Submitted:
14 June 2023
Posted:
15 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
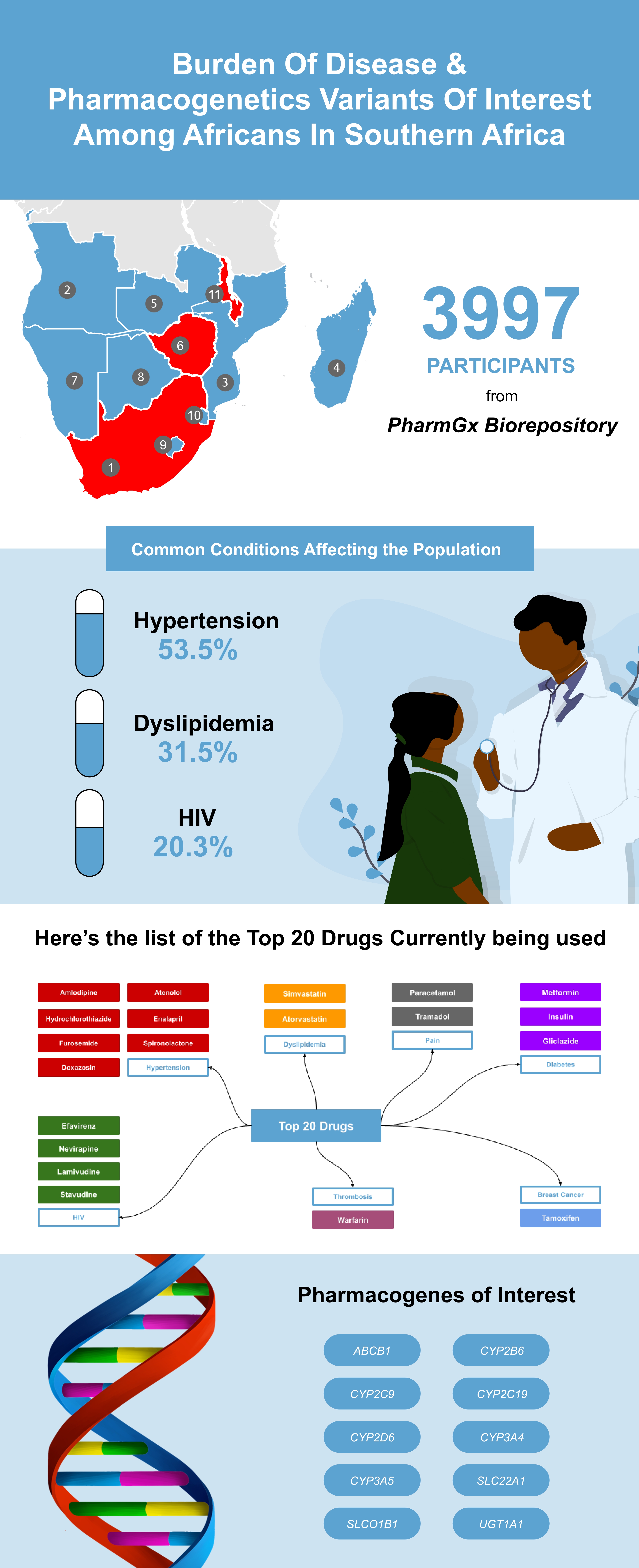
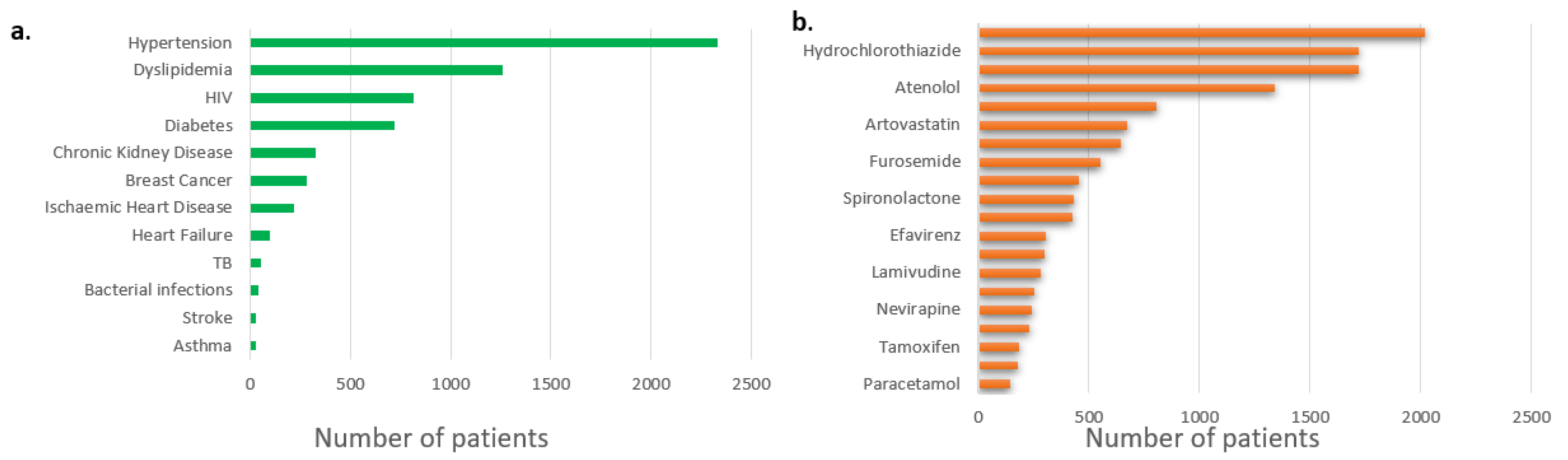
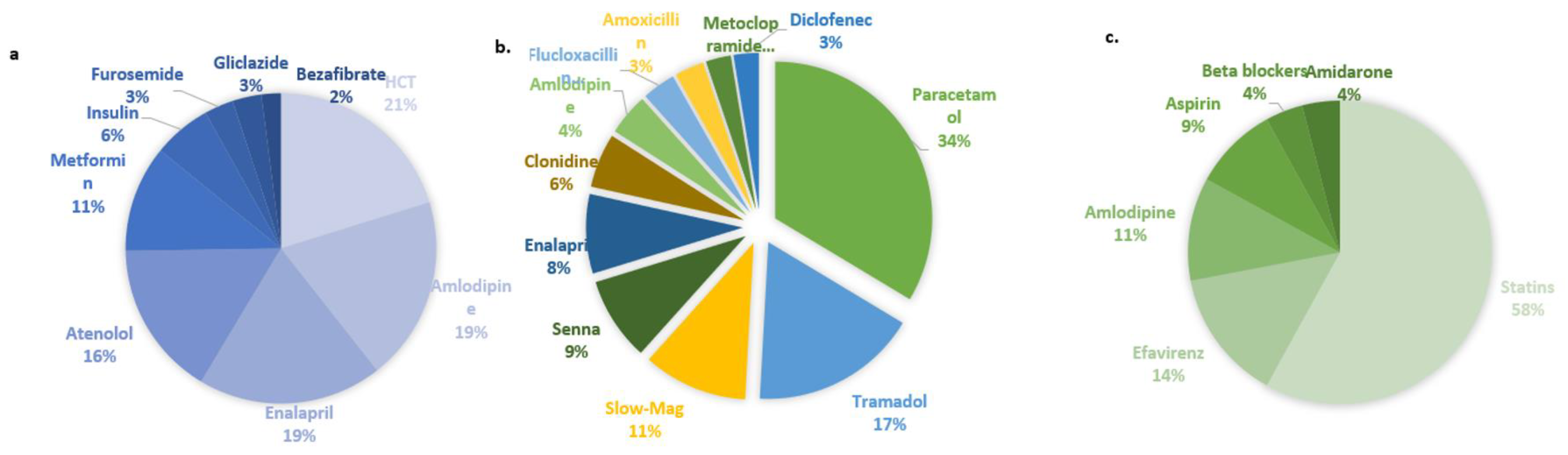
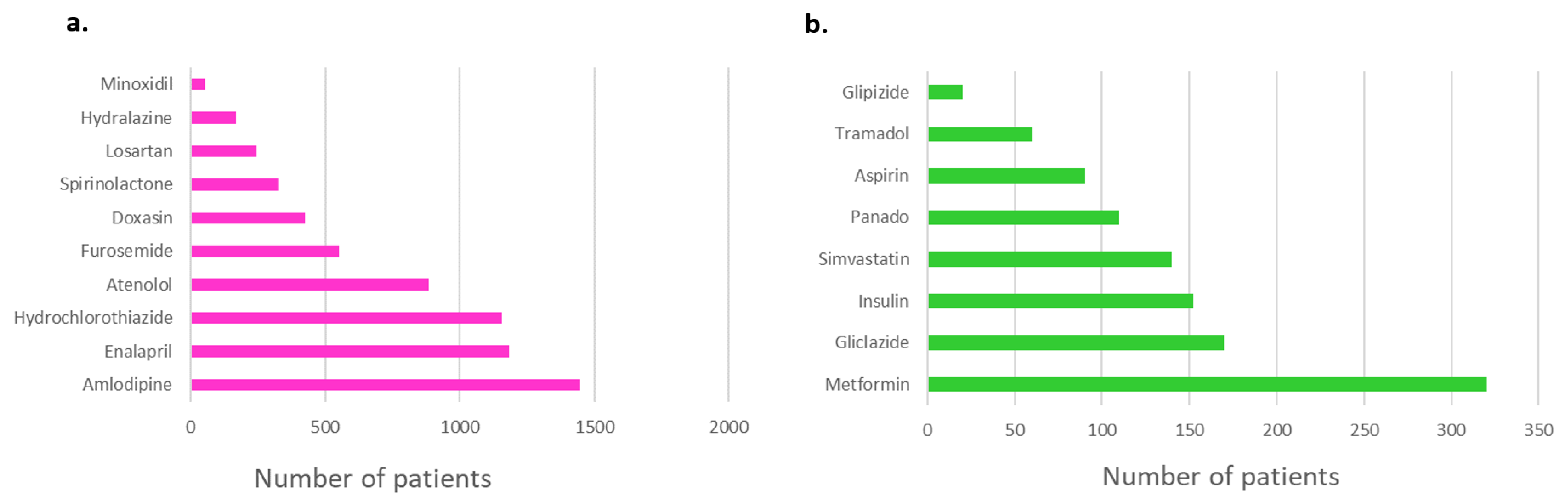
3. Results
3.1.1. Prescriptions and co-prescriptions per cohort
3.1.1.1. Breast cancer cohort
3.1.1.2. Dyslipidemia cohort
3.1.1.3. Hypertension cohort
3.1.1.4. Warfarin cohort
3.1.1.5. HIV cohort
3.1.2. Selected pharmacogenes and their variants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hockings, J.K.; Pasternak, A.L.; Erwin, A.L.; Mason, N.T.; Eng, C.; Hicks, J.K. Pharmacogenomics: An evolving clinical tool for precision medicine. Clevel. Clin. J. Med. 2020, 87, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.; Caudle, K.; Gong, L.; Whirl-Carrillo, M.; Stein, C.; Scott, S.; Lee, M.; Gage, B.; Kimmel, S.; Perera, M.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin. Pharmacol. Ther. 2017, 102, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Desta, Z.; Gammal, R.S.; Gong, L.; Whirl-Carrillo, M.; Gaur, A.H.; Sukasem, C.; Hockings, J.; Myers, A.; Swart, M.; Tyndale, R.F.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline forCYP2B6and Efavirenz-Containing Antiretroviral Therapy. Clin. Pharmacol. Ther. 2019, 106, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Crews, K.R.; Monte, A.A.; Huddart, R.; Caudle, K.E.; Kharasch, E.D.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Callaghan, J.T.; Samer, C.F.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin. Pharmacol. Ther. 2021, 110, 888–896. [Google Scholar] [CrossRef]
- Tata, E.B.; Ambele, M.A.; Pepper, M.S. Barriers to Implementing Clinical Pharmacogenetics Testing in Sub-Saharan Africa. A Critical Review. Pharmaceutics 2020, 12, 809. [Google Scholar] [CrossRef] [PubMed]
- Simkins, C. The Southern African development Community I-population [Internet]. Helen Suzman Foundation. 2022. Available online: https:/hsf.org.za/publications/hsf-briefs/the-southern-african-development-community-i-population (accessed on 7 December 2022).
- Gouda, H.N.; Charlson, F.; Sorsdahl, K.; Ahmadzada, S.; Ferrari, A.J.; Erskine, H.; Leung, J.; Santamauro, D.; Lund, C.; Aminde, L.N.; et al. Burden of non-communicable diseases in sub-Saharan Africa, 1990–2017: results from the Global Burden of Disease Study 2017. Lancet Glob. Health 2019, 7, e1375–e1387. [Google Scholar] [CrossRef]
- Gona, P.N.; Gona, C.M.; Ballout, S.; Rao, S.R.; Kimokoti, R.; Mapoma, C.C.; Mokdad, A.H. Burden and changes in HIV/AIDS morbidity and mortality in Southern Africa Development Community Countries, 1990–2017. BMC Public Health 2020, 20, 867. [Google Scholar] [CrossRef]
- Dwyer-Lindgren, L.; Cork, M.A.; Sligar, A.; Steuben, K.M.; Wilson, K.F.; Provost, N.R.; Mayala, B.K.; VanderHeide, J.D.; Collison, M.L.; Hall, J.B.; et al. Mapping HIV prevalence in sub-Saharan Africa between 2000 and 2017. Nature 2019, 570, 189–193. [Google Scholar] [CrossRef]
- Eichmeyer, J.; Munro, C. The building blocks of a pharmacogenomic program. From stakeholders to services, get the basics for crafting a program in this rising field of personalized medicine. Clinical Laboratory News. 2021. Available online: https://www.aacc.org/cln/articles/2021/april/the-building-blocks-of-a-pharmacogenomic-program.
- Whirl-Carrillo, M.; McDonagh, E.M.; Hebert, J.M.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Altman, R.B.; E Klein, T. Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2012, 92, 414–417. [Google Scholar] [CrossRef]
- Muyambo, S.; Ndadza, A.; Soko, N.D.; Kruger, B.; Kadzirange, G.; Chimusa, E.; Masimirembwa, C.M.; Ntsekhe, M.; Nhachi, C.F.; Dandara, C. Warfarin Pharmacogenomics for Precision Medicine in Real-Life Clinical Practice in Southern Africa: Harnessing 73 Variants in 29 Pharmacogenes. OMICS: A J. Integr. Biol. 2022, 26, 35–50. [Google Scholar] [CrossRef]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. CDC in South Africa. Available online: https://www.cdc.gov/globalhealth/countries/southafrica/default.htm#disease (accessed on 17 November 2022).
- Centers for Disease Control and Prevention. CDC in Zimbabwe. 2019. Available online: https://www.cdc.gov/globalhealth/countries/zimbabwe/pdf/zimbabwe-factsheet.pdf (accessed on 9 December 2022).
- Centers for Disease Control and Prevention. CDC in Malawi. 2019. Available online: https://www.cdc.gov/globalhealth/countries/malawi/pdf/Malawi_Factsheet-p.pdf (accessed on 9 December 2022).
- World Health Organization (WHO). Death from non communicable diseases on the rise in Africa. 2022. Available online: https://www.afro.who.int/news/deaths-noncommunicable-diseases-rise-africa (accessed on 18 January 2023).
- Fuchs, F.D.; Whelton, P.K. High Blood Pressure and Cardiovascular Disease. Hypertension 2020, 75, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Kjeldsen, S.E. Hypertension and cardiovascular risk: General aspects. Pharmacol. Res. 2018, 129, 95–99. [Google Scholar] [CrossRef]
- Noubiap, J.J.; Bigna, J.J.; Nansseu, J.R.; Nyaga, U.F.; Balti, E.V.; Echouffo-Tcheugui, J.B.; Kengne, A.P. Prevalence of dyslipidaemia among adults in Africa: a systematic review and meta-analysis. Lancet Glob. Heal. 2018, 6, e998–e1007. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation. Diabetes in Africa. IDF Diabetes Atlas 10th edition. 2021. Available online: https://www.idf.org/our-network/regions-members/africa/diabetes-in-africa.html (accessed on 19 January 2023).
- World Health Organization (WHO). Update of recommendations on first -and second-line antiretroviral regimens [Internet]. Geneva; 2019. Available online: https://www.who.int/publications/i/item/WHO-CDS-HIV-19.15.
- Makoni, M. The promise of paediatric dolutegravir in Zimbabwe. Lancet HIV 2022, 9, e603–e604. [Google Scholar] [CrossRef] [PubMed]
- National Department of Health SA. ART guidelines for the management of HIV in adults, pregnancy, adolescents, children, infants and neonates. Available online: https://www.health.gov.za/wp-content/uploads/2020/11/2019-art-guideline.pdf.
- Gong, L.; Goswami, S.; Giacomini, K.M.; Altman, R.B.; Klein, T.E. Metformin pathways. Pharmacogenetics Genom. 2012, 22, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Gong, L.; Giacomini, K.; Altman, R.B.; Klein, T.E. PharmGKB summary. Pharmacogenetics Genom. 2014, 24, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Tarasova, L.; Kalnina, I.; Geldnere, K.; Bumbure, A.; Ritenberga, R.; Nikitina-Zake, L.; Fridmanis, D.; Vaivade, I.; Pirags, V.; Klovins, J. Association of genetic variation in the organic cation transporters OCT1, OCT2 and multidrug and toxin extrusion 1 transporter protein genes with the gastrointestinal side effects and lower BMI in metformin-treated type 2 diabetes patients. Pharmacogenetics Genom. 2012, 22, 659–666. [Google Scholar] [CrossRef]
- Sajib, A.A.; Islam, T.; Paul, N.; Yeasmin, S. Interaction of rs316019 variants of SLC22A2 with metformin and other drugs- an in silico analysis. J. Genet. Eng. Biotechnol. 2018, 16, 769–775. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Johnson, S.G.; Caudle, K.E.; Haidar, C.E.; Voora, D.; Wilke, R.A.; Maxwell, W.D.; McLeod, H.L.; Krauss, R.M.; Roden, D.M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1 and Simvastatin-Induced Myopathy: 2014 Update. Clin. Pharmacol. Ther. 2014, 96, 423–428. [Google Scholar] [CrossRef]
- Niemi, M.; Pasanen, M.K.; Neuvonen, P.J. Organic Anion Transporting Polypeptide 1B1: a Genetically Polymorphic Transporter of Major Importance for Hepatic Drug Uptake. Pharmacol. Rev. 2011, 63, 157–181. [Google Scholar] [CrossRef] [PubMed]
- Pasanen, M.K.; Neuvonen, M.; Neuvonen, P.J.; Niemi, M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenetics Genom. 2006, 16, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Cooper-DeHoff, R.M.; Niemi, M.; Ramsey, L.B.; Luzum, J.A.; Tarkiainen, E.K.; Straka, R.J.; Gong, L.; Tuteja, S.; Wilke, R.A.; Wadelius, M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 2022, 111, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Ngaimisi, E.; Habtewold, A.; Minzi, O.; Makonnen, E.; Mugusi, S.; Amogne, W.; Yimer, G.; Riedel, K.-D.; Janabi, M.; Aderaye, G.; et al. Importance of Ethnicity, CYP2B6 and ABCB1 Genotype for Efavirenz Pharmacokinetics and Treatment Outcomes: A Parallel-Group Prospective Cohort Study in Two Sub-Saharan Africa Populations. PLOS ONE 2013, 8, e67946. [Google Scholar] [CrossRef]
- Soko, N.; Dandara, C.; Ramesar, R.; Kadzirange, G.; Masimirembwa, C. Pharmacokinetics of rosuvastatin in 30 healthy Zimbabwean individuals of African ancestry. Br. J. Clin. Pharmacol. 2016, 82, 326–328. [Google Scholar] [CrossRef]
- DeGorter, M.K.; Tirona, R.G.; Schwarz, U.I.; Choi, Y.-H.; Dresser, G.K.; Suskin, N.; Myers, K.; Zou, G.; Iwuchukwu, O.; Wei, W.-Q.; et al. Clinical and Pharmacogenetic Predictors of Circulating Atorvastatin and Rosuvastatin Concentrations in Routine Clinical Care. Circ. Cardiovasc. Genet. 2013, 6, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Soko, N.D.; Chimusa, E.; Masimirembwa, C.; Dandara, C. An African-specific profile of pharmacogene variants for rosuvastatin plasma variability: limited role for SLCO1B1 c.521T>C and ABCG2 c.421A>C. Pharmacogenomics J. 2018, 19, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Kiaco, K.; Rodrigues, A.S.; Rosário, V.D.; Gil, J.P.; Lopes, D. The drug transporter ABCB1 c.3435C>T SNP influences artemether–lumefantrine treatment outcome. Malar. J. 2017, 16, 383. [Google Scholar] [CrossRef]
- Tavares, L.C.; Marcatto, L.R.; Soares, R.A.G.; Krieger, J.E.; Pereira, A.C.; Santos, P.C.J.L. Association Between ABCB1 Polymorphism and Stable Warfarin Dose Requirements in Brazilian Patients. Front. Pharmacol. 2018, 9, 542. [Google Scholar] [CrossRef]
- Mittal, B.; Tulsyan, S.; Mittal, R.D. The effect of ABCB1 polymorphisms on the outcome of breast cancer treatment. Pharmacogenomics Pers. Med. 2016, ume 9, 47–58. [Google Scholar] [CrossRef]
- Finta, C.; Zaphiropoulos, P.G. The human cytochrome P450 3A locus. Gene evolution by capture of downstream exons. Gene 2000, 260, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Mpye, K.L.; Matimba, A.; Dzobo, K.; Chirikure, S.; Wonkam, A.; Dandara, C. Disease burden and the role of pharmacogenomics in African populations. Glob. Heal. Epidemiology Genom. 2017, 2, e1. [Google Scholar] [CrossRef] [PubMed]
- Bains, R.K.; Kovacevic, M.; A Plaster, C.; Tarekegn, A.; Bekele, E.; Bradman, N.N.; Thomas, M.G. Molecular diversity and population structure at the Cytochrome P450 3A5 gene in Africa. BMC Genet. 2013, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Birdwell, K.; Decker, B.; Barbarino, J.; Peterson, J.; Stein, C.; Sadee, W.; Wang, D.; Vinks, A.; He, Y.; Swen, J.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines forCYP3A5Genotype and Tacrolimus Dosing. Clin. Pharmacol. Ther. 2015, 98, 19–24. [Google Scholar] [CrossRef]
- Crewe, H.K.; Notley, L.M.; Wunsch, R.M.; Lennard, M.S.; Gillam, E.M.J. Metabolism of Tamoxifen by Recombinant Human Cytochrome P450 Enzymes: Formation of the 4-Hydroxy, 4′-Hydroxy andN-Desmethyl Metabolites and Isomerization oftrans-4-Hydroxytamoxifen. Drug Metab. Dispos. 2002, 30, 869–874. [Google Scholar] [CrossRef]
- Nyakutira, C.; Röshammar, D.; Chigutsa, E.; Chonzi, P.; Ashton, M.; Nhachi, C.; Masimirembwa, C. High prevalence of the CYP2B6 516G→T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur. J. Clin. Pharmacol. 2007, 64, 357–365. [Google Scholar] [CrossRef]
- Sinxadi, P.Z.; Leger, P.D.; McIlleron, H.M.; Smith, P.J.; Dave, J.A.; Levitt, N.S.; Maartens, G.; Haas, D.W. Pharmacogenetics of plasma efavirenz exposure in HIV-infected adults and children in South Africa. Br. J. Clin. Pharmacol. 2015, 80, 146–156. [Google Scholar] [CrossRef]
- Masimirembwa, C.; Persson, I.; Bertilsson, L.; Hasler, J.; Ingelman-Sundberg, M. A novel mutant variant of the CYP2D6 gene (CYP2D617) common in a black African population: association with diminished debrisoquine hydroxylase activity. Br. J. Clin. Pharmacol. 1996, 42, 713–719. [Google Scholar] [CrossRef]
- Theken, K.N.; Lee, C.R.; Gong, L.; Caudle, K.E.; Formea, C.M.; Gaedigk, A.; Klein, T.E.; Agúndez, J.A.; Grosser, T. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin. Pharmacol. Ther. 2020, 108, 191–200. [Google Scholar] [CrossRef]
- Li-Wan-Po, A.; Girard, T.; Farndon, P.; Cooley, C.; Lithgow, J. Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br. J. Clin. Pharmacol. 2010, 69, 222–230. [Google Scholar] [CrossRef]
- Gammal, R.S.; Court, M.H.; Haidar, C.E.; Iwuchukwu, O.F.; Gaur, A.H.; Alvarellos, M.; Guillemette, C.; Lennox, J.L.; Whirl-Carrillo, M.; Brummel, S.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline forUGT1A1and Atazanavir Prescribing. Clin. Pharmacol. Ther. 2015, 99, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Carrato, A. Precision Medicine: UGT1A1 Genotyping to Better Manage Irinotecan-Induced Toxicity. JCO Oncol. Pr. 2022, 18, 278–280. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food Drug Administration F. Prescribing information Camtosar. 1996.

| Drug | Pharmacogene |
|---|---|
| Amlodipine | CYP3A4 CYP3A5 CACNAIC ABCB1 ACE |
| Hydrochlorothiazide | ADD1 NEDD4L KCNJ1 WNK1 ACE |
| Enalapril | CES1 ACE VEGFA ABO ADRB2 |
| Atenolol | ADRB2 ADRB1 AGT GNB3 GRK4 |
| Simvastatin | SLCO1B1 ABCB1 CYP3A4 CYP3A5 CYP2C9 ABCG2 |
| Atorvastatin | SLCO1B1 ABCB1 CYP3A5 APOE CYP3A4 ABCG2 |
| Metformin | SLC22A1 SLC47A1 SLC47A2 SLC22A2 ATM |
| Furosemide | NPPA-ASI ACE ADD1 SCNN1G SLC12A3 |
| Warfarin | CYP2C9 VKORC1 CYP4F2 GGCX CYP2C19 |
| Spironolactone | ACE CYP4A11 ADRB1 ADRB2 ADD1 |
| Doxazosin | ADRA1B ADRA2A KCNH2 |
| Efavirenz | CYP2B6 NRI13 NRI12 UGT2B7 CYP2A6 ABCB1 |
| Insulin | G6PD SCNN1B SLC30A8 |
| Lamivudine | SLC22A1 OCT2 |
| Gliclazide | KCNJ11 CYP2C9 ABCC8 KCNQ1 |
| Nevirapine | CYP3A4 CYP2D6 CY2B6 ABCB1 |
| Stavudine | Discontinued |
| Tamoxifen | CYP2D6 CYP2C19 CYP3A5 ABCC2 CYP2B6 |
| Tramadol | CYP2D6 ABCB1 OPRM1 COMT SLC22A1 |
| Panado | SULT1A1 SULT1A3 UGT1A1 |
| Dolutegravir | CYP3A4 ABCG2 ABCB1 UGT1A1 UGT1A3 UGT1A9 SLC22A2 |
| The following polymorphism frequencies were obtained from Muyambo et al.; [12] | ||||||||
|---|---|---|---|---|---|---|---|---|
| Variant allele frequencies | ||||||||
| Gene | dbSNP | Africans (Southern Africa) | Mixed Ancestry (Southern Africa) |
West African (YRI) |
East African (LWK) |
African Americans | East Africans | Europeans |
| ABCB1 | rs1045642 | 0.09 | 0.40 | 0.13 | 0.14 | 0.23 | 0.40 | 0.52 |
| CYP2B6 | rs28399499 | 0.10 | 0.03 | 0.12 | 0.06 | 0.07 | 0.00 | 0.00 |
| rs3745274 (c.516G>T) | 0.35 | 0.32 | 0.40 | 0.36 | 0.35 | 0.22 | 0.24 | |
| CYP2C9 | rs1799853 (*2) | 0.01 | 0.04 | 0.00 | 0.00 | 0.07 | 0.001 | 0.12 |
| rs1057910 (*3) | 0.00 | 0.05 | 0.00 | 0.00 | 0.02 | 0.03 | 0.07 | |
| rs7900194 (*8) | 0.11 | 0.02 | 0.05 | 0.07 | 0.02 | 0.00 | 0.02 | |
| rs2256871 (*9) | 0.58 | ----- | 0.09 | 0.15 | 0.07 | 0.00 | 0.001 | |
| CYP2C19 | rs12248560 | 0.14 | 0.13 | 0.25 | 0.18 | 0.22 | 0.02 | 0.22 |
| rs4244285 | 0.17 | 0.22 | 0.17 | 0.21 | 0.18 | 0.31 | 0.15 | |
| CYP2D6 | rs1065852 | 0.07 | 0.10 | 0.11 | 0.04 | 0.19 | 0.57 | 0.20 |
| rs72549357 | 0.05 | 0.03 | ----- | ----- | ----- | ----- | ----- | |
| rs28371706 | 0.19 | 0.04 | 0.26 | 0.19 | 0.14 | 0.00 | 0.00 | |
| rs59421388 | 0.15 | 0.006 | 0.11 | 0.17 | 0.07 | 0.00 | 0.00 | |
| rs3892097 | 0.02 | 0.11 | 0.06 | 0.03 | 0.15 | 0.00 | 0.19 | |
| rs28371725 | 0.03 | 0.05 | 0.09 | 0.03 | 0.09 | 0.04 | 0.09 | |
| rs16947 | 0.12 | 0.23 | 0.56 | 0.65 | 0.46 | 0.14 | 0.34 | |
| CYP3A4 | rs35599367(*22) | 0.00 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.005 |
| CYP3A5 | rs776746 (*3) | 0.15 | 0.58 | 0.17 | 0.12 | 0.31 | 0.71 | 0.95 |
| rs10264272 (*6) | 0.24 | 0.05 | 0.17 | 0.24 | 0.12 | 0.00 | 0.0003 | |
| rs41303343 (*7) | 0.14 | 0.04 | 0.12 | 0.12 | 0.04 | 0.00 | 0.00 | |
| SLCO1B1 | rs4149056 | 0.005 | 0.08 | 0.009 | 0.02 | 0.04 | 0.12 | 0.16 |
| The following polymorphism frequencies were obtained from 1000 Genomes[13] | ||||||||
| Gene | Variant ID | African | East Asian | European | South Asian | American | ||
| CYP2B6 | rs4803419 | 0.08 | 0.44 | 0.32 | 0.34 | 0.35 | ||
| rs12208357 | 0.004 | 0.00 | 0.06 | 0.02 | 0.02 | |||
| rs34130495 | 0.003 | 0.00 | 0.02 | 0.01 | 0.07 | |||
| SLC22A1 | rs72552763 | 0.05 | 0.005 | 0.18 | 0.15 | 0.29 | ||
| rs34059508 | 0.00 | 0.00 | 0.02 | 0.00 | 0.02 | |||
| rs628031 | 0.73 | 0.74 | 0.59 | 0.61 | 0.78 | |||
| SLC22A2 | rs316019 | 0.19 | 0.14 | 0.11 | 0.13 | 0.09 | ||
| UGT1A1 | rs4148323 (*6) | 0.001 | 0.14 | 0.001 | 0.002 | 0.01 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





