Submitted:
12 June 2023
Posted:
14 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
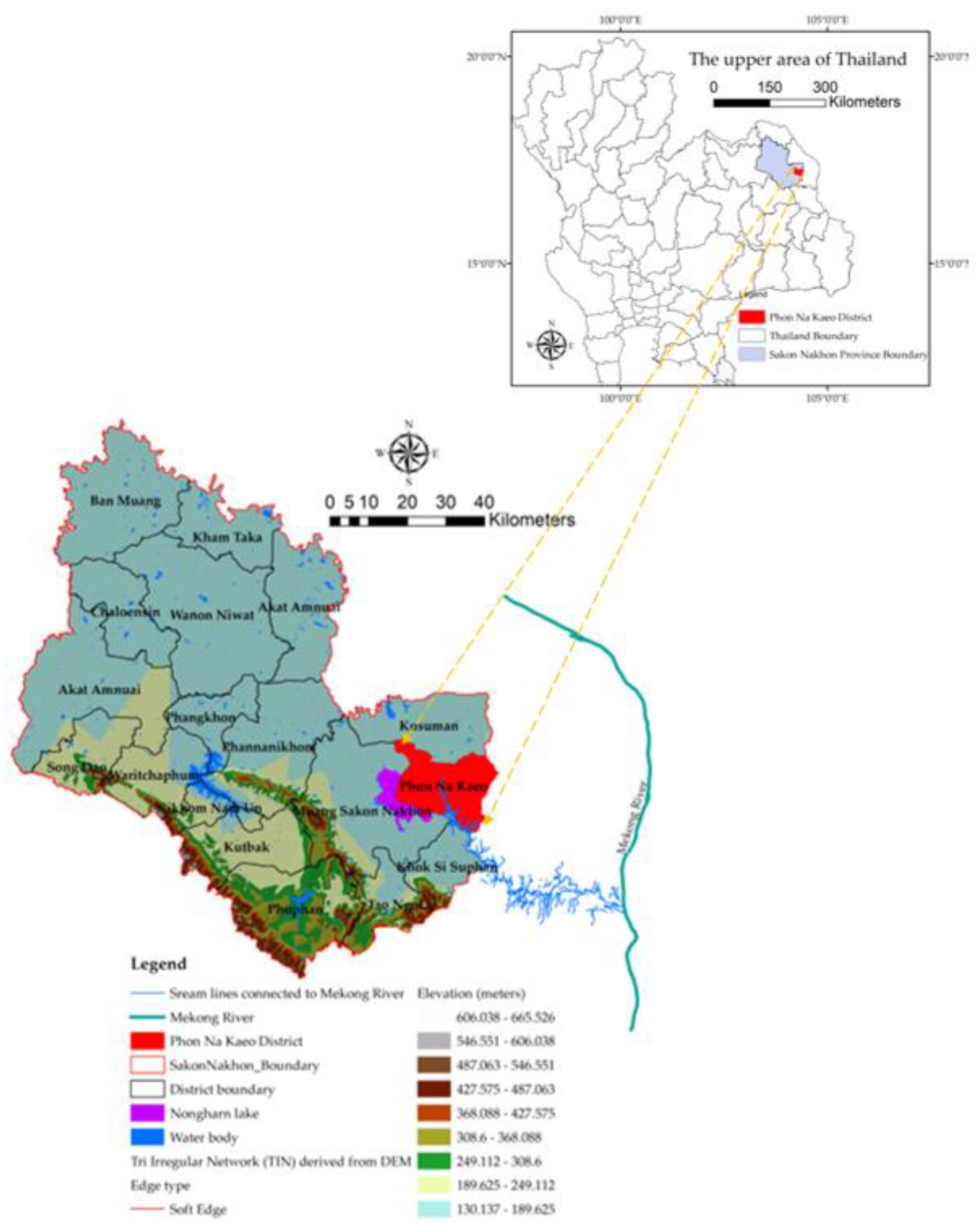
2.1. The Study Area
2.2. Datasets and Analyses
2.3. Independent variable modeling
2.4. GWR Modeling
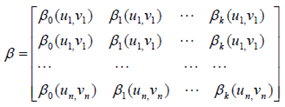
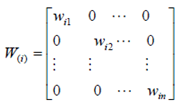
3. Results
3.1. Spatial Unit Design
3.2. Distribution of Independent Variables
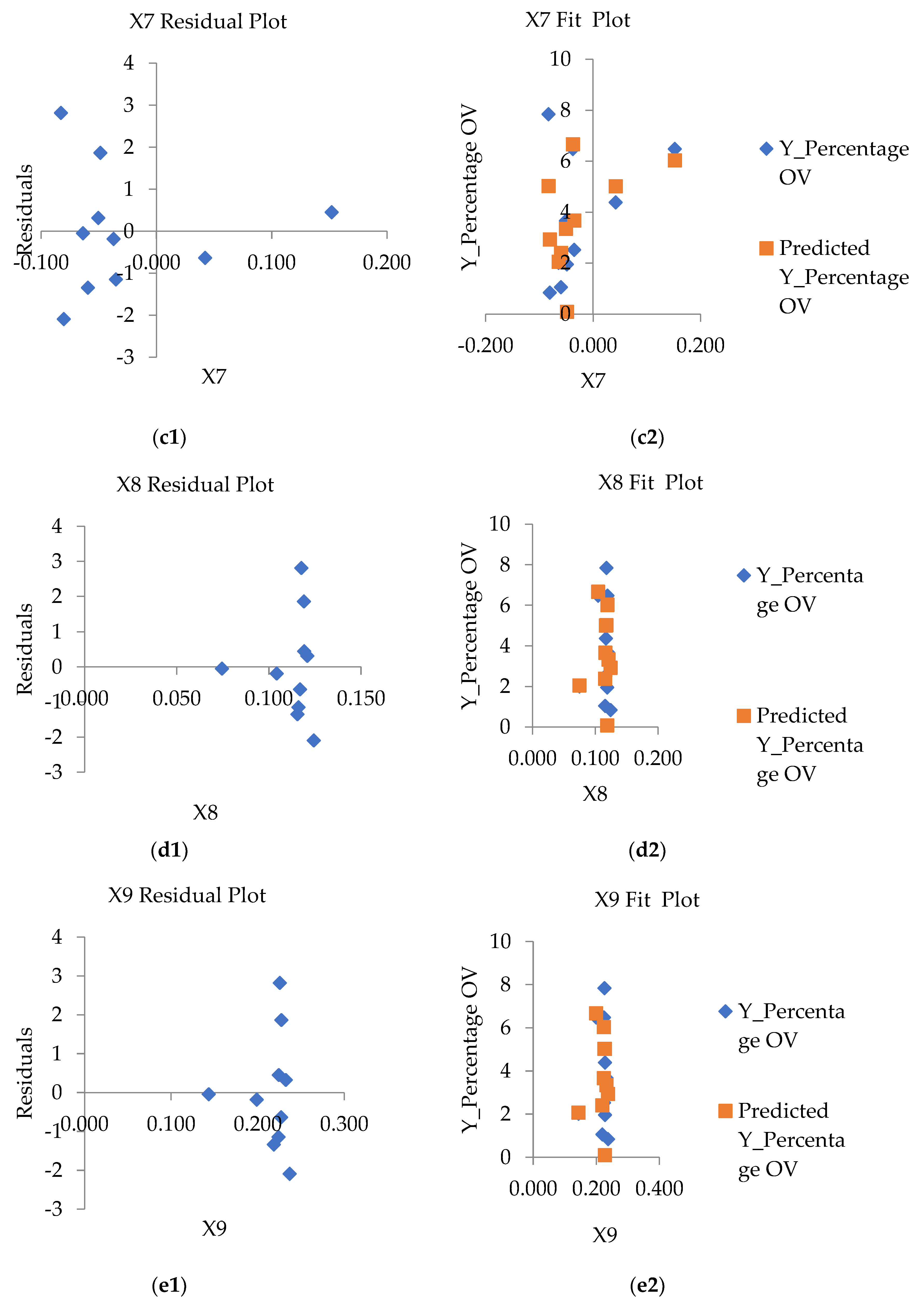
3.3. Selected the Influence Factors Associated with Spatial Liver Fluke (Opisthorchis viverrini) Infection
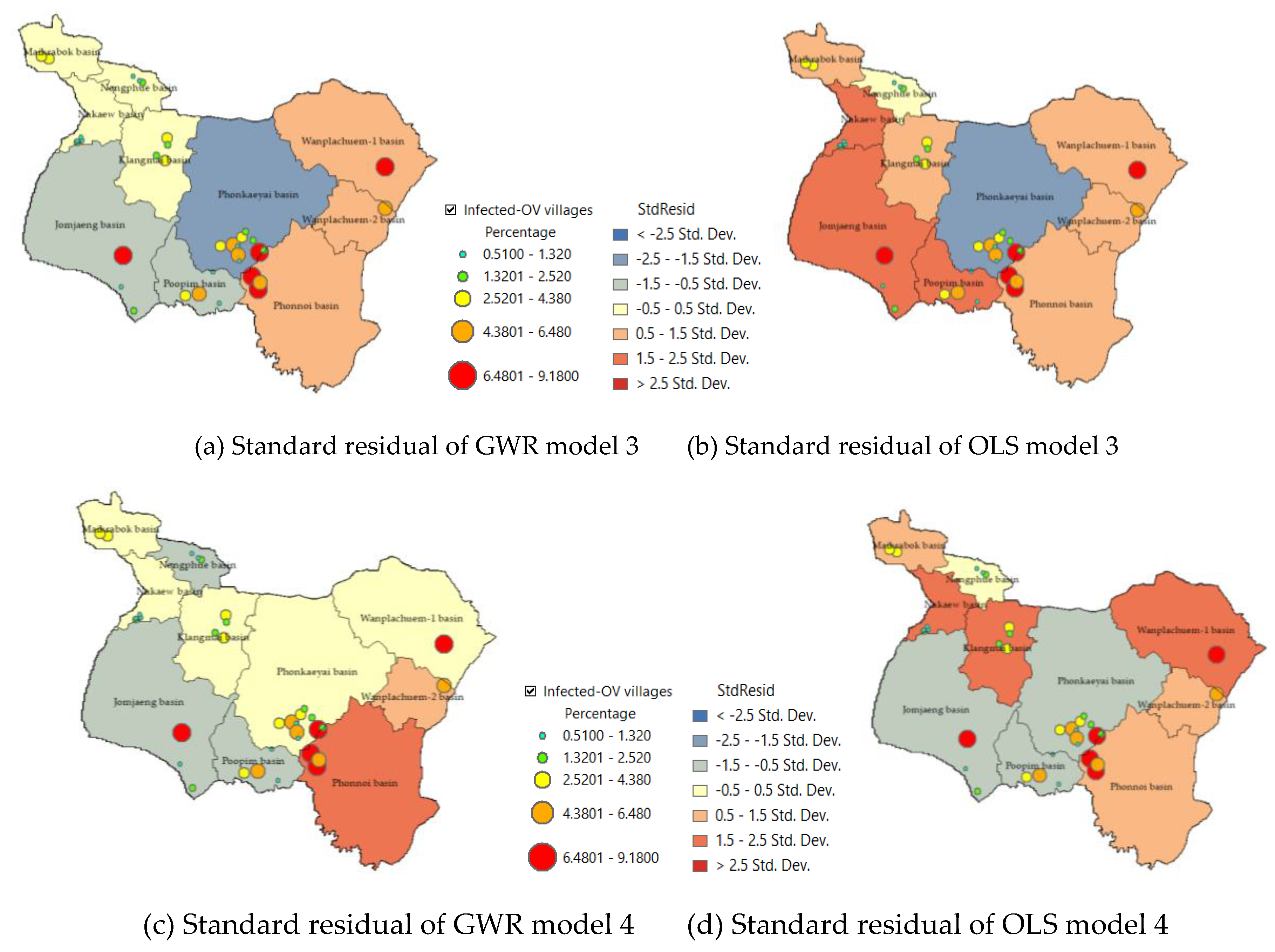
3.4. Optimal GWR Model for Predicted With Liver Fluke (Opisthorchis viverrini) Infection
4. Discussion
4.1. Redundancy of Independent Variable Sets
4.2. Model Capabilities and Development Approaches in Other Areas
4.3. Guidelines for Applying The Model to Provincial Public Health Policy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geadkaew-Krenc, A.; Krenc, D.; Thanongsaksrikul, J.; Grams, R.; Phadungsil, W.; Glab-ampai, K.; Chantree, P.; Martviset, P. Production and Immunological Characterization of ScFv Specific to Epitope of Opisthorchis Viverrini Rhophilin-Associated Tail Protein 1-like (OvROPN1L). Trop. Med. Infect. Dis. 2023, 8, 160. [Google Scholar] [CrossRef] [PubMed]
- Perakanya, P.; Ungcharoen, R.; Worrabannakorn, S.; Ongarj, P.; Artchayasawat, A.; Boonmars, T.; Boueroy, P. Prevalence and Risk Factors of Opisthorchis Viverrini Infection in Sakon Nakhon Province, Thailand. Trop. Med. Infect. Dis. 2022, 7, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Sadaow, L.; Rodpai, R.; Janwan, P.; Boonroumkaew, P.; Sanpool, O.; Thanchomnang, T.; Yamasaki, H.; Ittiprasert, W.; Mann, V.H.; Brindley, P.J.; et al. An Innovative Test for the Rapid Detection of Specific IgG Antibodies in Human Whole-Blood for the Diagnosis of Opisthorchis Viverrini Infection. Trop. Med. Infect. Dis. 2022, 7. [Google Scholar] [CrossRef]
- Boonjaraspinyo, S.; Boonmars, T.; Ekobol, N.; Artchayasawat, A.; Sriraj, P.; Aukkanimart, R.; Pumhirunroj, B.; Sripan, P.; Songsri, J.; Juasook, A.; et al. Prevalence and Associated Risk Factors of Intestinal Parasitic Infections: A Population-Based Study in Phra Lap Sub-District, Mueang Khon Kaen District, Khon Kaen Province, Northeastern Thailand. Trop. Med. Infect. Dis. 2023, 8. [Google Scholar] [CrossRef]
- Sripa, B.; Bethony, J.M.; Sithithaworn, P.; Kaewkes, S.; Mairiang, E.; Loukas, A.; Mulvenna, J.; Laha, T.; Hotez, P.J.; Brindley, P.J. Opisthorchiasis and Opisthorchis-Associated Cholangiocarcinoma in Thailand and Laos. Acta Trop. 2011, 120 Suppl, S158–S168. [Google Scholar] [CrossRef]
- Prasongwatana, J.; Laummaunwai, P.; Boonmars, T.; Pinlaor, S. Viable Metacercariae of Opisthorchis Viverrini in Northeastern Thai Cyprinid Fish Dishes--as Part of a Rational Program for Control of O. Viverrini-Associated Cholangiocarcinoma. Parasitol. Res. 2013, 112, 1323–1327. [Google Scholar] [CrossRef]
- Sripa, B.; Kaewkes, S.; Sithithaworn, P.; Mairiang, E.; Laha, T.; Smout, M.; Pairojkul, C.; Bhudhisawasdi, V.; Tesana, S.; Thinkamrop, B.; et al. Liver Fluke Induces Cholangiocarcinoma. PLOS Med. 2007, 4, e201. [Google Scholar] [CrossRef]
- Sripa, B.; Brindley, P.J.; Mulvenna, J.; Laha, T.; Smout, M.J.; Mairiang, E.; Bethony, J.M.; Loukas, A. The Tumorigenic Liver Fluke Opisthorchis Viverrini–Multiple Pathways to Cancer. Trends Parasitol. 2012, 28, 395–407. [Google Scholar] [CrossRef]
- Sripa, B.; Tangkawattana, S.; Laha, T.; Kaewkes, S.; Mallory, F.F.; Smith, J.F.; Wilcox, B.A. Toward Integrated Opisthorchiasis Control in Northeast Thailand: The Lawa Project. Acta Trop. 2015, 141, 361–367. [Google Scholar] [CrossRef]
- Haswell-elkins, M.R.; SAtarug, S.; Elkins, D.B. Opisthorchis Viverrini Infection in Northeast Thailand and Its Relationship to Cholangiocarcinoma. J. Gastroenterol. Hepatol. 1992, 7, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Mairiang, E.; Elkins, D.B.; Mairiang, P.; Chaiyakum, J.; Chamadol, N.; Loapaiboon, V.; Posri, S.; Sithithaworn, P.; Haswell-elkins, M. Relationship between Intensity of Opisthorchis Viverrini Infection and Hepatobiliary Disease Detected by Ultrasonography. J. Gastroenterol. Hepatol. 1992, 7, 17–21. [Google Scholar] [CrossRef]
- Pumhirunroj, B.; Aukkanimart, R. Liver Fluke-Infected Cyprinoid Fish in Northeastern Thailand (2016-2017). Southeast Asian J. Trop. Med. Public Health 2017, 51, 1–7. [Google Scholar]
- Pinlaor, S.; Onsurathum, S.; Boonmars, T.; Pinlaor, P.; Hongsrichan, N.; Chaidee, A.; Haonon, O.; Limviroj, W.; Tesana, S.; Kaewkes, S.; et al. Distribution and Abundance of Opisthorchis Viverrini Metacercariae in Cyprinid Fish in Northeastern Thailand. Korean J. Parasitol. 2013, 51, 703–710. [Google Scholar] [CrossRef]
- Suwannatrai, A.T.; Thinkhamrop, K.; Clements, A.C.A.; Kelly, M.; Suwannatrai, K.; Thinkhamrop, B.; Khuntikeo, N.; Gray, D.J.; Wangdi, K. Bayesian Spatial Analysis of Cholangiocarcinoma in Northeast Thailand. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Hasegawa, S.; Ikai, I.; Fujii, H.; Hatano, E.; Shimahara, Y. Surgical Resection of Hilar Cholangiocarcinoma: Analysis of Survival and Postoperative Complications. World J. Surg. 2007, 31, 1258–1265. [Google Scholar] [CrossRef]
- Thinkhamrop, K.; Suwannatrai, A.T.; Chamadol, N.; Khuntikeo, N.; Thinkhamrop, B.; Sarakarn, P.; Gray, D.J.; Wangdi, K.; Clements, A.C.A.; Kelly, M. Spatial Analysis of Hepatobiliary Abnormalities in a Population at High-Risk of Cholangiocarcinoma in Thailand. Sci. Rep. 2020, 10, 16855. [Google Scholar] [CrossRef]
- Pratumchart, K.; Suwannatrai, K.; Sereewong, C.; Thinkhamrop, K.; Chaiyos, J.; Boonmars, T.; Suwannatrai, A.T. Ecological Niche Model Based on Maximum Entropy for Mapping Distribution of Bithynia Siamensis Goniomphalos, First Intermediate Host Snail of Opisthorchis Viverrini in Thailand. Acta Trop. 2019, 193, 183–191. [Google Scholar] [CrossRef]
- Suwannatrai, A.T.; Thinkhamrop, K.; Clements, A.C.A.; Kelly, M.; Suwannatrai, K.; Thinkhamrop, B.; Khuntikeo, N.; Gray, D.J.; Wangdi, K. Bayesian Spatial Analysis of Cholangiocarcinoma in Northeast Thailand. Sci. Rep. 2019, 9, 14263. [Google Scholar] [CrossRef]
- Martviset, P.; Phadungsil, W.; Na-Bangchang, K.; Sungkhabut, W.; Panupornpong, T.; Prathaphan, P.; Torungkitmangmi, N.; Chaimon, S.; Wangboon, C.; Jamklang, M.; et al. Current Prevalence and Geographic Distribution of Helminth Infections in the Parasitic Endemic Areas of Rural Northeastern Thailand. BMC Public Health 2023, 23, 448. [Google Scholar] [CrossRef]
- Littidej, P.; Buasri, N. Built-up Growth Impacts on Digital Elevation Model and Flood Risk Susceptibility Prediction in Muaeng District, Nakhon Ratchasima (Thailand). Water (Switzerland) 2019, 11. [Google Scholar] [CrossRef]
- Littidej, P.; Uttha, T.; Pumhirunroj, B. Spatial Predictive Modeling of the Burning of Sugarcane Plots in Northeast Thailand with Selection of Factor Sets Using a GWR Model and Machine Learning Based on an ANN-CA. Symmetry (Basel). 2022, 14. [Google Scholar] [CrossRef]
- Prasertsri, N.; Littidej, P. Spatial Environmental Modeling for Wildfire Progression Accelerating Extent Analysis Using Geo-Informatics. Polish J. Environ. Stud. 2020, 29, 3249–3261. [Google Scholar] [CrossRef]
- Lu, B.; Charlton, M.; Fotheringham, A.S. Geographically Weighted Regression Using a Non-Euclidean Distance Metric with a Study on London House Price Data. Procedia Environ. Sci. 2011, 7, 92–97. [Google Scholar] [CrossRef]
- Lu, B.; Charlton, M.; Harris, P.; Fotheringham, A.S. Geographically Weighted Regression with a Non-Euclidean Distance Metric: A Case Study Using Hedonic House Price Data. Int. J. Geogr. Inf. Sci. 2014, 28, 660–681. [Google Scholar] [CrossRef]
- Fotheringham, A.; Charlton, M. Geographically Geographically Weighted Weighted Regression Regression A Stewart Fotheringham. 2014.
- Suwannahitatorn, P.; Webster, J.; Riley, S.; Mungthin, M.; Donnelly, C.A. Uncooked Fish Consumption among Those at Risk of Opisthorchis Viverrini Infection in Central Thailand. PLoS One 2019, 14, e0211540. [Google Scholar] [CrossRef]
- Sripa, B.; Kaewkes, S.; Intapan, P.M.; Maleewong, W.; Brindley, P.J. Chapter 11 - Food-Borne Trematodiases in Southeast Asia: Epidemiology, Pathology, Clinical Manifestation and Control. In Important Helminth Infections in Southeast Asia: Diversity and Potential for Control and Elimination, Part A; Zhou, X.-N., Bergquist, R., Olveda, R., Utzinger, J.B.T.-A. in P., Eds.; Academic Press, 2010; Vol. 72, pp. 305–350 ISBN 0065-308X.
- Qian, M.-B.; Utzinger, J.; Keiser, J.; Zhou, X.-N. Clonorchiasis. Lancet 2016, 387, 800–810. [Google Scholar] [CrossRef]
- Brindley, P.J.; Bachini, M.; Ilyas, S.I.; Khan, S.A.; Loukas, A.; Sirica, A.E.; Teh, B.T.; Wongkham, S.; Gores, G.J. Cholangiocarcinoma. Nat. Rev. Dis. Prim. 2021, 7. [Google Scholar] [CrossRef]
- Sakon Nakhon Provincial Public Health Office (SKKO). Annual Report 2023. 2023. Available online: https://skko.moph.go.th/dward/web/index.php?module=skko (accessed on 1 June 2022).
- Dao, T.T.H.; Van Bui, T.; Abatih, E.N.; Gabriël, S.; Nguyen, T.T.G.; Huynh, Q.H.; Van Nguyen, C.; Dorny, P. Opisthorchis Viverrini Infections and Associated Risk Factors in a Lowland Area of Binh Dinh Province, Central Vietnam. Acta Trop. 2016, 157, 151–157. [Google Scholar] [CrossRef]
- Ruantip, S.; Eamudomkarn, C.; Kopolrat, K.Y.; Sithithaworn, J.; Laha, T.; Sithithaworn, P. Analysis of Daily Variation for 3 and for 30 Days of Parasite-Specific IgG in Urine for Diagnosis of Strongyloidiasis by Enzyme-Linked Immunosorbent Assay. Acta Trop. 2021, 218, 105896. [Google Scholar] [CrossRef]
- The 8th Health District Office (Region, (R8)) Annual Report 2021. 2021. Available online: https://r8way.moph.go.th/r8way/index (accessed on 3 June 2022).
- Honjo, S.; Srivatanakul, P.; Sriplung, H.; Kikukawa, H.; Hanai, S.; Uchida, K.; Todoroki, T.; Jedpiyawongse, A.; Kittiwatanachot, P.; Sripa, B.; et al. Genetic and Environmental Determinants of Risk for Cholangiocarcinoma via Opisthorchis Viverrini in a Densely Infested Area in Nakhon Phanom, Northeast Thailand. Int. J. Cancer 2005, 117, 854–860. [Google Scholar] [CrossRef]
- Zhao, T.-T.; Feng, Y.-J.; Doanh, P.N.; Sayasone, S.; Khieu, V.; Nithikathkul, C.; Qian, M.-B.; Hao, Y.-T.; Lai, Y.-S. Model-Based Spatial-Temporal Mapping of Opisthorchiasis in Endemic Countries of Southeast Asia. Elife 2021, 10, e59755. [Google Scholar] [CrossRef]
- Forrer, A.; Sayasone, S.; Vounatsou, P.; Vonghachack, Y.; Bouakhasith, D.; Vogt, S.; Glaser, R.; Utzinger, J.; Akkhavong, K.; Odermatt, P. Spatial Distribution of, and Risk Factors for, Opisthorchis Viverrini Infection in Southern Lao PDR. PLoS Negl. Trop. Dis. 2012, 6, e1481. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Jiang, S.; Peng, H.-J. Association between Liver Fluke Infection and Hepatobiliary Pathological Changes: A Systematic Review and Meta-Analysis. PLoS One 2015, 10, e0132673. [Google Scholar] [CrossRef] [PubMed]
- Brunton, L.A.; Alexander, N.; Wint, W.; Ashton, A.; Broughan, J.M. Using Geographically Weighted Regression to Explore the Spatially Heterogeneous Spread of Bovine Tuberculosis in England and Wales. Stoch. Environ. Res. Risk Assess. 2017, 31, 339–352. [Google Scholar] [CrossRef]
- Rujirakul, R.; Ueng-arporn, N.; Kaewpitoon, S.; Loyd, R.J.; Kaewthani, S.; Kaewpitoon, N. GIS-Based Spatial Statistical Analysis of Risk Areas for Liver Flukes in Surin Province of Thailand. Asian Pac. J. Cancer Prev. 2015, 16, 2323–2326. [Google Scholar] [CrossRef]
- Brunsdon, C.; Fotheringham, S.; Charlton, M. Geographically Weighted Regression-Modelling Spatial Non-Stationarity. J. R. Stat. Soc. Ser. D (The Stat. 1998, 47, 431–443. [Google Scholar]
- Comber, A.; Brunsdon, C.; Charlton, M.; Dong, G.; Harris, R.; Lu, B.; Lü, Y.; Murakami, D.; Nakaya, T.; Wang, Y.; et al. A Route Map for Successful Applications of Geographically Weighted Regression. 2023, 155–178. [CrossRef]
- Lu, B.; Hu, Y.; Murakami, D.; Brunsdon, C.; Comber, A.; Charlton, M.; Harris, P. High-Performance Solutions of Geographically Weighted Regression in R. Geo-spatial Inf. Sci. 2022, 25, 536–549. [Google Scholar] [CrossRef]
- Leong, Y.Y.; Yue, J.C. A Modification to Geographically Weighted Regression. Int. J. Health Geogr. 2017, 1–18. [Google Scholar] [CrossRef]
- Isazade, V.; Qasimi, A.B.; Dong, P.; Kaplan, G.; Isazade, E. Integration of Moran’s I, Geographically Weighted Regression (GWR), and Ordinary Least Square (OLS) Models in Spatiotemporal Modeling of COVID-19 Outbreak in Qom and Mazandaran Provinces, Iran. Model. Earth Syst. Environ. 2023. [Google Scholar] [CrossRef]
- Düzgün, H.S.; Kemeç, S. Spatial and Geographically Weighted Regression BT - Encyclopedia of GIS, Shekhar, S., Xiong, H., Eds.; Springer US: Boston, MA, 2008; ISBN 978-0-387-35973-1.
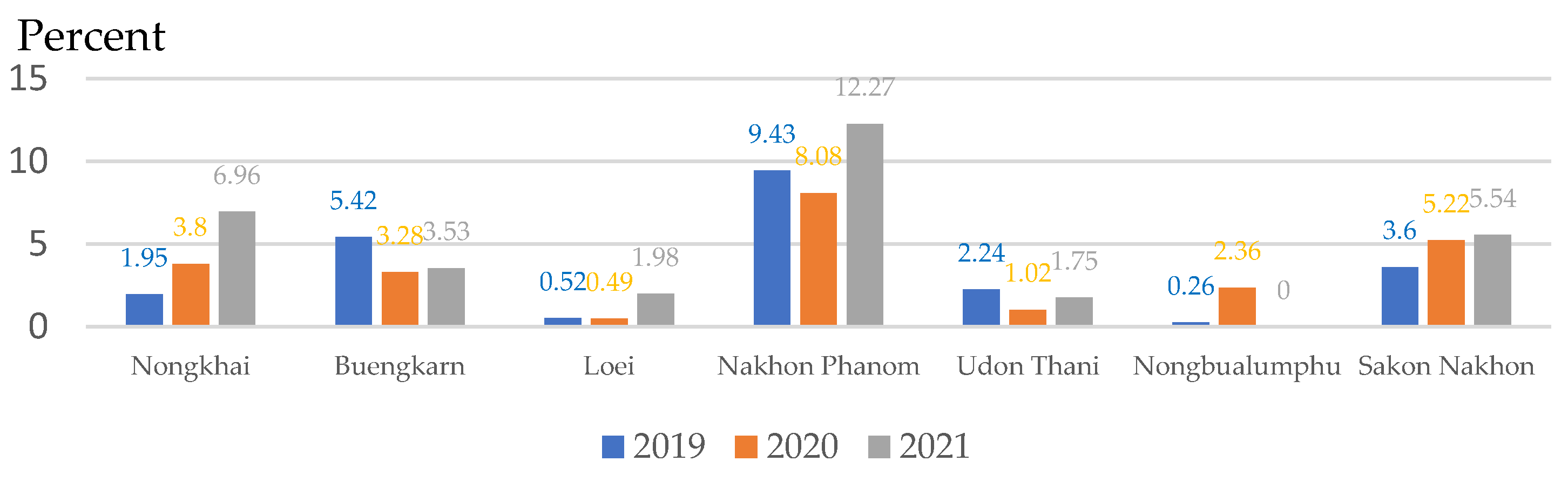
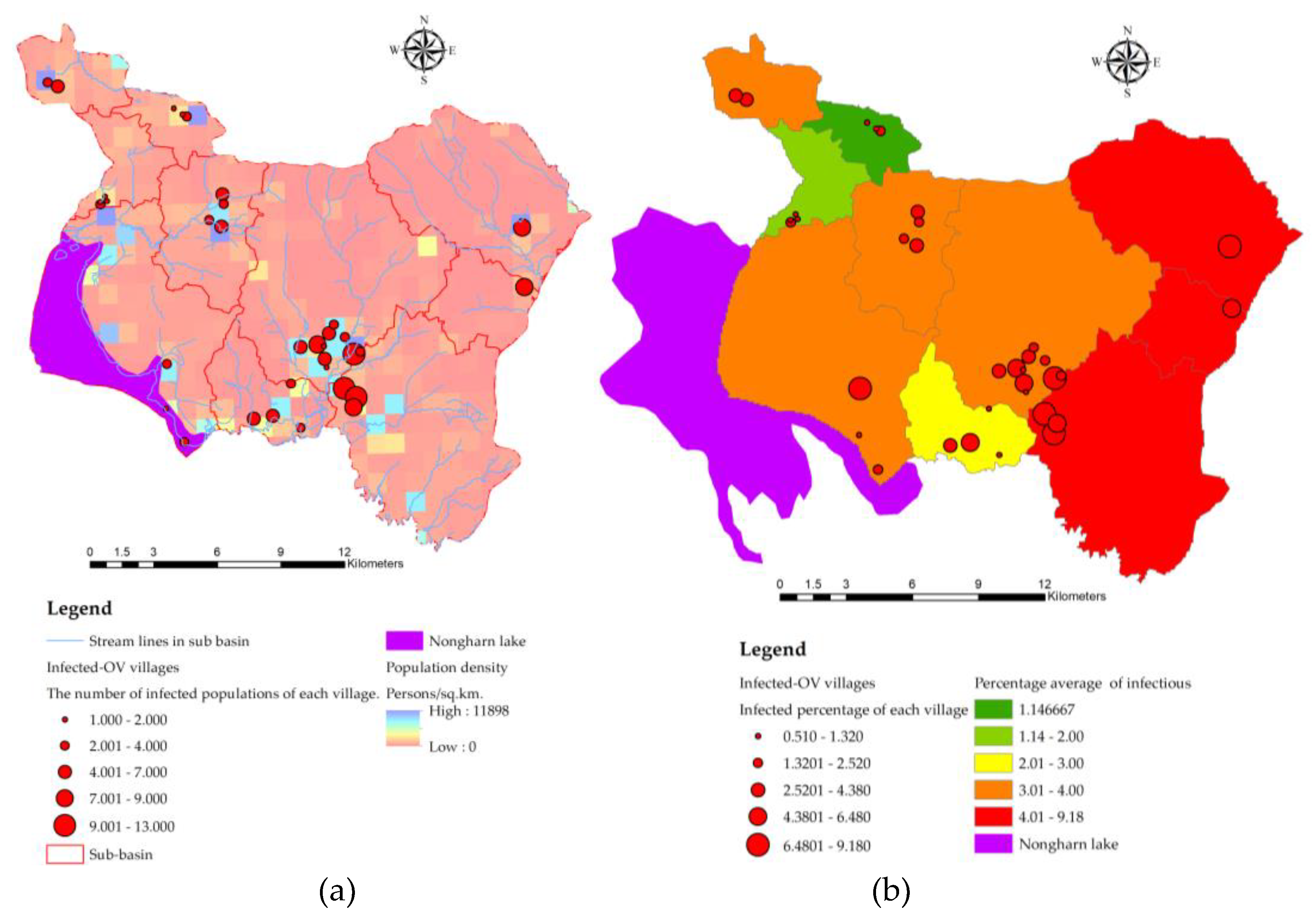

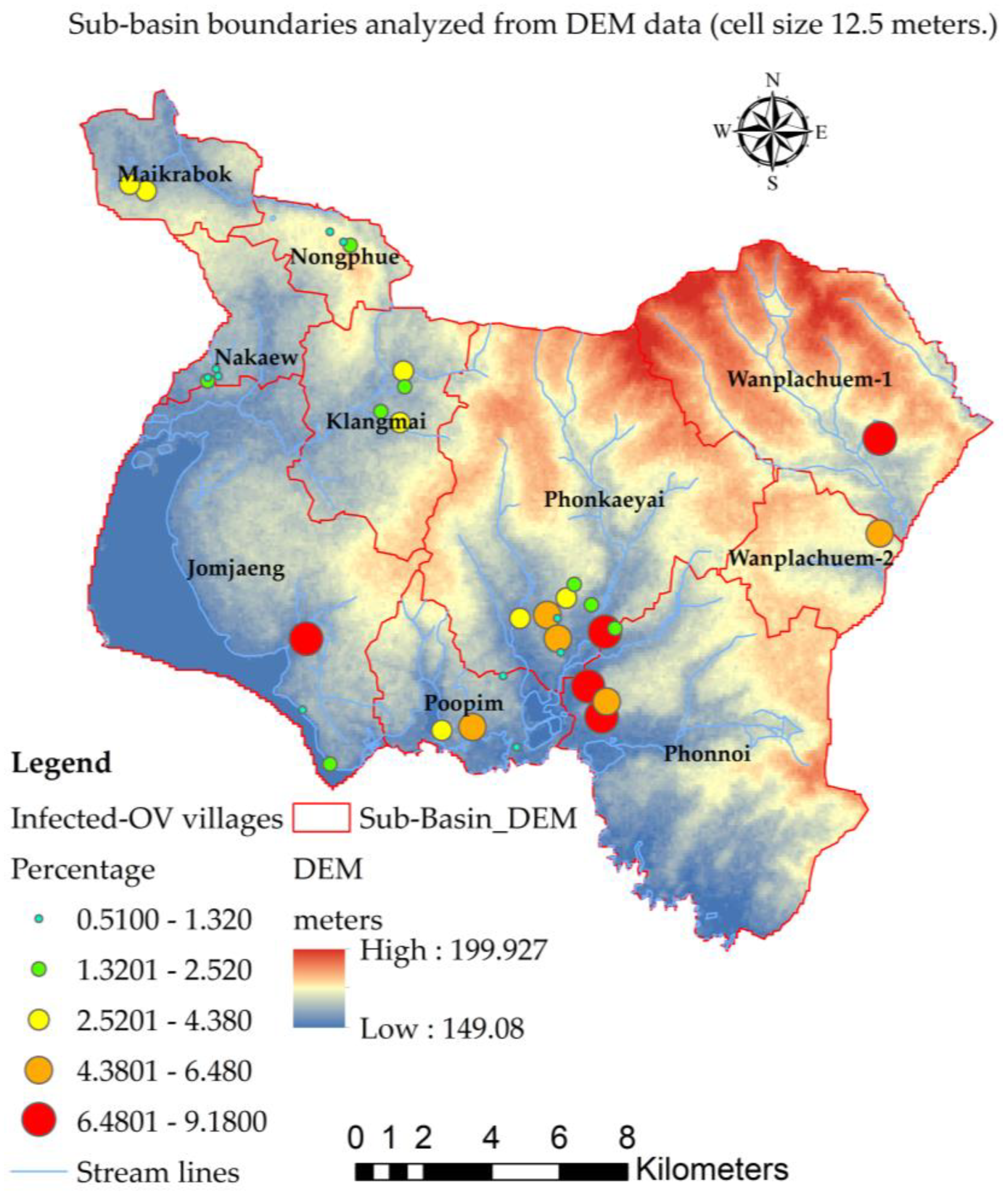

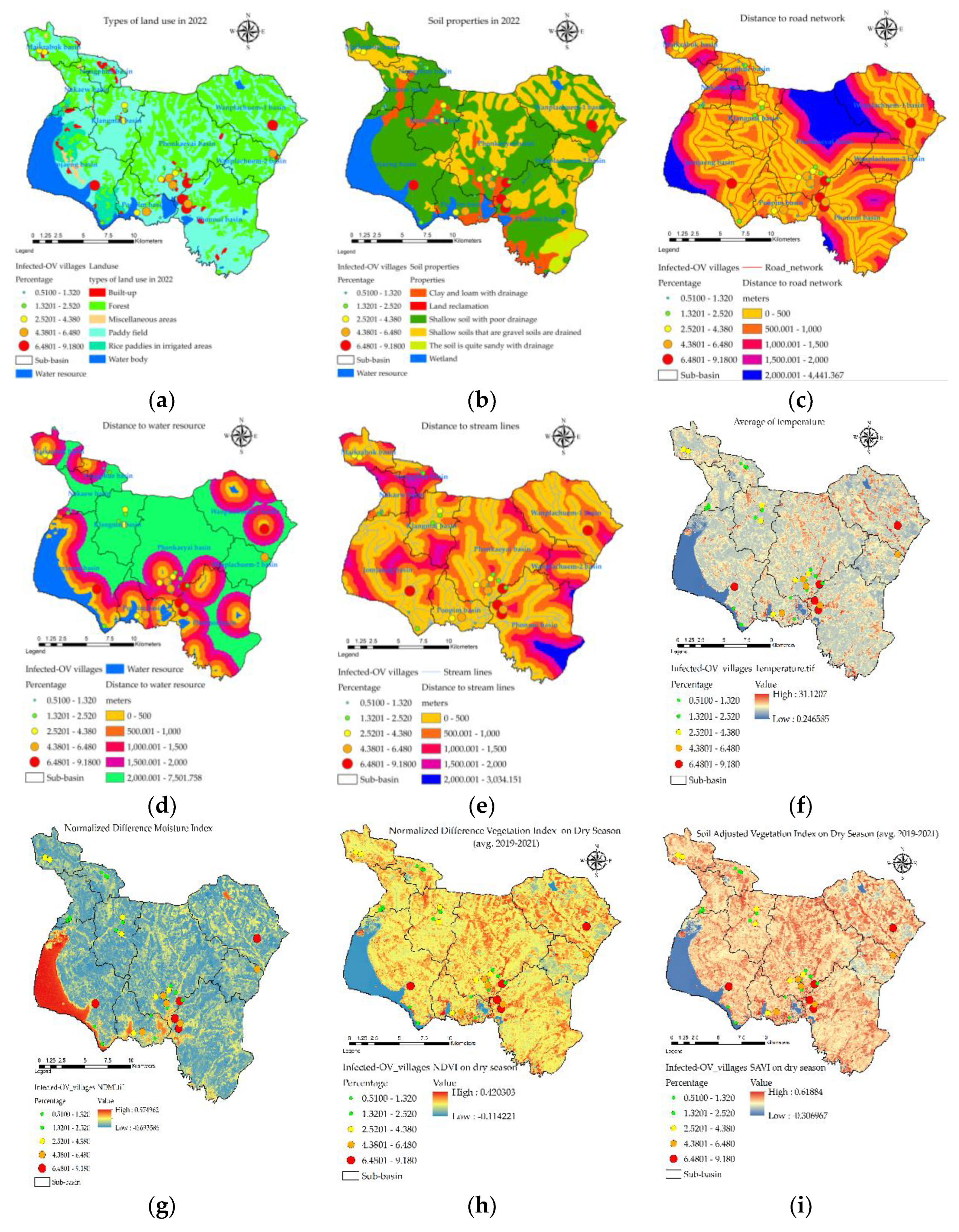
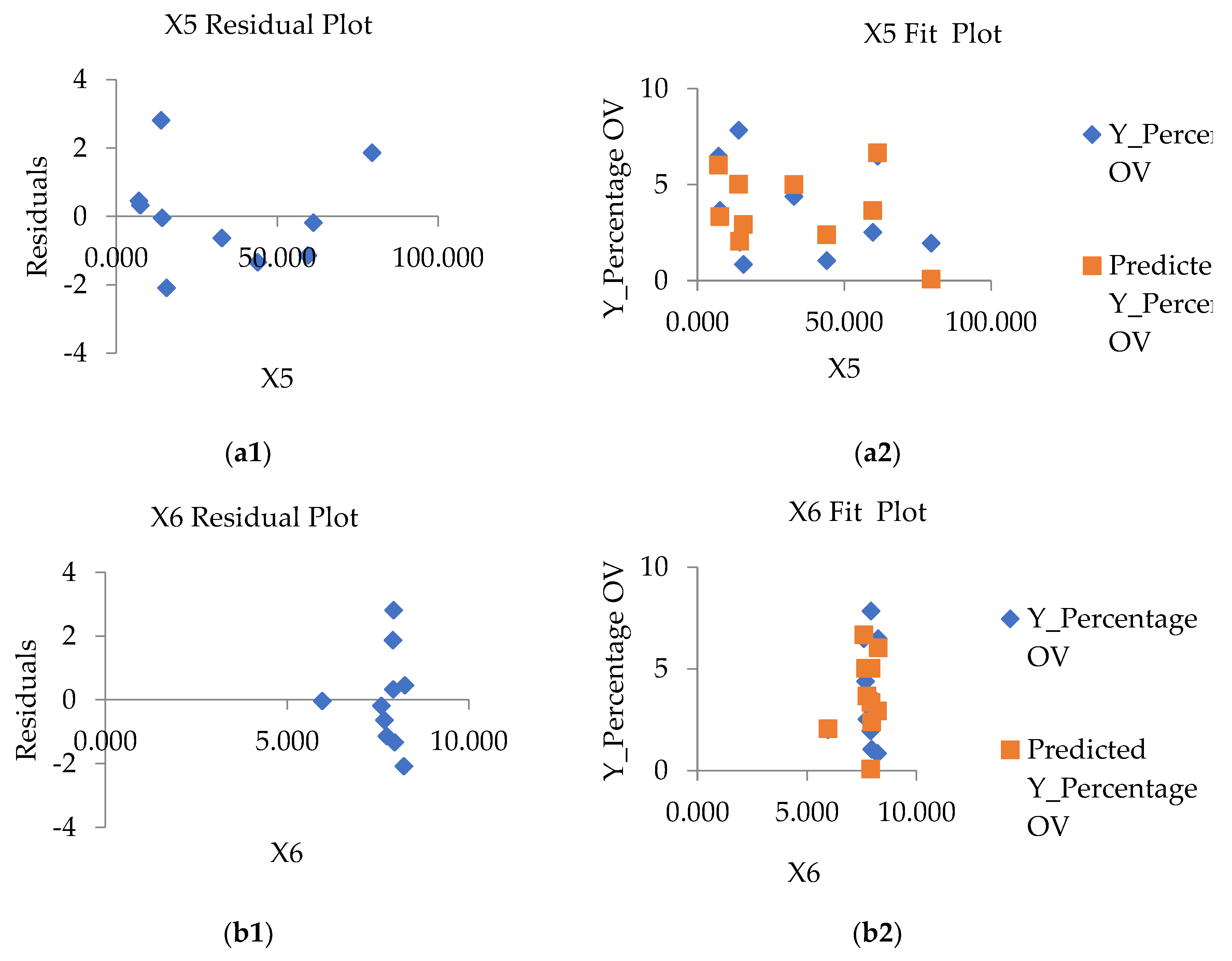
| Provinces | Number of people with | Number of people with |
|---|---|---|
| cholangiocarcinoma in 2019 | cholangiocarcinoma in 2020 | |
| Nongkhai | 22 | 37 |
| Buengkarn | 8 | 7 |
| Loei | 54 | 84 |
| Nakhon Phanom | 7 | 10 |
| Udon Thani | 50 | 88 |
| Nongbualumphu | 19 | 12 |
| Sakon Nakhon | 161 | 130 |
| Sub-basin name | Areas (sq.km.) | Perimeters (km.) | Average of DEM (meters) |
|---|---|---|---|
| Jomjaeng | 66.183 | 46.434 | 160.445 |
| Poopim | 18.432 | 23.986 | 161.624 |
| Phonnoi | 64.081 | 46.779 | 166.147 |
| Phonkaeyai | 72.794 | 43.279 | 172.894 |
| Wanplachuem-1 | 51.264 | 36.087 | 180.397 |
| Wanplachuem-2 | 16.071 | 20.354 | 174.412 |
| Klangmai | 27.738 | 28.053 | 167.640 |
| Nakaew | 12.708 | 24.316 | 164.155 |
| Nongphue | 10.040 | 18.698 | 170.894 |
| Maikrabok | 14.259 | 19.312 | 163.884 |
| Sub-basin | Y(% of OV) | X1(lu) | X2(soil) | X3(road) | X4(water) | X5(stream) | X6(temp) | X7(ndmi) | X8(ndvi) | X9(savi) |
|---|---|---|---|---|---|---|---|---|---|---|
| Jomjaeng | 2.01 | 14.773 | 9.144 | 14.755 | 7.361 | 14.293 | 5.966 | -0.064 | 0.075 | 0.143 |
| Poopim | 1.05 | 49.947 | 33.838 | 47.748 | 29.494 | 43.984 | 7.954 | -0.060 | 0.115 | 0.218 |
| Phonnoi | 7.84 | 17.688 | 6.252 | 15.376 | 6.922 | 13.931 | 7.925 | -0.083 | 0.118 | 0.225 |
| Phonkaeyai | 0.84 | 14.279 | 11.576 | 14.489 | 6.149 | 15.661 | 8.210 | -0.081 | 0.124 | 0.237 |
| Wanplachuem-1 | 9.18 | 20.042 | 8.565 | 19.993 | 5.129 | 7.122 | 8.241 | 0.152 | 0.119 | 0.224 |
| Wanplachuem-2 | 6.48 | 60.884 | 24.128 | 64.132 | 14.349 | 61.311 | 7.593 | -0.037 | 0.104 | 0.199 |
| Klangmai | 4.38 | 37.048 | 24.577 | 37.011 | 5.862 | 32.838 | 7.677 | 0.042 | 0.117 | 0.227 |
| Nakaew | 2.52 | 60.758 | 29.858 | 74.811 | 40.229 | 59.603 | 7.740 | -0.035 | 0.116 | 0.224 |
| Nongphue | 1.95 | 80.795 | 34.581 | 90.847 | 18.963 | 79.482 | 7.909 | -0.049 | 0.119 | 0.227 |
| Maikrabok | 3.66 | 5.235 | 19.740 | 4.753 | 15.510 | 7.539 | 7.920 | -0.050 | 0.121 | 0.232 |
| Y(% of OV) | X1(lu) | X2(soil) | X3(road) | X4(water) | X5(stream) | X6(Temp) | X7(ndmi) | X8(ndvi) | X9(savi) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Y(% of OV) | 1.000 | - | - | - | - | - | - | - | - | - |
| X1(land use) | -0.167 | 1.000 | - | - | - | - | - | - | - | |
| X2(soil) | -0.437 | 0.826 | 1.000 | - | - | - | - | - | - | - |
| X3(road) | -0.189 | 0.992 | 0.813 | 1.000 | - | - | - | - | - | - |
| X4(water) | -0.402 | 0.599 | 0.739 | 0.635 | 1.000 | - | - | - | - | - |
| X5(stream) | -0.226 | 0.985 | 0.838 | 0.984 | 0.612 | 1.000 | - | - | - | - |
| X6(temp) | 0.173 | 0.116 | 0.184 | 0.106 | 0.109 | 0.067 | 1.000 | - | - | - |
| X7(ndmi) | 0.395 | 0.060 | -0.143 | -0.061 | -0.258 | -0.193 | 0.243 | 1.000 | - | - |
| X8(ndvi) | 0.082 | 0.092 | 0.227 | 0.095 | 0.134 | 0.062 | 0.969 | 0.171 | 1.000 | - |
| X9(savi) | 0.079 | 0.097 | 0.242 | 0.103 | 0.144 | 0.074 | 0.950 | 0.150 | 0.997 | 1.000 |
| GWR models | Independent Variables |
coefficients | t-Stat |
p-Valuea GWR |
R2 GWR |
R2 OLS |
|---|---|---|---|---|---|---|
| Y%ov1=0.475+1.525(X8ndvi)+ 6.021(X9savi) |
Intercept | 0.475 | 4.573*** | 0.000*** | 0.463 | 0.445 |
| X8ndvi | 1.525 | 0.918 n/s | 0.236 n/s | |||
| X9savi | 6.021 | 2.152 n/s | 0.135 n/s | |||
| Y%ov2=4.528+1.125(X7ndmi)+ 3.116(X8ndvi)-9.852(X9savi) |
Intercept | 4.528 | 1.975*** | 0.000*** | 0.521 | 0.483 |
| X7ndmi | 1.125 | 0.799 n/s | 1.154 n/s | |||
| X8ndvi | 3.116 | 0.890 n/s | 2.021 n/s | |||
| X9savi | -9.852 | -2.326*** | 0.038*** | |||
| Y%ov3=62.042-5.047(X5stream)+ 4.246 (X7ndmi)-9.874(X9savi) |
Intercept | 62.042 | 3.031*** | 0.000*** | 0.624 | 0.576 |
| X5stream | -5.047 | -2.068*** | 0.048*** | |||
| X7ndmi | 4.246 | 1.875 *** | 0.034 *** | |||
| X9savi | -9.874 | -2.661*** | 0.021*** | |||
| Y%ov4=59.410.039(X5stream)+21.21(X6temp)+7.23(X7ndmi)-3752.16(X8ndvi+1503.27(X9savi) | Intercept | 59.410 | 0.999*** | 0.000*** | 0.646 | 0.591 |
| X5stream | -0.0390 | -3.561*** | 0.041*** | |||
| X6temp | 21.210 | 0.774 n/s | 1.243 n/s | |||
| X7ndmi | 7.230 | 0.550 n/s | 0.764 n/s | |||
| X8ndvi | 1503.270 | 0.678 n/s | 0.689 n/s | |||
| X9savi | -2752.160 | -2.156*** | 0.037*** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





