Submitted:
13 June 2023
Posted:
14 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
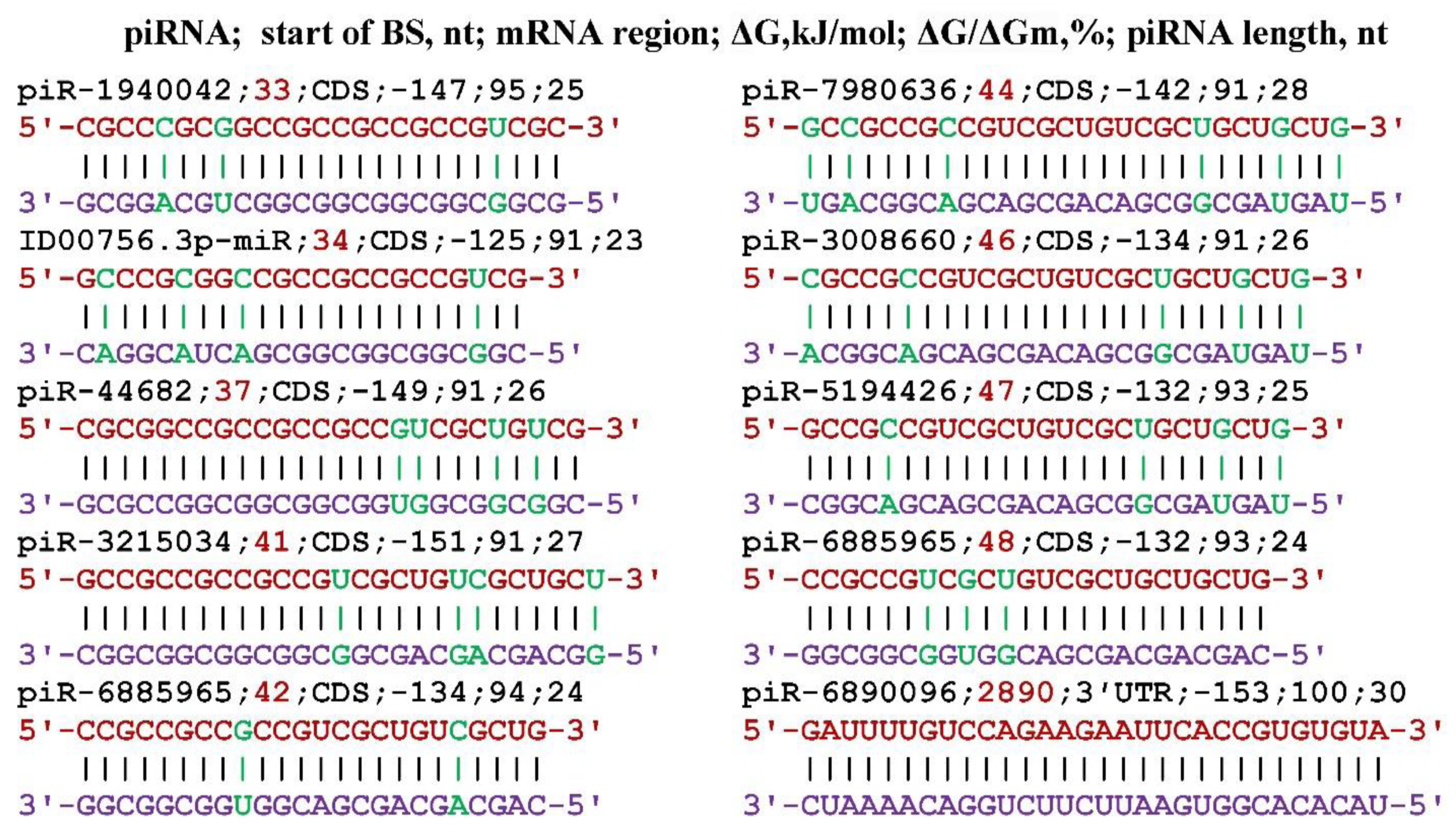
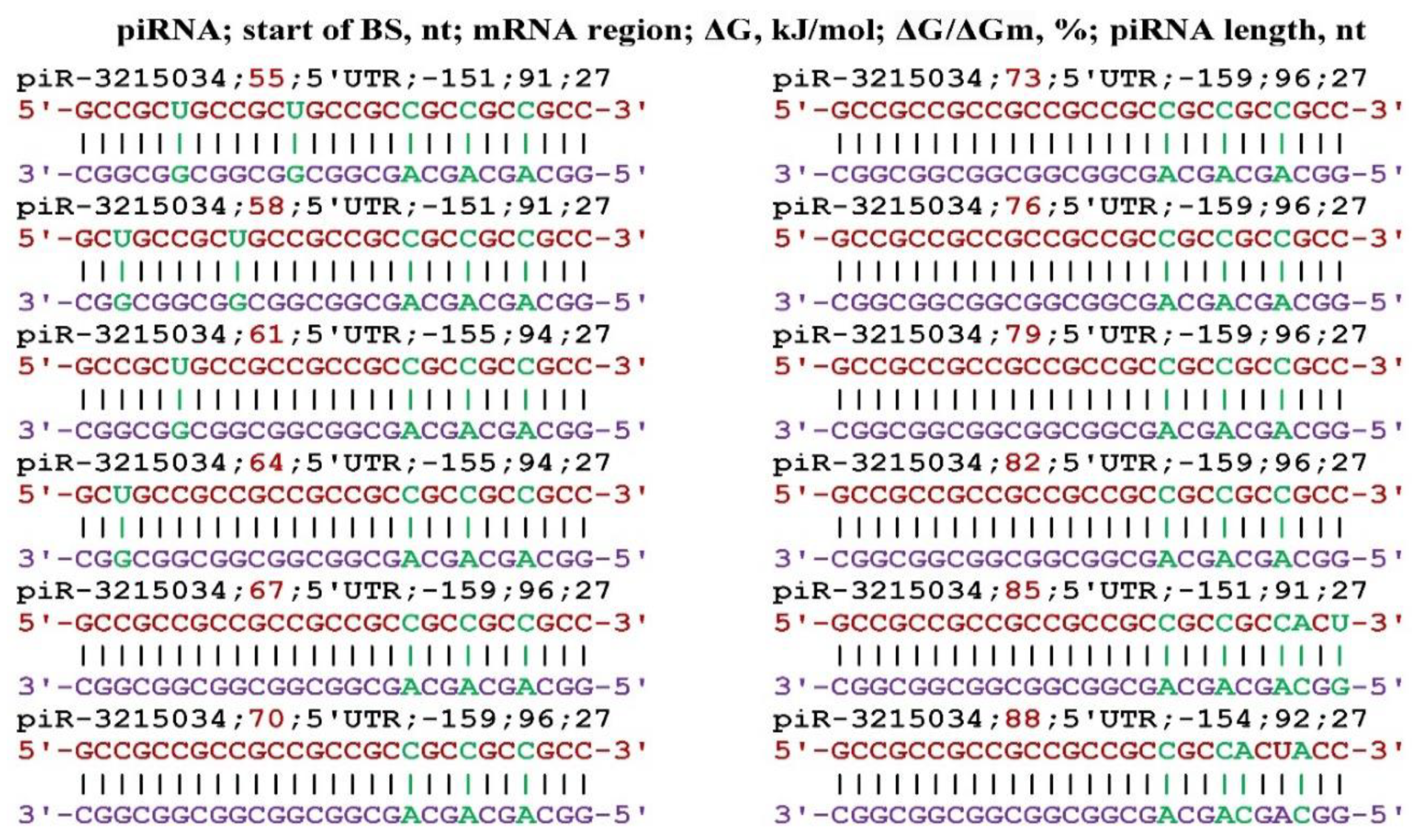
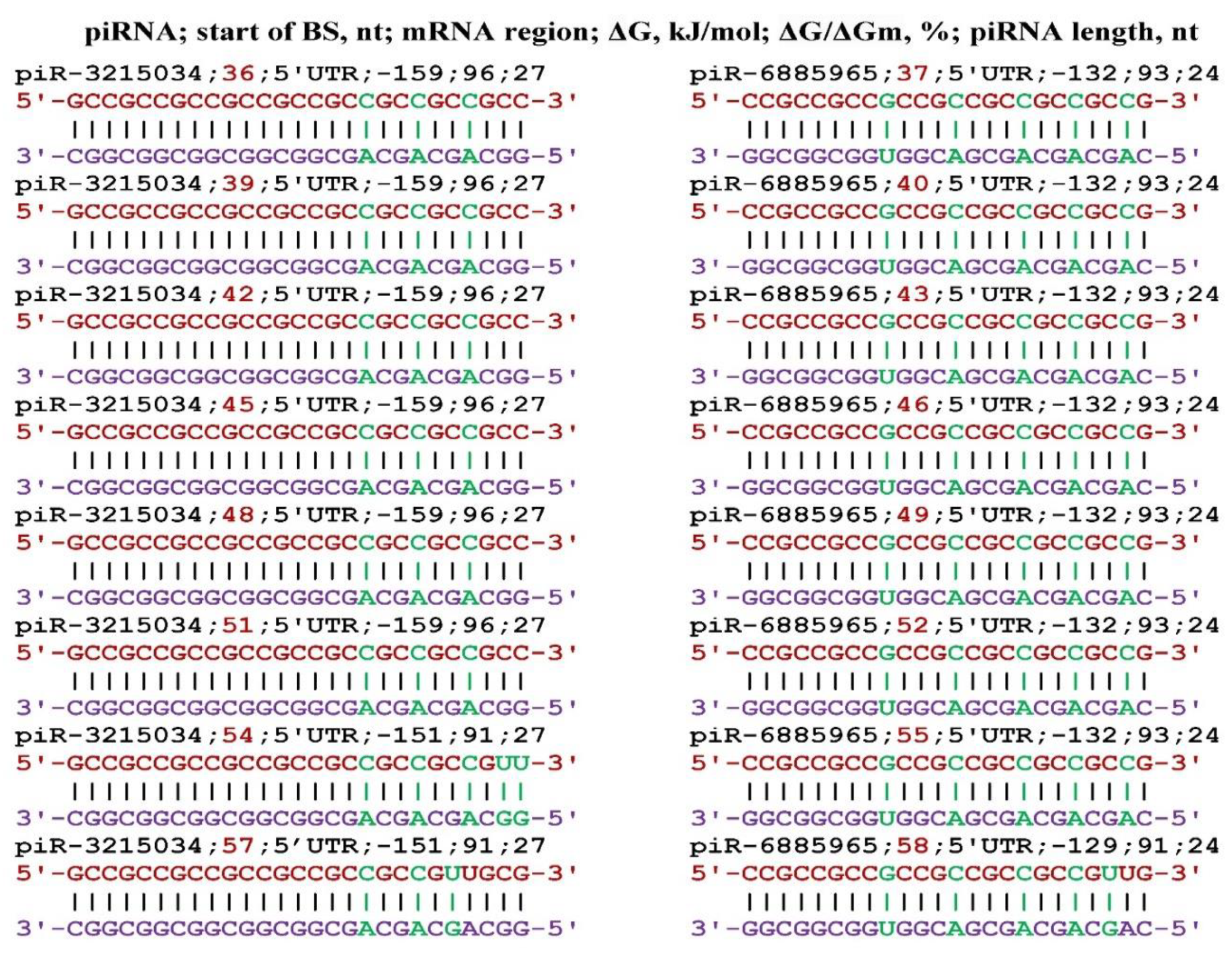
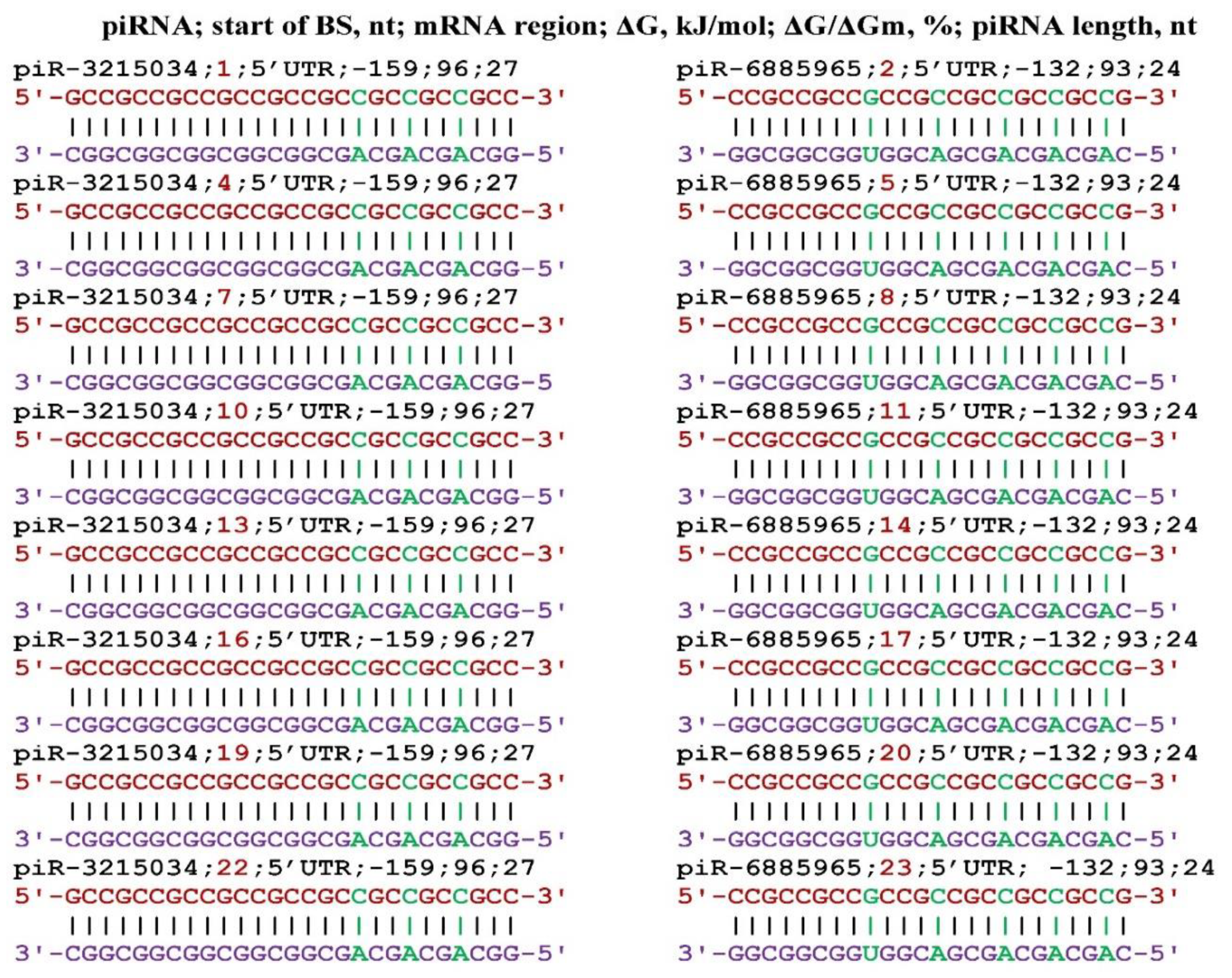
3. Results
4. Discussion
References
- Kuro-o, M.; Matsumura, Y.; Aizawa, H.; Kawaguchi, H.; Suga, T.; Utsugi, T.; Ohyama, Y.; Kurabayashi, M.; Kaname, T.; Kume, E.; et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997, 390, 45–51. [Google Scholar] [CrossRef]
- Li, S.S.; Sheng, M.J.; Sun, Z.Y.; Liang. Y.; Yu, L.X.; Liu, Q.F. Upstream and downstream regulators of Klotho expression in chronic kidney disease. Metabolism 2023, 142, 155530. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Zhang, Y.; Yin, Z. The protective mechanism of Klotho gene-modified bone marrow mesenchymal stem cells on acute kidney injury induced by rhabdomyolysis. Regen Ther. 2021, 13, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jiang, W.; Li, Y.; Gu, W.; Huang, H.; Wei, Q.; Bai, G.; Wang, J.; Rizak, J.D.; Zhou, Z. Evaluation of Klotho gene expression and NGAL levels following acute kidney injury during pregnancy hypertensive disorders. Pregnancy Hypertens 2022, 30, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Cao, W. Epigenetic modifications of Klotho expression in kidney diseases. J Mol Med (Berl) 2021, 99, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.S.; Aamir, A.; Khan, A.; Khan, Z,; Shah, S. Q. et al. Investigation of Klotho G395A and C1818T Polymorphisms and Their Association with Serum Glucose Level and Risk of Type 2 Diabetes Mellitus. Genes (Basel) 2022, 13, 1532. [Google Scholar] [CrossRef] [PubMed]
- Speer, T.; Schunk, S.J. Klotho in diabetic kidney disease: more than dust in the Wnt. Kidney Int 2022, 102, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Banchs, P.A.P.; Liu, Y.; Fu, H.; Arena, V.C.; Forno, E.; Libman, I.; Ho, J.; Muzumdar, R. Serum α-KL, a potential early marker of diabetes complications in youth with T1D, is regulated by miRNA 192. Front Endocrinol (Lausanne) 2022, 13, 937093. [Google Scholar] [CrossRef]
- Typiak, M, Piwkowska, A. Antiinflammatory Actions of Klotho: Implications for Therapy of Diabetic Nephropathy. Int J Mol Sci. 2021, 22, 956. [Google Scholar] [CrossRef]
- Iijima, H.; Gilmer, G.; et al. Age-related matrix stiffening epigenetically regulates α-Klotho expression and compromises chondrocyte integrity. Nat Commun 2023, 14, 18. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, J.; Kalantar-Zadeh, K.; Chen, J. Focusing on Phosphorus Loads: From Healthy People to Chronic Kidney Disease. Nutrients 2023, 15, 1236. [Google Scholar] [CrossRef]
- Liu, S.; Yao, W. Prediction of lung cancer using gene expression and deep learning with KL divergence gene selection. BMC Bioinformatics 2022, 23, 175. [Google Scholar] [CrossRef]
- Xie, B.; Chen, J.; Liu, B.; Zhan, J. Klotho acts as a tumor suppressor in cancers. Pathol Oncol Res 2013, 19, 611–617. [Google Scholar] [CrossRef]
- Kim, G.; Chung, H.; Lee, S.; Kim, W.H. Reduced Klotho expression and its prognostic significance in canine hepatocellular carcinoma. Vet Comp Oncol 2023, 21, 91–99. [Google Scholar] [CrossRef]
- Chung, H.; Lee, S.; Kim, G.A.; Kim, W.H. Down-expression of klotho in canine mammary gland tumors and its prognostic significance. PLoS One 2022, 17, e0265248. [Google Scholar] [CrossRef]
- Xie, H.; Li, N.; Zhou, G.; Liu, Q.; Wang, H.; Han, J.; Shen, L.; Yu, P.; Chen, J.; Chen, X. Plasma S-Klotho level affects the risk of hyperuricemia in the middle-aged and elderly people. Eur J Med Res 2022, 27, 262. [Google Scholar] [CrossRef]
- Abraham, C.R.; Li, A. Aging-suppressor Klotho: Prospects in diagnostics and therapeuticAgeing Res Rev 2022, 82, 101766. 82. [CrossRef]
- Donate-Correa, J.; Matos-Perdomo, E.; González-Luis, A.; Martín-Olivera, A.; Ortiz, A.; Mora-Fernández, C.; Navarro-González, J.F. The Value of Klotho in Kidney Transplantation. Transplantation 2023, 107, 616–627. [Google Scholar] [CrossRef]
- Topal, M.; Guney, I. The association of soluble Klotho levels with anemia and hemoglobin variability in hemodialysis patients. Semin Dial 2023, 36, 142–146. [Google Scholar] [CrossRef]
- Prud’homme, G.J.; Kurt, M.; Wang, Q. Pathobiology of the Klotho Antiaging Protein and Therapeutic Considerations. Front Aging 2022, 3, 931331. [Google Scholar] [CrossRef]
- Xu, J.P.; Zeng, R.X.; He, M.H.; Lin, S.S.; Guo, L.H.; Zhang, M.Z. Associations Between Serum Soluble α-Klotho and the Prevalence of Specific Cardiovascular Disease. Front Cardiovasc Med 2022, 9, 899307. [Google Scholar] [CrossRef]
- Orces, C.H. The association between metabolic syndrome and the anti-aging humoral factor klotho in middle-aged and older adults. Diabetes Metab Syndr 2022, 16, 102522. [Google Scholar] [CrossRef]
- Kim, H.J; Kim, Y.; Kang, M.; Kim, S.; Park, S.K; Sung, S.; Hyun, Y.Y.; Jung, J.Y.; Ahn, C.; Oh, K.H. Low Klotho/Fibroblast Growth Factor 23 Ratio Is an Independent Risk Factor for Renal Progression in Chronic Kidney Disease: Finding From KNOW-CKD. Front Med (Lausanne) 2022, 9, 904963. [Google Scholar] [CrossRef]
- Saxena, A.; Sachan, T.; Gupta, A.; Kapoor, V. Effect of Dietary Phosphorous Restriction on Fibroblast Growth 2 Factor-23 and sKlotho Levels in Patients with Stages 1-2 Chronic Kidney Disease. Nutrients 2022, 14, 3302. [Google Scholar] [CrossRef]
- Portales-Castillo, I.; Simic, P. PTH, FGF-23, Klotho and Vitamin D as regulators of calcium and phosphorus: Genetics, epigenetics and beyond. Front Endocrinol (Lausanne) 2022, 13, 992666. [Google Scholar] [CrossRef]
- Terzi Demirsoy, E.; Mehtap, O.; Birtas Atesoglu, E.; Tarkun, P.; Gedük, A.; Eren, N.; Hacihanefioglu, A. Prognostic Value of Serum Soluble Klotho and Fibroblast Growth Factor-23 in Multiple Myeloma Patients. Indian J Hematol Blood Transfus 2022, 38, 454–463. [Google Scholar] [CrossRef]
- Biscetti, F.; Rando, M.M.; Cecchini, A.L.; Nicolazzi, M.A.; Rossini, E.; Angelini, F.; Iezzi, R.; Eraso, L.H.; Dimuzio, P.J.; Pitocco, D.; Gasbarrini, A.; Massetti, M.; Flex, A. The role of Klotho and FGF23 in cardiovascular outcomes of diabetic patients with chronic limb threatening ischemia: a prospective study. Sci Rep 2023, 13, 6150. [Google Scholar] [CrossRef]
- Mizuno, Y.; Ishida, T.; Kugimiya, F.; Takai, S.; Nakayama, Y.; Yonemitsu, K.; Harada, E. Deterioration of Phosphate Homeostasis Is a Trigger for Cardiac Afterload- Clinical Importance of Fibroblast Growth Factor 23 for Accelerated. Aging Circ Rep 2022, 5, 4–12. [Google Scholar] [CrossRef]
- Castelblanco, E.; Hernández, M.; Alonso, N.; Ribes-Betriu, A.; Real, J. , Granado-Casas, M.; Rossell, J.; Rojo-López M.I.; Dusso, A.S.; Julve, J.; Mauricio, D. Association of α-klotho with subclinical carotid atherosclerosis in subjects with type 1 diabetes mellitus. Cardiovasc Diabetol 2022, 21, 207. [Google Scholar] [CrossRef]
- Reggiani, F.; Moroni, G.; Ponticelli, C. Cardiovascular Risk after Kidney Transplantation: Causes and Current Approaches to a Relevant Burden J Pers Med 2022, 12, 1200. 12. [CrossRef]
- Liu, S.H.; Xiao, Z.; Mishra, S.K.; Mitchell, J.C.; Smith, J.C.; Quarles, L.D.; Petridis, L. Identification of Small-Molecule Inhibitors of Fibroblast Growth Factor 23 Signaling via In Silico Hot Spot Prediction and Molecular Docking to α-Klotho. J Chem Inf Model 2022, 62, 3627–3637. [Google Scholar] [CrossRef]
- Abiola, B.I.; Raji, Y.R.; Ajayi, S.; Adeoye, A.M.; Salako, B.L.; Arije, A.; Kadiri, S. Comparative analysis of fibroblast growth Factor-23 as a correlate of cardiovascular disease among individuals with chronic kidney disease, hypertensives, and healthy controls. Niger J Clin Pract 2022, 25, 1247–1255. [Google Scholar] [CrossRef]
- Golüke, N.M.S.; Schoffelmeer, M.A.; De Jonghe, A.; Emmelot-Vonk, M.H.; De Jong, P.A.; Koek, H.L. Serum biomarkers for arterial calcification in humans: A systematic review Bone Rep, 2022, 17, 101599. 17. [CrossRef]
- Biscetti, F.; Rando, M.M.; Cecchini, A.L.; Nicolazzi, M.A.; Rossini, E.; Angelini, F.; Iezzi, R.; Eraso, L.H.; Dimuzio, P.J.; Pitocco, D.; Gasbarrini, A.; Massetti, M.; Flex, A. The role of Klotho and FGF23 in cardiovascular outcomes of diabetic patients with chronic limb threatening ischemia: a prospective study. Sci Rep 2023, 13, 6150. [Google Scholar] [CrossRef]
- Wang, Y.P.; Sidibé, A.; Fortier, C.; Desjardins, M.P.; Ung, R.V.; Kremer, R.; Agharazii, M.; Mac-Way, F. Wnt/β-catenin pathway inhibitors, bone metabolism and vascular health in kidney transplant patients. J Nephrol 2023. [Google Scholar] [CrossRef]
- Bishop, N.C.; Burton, J.O.; Graham-Brown, M.P.M.; Stensel, D.J.; Viana, J.L.; Watson, E.L. Exercise and chronic kidney disease: potential mechanisms underlying the physiological benefits. Nat Rev Nephrol 2023, 19, 244–256. [Google Scholar] [CrossRef]
- Yanucil, C.; Kentrup, D.; Campos, I.; Czaya, B.; Heitman. K.; Westbrook, D.; Osis, G.; Grabner, A.; Wende, A.R.; Vallejo, J.; Wacker, M.J.; Navarro-Garcia, J.A.; Ruiz-Hurtado, G.; Zhang, F.; Song, Y.; Linhardt, R.J.; White, K.; Kapiloff, M.S.; Faul, C. Kidney Int 2022, 102, 261–279. [Google Scholar] [CrossRef]
- Kužmová, Z.; Kužma, M.; Gažová, A.; Kovářová, M.; Jackuliak, P.; Killinger, Z.; Kyselovič, J.; Payer, J. Fibroblast Growth Factor 23 and Klotho Are Associated With Trabecular Bone Score but Not Bone Mineral Density in the Early Stages of Chronic Kidney Disease: Results of the Cross-Sectional Study. Physiol Res 2021, 70, S43–S51. [Google Scholar] [CrossRef]
- Wang, J.; Shi, Y.; Zhou, H.; Zhang, P.; Song, T.; Ying, Z.; Yu, H.; Li, Y.; Zhao, Y.; Zeng, X.; He, S.; Chen, R. piRBase: integrating piRNA annotation in all aspects. Nucleic Acids Res 2021, 50, 265–272. [Google Scholar] [CrossRef]
- Londin, E.; Loher, P.; Telonis, A.G.; et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci USA 2015, 112, E1106–E1115. [Google Scholar] [CrossRef]
- Backes, C.; Meder, B.; Hart, M.; Ludwig, N.; Leidinger, P.; Vogel, B.; Galata, V.; Roth, P.; Menegatti, J.; Grasser, F.A.; et al. Prioritizing and selecting likely novel miRNAs from NGS data. Nucleic Acids Res 2015, 44, e53. [Google Scholar] [CrossRef]
- Ivashchenko, A.; Berillo, O.; Pyrkova, A.; Niyazova, R.; Atambayeva, S. MiR-3960 binding sites with mRNA of human genes. Bioinformation 2014, 10, 423–427. [Google Scholar] [CrossRef]
- Friedman RA, Honig BA. Free Energy Analysis of Nucleic Acid Base Stacking in Aqueous Solution. Biophys. J 1995, 69, 1528–1535. [Google Scholar] [CrossRef]
- Garg, A.; Heinemann, U.A. Novel Form of RNA Double Helix Based on G·U and C·A+ Wobble Base Pairing. RNA, 2018, 24, 209–218. [Google Scholar] [CrossRef]
- Leontis, N.B.; Stombaugh, J.; Westhof, E. The Non-watson-crick Base Pairs and Their Associated Isostericity Matrices. Nucleic Acids Res 2002, 30, 3497–3531. [Google Scholar] [CrossRef]
- Kool, E.T. Hydrogen Bonding, Base Stacking, and Steric Effects in DNA Replication. Annu. Rev. Biophys. Biomol. Struct 2001, 30, 1–22. [Google Scholar] [CrossRef]
- Davis, E.; Caiment, F.; Tordoir, X.; Cavaillé, J.; Ferguson-Smith, A.; Cockett, N.; Georges, M.; Charlier, C. RNAi-Mediated Allelic Trans-interaction at the Imprinted Rtl1/ Peg11 Locus. Curr. Biol 2005, 15, 743–749. [Google Scholar] [CrossRef]
- Atambayeva, S.; Niyazova, R.; Ivashchenko, A.; Pyrkova, A.; Pinsky, I.; Akimniyazova, A.; Labeit, S. The Binding Sites of miR-619-5p in the mRNAs of Human and Orthologous Genes. BMC Genom 2017, 18, 428. [Google Scholar] [CrossRef]
- Belkozhayev, A.; Niyazova, R.; Wilson, C.; Jainakbayev, N.; Pyrkova, A.; Ashirbekov, Y.; Akimniyazova, A.; Sharipov, K.; Ivashchenko, A. Bioinformatics Analysis of the Interaction of miRNAs and piRNAs with Human mRNA Genes Having di- and Trinucleotide Repeats. Genes, 2022, 13, 800. [Google Scholar] [CrossRef]
- Kondybayeva, А.М.; Akimniyazova, A.N.; Kamenova, S.U.; Ivashchenko, А.Т. The characteristics of miRNA binding sites in mRNA of ZFHX3 gene and its orthologs. Vavilov journal of genetics and breeding 2018, 22, p–438. [Google Scholar] [CrossRef]
- Kamenova, S.; Sharapkhanova, A.; Akimniyazova, A.; Kuzhybayeva, K.; Kondybayeva, A.; Rakhmetullina, A.; Pyrkova, A.; Ivashchenko, A. piRNA and miRNA Can Suppress the Expression of Multiple Sclerosis Candidate Genes. Nanomaterials (Basel) 2023, 13, 22. [Google Scholar] [CrossRef]
- Kuo, Y.J.; Lewis, J.S.; Zhai, C.; Chen, Y.A.; Chernock, R.D.; Hsieh, M.S.; Lan, M.Y.; Lee, C.K.; Weinreb, I.; Hang, J.F. DEK-AFF2 fusion-associated papillary squamous cell carcinoma of the sinonasal tract: clinicopathologic characterization of seven cases with deceptively bland morphology. Mod Pathol, 2021, 34, 1820–1830. [Google Scholar] [CrossRef]
- Kuo, Y.J.; Lewis, J.S.Jr.; Truong, T.; Yeh, Y.C.; Chernock, R.D.; Zhai, C.; Chen, Y.A.; Hongo, T.; Lee, C.K.; Shi, Q.; Velez Torres, J.M.; Geromes, A.B.; Chu, Y.H.; Hsieh, M.S.; Yamamoto, H.; Weinreb, I.; Hang, J.F. Nuclear expression of AFF2 C-terminus is a sensitive and specific ancillary marker for DEK: AFF2 carcinoma of the sinonasal tract. Mod Pathol 2022, 35, 1587–1595. [Google Scholar] [CrossRef]
- Savari, O.; Chang, J.C.; Bishop, J.A.; Sakthivel, M.K.; Askin, F.B.; Rekhtman, N. First Report of Thoracic Carcinoma With DEK: AFF2 Rearrangement: A Case Report. J Thorac Oncol 2022, 17, 1050–1053. [Google Scholar] [CrossRef]
- Ruangritchankul, K.; Sandison, A. DEK: AFF2 Fusion Carcinomas of Head and Neck. Adv Anat Pathol 2023, 30, 86–94. [Google Scholar] [CrossRef]
- Ji, Z.; Lu, R.; Wu, T.; Chen, Z.; Wen, Z.; Li, Z.; Zheng, X.; Tang, J.; Chen, X.; Yang. Y.; Zheng. Q. Expression profiling of circular RNA reveals a potential miR-145-5p sponge function of circ-AFF2 and circ-ASAP1 in renal cell carcinoma. Am J Transl Res 2023, 15, 82–98. [Google Scholar]
- Ji, Z.; Lu, R.; Wu, T. , Chen, Z., Wen, Z., Li, Z., Zheng, X., Tang, J., Chen, X., Yang, Y., Zheng, Q. Expression profiling of circular RNA reveals a potential miR-145-5p sponge function of circ-AFF2 and circ-ASAP1 in renal cell carcinoma. Am J Transl Res 2023, 15, 82–98. [Google Scholar]
- Taherkhani, A.; Dehto, S.S.; Jamshidi, S.; Shojaei, S. Pathogenesis and prognosis of primary oral squamous cell carcinoma based on microRNAs target genes: a systems biology approach. Genomics Inform 2022, 20, e27. [Google Scholar] [CrossRef]
- Flores, D.; Lopez, A.; Udawant, S.; Gunn, B.; Keniry, M. The FOXO1 inhibitor AS1842856 triggers apoptosis in glioblastoma multiforme and basal-like breast cancer cells. FEBS Open Bio 2023, 13, 352–362. [Google Scholar] [CrossRef]
- Rose, M.M.; Espinoza, V.L.; Hoff, K.J.; Pike, L.A.; Sharma, V.; Hofmann, M.C.; Tan, A.C.; Pozdeyev, N. Schweppe RE. BCL2L11 Induction Mediates Sensitivity to Src and MEK1/2 Inhibition in Thyroid Cancer. Cancers (Basel) 2023, 15, 378. [Google Scholar] [CrossRef]
- Ranapour, S.; Motamed, N. Effect of Silibinin on the Expression of Mir-20b, Bcl2L11, and Erbb2 in Breast Cancer Cell Lines. Mol Biotechnol 2023. [Google Scholar] [CrossRef]
- Li, J.; Zheng, W.; Wu, J.; Zhang, J.; Lv, B.; Li, W.; Liu, J.; Zhang, X.; Huang, T.; Luo, Z. CPT1C-mediated fatty acid oxidation facilitates colorectal cancer cell proliferation and metastasis. Acta Biochim Biophys Sin (Shanghai) 2023. [Google Scholar] [CrossRef]
- Liao, L.; Zhang, F.; Zhuo, Z.; Huang, C.; Zhang, X.; Liu, R.; Gao, B.; Ding, S. Regulation of Fatty Acid Metabolism and Inhibition of Colorectal Cancer Progression by Erchen Decoction Evid Based Complement Alternat Med 2023, 9557720. [CrossRef]
- Xiong, L.; He, T.; Liu, C.; Qin, S.; Xiao, T.; Xin, W.; Wang, Y.; Ran, L.; Zhang, B.; Zhao, J. IL-37 Ameliorates Renal Fibrosis by Restoring CPT1A-Mediated Fatty Acid Oxidation in Diabetic Kidney Disease. Kidney Dis (Basel) 2023, 9, 104–117. [Google Scholar] [CrossRef]
- Li, S.; Liu, M.; Chen, J.; Chen, Y.; Yin, M.; Zhou, Y.; Li. Q.; Xu, F.; Li, Y.; Yan, X.; Xia, Y.; Chen, A.; Lu, D.; Li, C.; Shen, L.; Chen, Z.; Qian, J.; Ge, J. L-carnitine alleviates cardiac microvascular dysfunction in diabetic cardiomyopathy by enhancing PINK1-Parkin-dependent mitophagy through the CPT1a-PHB2-PARL pathways. Acta Physiol (Oxf) 2023, 12, e13975. [Google Scholar] [CrossRef]
- Tian, T.; Lu, Y.; Lin, J.; Chen, M.; Qiu, H.; Zhu, W.; Sun, H.; Huang, J.; Yang, H.; Deng, W. CPT1A promotes anoikis resistance in esophageal squamous cell carcinoma via redox homeostasis. Redox Biol. 2022, 58, 102544. [Google Scholar] [CrossRef]
- Bernard, J.N.; Chinnaiyan, V.; Andl, T.; Le Bras, G.F.; Qureshi, M.N.; Altomare, D.A.; Andl, C.D. Аugmented CPT1A Expression Is Associated with Proliferation and Colony Formation during Barrett’s Tumorigenesis. Int J Mol Sci 2022, 23, 11745. [Google Scholar] [CrossRef]
- Deng, J.J.; Li, G.P.; Lu, W.; Yan, Z.; Wang, Y. DAZAP1 overexpression promotes growth of HCC cell lines: a primary study using CEUS. Clin Transl Oncol 2022, 24, 1168–1176. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Wang, W.; Liu, B.; Yang, G.; Xu, Z.; Li, J.; Liu, Z. RNA binding protein DAZAP1 promotes HCC progression and regulates ferroptosis by interacting with SLC7A11 mRNA. Exp Cell Res 2021, 399, 112453. [Google Scholar] [CrossRef]
- Kim, M.C.; Park, M.H.; Kang, S.H.; Bae, Y.K. NDRG3 protein expression is associated with aggressive biologic phenotype and unfavorable outcome in patients with invasive breast cancer. Int J Clin Exp Pathol 2019, 12, 3886–3893. [Google Scholar]
- Liu, Y.; Xia, J.; Zhou, Y.; Shao, S. High expression of NDRG3 correlates with poor prognosis in gastric cancer patients. Rev Esp Enferm Dig 2021, 113, 524–528. [Google Scholar] [CrossRef]
- Ma, W.; Zhao, X.; Xue, N.; Gao, Y.; Xu, Q. The LINC01410/miR-122-5p/NDRG3 axis is involved in the proliferation and migration of osteosarcoma cells. IUBMB Life 2021, 73, 705–717. [Google Scholar] [CrossRef]
- Pappula, A.L.; Rasheed, S.; Mirzaei, G.; Petreaca, R.C.; Bouley, R.A. A Genome-Wide Profiling of Glioma Patients with an IDH1 Mutation Using the Catalogue of Somatic Mutations in Cancer Database. Cancers (Basel) 2021, 13, 4299. [Google Scholar] [CrossRef]
- Yin, X.; Yu, H.; He, X.K.; Yan, S.X. Prognostic and biological role of the N-Myc downstream-regulated gene family in hepatocellular carcinoma. World J Clin Cases 2022, 10, 2072–2086. [Google Scholar] [CrossRef]
- Liu, Y.; Xia, J.; Zheng, R.; Shao, S. High expression of NDRG3 suppresses cell apoptosis and promotes the cell proliferation and migration in gastric cancer. Asian J Surg 2022, 45, 2019–2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Quan, J.; Liu, J.; Tian, L.; Dong, C. Relationship between serum NDRG3 and papillary thyroid carcinoma. Front Endocrinol (Lausanne) 2022, 13, 1091462. [Google Scholar] [CrossRef]
- Zhang, H.; Ge, Z.; Wang, Z.; Gao, Y.; Wang, Y.; Qu, X. Circular RNA RHOT1 promotes progression and inhibits ferroptosis via mir-106a-5p/STAT3 axis in breast cancer. Aging (Albany NY) 2021, 13, 8115–8126. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yao, L.; Wei, Y.; Geng, S.; He, C.; Jiang, H. Role of RHOT1 on migration and proliferation of pancreatic cancer. Am J Cancer Res 2015, 5, 1460–70. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; He, C.; Geng, S.; Sheng, H.; Shen, X.; Zhang, X.; Li, H.; Zhu, S.; Chen, X.; Yang, C.; Gao, H. RhoT1 and Smad4 are correlated with lymph node metastasis and overall survival in pancreatic cancer. PLoS One. 2012, 7, e42234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cheng, F.; Zhang, Z.; Wang, B.; Zhang, X. Propofol suppresses non-small cell lung cancer tumorigenesis by regulation of circ-RHOT1/miR-326/FOXM1 axis. Life Sci. 2021, 119042. [Google Scholar] [CrossRef]
- Wang, L.; Long, H.; Zheng, Q.; Bo, X.; Xiao, X.; Li, B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer 2019, 18, 119. [Google Scholar] [CrossRef]
- Periñán, M.T.; Gómez-Garre, P.; Blauwendraat, C.; Mir, P.; Bandres-Ciga, S.; International Parkinson’s Disease Genomics Consortium (IPDGC). The role of RHOT1 and RHOT2 genetic variation on Parkinson disease risk and onset. Neurobiol Aging 2021, 97, 144–e1. [Google Scholar] [CrossRef]
- Sun, X.; Luo, Zh.; Gong, L.; Tan, X.; Chen, J.; Liang, X.; Cai, M. Identification of significant genes and therapeutic agents for breast cancer by integrated genomics. Bioengineered 2021, 12, 2140–2154. [Google Scholar] [CrossRef]
- Xing, Q.; Liu, S.; Luan, J.; Wang, Y.; Ma, L. A novel 13 RNA binding proteins (RBPs) signature could predict prostate cancer biochemical recurrence. Pathol Res Pract, 1: 225, 1535. [Google Scholar] [CrossRef]
- Henning, L.M.; Santos, K.F.; Sticht, J.; Jehle, S.; Lee, C.T.; Wittwer, M.; Urlaub, H.; Stelzl, U.; Wahl, M.C.; Freund, C. A new role for FBP21 as regulator of Brr2 helicase activity. Nucleic Acids Res 2017, 45, 7922–7937. [Google Scholar] [CrossRef]
- Henning, L.M.; Bhatia, S.; Bertazzon, M.; Marczynke, M.; Seitz, O.; Volkmer, R.; Haag, R.; Freund, C. Exploring monovalent and multivalent peptides for the inhibition of FBP21-tWW. Beilstein, J. Org Chem. 2015, 11, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Song, J.; Xu, T.; Liu, J.; Chai, J.; Yang, Y.; Li, L.; Li, M.; Yang, X. ZIC5 promotes aggressiveness and cancer stemness in cervical squamous cell carcinoma. Pathol Res Pract 2023, 241, 154268. [Google Scholar] [CrossRef] [PubMed]
- Satow, R.; Watanabe, T.; Nomura, M.; Inagaki, S.; Yoneda, A.; Fukami, K. Patulin and LL-Z1640-2 induce apoptosis of cancer cells by decreasing endogenous protein levels of Zic family member 5. J Cell Mol Med 2022, 26, 5680–5689. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.F.; Zhang, Y.; Ge, S.Y.; Zhong, F.; Sun, C.Y.; Xia, G.W. AR-regulated ZIC5 contributes to the aggressiveness of prostate cancer. Cell Death Discov 2022, 8, 393. [Google Scholar] [CrossRef]
- Song, W.; Yu, W.; Li, D.; Cheng, C.; Wu, X.; Chen, J.; Zhang, W. ZIC5 promotes human hepatocellular carcinoma cell proliferation through upregulating COL1A1. J Gastrointest Oncol 2022, 13, 1237–1247. [Google Scholar] [CrossRef]
- Chang, C.C.; Kuo, H.Y.; Chen, S.Y.; Lin, W.T.; Lu, K.M.; Saito, T.; Liu, F.C. Developmental Characterization of Schizophrenia-Associated Gene Zswim6 in Mouse Forebrain. Front Neuroanat 2021, 15, 669631. [Google Scholar] [CrossRef]
- Yanagishita, T.; Eto, K.; Yamamoto-Shimojima, K.; Segawa, O.; Nagata, M.; Ishihara, Y.; Miyashita, Y.; Asano, Y.; Sakata, Y.; Nagata, S.; Yamamoto, T. A recurrent de novo ZSWIM6 variant in a Japanese patient with severe neurodevelopmental delay and frequent vomiting. Hum Genome Var 2021, 8, 16. [Google Scholar] [CrossRef]
- Tischfield, D.J.; Saraswat, D.K.; Furash, A.; Fowler, S.C.; Fuccillo, M.V.; Anderson, S.A. Loss of the neurodevelopmental gene Zswim6 alters striatal morphology and motor regulation. Neurobiol Dis 2017, 103, 174–183. [Google Scholar] [CrossRef]
- Bogdanova, E.; Sadykov, A.; Ivanova, G.; Zubina, I.; Beresneva, O.; Semenova, N.; Galkina, O.; Parastaeva, M.; Sharoyko, V.; Dobronravov, V. Mild Chronic Kidney Disease Associated with Low Bone Formation and Decrease in Phosphate Transporters and Signaling Pathways Gene Expression. Int J Mol Sci 2023, 24, 7270. [Google Scholar] [CrossRef]
- Biscetti, F.; Rando, M.M.; Cecchini, A.L.; Nicolazzi, M.A.; Rossini, E.; Angelini, F.; Iezzi, R.; Eraso, L.H.; Dimuzio, P.J.; Pitocco, D.; Gasbarrini, A.; Massetti, M.; Flex, A. The role of Klotho and FGF23 in cardiovascular outcomes of diabetic patients with chronic limb threatening ischemia: a prospective study. Sci Rep 2023, 13, 6150. [Google Scholar] [CrossRef]
- Kubota, M.; Hamasaki, Y.; Hashimoto, J.; Aoki, Y.; Kawamura, T.; Saito, A.; Yuasa, R.; Muramatsu, M.; Komaba, H.; Toyoda, M.; Fukagawa, M.; Shishido, S.; Sakai, K. Fibroblast growth factor 23-Klotho and mineral metabolism in the first year after pediatric kidney transplantation: A single-center prospective study. Pediatr Transplant 2023, 27, e14440. [Google Scholar] [CrossRef] [PubMed]
- Balcázar-Hernández, L.; Manuel-Apolinar, L.; Vargas Ortega, G.; González-Virla, B.; Reza-Albarrán, A.A.; Jiménez Martínez, M.D.C.; Martínez Ordaz, J.L.; Mendoza-Zubieta, V.; Basurto, L. Vitamin D and its positive effect on the PTH/vitamin D/calcium-FGF23/klotho/phosphorus axis in kidney transplant recipients. Nutr Hosp 2023, 40, 428–435. [Google Scholar] [CrossRef]
- Nakano, T.; Kishimoto, H.; Tokumoto, M. Direct and indirect effects of fibroblast growth factor 23 on the heart. Front Endocrinol (Lausanne) 2023, 14, 1059179. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, J.; Kalantar-Zadeh, K.; Chen, J. Focusing on Phosphorus Loads: From Healthy People to Chronic Kidney Disease. Nutrients 2023, 15, 1236. [Google Scholar] [CrossRef]
- Karava, V.; Dotis, J.; Kondou, A.; Christoforidis, A.; Taparkou, A.; Farmaki, E.; Economou, M.; Printza, N. Fibroblast growth-factor 23 and vitamin D are associated with iron deficiency and anemia in children with chronic kidney disease. Pediatr Nephrol 2023. [Google Scholar] [CrossRef]
- Kaplan, J.; Tommasini, S.; Yao, G.Q.; Zhu, M.; Nishimura, S.; Ghazarian, S.; Louvi, A.; Insogna, K. Altered Expression of Several Molecular Mediators of Cerebrospinal Fluid Production in Hyp Mice. J Endocr Soc 2023, 7, bvad022. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Sidibé, A.; Fortier, C.; Desjardins, M.P.; Ung, R.V.; Kremer, R.; Agharazii, M.; Mac-Way, F. Wnt/β-catenin pathway inhibitors, bone metabolism and vascular health in kidney transplant patients. J Nephrol 2023. [Google Scholar] [CrossRef] [PubMed]
- Bishop, N.C.; Burton, J.O.; Graham-Brown, M.P.M.; Stensel, D.J.; Viana, J.L.; Watson, E.L. Exercise and chronic kidney disease: potential mechanisms underlying the physiological benefits. Nat Rev Nephrol 2023, 19, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, Y.; Ishida, T.; Kugimiya, F.; Takai, S.; Nakayama, Y.; Yonemitsu, K.; Harada, E. Deterioration of Phosphate Homeostasis Is a Trigger for Cardiac Afterload- Clinical Importance of Fibroblast Growth Factor 23 for Accelerated Aging. Circ Rep. 2022, 5, 4–12. [Google Scholar] [CrossRef] [PubMed]



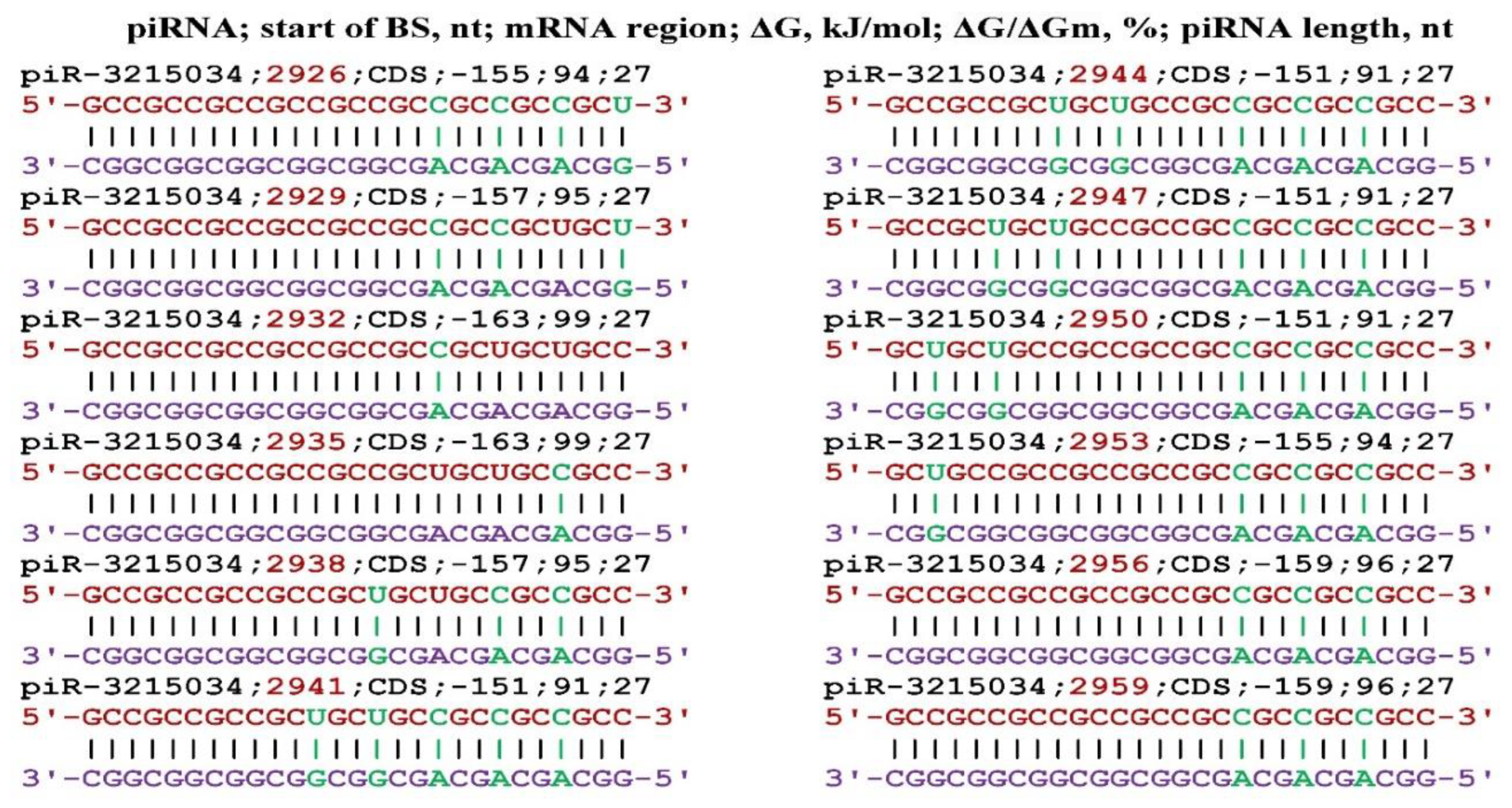
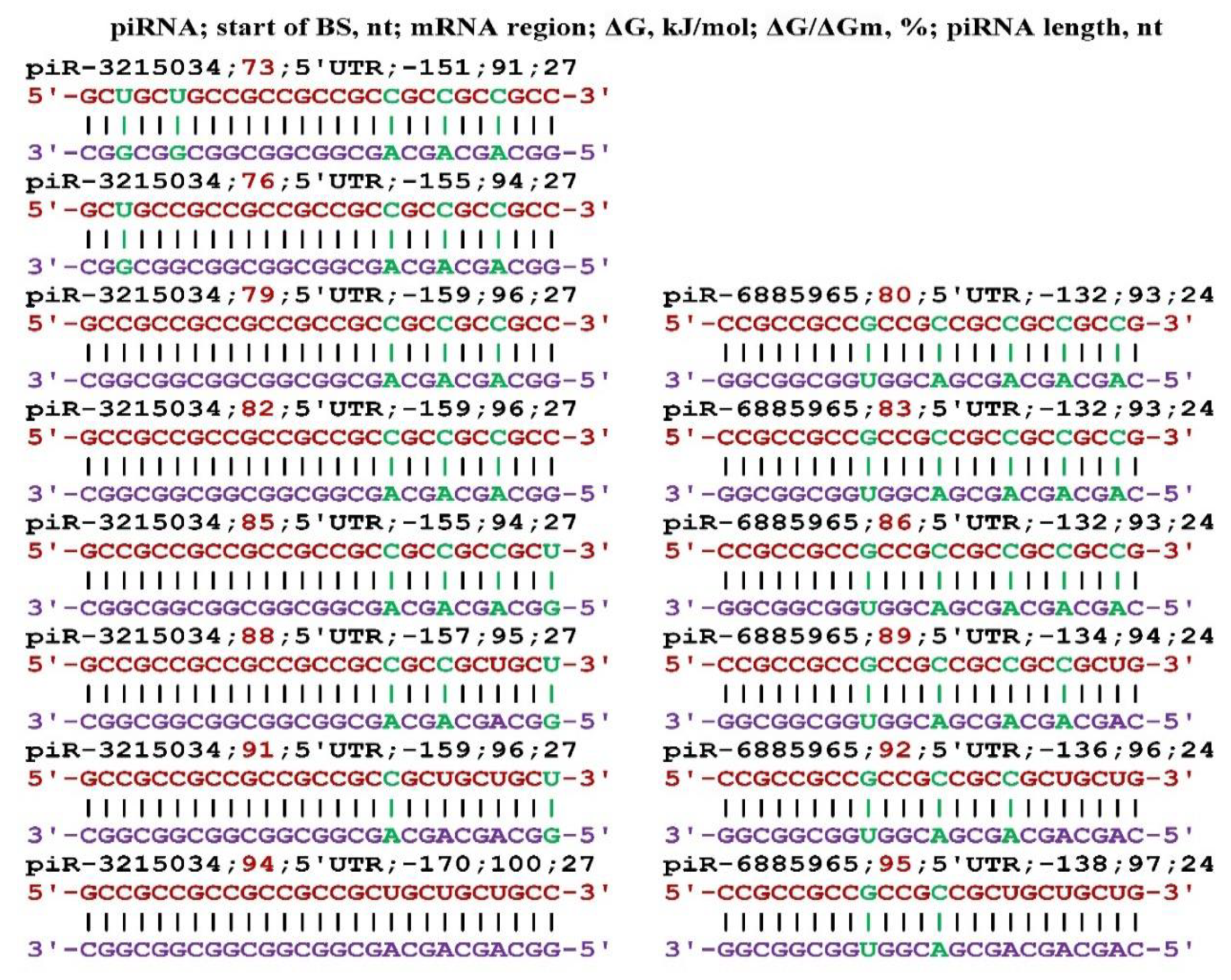


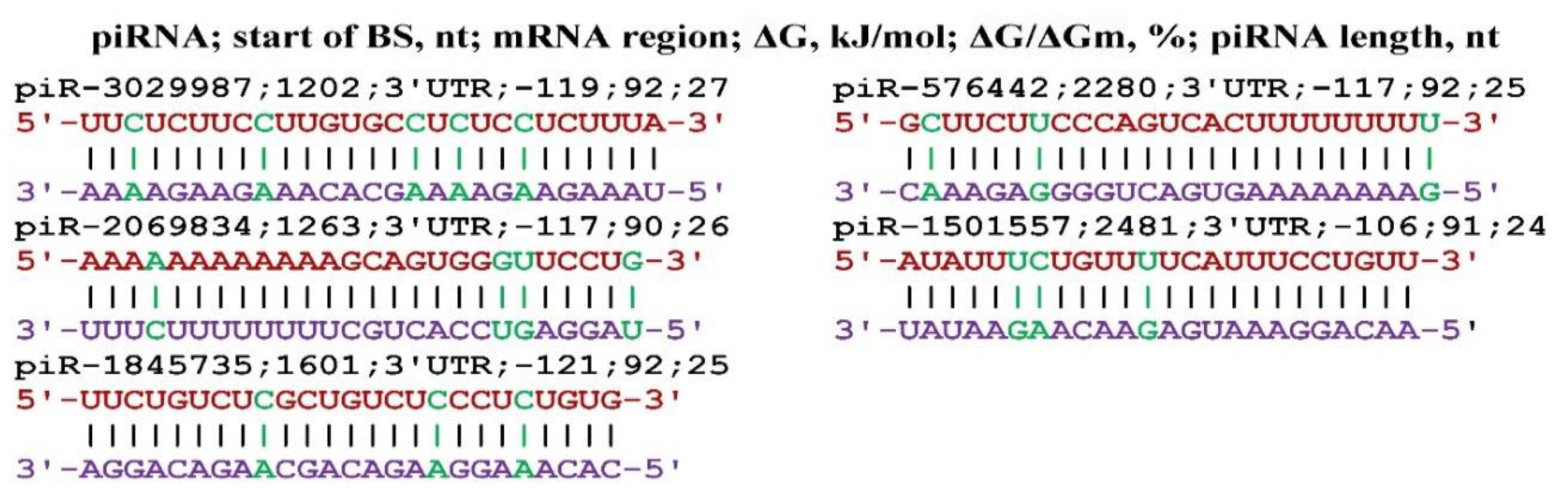
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




