (24:1) were positively correlated with clinical and inflammatory indicators in COVID-19, including the clinical score, hospitalization days, neutrophil count, as well as the production of IL-10, IL-6, and IL-8. On the other hand, the correlation between Cer (24:1) and these indicators was weaker. These systemic SM species hold promise as potential indicators or targets for preventing disease progression and exploring new treatments. They may have direct effects on regulating inflammation, coagulopathy, and the cytokine storm, which are defining characteristics of severe COVID-19 (58,59). Furthermore, the combination of SL species production and expression of enzymes involved in SL metabolism shows diagnostic potential and can assist in prioritizing individuals for therapy with newly developed antiviral medications (60,61).
Our study distinguishes itself from previous research by including a larger and more extensively characterized cohort of COVID-19 patients, encompassing healthy controls and individuals in the convalescent phase. However, it is crucial to acknowledge certain limitations that should be addressed in future investigations. Firstly, we have not accounted for the impact of chronic disorders and secondary infections, which could potentially contribute to the dysregulation of SL metabolism. These factors may influence the observed SL patterns and should be considered in future studies. Additionally, it is important to note that our sample is regionally limited, which may introduce geographic and demographic biases. To establish the predictive effect of SL biomarkers in critically ill individuals with COVID-19, it is necessary to conduct cohort studies with broader representation and diverse populations. Furthermore, to gain a comprehensive understanding of SL dynamics, it would be advantageous to assess the SL profile over the course of the disease. This longitudinal analysis would allow us to examine how SL patterns evolve and whether early alterations can serve as predictive markers for disease progression and outcome. Such investigations could provide valuable insights into the pathogenesis of COVID-19 and aid in the development of targeted interventions. By considering additional factors, expanding the scope of the study population, and conducting longitudinal analyses, we can enhance our understanding of the predictive potential of SL patterns and their implications for patient management and therapeutic interventions.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Study conceptualization, D.M.T. and C.A.S; Coordinated the samples collection and ethics committee approval, C.A.S., C.R.B.C., E.M.S.R., A.P.M.F., A.L.V; Supervised the collection of domiciliary and healthy individuals, C.A.S; Data management, collection and analysis from hospitalized patients, M.M.P., V.E.P., L.F.C., A.M.D., O.F., F.A.M.O., M.R.F., R.S.P., F.C.V., G.G.G., J.J.R.d.R., O.F; Applied the clinical questionnaire and collected samples, P.V.d.S.-N., J.C.S.d.C., V.E.P., C.N.S.O., M.M.P., D.M.T., I.C.-G., J.G.M.A., T.M.F., G.P.C; Collected demographic/clinical data and classified patient clinical scores, M.M.P., V.E.P., P.V.d.S.-N., M.D.-B., D.M.T., N.T.N.; Performed sample processing and storage, P.V.d.S.-N., T.F.C.F.-S., D.M.T., V.E.P., J.C.S.d.C., C.N.S.O., J.G.M.A.,G.R.C.,L.C.R; Measured cytokine/protein, P.V.d.S.-N., C.N.S.O., T.F.C.F.-S., V.E.P., M.M.P., D.M.T., and C.R.B.C; Bioinformatic analyses of the transcriptome, S.R.M., T.F.C.F.-S., C.A.F.; Measured sphingolipids, D.M.T., G.R.C., J.C.S.d.C., P.V.d.S.-N., P.N.A, T.V.D.F; Statistical analysis, P.V.d.S.-N., D.M.T., C.A.F.; Drafted the manuscript, D.M.T., P.V.d.S.-N., A.F.L.V, P.N.A and C.A.S; Provided materials, resources and equipment, L.H.F., V.L.D.B., C.R.B.C., R.T.S.; Coordination and idealization of the ImmunoCovid project and revision of the manuscript, C.A.S., S.R.C.M., E.M.S.R., A.P.M.F., I.K.F.M.S., V.L.D.B., C.R.B.C., M.D.-B., A.M., R.T.S. and L.H.F.; Supervised the project, C.A.S. and L.H.F.; Coordination and idealization of the AEROBICOVID project for convalescent samples collection, A.A.T., C.A.S.; All authors revised the manuscript and agreed with the published version of the manuscript.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo–FAPESP (grants #2020/05207-6, #2014/07125-6, and #2015/00658-1 for L.H.F., #2020/08534-8 for M.M.P.; grant #2020/05270-0 for V.L.D.B and grant #2021/04590-3 for C.A.S.); and grant #R01 HL144478 NIH/NHLBI and grant #I01 BX001786 Department of Veterans Affairs for R.T.S. Additional support was provided by the National Council for Scientific and Technological Development (CNPq), the Coordination for the Improvement of Higher Educational Personnel (CAPES-Finance Code 001)), Fundação de apoio à Universidade de São Paulo–FUSP by USP VIDA program. RES. N. 006/2020 - POSGRAD 2020 FAPEAM, Fundação de Amparo à Pesquisa do Estado do Amazonas-FAPEAM for D.M.T and C.A.S, and from Conselho Nacional de Desenvolvimento Científico e Tecnológico CNPq grant numbers: (CNPq grants 312606/2019-2 for M.D.B, 303259/2020-5 for L.H.F., and #309583/2019-5 for C.R.B.C.) and Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM) POSGRAD (Resolution 006/2020). The AEROBICOVID project received funding from the 'USP Vida' Project (code – 3518/2020) and Integrated Research Projects in Strategic Areas (PIPAE) from the Dean of Research-USP (2021.1.10424.1.9).
Data Availability
Normalized transcriptomic data from the participants' whole blood leukocytes and associated metadata were deposited on the ArrayExpress database at EMBL-EBI (
www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-11240.
Acknowledgments
The authors acknowledge the support of the ICU team of doctors, nurses, physiotherapists, and the collaboration of Hospital Santa Casa de Misericórdia of Ribeirão Preto and Hospital São Paulo of Ribeirão Preto; the laboratory support from Supera Parque-Innovation and Technology Park-Ribeirão Preto/SP for testing infection by SARS-CoV-2 in healthy volunteers; and the valuable contribution by Municipal Health Department of Ribeirão Preto city and Analysis Service Clinics (SAC) from Faculdade de Ciências Farmacêuticas de Ribeirão Preto–USP. We are grateful to Fabiana R. de Moraes for helping with flow cytometry acquisition and data analysis; and Professors Victor Hugo Aquino Quintana, Márcia Regina von Zeska Kress, and Marcia Eliana da Silva Ferreira for sharing the viral BS-2 lab.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ImmunoCovid Consortium group
Angelina L Viana1, Ingryd Carmona-Garcia1, Nicola T Neto1, Ana C. Xavier1, Giovanna da S. Porcel1, Isabelle C. Guarneri1, Kamila Zaparoli1, Caroline T. Garbato1, Ângelo A.F. Júnior1; Alessandro P. de Amorim2, Leticia F Constant2, Dayane P. da Silva2, Rafael C. da Silva2, Rita de C.C. Barbieri3, Cristiane M. Milanezi4, Cassia F.S.L. Dias5, Lilian C. Rodrigues5 and Talita M. Fernandes1
1Escola de Enfermagem de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, SP, Brazil
2Hospital Santa Casa de Misericórdia de Ribeirão Preto, Ribeirão Preto, SP, Brazil.
3Hospital São Paulo, Ribeirão Preto, SP, Brazil.
4Departamento de Bioquímica e Imunologia. Faculdade de Medicina de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, SP, Brazil.
5Faculdade de Ciências Farmacêuticas de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, SP, Brazil.
References
- Gandhi RT, Lynch JB, del Rio C. Mild or Moderate Covid-19. New England Journal of Medicine (2020) 383:1757–1766. [CrossRef]
- Murthy S, Gomersall CD, Fowler RA. Care for Critically Ill Patients With COVID-19. JAMA (2020) 323:1499–1500. [CrossRef]
- Erdinc B, Sahni S, Gotlieb V. Hematological manifestations and complications of COVID-19. Adv Clin Exp Med (2021) 30:101—107. [CrossRef]
- Costanzo M, de Giglio M, Roviello G. SARS-CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other Drugs for the Treatment of the New Coronavirus. Curr Med Chem (2020) 27:.
- Carpinteiro A, Edwards MJ, Hoffmann M, Kochs G, Gripp B, Weigang S, Adams C, Carpinteiro E, Gulbins A, Keitsch S, et al. Pharmacological Inhibition of Acid Sphingomyelinase Prevents Uptake of SARS-CoV-2 by Epithelial Cells. Cell Rep Med (2020) 1:100142. [CrossRef]
- Schneider-Schaulies J, Schneider-Schaulies S. Sphingolipids in viral infection. Biol Chem (2015) 396:585–595. doi: doi:10.1515/hsz-2014-0273.
- Simonis A, Schubert-Unkmeir A. The role of acid sphingomyelinase and modulation of sphingolipid metabolism in bacterial infection. Biol Chem (2018) 399:1135–1146. doi: doi:10.1515/hsz-2018-0200.
- Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol (2018) 19:175–191. [CrossRef]
- Gomez-Larrauri A, Presa N, Dominguez-Herrera A, Ouro A, Trueba M, Gomez-Muñoz A. Role of bioactive sphingolipids in physiology and pathology. Essays Biochem (2020) 64:579–589. [CrossRef]
- Hussain G, Wang J, Rasul A, Anwar H, Imran A, Qasim M, Zafar S, Kamran SKS, Razzaq A, Aziz N, et al. Role of cholesterol and sphingolipids in brain development and neurological diseases. Lipids Health Dis (2019) 18:26. [CrossRef]
- Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer (2018) 18:33–50. [CrossRef]
- Vitner EB, Achdout H, Avraham R, Politi B, Cherry L, Tamir H, Yahalom-Ronen Y, Paran N, Melamed S, Erez N, et al. Glucosylceramide synthase inhibitors prevent replication of SARS-CoV-2 and influenza virus. Journal of Biological Chemistry (2021) 296:100470. [CrossRef]
- Bezgovsek J, Gulbins E, Friedrich S-K, Lang KS, Duhan V. Sphingolipids in early viral replication and innate immune activation. Biol Chem (2018) 399:1115–1123. doi: doi:10.1515/hsz-2018-0181.
- Wigger D, Gulbins E, Kleuser B, Schumacher F. Monitoring the Sphingolipid de novo Synthesis by Stable-Isotope Labeling and Liquid Chromatography-Mass Spectrometry. Front Cell Dev Biol (2019) 7:. [CrossRef]
- Kurz J, Parnham MJ, Geisslinger G, Schiffmann S. Ceramides as Novel Disease Biomarkers. Trends Mol Med (2019) 25:20–32. [CrossRef]
- Gandy KAO, Obeid LM. “Regulation of the Sphingosine Kinase/Sphingosine 1-Phosphate Pathway BT - Sphingolipids in Disease.,” In: Gulbins E, Petrache I, editors. Vienna: Springer Vienna (2013). p. 275–303. [CrossRef]
- Pyne S, Adams DR, Pyne NJ. Sphingosine 1-phosphate and sphingosine kinases in health and disease: Recent advances. Prog Lipid Res (2016) 62:93–106. [CrossRef]
- Törnquist K, Asghar MY, Srinivasan V, Korhonen L, Lindholm D. Sphingolipids as Modulators of SARS-CoV-2 Infection. Front Cell Dev Biol (2021) 9:. [CrossRef]
- Vitner EB, Avraham R, Politi B, Melamed S, Israely T. Elevation in sphingolipid upon SARS-CoV-2 infection: possible implications for COVID-19 pathology. Life Sci Alliance (2022) 5:e202101168. [CrossRef]
- Marín-Corral J, Rodríguez-Morató J, Gomez-Gomez A, Pascual-Guardia S, Muñoz-Bermúdez R, Salazar-Degracia A, Pérez-Terán P, Restrepo MI, Khymenets O, Haro N, et al. Metabolic Signatures Associated with Severity in Hospitalized COVID-19 Patients. Int J Mol Sci (2021) 22:. [CrossRef]
- Lee JW, Su Y, Baloni P, Chen D, Pavlovitch-Bedzyk AJ, Yuan D, Duvvuri VR, Ng RH, Choi J, Xie J, et al. Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nat Biotechnol (2022) 40:110–120. [CrossRef]
- Torretta E, Garziano M, Poliseno M, Capitanio D, Biasin M, Santantonio TA, Clerici M, Lo Caputo S, Trabattoni D, Gelfi C. Severity of COVID-19 Patients Predicted by Serum Sphingolipids Signature. Int J Mol Sci (2021) 22:. [CrossRef]
- Janneh AH, Kassir MF, Dwyer CJ, Chakraborty P, Pierce JS, Flume PA, Li H, Nadig SN, Mehrotra S, Ogretmen B. Alterations of lipid metabolism provide serologic biomarkers for the detection of asymptomatic versus symptomatic COVID-19 patients. Sci Rep (2021) 11:14232. [CrossRef]
- Diagnosis and treatment protocol for novel coronavirus pneumonia (Trial version 7). Chin Med J (Engl) (2020) 133:. [CrossRef]
- Wan S, Xiang Y, Fang W, Zheng Y, Li B, Hu Y, Lang C, Huang D, Sun Q, Xiong Y, et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. J Med Virol (2020) 92:797–806. [CrossRef]
- Reimann C-M, Gräler M. Extraction and Quantification of Sphingosine 1-Phosphate (S1P). Bio Protoc (2016) 6:. [CrossRef]
- Andréani P, Gräler MH. Comparative quantification of sphingolipids and analogs in biological samples by high-performance liquid chromatography after chloroform extraction. Anal Biochem (2006) 358:. [CrossRef]
- Shaner RL, Allegood JC, Park H, Wang E, Kelly S, Haynes CA, Sullards MC, Merrill AH. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J Lipid Res (2009) 50:. [CrossRef]
- R Development Core Team. R a language and environment for statistical computing : reference index. R Foundation for Statistical Computing (2010).
- RStudio Team. RStudio: Integrated development environment for R. RStudio: Integrated Development Environment for R (2019).
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol (2004). [CrossRef]
- Benjaminit Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. (1995). 289–300 p.
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. Journal of Computational Biology (2005) 12:. [CrossRef]
- Hellemans J, Mortier G, de Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol (2008) 8:. [CrossRef]
- Epskamp S, Cramer AOJ, Waldorp LJ, Schmittmann VD, Borsboom D. qgraph: Network Visualizations of Relationships in Psychometric Data. (2012). http://www.jstatsoft.org/.
- Szymańska E, Saccenti E, Smilde AK, Westerhuis JA. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics (2012) 8:. [CrossRef]
- Szymańska E, Saccenti E, Smilde AK, Westerhuis JA. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics (2012) 8:3–16. [CrossRef]
- Mühle C, Bilbao Canalejas RD, Kornhuber J. Sphingomyelin Synthases in Neuropsychiatric Health and Disease. Neurosignals (2019) 27:54—76. [CrossRef]
- Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind JS, Spiegel S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature (1996) 381:800–803. [CrossRef]
- Sassa T, Hirayama T, Kihara A. Enzyme Activities of the Ceramide Synthases CERS2–6 Are Regulated by Phosphorylation in the C-terminal Region*. Journal of Biological Chemistry (2016) 291:7477–7487. [CrossRef]
- Coant N, Sakamoto W, Mao C, Hannun YA. Ceramidases, roles in sphingolipid metabolism and in health and disease. Adv Biol Regul (2017) 63:122–131. [CrossRef]
- Prakash H, Upadhyay D, Bandapalli OR, Jain A, Kleuser B. Host sphingolipids: Perspective immune adjuvant for controlling SARS-CoV-2 infection for managing COVID-19 disease. Prostaglandins Other Lipid Mediat (2021) 152:106504. [CrossRef]
- Harvald EB, Olsen ASB, Færgeman NJ. Autophagy in the light of sphingolipid metabolism. Apoptosis (2015) 20:658–670. [CrossRef]
- Ghidoni R, Caretti A, Signorelli P. Role of Sphingolipids in the Pathobiology of Lung Inflammation. Mediators Inflamm (2015) 2015:487508. [CrossRef]
- BLITTERSWIJK WJ van, LUIT AH van der, VELDMAN RJ, VERHEIJ M, BORST J. Ceramide: second messenger or modulator of membrane structure and dynamics? Biochemical Journal (2003) 369:199–211. [CrossRef]
- Spadaro F, Cecchetti S, Fantuzzi L. Macrophages and Phospholipases at the Intersection between Inflammation and the Pathogenesis of HIV-1 Infection. Int J Mol Sci (2017) 18:. [CrossRef]
- Chan CJ, Smyth MJ, Martinet L. Molecular mechanisms of natural killer cell activation in response to cellular stress. Cell Death Differ (2014) 21:5–14. [CrossRef]
- Bai A, Guo Y. Acid sphingomyelinase mediates human CD4+ T-cell signaling: potential roles in T-cell responses and diseases. Cell death & disease (2017) 8:e2963. [CrossRef]
- Peng H, Li C, Kadow S, Henry BD, Steinmann J, Becker KA, Riehle A, Beckmann N, Wilker B, Li P-L, et al. Acid sphingomyelinase inhibition protects mice from lung edema and lethal Staphylococcus aureus sepsis. J Mol Med (2015) 93:675–689. [CrossRef]
- Chung H-Y, Witt CJ, Jbeily N, Hurtado-Oliveros J, Giszas B, Lupp A, Gräler MH, Bruns T, Stallmach A, Gonnert FA, et al. Acid Sphingomyelinase Inhibition Prevents Development of Sepsis Sequelae in the Murine Liver. Sci Rep (2017) 7:12348. [CrossRef]
- Wigger D, Schumacher F, Schneider-Schaulies S, Kleuser B. Sphingosine 1-phosphate metabolism and insulin signaling. Cell Signal (2021) 82:109959. [CrossRef]
- Chaurasia B, Summers SA. Ceramides in Metabolism: Key Lipotoxic Players. Annu Rev Physiol (2021) 83:303–330. [CrossRef]
- Lu Y, Liu DX, Tam JP. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem Biophys Res Commun (2008) 369:344–349. [CrossRef]
- Noriyuki O, Masafumi S, Kyoko S, Kiyoko O, Yoshio M, Kentaro H, Makoto T. Both Sphingomyelin and Cholesterol in the Host Cell Membrane Are Essential for Rubella Virus Entry. J Virol (2017) 92:10.1128/jvi.01130-17. [CrossRef]
- Radenkovic D, Chawla S, Pirro M, Sahebkar A, Banach M. Cholesterol in Relation to COVID-19: Should We Care about It? J Clin Med (2020) 9:. [CrossRef]
- Audi A, Soudani N, Dbaibo G, Zaraket H. Depletion of Host and Viral Sphingomyelin Impairs Influenza Virus Infection. Front Microbiol (2020) 11:. [CrossRef]
- Abusukhun M, Winkler MS, Pöhlmann S, Moerer O, Meissner K, Tampe B, Hofmann-Winkler H, Bauer M, Gräler MH, Claus RA. Activation of Sphingomyelinase-Ceramide-Pathway in COVID-19 Purposes Its Inhibition for Therapeutic Strategies. Front Immunol (2021) 12:. [CrossRef]
- Creeden JF, Imami AS, Eby HM, Gillman C, Becker KN, Reigle J, Andari E, Pan ZK, O’Donovan SM, McCullumsmith RE, et al. Fluoxetine as an anti-inflammatory therapy in SARS-CoV-2 infection. Biomedicine & Pharmacotherapy (2021) 138:111437. [CrossRef]
- Sukhatme VP, Reiersen AM, Vayttaden SJ, Sukhatme V V. Fluvoxamine: A Review of Its Mechanism of Action and Its Role in COVID-19. Front Pharmacol (2021) 12:. [CrossRef]
- Whitley R. Molnupiravir — A Step toward Orally Bioavailable Therapies for Covid-19. New England Journal of Medicine (2021) 386:592–593. [CrossRef]
- Wang Z, Yang L. In the age of Omicron variant: Paxlovid raises new hopes of COVID-19 recovery. J Med Virol (2022) 94:1766–1767. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).