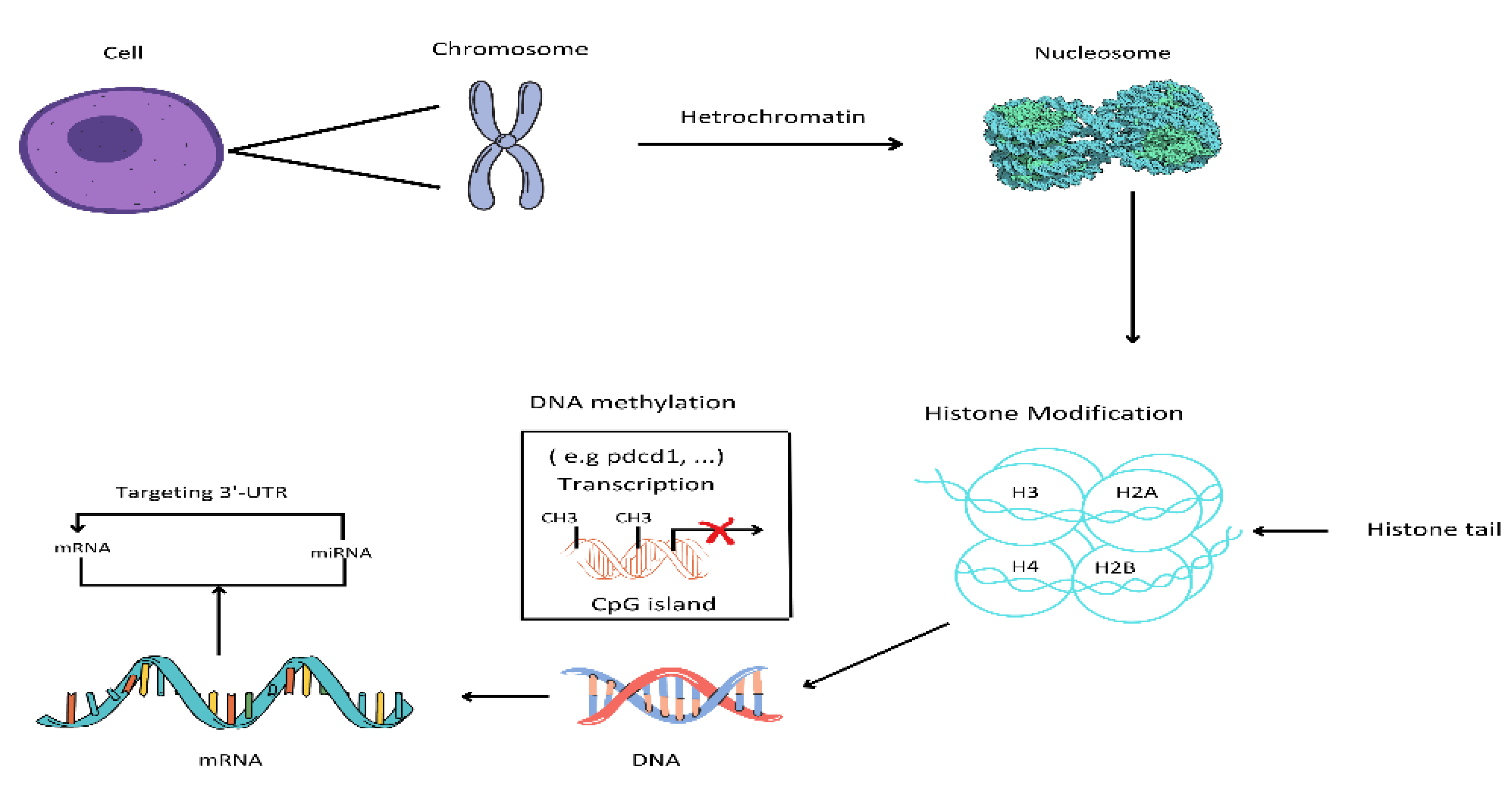
1. Introduction
Epigenetic alterations are heritable and stable changes in the gene expression that are independent of changes in the underlying DNA sequence [
1]. This phenomenon underlies the inheritance of cellular phenotypes and the ability of cells with identical genetic material to differentiate into various types of cells. Epigenetics is an important phenomenon for controlling expression patterns during development, cell cycle, and in response to biological or environmental changes [
2]. These alterations mediate environmental effects on gene expression and influence genetic disease risks, providing a valuable tool for understanding how genetic and environmental factors interact to confer disease risk [
3]. Environmental factors including pollution, toxicants, inflammation, and nutrients can encourage epigenetic alterations, which in turn can lead to various disorders, including autoimmune disorders, diabetes, and cancers [
2,
4]. Epigenetic alterations include noncoding RNAs, histone modifications, and DNA methylation.
DNA methylation is an important epigenetic mechanism in mammals, which involves the formation of 5-methylcytosine through the transfer of methyl group onto the cytosine C5 position. The main fundtion of DNA methylation is the regulation of gene expression by employing proteins involved in the repression of genes or through inhibition of transcription factor bindings to the DNA [
5]. These modifications play a significant role in sepsis and viral infections. Recent studies have shown that epigenetic alterations have an impact on immune responses [
6,
7]. The epigenome plays a critical role in the normal development of lymphocytes and the regulation of immune responses [
8]. Immune evasion is an approach used by tumors and pathogens to avoid the immune response of the host, promoting their growth and transmission to new hosts [
9]. The mechanisms of immune evasion in tumors involve multiple factors that promote antiapoptotic processes, as well as the secretion of immune-suppressive factors in the tumor micro-environment. These factors include transforming growth factor (TGF)-β, interleukin (IL)-10, indoleamine 2,3-dioxygenase, and vascular endothelial growth factor (VEGF), which contribute to the loss of tumor antigen expression and the potentiation of immune-suppressive lymphocytes such as tumor-associated macrophages, regulatory T-cells, and myeloid suppressive cells. Additionally, the expression of inhibitory molecules plays a role in immune evasion [
10].
T-cell malignancies are highly malignant and aggressive diseases with poor clinical outcomes. Epigenetic modifications play a critical role in their development and pathogenesis by regulating gene expression and signal transduction [
11]. Epigenetic alteration leads to immune evasion in several ways, including changes in DNA methylation patterns. DNA methylation involves the addition of a methyl group to the cytosine base of DNA. Alterations in DNA methylation such as hypomethylation or hypermethylation can lead to immune evasion. Furthermore, the addition or removal of chemical groups to histone proteins can alter their structure and gene expression, leading to changes in immune responses. Histone deacetylation and altered gene expression also contribute to immune evasion [
12]. Non-coding RNAs have also been found to play a key role in the alteration of immune responses. Epigenetic changes, inherited across generations can contribute significantly to the alteration of immune responses. The term used for this purpose is transgenerational inheritance [
13]. Epigenetic alterations in tumor suppressor genes and oncogenesis contribute towards carcinogenesis. Understanding the genetic and epigenetic components of immune evasion is crucial. Studies on the modulation of epigenetics and immune checkpoints have shown an interaction between immune modulation and epigenetic alterations [
14]. This review article provides a comprehensive summary of the latest research on the epigenetic modifications that can result in immune evasion and the development of T-cell malignancies. The article is of great significance as it highlights the emerging importance of epigenetic mechanisms in T-cell malignancies and their potential role in the development of novel therapeutic approaches.
2. Epigenetic Alterations
In the modern era, Human Genome Project is one of the most remarkable accomplishments which sheds light on various surprising mechanisms encoded within the genome. Among these mechanisms, epigenetics has emerged as a crucial factor which represent the chemical interactions and regulatory systems governing genetic code expression. [
15]. Currently, regulation of tissue-specific expressions, X chromosome inactivation, or genomic imprinting are the main roles of epigenetic modifications. Additionally, the variability of epigenetic alterations in human disorders particularly cancers has further highlighted the significance of epigenetic regulation mechanisms [
16]. Recent breakthroughs in understanding these mechanisms have refined the definition of epigenetics from a process of the development of a fertilized zygote into mature organisms, to an emphasis on the heritability of traits and gene expression mechanisms [
17]. Epigenetics is now understood to control both normal and abnormal events in organisms [
18]. One of the main activators of epigenetic alterations is environmental stimuli. Chronic inflammation and aging has been identified as activator of aberrant DNA methylation [
19,
20]. Cigarette smoking is also known to induce invitro abnormal DNA methylation [
21]. In the case of cultured epithelial cells of mammals, estrogen treatment is also a well-known abnormal DNA methylation accelerator [
22].
3. Epigenetics of Normal Cells
In mammals, epigenetics is an essential mechanism for the development of normal cells and the maintenance of gene expression (tissue-specific) patterns [
23]. Chromatin is a set of nucleosomes, that consist of 146 base pairs of wrapped DNA around an octant of 4 core histone proteins. these are H2A, H2B, H3 and H4 [
24]. Epigenetics that modify the structure of chromatin can be classified into 4 classes. These categories include DNA methylation, covalent histone modifications, non-covalent mechanisms, and the non-coding RNAs [
25].
4. Epigenetics of Abnormal Cells
4.1. Changes in DNA Methylation
The epigenetic profile of cancer cells is different from that of normal cells. There are two types of changes in DNA methylation. Hypomethylation (DNA with less amount of DNA methylation than normal cells) and hypermethylation ( DNA with more amounts of DNA methylation). In cancer cells, across much of the genome, a decreased DNA methylation occurs [
27]. This decreased DNA methylation consequences in the alteration of many of the activities of the genes. This is because methylation is linked to low gene activities but in the case of hypomethylation the activity of genes affected is increased [
28].
Genes that control cell differentiation and growth typically display less methylation and higher levels of activity, making them prime candidates for the development of cancer [
29]. On the other hand, hypermethylation-associated silencing of tumor suppressor genes is a more limited phenomenon that can occur at specific points, or hotspots in the genome. In cancer cells, DNA hypermethylation primarily affects tumor suppressor genes which lead to decreased gene activity and subsequent cancer development [
30]. Such hypomethylated and hypermethylated cells tend to develop and grow faster than the normally methylated cells. They grow in abundance and take over the whole population [
31]. However, the specific DNA methylation profiles that lead to cancer development can vary greatly between different types of cancer. For instance, while the BRCA1 gene is hypermethylated in ovarian and breast tumors, it is demethylated in other types of tumors [
32]. DNA methylation profile in both normal and tumor cells is shown in
Figure 1.
5. Immune Evasion Due To DNA Methylation
DNA methylation leads to the silencing of transcription genes, either by recruiting the chromatin-modifying protein or directly inhibiting the transcription factors binding to suppress the gene expression and chromatin structure [
34]. Consequently, malignant cells evade detection and attack by natural killer cells and T-cells of the immune system [
35]
. One of the main immune evasion mechanisms through DNA methylation is the suppression of genes encoding cancer antigens. These antigens are recognized by the immune cells as a foreign agent, after being expressed by the tumor cells [
36]. These foreign agents trigger immune responses. However, the silencing or suppression of these antigen-coding genes through DNA methylation renders tumor cells invisible to the immune system, as these genes do not express antigens that can activate the immune system [
37]
.
In a study by Jung et al., they proposed that DNA methylation aberrations play a crucial role in determining how tumors respond to the host immune system. They suggested that such aberrations help highly mutated and rapidly dividing cancer cells to evade the immune system and resist immune therapies. The strategic mechanism involves the heterochromatin formation which is then coupled with an advanced level of methylation loss. This in turn provides support to cancer evolution as they help the cancer cells to evade the immune cells and aid in the fitness of such cells [
38]. In another study by Li et al., they reported that the inactivation of histone H3k36, methyltransferase NSD1 induces DNA hypomethylation. This results in reduced immune infiltration of tumor cells. Silencing of genes if innate immunity occurs upon loss of NSD1, these include Type 3 interferon IFNLR1 receptor, through H3K36diemethylation depletion gain of tri methyl H3K27 [
39].
6. Downregulation of Antigen Processing
DNA methylation also leads to the deregulation of genes responsible for the presentation and processing of genes. Macrophages and dendritic cells rely on the genes to present and process cancerous antigens to T-cells [
5]. On silencing of these genes, cancer cells become resistant to the immune elimination and recognition system. DNA methylation also causes a suppression of chemokines and cytokines expression, which are a necessity of immune cell recruiting to the site of tumor growth or infection [
40]. If these signals are not provided to the immune system, it is unable to locate and recognize the tumor cells in the body. This eventually leads to cancer progression and immune evasion [
41]. This powerful mechanism of DNA methylation is used by cancer cells to evade the immune destruction and recognition process. It is important to identify and target the immune evasion mediated by DNA methylation, this might help in developing new therapies for cancer and the immune system's power to fight against such cells [
42].
Li et al., in 2021, identified that aberrant DNA methylation of PPP2R2B results in tumor suppression in triple-negative breast cancer (TNBC). Analysis was done schematically through bioinformatics. Pieces of evidence obtained through transcriptome, in-vitro experiments, and genome supported that downregulation of PPP2R2B could assist TNBC cells in immune evasion via suppressing the immune response against tumors. Inclusively, PPP2R2B could be a favorable biomarker in the case of TNBC. It also helps in predicting responses to immunotherapies and direct modified TNBC treatment strategies [
39].
7. Modification in Histone
Histone methyltransferases (HMTs) carry out the methyl group addition to the histones. The histone demethylase (HDMs) function is opposite to the HMTs. In case of abnormal cell development, methyl groups are placed at the wrong spot when HMT functions are altered, this will lead to the silencing of tumor-suppressing genes[
43]. In the same way, HDMS activity is also affected and led to increased oncogenic activity. Histone acetyl markers are lost in the epigenetics of cancer cells because of increased deacetylation of histone [
44]. The change of this protein will modify the link between DNA and histones and the shape of complexes of DNA and histones. The methylation effect on the activities of genes varies in according to the amino acid variability. Methyl marks are either regarded as repressing or activating based on their dependence on gene activity. But there is an interesting turnover that few HDMs can eradicate both repressing and activating marks [
45].
8. Modification of Histones that Cause Immune Evasion
Immune evasion is also affected by histone modification through an alteration of gene expression involved in immune evasion. These include chemokines, antigen-presenting molecules, and encoding cytokines. Changing the DNA accessibility and transcription binding factors to a certain genetic point. The expression of genes is regulated by the histone modifications in two forms: either suppression of immune-responsive genes or their promotion [
46].
Steinbach and Riemer have narrated about the human papillomavirus (HPV) immune evasion mechanism. HPV active immune evasion is mediated intracellularly through disturbed functions of proteins and altered gene expression and interfering extracellularly with immune networks from antigen-presenting cells to T cells. Suppressed IFN and cGAS-STING reaction inhibits the antiviral state induction. Downregulation of adhesion molecules and TLRs plus decreased chemokine production by infected keratinocytes pave the path of reduced antigen-presenting cell attraction and thus consequence in a delayed immune response to Anti-HPV. Interference with antigen processing and low protein expressions add a lot to decreased HPV epitope performance. All these mechanisms help in the persistence of HPV for a long time until it completes its life cycle. But this in turn increases lesion persistence risk and malignant transformation onset [
47]
.
9. Inhibition of Immune Responses
Histone modification can inhibit immune responses by promoting the gene expression involved in inhibiting the immune mechanism [
48]. For example, tumor cells can alter histones to overpower the gene expression which encodes the major histocompatibility complex molecules (MHC), which are crucial for presenting the foreign antigen to the T-cells [
49]. Histone modification will lead to a suppression of MHC encoding gene expression, so tumor cells will escape recognition from the immune system, and will be avoided by the T-cell destructive mechanism. In addition to this, modification in histones by tumors for the promotion of expression of immune response molecules. For example, PD-L1 inhibits the activation of T cell and hence prevent the immune response [
48]. A blockage of T-cell functionality can help lymphomas to evade the recognition and destruction of the immune system [
50].
10. Inhibition of Immune Activation
Histone modifications can also evade immune responses by suppressing the immune-activating gene expressions. Such as, in the case o autoimmune disorders, histone modifications can overpower the chemokines and cytokines expressions, which are responsible for promoting immune responses [
51]. This will result in a reduction of immune responses and tissue damage will also be reduced, which allows the disorder to persist in the body. Moreover, in case of viral infections, viruses alter histone and this alteration will suppress interferon-stimulated expression which is crucial for the antiviral response of the immune system. Thus viruses evade the immune responses, detection, and destruction by the T-cells [
48].
Histone deacetylases (HDACs eliminate the acetyl group from non-histone and histone proteins and play the role of transcriptional repressor. Yeon et al., in 2020 in their study predicted that HDACs are frequently dysregulated in malignancies, regulation of MAPK signaling, progression of cancer cells, and reaction to several anti-tumor drugs. HDACs have been known to regulate the PD-1/PD-L1 expression and genes that contribute towards immune evasion [
52]. In short, immune inhibition, and activation-related gene expression can be regulated by histone modifications. Considering the contribution of histone modifications to immune evasion is an important issue and this will contribute a lot to the development of new autoimmune and cancer therapies plus mechanism enhances host and virus interactions [
53].
11. Micro RNAs (mRNAs) and Immune Evasion
miRNA are short, endogenous 19-25 nucleotide long, non-coding RNAs that perfectly or partially match the target messenger RNA 3′ untranslated regions (3′UTR) for the regulation of expression of genes through post-transcriptional silencing and degradation of targeted mRNAs [
54]. miRNAs have a role in all the processes of life such as cell growth, apoptosis, regulation of cell cycle, stress reaction, and cell differentiation as 30 percent of the human genes are directly targeted by miRNAs, this has been confirmed by the experimental studies and bioinformatics. Sanger miRNA's latest registry annotates that there are more than 800 miRNAs in humans and several more miRNAs are surely be identified in the future[
55].
In normal cells, just like other protein-coding genes, miRNAs are tightly regulated for their contributions to the normal cell transcriptome. While in the case of lymphomas, they were found to be very deregulated and massive. The interaction of epigenetic mechanisms and miRNA is a complex regulatory system [
56]. There are also pieces of evidence that miRNA is tissue-specific and can affect epigenetic mechanisms such as histone modifications and DNA methylation, and regulate gene transcription and post-transcriptional silencing of genes [
56,
57]
.
12. T-Cell malignancies and Epigenetic Alterations
T-cell Lymphoma is a malignancy of T-cells and mature CD+4 cells. This is mainly caused by the T-cell leukemia virus Type 1 (HTLV-1) [
58,
59]. In comparison with the human immune deficiency virus (HIV) (retrovirus and pathogenic), in vivo HTLV-1 replication level is low, and virions of HTLV-1 transmission are not very efficient [
60]. Suppression of infected cell death and clonal proliferation is done by the HTLV-1 for a persistent infection. Immune evasion is also done by this virus through the exceptional function of its accessory and regulatory genes. The survival and proliferation of infected cells lead to the accumulation of epigenetic and genetic aberrations in the genes of the host cells [
60].
13. Immune Evasion of T-Lymphoma
Host immunity prevents the development of adult T-cell lymphoma (ATL), due to the immune response towards the HLTV-1. In approximately 90 percent of the ATL cases, immune responses by the host immunity are seen [
59]. Strong T-cell responses are recorded in patients who receive a very stressful treatment such as hematopoietic cells[
61,
62]. Reported results indicated the anti-tumor or anti-viral immune response against the development of T- cell Lymphomas [
59]. Despite the immune responses to this viral disease, it has been reported in a recent study that cells of ATL can escape from natural killer cells (NK) mediated immunity through the deregulation of CD48 [
63]. This observation suggests that gene silencing for viral expression in cells of ATL and defects in the anti-viral immune responses permits the HTLV-1 infected cells to do immune evasion, survive and transform eventually into clones [
59].
14. Epigenetic Regulators of T Lymphocytes
Epigenetic aberrations can lead to transcriptional dysregulations in all types of lymphomas. Epigenetic changes result in the silencing of tumor suppressor genes in their promotor regions. This is an important mechanism in the case of oncogenesis and several other such genes [
64]. These include miR-31, p16
INK4A, NDRG
2, and TCF-8, these greens are well known for their deregulation in the ALT cells by the repressive histone modifications or CpG hypermethylation. [
65]. In this way, ALT cells evade the host's immune response and survive [
59]. Other main epigenetic regulators of lymphomas include EZH2, KMT2, CREBBP, ARID1A, DNMTA, TET2, and IDH2 [
66].
15. Immunotherapy challenges
Identification of biomarkers that envisage clinical responses to PD-1/PDL1 and CTLA-4 blockade is one of the major challenges faced by current immunotherapies[
38]. Somatic copy number alterations (SCNAs), genetic alterations of certain types of genes, or in pathways, and tumor heterogeneity have been recognized as the resistance factors of immunotherapies [
67,
68]. Global methylation also has adverse effects on the checkpoint blockade clinical advantages. On the other hand neoantigen or mutational load and existing T-cell infiltrations are thought to be pointers of the clinical advantage of blockade checkpoints [
69]. Chen et al., 2020 hypothesize that treatment with a hypomethylating (HMA) agent would help in the induction of an antitumor immune response to sensitize people suffering from ovarian cancer to the anti-PD-1, immunotherapy. Phase 2 clinical trial was performed by the authors to test the combination of a second-generation HMA, guadecitabine, with an immune PD-1 checkpoint inhibitor, pembrolizumab.
The clinical trial was performed on 35 platinum-resistant patients with ovarian cancer. The hoped result was not attained from the immune checkpoint blockade and HMA but correlation analysis gave information about which immune therapy will be beneficial for people with ovarian cancer [
70]. Cellular adoptive therapy, dendritic cell vaccines, and some other strategies have yet to display success for extensive types of tumor cells. As we have talked about above that immune therapy becomes resistant to cancer cells by intrinsic and extrinsic factors in tumor cells that lead to immune evasion. Extrinsic factors are immune suppressive cells such as T regulatory cells, myeloid-derived suppressor cells, and tumor-associated macrophages. These cells secrete and produce immune suppressive aspects and show inhibitory ligands for interaction with receptors of T-cells such as CTLA-4 and PD-1. In this type of therapy, both acquired and primary resistance are a problem. But these PD-1 and CTLA-4 immune checkpoint blockade therapies are known to have some success in immune activation enhancement [
35].
16. Prospects of Epigenetic Treatment of Lymphomas
From all the above discussion, it is clear that several studies have shown that epigenetic aberrations are a leading cause of the development and spread of lymphomas. But epigenetic therapies in contrast to chemotherapy which persuades cytotoxicity have a greater effect on cellular processes. For the achievement of a therapeutic effect in the case of lymphomas, reprogramming of cells is done [
71]. Epigenetic monotherapy has been known to have auspicious results in recent clinical trials. In this way, many epigenetic agents were approved for use in the lymphoma treatment. Nowadays, immunotherapies and chemotherapies are being used in combination with epigenetic therapies in several clinical trials [
72].
This combination therapy is being used to overcome the confrontation with a single agent by cell signaling inhibition bypass pathways and is extensively studied preclinically. Such as BET inhibitors are used in combination with an assemblage of tiny inhibitor molecules including HDACs, EZH2, ATR, BTK, P13K, mTOR plus lenalidomide [
73,
74].In the same way, decitabine was combined with BET143, AKT, JAK-STAT, and BCL2 inhibitors. Several such combinations are being studied at the preclinical level and some of them are being tried on mice, before their application in humans. The most thrilling avenue for future research studies is the combination of immunotherapy with epigenetic modulating agents [
72].
17. Conclusion
Epigenetic alterations are the backbone of several immune evasions mechanism. Alteration in epigenetics leads to several autoimmune disorders diabetes and lymphomas. The normal epigenetic mechanism includes DNA methylation, covalent histone modifications, non-covalent mechanisms, and the non-coding RNAs. Immune evasion results due to alteration in this mechanism: hypomethylation and hypermethylation of DNA suppress the gene expressions that are responsible for detecting the foreign cells and inducing an immune response. Downregulation antigen is also seen in the altered DNA methylation process. Similarly, epigenetic changes in the modifications lead to the suppression or activation of chemokines and cytokines expressions, that alter the actual activity of these genes resulting in the survival and development of tumor cells. T-cell lymphomas are also capable of immune evasion due to antiviral gene suppression in the immune system. Epigenetic therapies are also emerging in addition to the traditional immunotherapy and chemotherapy regimes as a striking add-on. These merging therapies work in synergy with other treatments and offer low toxicity. Further research is required on the novel combinations of inhibiting agents, therapeutic roles, and finding new epigenetic pathways.
Author Contributions
Conceptualization, M.A.; methodology, M.A..; validation, M.A.; formal analysis, M.A..; investigation, M.A..;. resources, M.A.; data curation M.A. ; writing—original draft preparation, M.A.; writing—review and editing M.A.; visualization, M.A.; supervision,M.A..
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kiers, T. Current Biology 2022, 32, R147–R148.
- El-Sayed, A.; Aleya, L.; Kamel, M. Microbiota and epigenetics: promising therapeutic approaches? Environmental Science and Pollution Research 2021, 28, 49343–49361. [Google Scholar]
- Zhou, Z.; Rajasingh, S.; Barani, B.; Samanta, S.; Dawn, B.; Wang, R.; Rajasingh, J. Therapy of Infectious Diseases Using Epigenetic Approaches. Epigenetics in Human Disease 2018, 689–715. [Google Scholar]
- Mongan, A.E.; Tuda, J.S.B.; Runtuwene, L.R. Portable sequencer in the fight against infectious disease. Journal of human genetics 2020, 65, 35–40. [Google Scholar]
- Skvortsova, K.; Stirzaker, C.; Taberlay, P. The DNA methylation landscape in cancer. Essays in biochemistry 2019, 63, 797–811. [Google Scholar]
- Lamarche, C.; Iliuta, I.-A.; Kitzler, T. Infectious disease risk in dialysis patients: a transdisciplinary approach. Canadian Journal of Kidney Health and Disease 2019, 6, 2054358119839080. [Google Scholar]
- Tang, C.; Chen, H.; Jiang, L.; Liu, L. Liver regeneration: changes in oxidative stress, immune system, cytokines, and epigenetic modifications associated with aging. Oxidative Medicine and Cellular Longevity 2022, 2022. [Google Scholar]
- Rosikiewicz, W.; Chen, X.; Dominguez, P.M.; Ghamlouch, H.; Aoufouchi, S.; Bernard, O.A.; Melnick, A.; Li, S. TET2 deficiency reprograms the germinal center B cell epigenome and silences genes linked to lymphomagenesis. Science advances 2020, 6, eaay5872. [Google Scholar]
- Mir, M.A.; Mir, B.; Kumawat, M.; Alkhanani, M.; Jan, U. Manipulation and exploitation of host immune system by pathogenic Mycobacterium tuberculosis for its advantage. Future Microbiology 2022, 17, 1171–1198. [Google Scholar]
- Crispen, P.L.; Kusmartsev, S. Mechanisms of immune evasion in bladder cancer. Cancer Immunology, Immunotherapy 2020, 69, 3–14. [Google Scholar]
- Zhang, P.; Zhang, M. Epigenetic alterations and advancement of treatment in peripheral T-cell lymphoma. Clinical epigenetics 2020, 12, 169. [Google Scholar]
- Zhang, P.; Zhang, M. Epigenetic alterations and advancement of treatment in peripheral T-cell lymphoma. Clinical epigenetics. 2020 Dec; 12 (1): 1-7.
- Gillson, S.L.; Ross, D.A. From generation to generation: Rethinking “soul wounds” and historical trauma. Biological psychiatry 2019, 86, e19–e20. [Google Scholar]
- Liu, J.; Li, J.-n.; Wu, H.; Liu, P. The status and prospects of epigenetics in the treatment of lymphoma. Frontiers in Oncology 2022, 12, 874645. [Google Scholar]
- Lu, Q. The critical importance of epigenetics in autoimmunity. Journal of autoimmunity 2013, 41, 1–5. [Google Scholar]
- Ilango, S.; Paital, B.; Jayachandran, P.; Padma, P.R.; Nirmaladevi, R. Epigenetic alterations in cancer. Frontiers in Bioscience-Landmark 2020, 25, 1058–1109. [Google Scholar]
- Felsenfeld, G. Cold Spring Harbor Perspect. Biol 2014, 6, a018200. [Google Scholar]
- Johnson, C.; Warmoes, M.O.; Shen, X.; Locasale, J.W. Epigenetics and cancer metabolism. Cancer letters 2015, 356, 309–314. [Google Scholar]
- Sun, A.; Cole, L.; Maegawa, S.; Park, P.H.; Keith, K.; Madzo, J.; Jelinek, J.; Jobin, C.; Issa, J.-P.J. Microbiota Accelerates Age-Related CpG Island Methylation in Colonic Mucosa. bioRxiv, 2020; 2020.2008. 2028.268367. [Google Scholar]
- Keith, K.; Issa, J.-P.J.; Panjarian, S. Age-Related Variation in DNA Methylation. In Epigenetic Epidemiology; Springer: 2022; pp. 235-259.
- Shirvaliloo, M. The unfavorable clinical outcome of COVID-19 in smokers is mediated by H3K4me3, H3K9me3 and H3K27me3 histone marks. Epigenomics 2022, 14, 153–162. [Google Scholar]
- Gadaleta, E.; Thorn, G.J.; Ross-Adams, H.; Jones, L.J.; Chelala, C. Field cancerization in breast cancer. The Journal of Pathology 2022, 257, 561–574. [Google Scholar]
- Wang, Z.; Yang, C. Metal carcinogen exposure induces cancer stem cell-like property through epigenetic reprograming: A novel mechanism of metal carcinogenesis. In Proceedings of the Seminars in cancer biology; 2019; pp. 95–104. [Google Scholar]
- Sato, S.; Takizawa, Y.; Hoshikawa, F.; Dacher, M.; Tanaka, H.; Tachiwana, H.; Kujirai, T.; Iikura, Y.; Ho, C.-H.; Adachi, N. Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones. Nucleic Acids Research 2021, 49, 8934–8946. [Google Scholar]
- De Sarkar, N.; Patton, R.D.; Doebley, A.-L.; Hanratty, B.; Adil, M.; Kreitzman, A.J.; Sarthy, J.F.; Ko, M.; Brahma, S.; Meers, M.P. Nucleosome patterns in circulating tumor DNA reveal transcriptional regulation of advanced prostate cancer phenotypes. Cancer Discovery 2023, 13, 632–653. [Google Scholar]
- Chen, Q.W.; Zhu, X.; Li, Y.; Meng, Z. Epigenetic regulation and cancer. Oncology reports 2014, 31, 523–532. [Google Scholar]
- Ghasemi, S. Cancer's epigenetic drugs: where are they in the cancer medicines? The pharmacogenomics journal 2020, 20, 367–379. [Google Scholar]
- Rugo, H.S.; Jacobs, I.; Sharma, S.; Scappaticci, F.; Paul, T.A.; Jensen-Pergakes, K.; Malouf, G.G. The promise for histone methyltransferase inhibitors for epigenetic therapy in clinical oncology: a narrative review. Advances in therapy 2020, 37, 3059–3082. [Google Scholar]
- Pop, S.; Enciu, A.M.; Tarcomnicu, I.; Gille, E.; Tanase, C. Phytochemicals in cancer prevention: modulating epigenetic alterations of DNA methylation. Phytochemistry Reviews 2019, 18, 1005–1024. [Google Scholar]
- Park, J.W.; Han, J.-W. Targeting epigenetics for cancer therapy. Archives of pharmacal research 2019, 42, 159–170. [Google Scholar]
- Chen, Z.; Fan, Y.; Liu, X.; Shang, X.; Qi, K.; Zhang, S. Clinicopathological significance of DAPK gene promoter hypermethylation in non-small cell lung cancer: A meta-analysis. The International Journal of Biological Markers 2022, 37, 47–57. [Google Scholar]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nature Reviews Molecular Cell Biology 2020, 21, 284–299. [Google Scholar]
- O'Connor, K.M.; Das, A.B.; Winterbourn, C.C.; Hampton, M.B. Inhibition of DNA methylation in proliferating human lymphoma cells by immune cell oxidants. Journal of Biological Chemistry 2020, 295, 7839–7848. [Google Scholar]
- Wu, R.; Ivan, E.; Sahasrabuddhe, A.A.; Shaw, T.; Mullighan, C.G.; Leventaki, V.; Elenitoba-Johnson, K.S.; Lim, M.S. Epigenetic modulation of CD48 By NPM-ALK promotes immune evasion in ALK+ ALCL. Blood 2019, 134, 1510. [Google Scholar]
- Gomez, S.; Tabernacki, T.; Kobyra, J.; Roberts, P.; Chiappinelli, K.B. Combining epigenetic and immune therapy to overcome cancer resistance. In Proceedings of the Seminars in cancer biology; 2020; pp. 99–113. [Google Scholar]
- Wei, W.L.; Ee, C.J.; Ying, L.J.; Miaomiao, L. Decitabine as a Latency Perturbing Agent in Epstein-Barr Virus (EBV) Positive Natural Killer/T-Cell Lymphoma (NKTL). In Proceedings of the IRC-SET 2021: Proceedings of the 7th IRC Conference on Science, Engineering and Technology, August 2021, Singapore, 2022; pp. 485–495.
- Greenberg, M.V.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nature reviews Molecular cell biology 2019, 20, 590–607. [Google Scholar]
- Jung, H.; Kim, H.S.; Kim, J.Y.; Sun, J.-M.; Ahn, J.S.; Ahn, M.-J.; Park, K.; Esteller, M.; Lee, S.-H.; Choi, J.K. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nature communications 2019, 10, 4278. [Google Scholar]
- Li, Y.; Goldberg, E.M.; Chen, X.; Xu, X.; McGuire, J.T.; Leuzzi, G.; Karagiannis, D.; Tate, T.; Farhangdoost, N.; Horth, C. Histone methylation antagonism drives tumor immune evasion in squamous cell carcinomas. Molecular Cell 2022, 82, 3901–3918.e3907. [Google Scholar]
- Zhao, S.G.; Chen, W.S.; Li, H.; Foye, A.; Zhang, M.; Sjöström, M.; Aggarwal, R.; Playdle, D.; Liao, A.; Alumkal, J.J. The DNA methylation landscape of advanced prostate cancer. Nature genetics 2020, 52, 778–789. [Google Scholar]
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.; Ross, J.P. DNA methylation cancer biomarkers: translation to the clinic. Frontiers in genetics 2019, 10, 1150. [Google Scholar]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA methylation. PLoS genetics 2020, 16, e1009034. [Google Scholar]
- Jeusset, L.M.; McManus, K.J. Developing targeted therapies that exploit aberrant histone ubiquitination in cancer. Cells 2019, 8, 165. [Google Scholar]
- Chandrasekaran, B.; Tapadar, S.; Wu, B.; Saran, U.; Tyagi, A.; Johnston, A.; Gaul, D.A.; Oyelere, A.K.; Damodaran, C. Antiandrogen-Equipped Histone Deacetylase Inhibitors Selectively Inhibit Androgen Receptor (AR) and AR-Splice Variant (AR-SV) in Castration-Resistant Prostate Cancer (CRPC). Cancers 2023, 15, 1769. [Google Scholar]
- Cao, J.; Yan, Q. Cancer epigenetics, tumor immunity, and immunotherapy. Trends in cancer 2020, 6, 580–592. [Google Scholar]
- Sim, W.; Lim, W.-M.; Hii, L.-W.; Leong, C.-O.; Mai, C.-W. Targeting pancreatic cancer immune evasion by inhibiting histone deacetylases. World Journal of Gastroenterology 2022, 28, 1934. [Google Scholar]
- Steinbach, A.; Riemer, A.B. Immune evasion mechanisms of human papillomavirus: An update. International journal of cancer 2018, 142, 224–229. [Google Scholar]
- Darvin, P.; Sasidharan Nair, V.; Elkord, E. PD-L1 expression in human breast cancer stem cells is epigenetically regulated through posttranslational histone modifications. Journal of oncology 2019, 2019. [Google Scholar]
- Burr, M.L.; Sparbier, C.E.; Chan, K.L.; Chan, Y.-C.; Kersbergen, A.; Lam, E.Y.; Azidis-Yates, E.; Vassiliadis, D.; Bell, C.C.; Gilan, O. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer cell 2019, 36, 385–401.e388. [Google Scholar]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer immune evasion through loss of MHC class I antigen presentation. Frontiers in immunology 2021, 12, 636568. [Google Scholar]
- Zhang, Q.; Cao, X. Epigenetic regulation of the innate immune response to infection. Nature Reviews Immunology 2019, 19, 417–432. [Google Scholar]
- Yeon, M.; Kim, Y.; Jung, H.S.; Jeoung, D. Histone deacetylase inhibitors to overcome resistance to targeted and immuno therapy in metastatic melanoma. Frontiers in cell and developmental biology 2020, 8, 486. [Google Scholar]
- Qin, J.; Wen, B.; Liang, Y.; Yu, W.; Li, H. Histone modifications and their role in colorectal cancer. Pathology & Oncology Research 2020, 26, 2023–2033. [Google Scholar]
- Ng, K.-L.; Taguchi, Y.-H. Identification of miRNA signatures for kidney renal clear cell carcinoma using the tensor-decomposition method. Scientific Reports 2020, 10, 15149. [Google Scholar]
- Arif, K.T.; Elliott, E.K.; Haupt, L.M.; Griffiths, L.R. Regulatory mechanisms of epigenetic miRNA relationships in human cancer and potential as therapeutic targets. Cancers 2020, 12, 2922. [Google Scholar]
- Sadakierska-Chudy, A. MicroRNAs: diverse mechanisms of action and their potential applications as cancer epi-therapeutics. Biomolecules 2020, 10, 1285. [Google Scholar]
- Zhang, S.; Cheng, Z.; Wang, Y.; Han, T. The risks of miRNA therapeutics: in a drug target perspective. Drug design, development and therapy, 2021; 721–733. [Google Scholar]
- Hedayati-Moghaddam, M.R.; Fathimoghadam, F.; Soghandi, L.; Darrudi, A. High prevalence of HTLV-1 infection among hemodialysis patients in Neyshabur, northeast of Iran. International Journal of Infection 2019, 6. [Google Scholar]
- Yasunaga, J.-i. Viral, genetic, and immune factors in the oncogenesis of adult T-cell leukemia/lymphoma. International Journal of Hematology 2023, 117, 504–511. [Google Scholar]
- Yasunaga, J.-i. Strategies of human T-cell leukemia virus Type 1 for persistent infection: Implications for leukemogenesis of adult T-cell leukemia-lymphoma. Frontiers in microbiology 2020, 11, 979. [Google Scholar]
- Utsunomiya, A. Progress in allogeneic hematopoietic cell transplantation in adult T-cell leukemia-lymphoma. Frontiers in microbiology 2019, 10, 2235. [Google Scholar]
- Tanaka, A.; Nishikawa, H.; Noguchi, S.; Sugiyama, D.; Morikawa, H.; Takeuchi, Y.; Ha, D.; Shigeta, N.; Kitawaki, T.; Maeda, Y. Tyrosine kinase inhibitor imatinib augments tumor immunity by depleting effector regulatory T cells. Journal of Experimental Medicine 2020, 217. [Google Scholar]
- Maruoka, Y.; Furusawa, A.; Okada, R.; Inagaki, F.; Fujimura, D.; Wakiyama, H.; Kato, T.; Nagaya, T.; Choyke, P.L.; Kobayashi, H. Combined CD44-and CD25-Targeted Near-Infrared Photoimmunotherapy Selectively Kills Cancer and Regulatory T Cells in Syngeneic Mouse Cancer ModelsCombined CD44-and CD25-Targeted NIR-PIT. Cancer immunology research 2020, 8, 345–355. [Google Scholar]
- Muranushi, H.; Shindo, T.; Hishizawa, M.; Tokunaga, M.; Wake, A.; Nakano, N.; Eto, T.; Hidaka, M.; Choi, I.; Miyamoto, T. GVHD-free, relapse-free survival provides novel clues for optimizing allogeneic-HSCT for adult T-cell leukemia/lymphoma. Bone Marrow Transplantation 2021, 56, 155–166. [Google Scholar]
- Ratner, L. Epigenetic regulation of human t-cell leukemia virus gene expression. Microorganisms 2021, 10, 84. [Google Scholar]
- Harrop, S.; Yannakou, C.K.; Van Der Weyden, C.; Prince, H.M. Epigenetic Modifications in Lymphoma and Their Role in the Classification of Lymphomas. Hemato 2022, 3, 174–187. [Google Scholar]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Geukes Foppen, M.H.; Goldinger, S.M. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar]
- Ock, C.-Y.; Hwang, J.-E.; Keam, B.; Kim, S.-B.; Shim, J.-J.; Jang, H.-J.; Park, S.; Sohn, B.H.; Cha, M.; Ajani, J.A. Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nature communications 2017, 8, 1050. [Google Scholar]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar]
- Chiappinelli, K.B.; Baylin, S.B. Inhibiting DNA methylation improves antitumor immunity in ovarian cancer. The Journal of clinical investigation 2022, 132. [Google Scholar]
- Bates, S.E. Epigenetic therapies for cancer. New England Journal of Medicine 2020, 383, 650–663. [Google Scholar]
- Sermer, D.; Pasqualucci, L.; Wendel, H.-G.; Melnick, A.; Younes, A. Emerging epigenetic-modulating therapies in lymphoma. Nature reviews Clinical oncology 2019, 16, 494–507. [Google Scholar]
- Sarnik, J.; Popławski, T.; Tokarz, P. BET proteins as attractive targets for cancer therapeutics. International Journal of Molecular Sciences 2021, 22, 11102. [Google Scholar]
- Sender, S.; Sultan, A.; Palmer, D.; Koczan, D.; Sekora, A.; Beck, J.; Schuetz, E.; Taher, L.; Brenig, B.; Fuellen, G. Evaluation of the Synergistic Potential of Simultaneous Pan-or Isoform-Specific BET and SYK Inhibition in B-Cell Lymphoma: An In Vitro Approach. Cancers 2022, 14, 4691. 2022.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).