Submitted:
15 June 2023
Posted:
16 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patient and tissue samples
2.2. Slide digitisation and viewing
2.3. Preparation of TMA
2.4. On-slide biomarkers
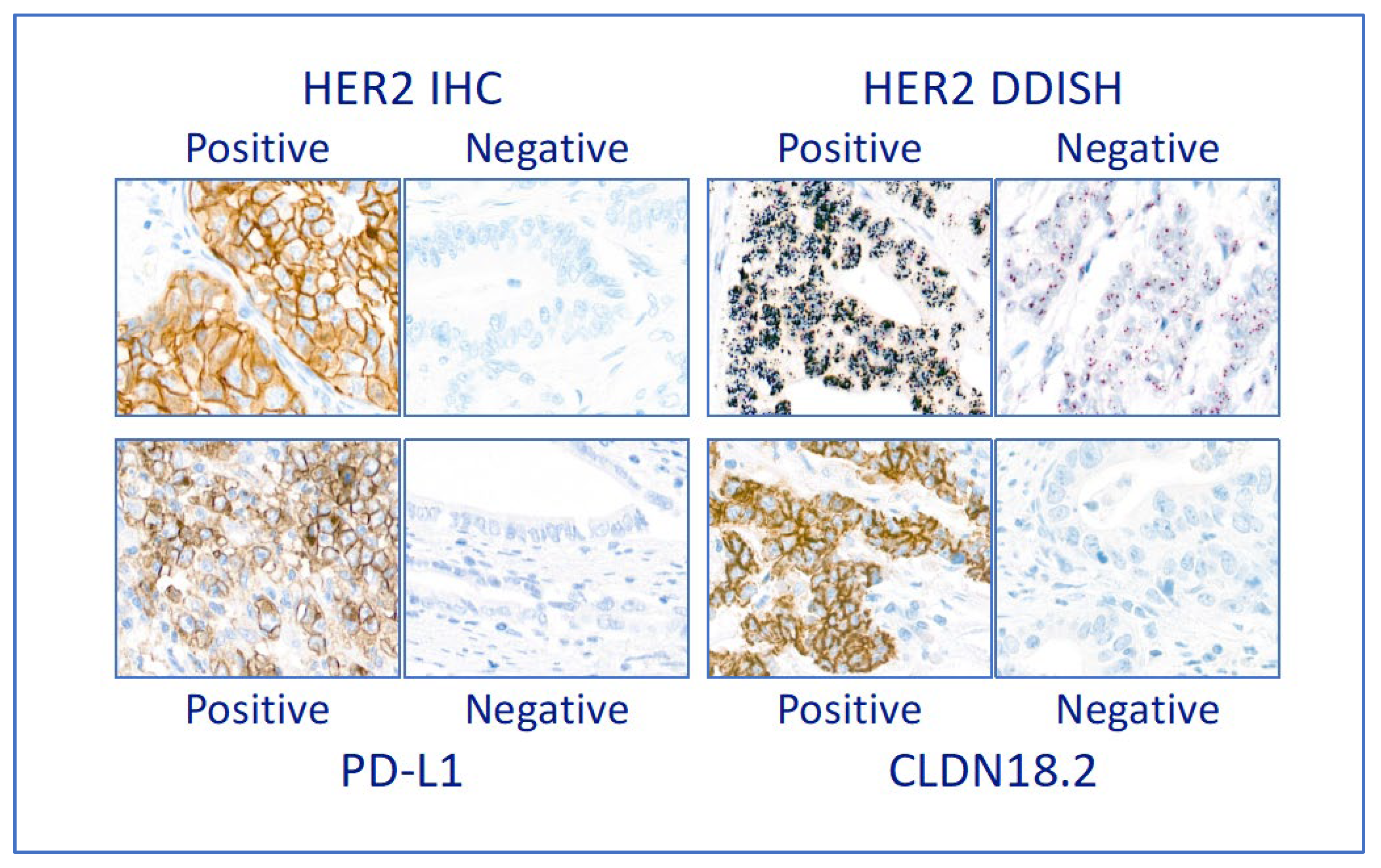
2.5. Evaluation of ISH and IHC staining
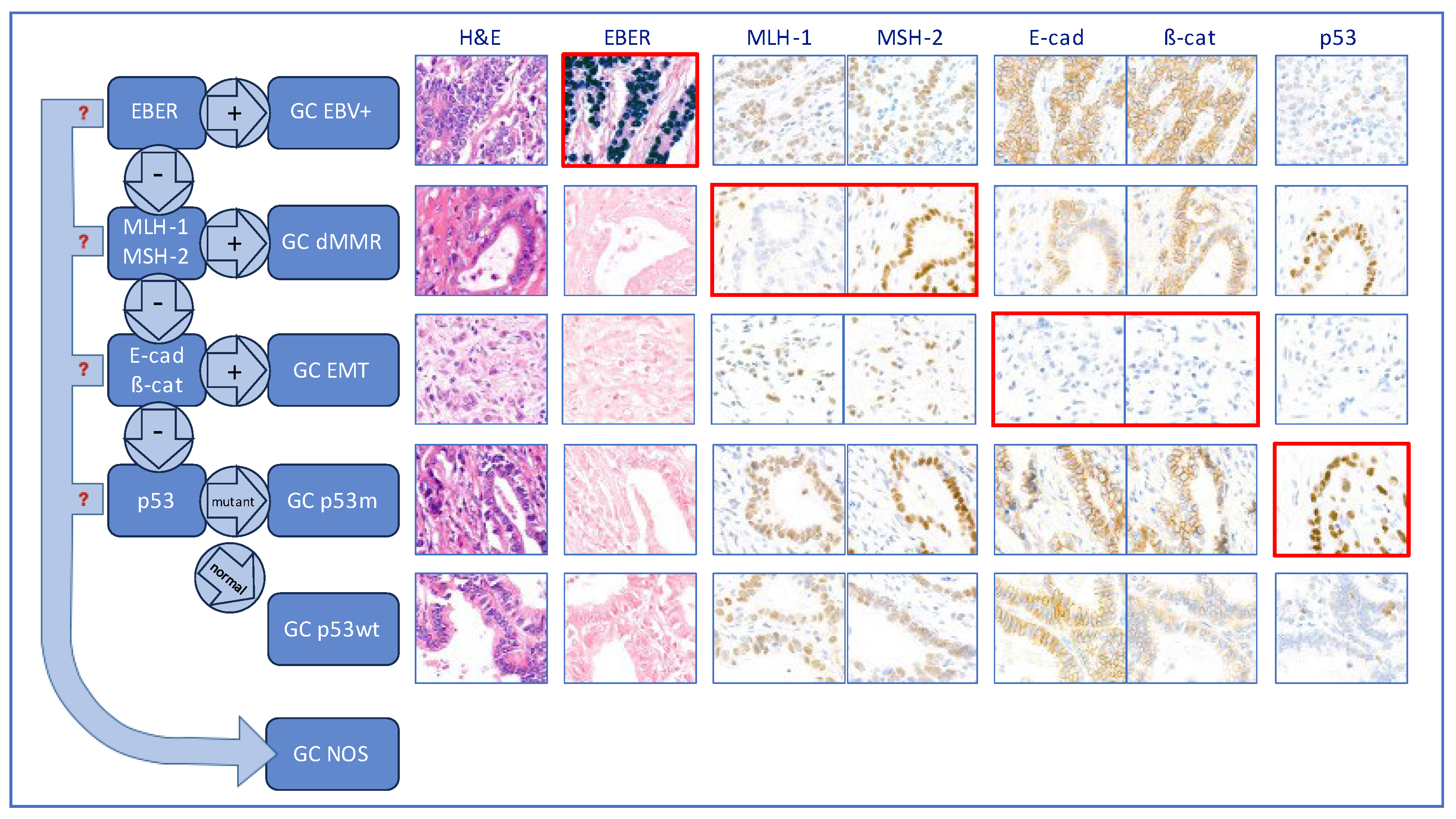
2.6. Identification of the molecular subtypes using a hierarchical approach
2.7. Assessment of Laurén type
3. Results
3.1. Cohort description
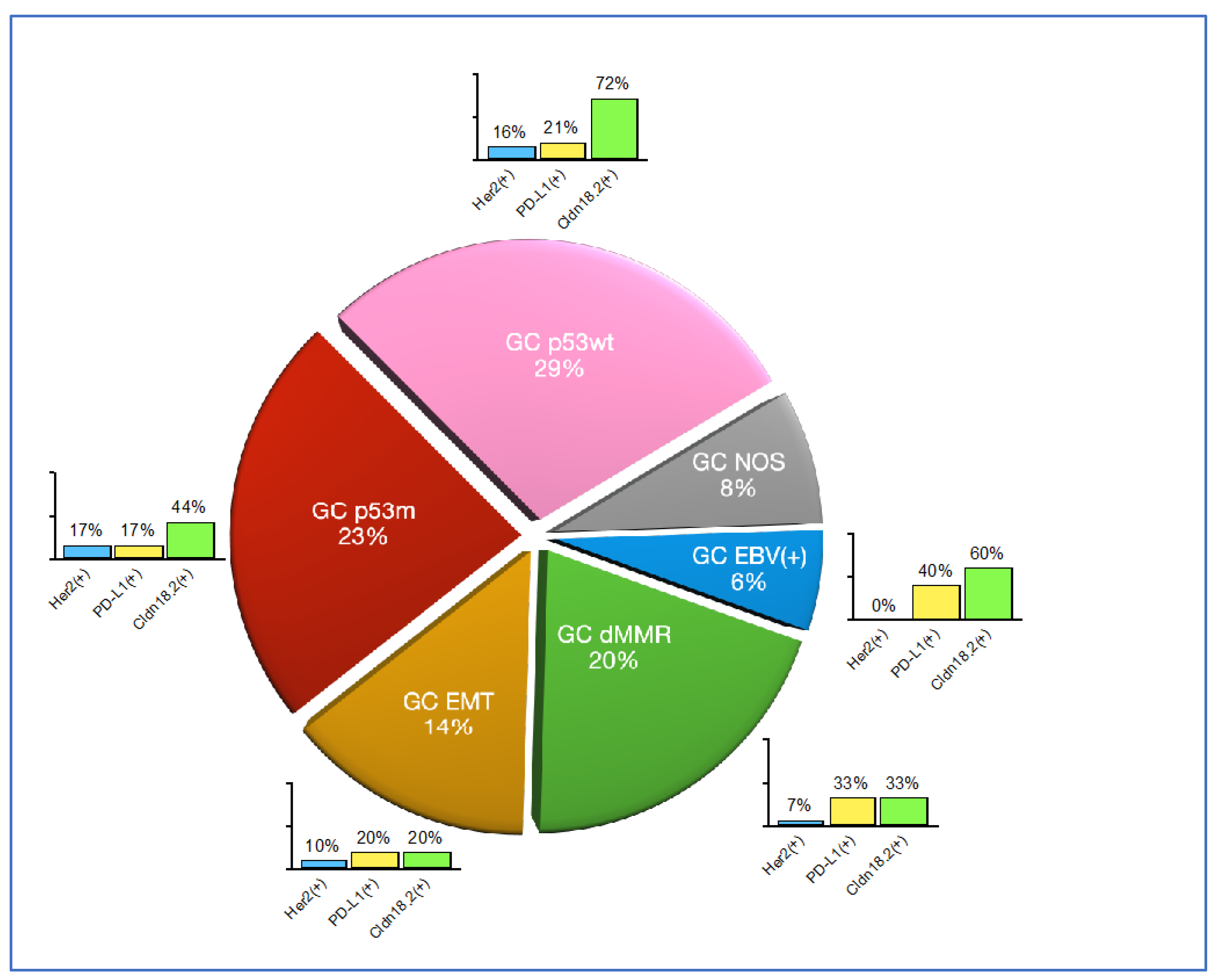
3.2. Molecular classification
3.3. Companion diagnostic predictive biomarkers
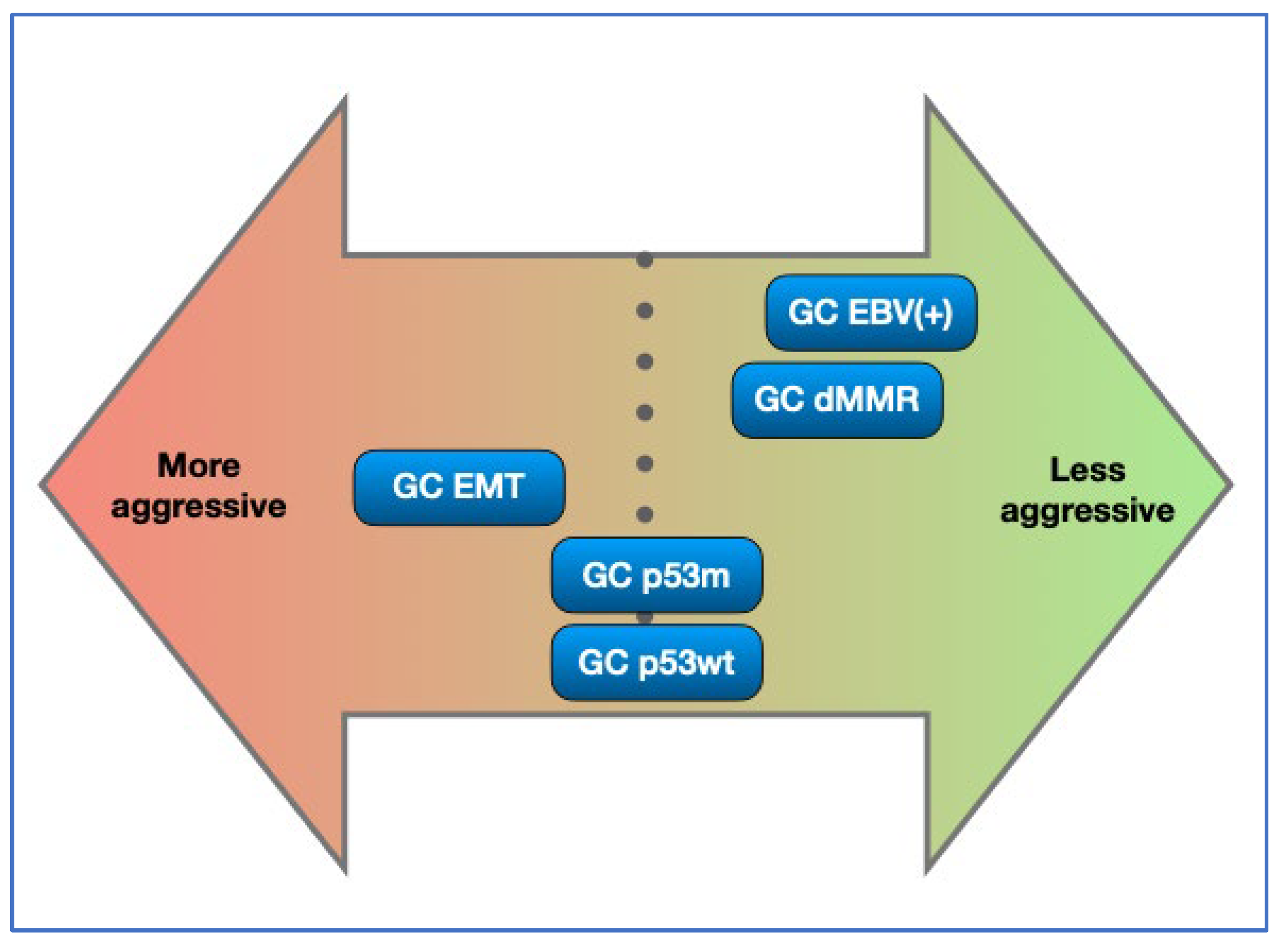
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ichikawa, H.; Nagahashi, M.; Shimada, Y.; Hanyu, T.; Ishikawa, T.; Kameyama, H.; Kobayashi, T.; Sakata, J.; Yabusaki, H.; Nakagawa, S.; et al. Actionable Gene-Based Classification toward Precision Medicine in Gastric Cancer. Genome Med 2017, 9, 93. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular Analysis of Gastric Cancer Identifies Subtypes Associated with Distinct Clinical Outcomes. Nat Med 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Wong, S.S.; Kim, K.-M.; Ting, J.C.; Yu, K.; Fu, J.; Liu, S.; Cristescu, R.; Nebozhyn, M.; Gong, L.; Yue, Y.G.; et al. Genomic Landscape and Genetic Heterogeneity in Gastric Adenocarcinoma Revealed by Whole-Genome Sequencing. Nat Commun 2014, 5, 5477. [Google Scholar] [CrossRef]
- Setia, N.; Agoston, A.T.; Han, H.S.; Mullen, J.T.; Duda, D.G.; Clark, J.W.; Deshpande, V.; Mino-Kenudson, M.; Srivastava, A.; Lennerz, J.K.; et al. A Protein and MRNA Expression-Based Classification of Gastric Cancer. Modern Pathology 2016, 29, 772–784. [Google Scholar] [CrossRef]
- Ahn, S.; Lee, S.-J.; Kim, Y.; Kim, A.; Shin, N.; Choi, K.U.; Lee, C.-H.; Huh, G.Y.; Kim, K.-M.; Setia, N.; et al. High-Throughput Protein and MRNA Expression–Based Classification of Gastric Cancers Can Identify Clinically Distinct Subtypes, Concordant With Recent Molecular Classifications. American Journal of Surgical Pathology 2017, 41, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Feng, Z.; He, H.; Zang, D.; Du, H.; Huang, H.; Du, Y.; He, J.; Zhou, Y.; Nie, Y. Protein Expression-Based Classification of Gastric Cancer by Immunohistochemistry of Tissue Microarray. PLoS ONE 2020, 15, e0238836. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.F.K.P.; Pereira, M.A.; Amorim, L.C.; Mello, E.S.; Faraj, S.F.; Ribeiro, U.; Hoff, P.M.G.; Cecconello, I.; Castria, T.B. Gastric Cancer Molecular Classification and Adjuvant Therapy: Is There a Different Benefit According to the Subtype? J Surg Oncol 2019, 5792. [Google Scholar] [CrossRef] [PubMed]
- Alexa, M.; Hasenburg, A.; Battista, M.J. The TCGA Molecular Classification of Endometrial Cancer and Its Possible Impact on Adjuvant Treatment Decisions. Cancers 2021, 13, 1478. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; McAlpine, J.N. New Classification of Endometrial Cancers: The Development and Potential Applications of Genomic-Based Classification in Research and Clinical Care. gynaecol oncol res pract 2016, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Female Genital Tumours; Organisation mondiale de la santé, Centre international de recherche sur le cancer, Eds.; World health organization classification of tumours; 5th ed.; International agency for research on cancer: Lyon, 2020; ISBN 978-92-832-4504-9.
- Costache, S.; Sajin, M.; Wedden, S.; D’Arrigo, C. A Consolidated Working Classification of Gastric Cancer for Histopathologists (in Press.).
- Nakamura, Y.; Kawazoe, A.; Lordick, F.; Janjigian, Y.Y.; Shitara, K. Biomarker-Targeted Therapies for Advanced-Stage Gastric and Gastro-Oesophageal Junction Cancers: An Emerging Paradigm. Nat Rev Clin Oncol 2021, 18, 473–487. [Google Scholar] [CrossRef]
- Digestive System Tumours; Organisation mondiale de la santé, Centre international de recherche sur le cancer, Eds.; World health organization classification of tumours; 5th ed.; International agency for research on cancer: Lyon, 2019; ISBN 978-92-832-4499-8.
- Laurén, P. THE TWO HISTOLOGICAL MAIN TYPES OF GASTRIC CARCINOMA: DIFFUSE AND SO-CALLED INTESTINAL-TYPE CARCINOMA: An Attempt at a Histo-Clinical Classification. Acta Pathologica Microbiologica Scandinavica 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to Build a Bridge from a Population-Based to a More “Personalized” Approach to Cancer Staging. CA Cancer J Clin 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Gulley, M.L.; Glaser, S.L.; Craig, F.E.; Borowitz, M.; Mann, R.B.; Shema, S.J.; Ambinder, R.F. Guidelines for Interpreting EBER In Situ Hybridization and LMP1 Immunohistochemical Tests for Detecting Epstein-Barr Virus in Hodgkin Lymphoma. Am J Clin Pathol 2002, 117, 259–267. [Google Scholar] [CrossRef]
- McCarthy, A.J.; Capo-Chichi, J.; Spence, T.; Grenier, S.; Stockley, T.; Kamel-Reid, S.; Serra, S.; Sabatini, P.; Chetty, R. Heterogenous Loss of Mismatch Repair (MMR) Protein Expression: A Challenge for Immunohistochemical Interpretation and Microsatellite Instability (MSI) Evaluation. J Pathol Clin Res 2019, 5, 115–129. [Google Scholar] [CrossRef]
- Burandt, E.; Lübbersmeyer, F.; Gorbokon, N.; Büscheck, F.; Luebke, A.M.; Menz, A.; Kluth, M.; Hube-Magg, C.; Hinsch, A.; Höflmayer, D.; et al. E-Cadherin Expression in Human Tumors: A Tissue Microarray Study on 10,851 Tumors. Biomark Res 2021, 9, 44. [Google Scholar] [CrossRef]
- Montgomery, E.; Folpe, A.L. The Diagnostic Value of β-Catenin Immunohistochemistry. Advances in Anatomic Pathology 2005, 12, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Köbel, M.; Ronnett, B.M.; Singh, N.; Soslow, R.A.; Gilks, C.B.; McCluggage, W.G. Interpretation of P53 Immunohistochemistry in Endometrial Carcinomas: Toward Increased Reproducibility. International Journal of Gynecological Pathology 2019, 38, S123–S131. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Stoss, O.; Shi, D.; Büttner, R.; Van De Vijver, M.; Kim, W.; Ochiai, A.; Rüschoff, J.; Henkel, T. Assessment of a HER2 Scoring System for Gastric Cancer: Results from a Validation Study. Histopathology 2008, 52, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Abrahao-Machado, L.F.; Scapulatempo-Neto, C. HER2 Testing in Gastric Cancer: An Update. WJG 2016, 22, 4619. [Google Scholar] [CrossRef] [PubMed]
- Abrahao-Machado, L.F.; Scapulatempo-Neto, C. HER2 Testing in Gastric Cancer: An Update. WJG 2016, 22, 4619. [Google Scholar] [CrossRef]
- Scheel, A.H.; Penault-Llorca, F.; Hanna, W.; Baretton, G.; Middel, P.; Burchhardt, J.; Hofmann, M.; Jasani, B.; Rüschoff, J. Physical Basis of the ‘Magnification Rule’ for Standardized Immunohistochemical Scoring of HER2 in Breast and Gastric Cancer. Diagn Pathol 2018, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Kulangara, K.; Hanks, D.A.; Waldroup, S.; Peltz, L.; Shah, S.; Roach, C.; Juco, J.W.; Emancipator, K.; Stanforth, D. Development of the Combined Positive Score (CPS) for the Evaluation of PD-L1 in Solid Tumors with the Immunohistochemistry Assay PD-L1 IHC 22C3 PharmDx. JCO 2017, 35, e14589–e14589. [Google Scholar] [CrossRef]
- Carneiro, F.; Seixas, M.; Sobrinho-Simões, M. New Elements for an Updated Classification of the Carcinomas of the Stomach. Pathology - Research and Practice 1995, 191, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Grogg, K.L.; Lohse, C.M.; Pankratz, V.S.; Halling, K.C.; Smyrk, T.C. Lymphocyte-Rich Gastric Cancer: Associations with Epstein-Barr Virus, Microsatellite Instability, Histology, and Survival. Modern Pathology 2003, 16, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, X.; Wang, X.; Luo, Y.; Zhou, W.; Luo, H.; Bianba, Z.; Nima, Z.; Wang, Q.; Wang, H.; et al. Prevalence of Epstein–Barr Virus Infection and Mismatch Repair Protein Deficiency and the Correlation of Immune Markers in Tibetan Patients with Gastric Cancer. BioMed Research International 2022, 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fanaian, N.K.; Cohen, C.; Waldrop, S.; Wang, J.; Shehata, B.M. Epstein-Barr Virus (EBV)-Encoded RNA: Automated In-Situ Hybridization (ISH) Compared with Manual ISH and Immunohistochemistry for Detection of EBV in Pediatric Lymphoproliferative Disorders. Pediatr Dev Pathol 2009, 12, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Siciliano, M.C.; Tornambè, S.; Cevenini, G.; Sorrentino, E.; Granai, M.; Giovannoni, G.; Marrelli, D.; Biviano, I.; Roviello, F.; Yoshiyama, H.; et al. EBV Persistence in Gastric Cancer Cases Conventionally Classified as EBER-ISH Negative. Infect Agents Cancer 2022, 17, 57. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Gulley, M.L. Molecular Methods for Detecting Epstein-Barr Virus (Part I): In Situ Hybridization to Epstein-Barr Virus-Encoded RNA (EBER) Transcripts. In Molecular Pathology Protocols; Humana Press: New Jersey, 2000; ISBN 978-1-59259-081-0. [Google Scholar]
- Tavakoli, A.; Monavari, S.H.; Solaymani Mohammadi, F.; Kiani, S.J.; Armat, S.; Farahmand, M. Association between Epstein-Barr Virus Infection and Gastric Cancer: A Systematic Review and Meta-Analysis. BMC Cancer 2020, 20, 493. [Google Scholar] [CrossRef]
- Mundo, L.; Ambrosio, M.R.; Picciolini, M.; Lo Bello, G.; Gazaneo, S.; Del Porro, L.; Lazzi, S.; Navari, M.; Onyango, N.; Granai, M.; et al. Unveiling Another Missing Piece in EBV-Driven Lymphomagenesis: EBV-Encoded MicroRNAs Expression in EBER-Negative Burkitt Lymphoma Cases. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Sun, K.; Jia, K.; Lv, H.; Wang, S.-Q.; Wu, Y.; Lei, H.; Chen, X. EBV-Positive Gastric Cancer: Current Knowledge and Future Perspectives. Front. Oncol. 2020, 10, 583463. [Google Scholar] [CrossRef]
- Li, G.; Zhou, Z.; Wang, Z.; Wang, Z. Assessing Epstein–Barr Virus in Gastric Cancer: Clinicopathological Features and Prognostic Implications. Infect Agents Cancer 2023, 18, 11. [Google Scholar] [CrossRef]
- Teodoridis, J.M.; Hardie, C.; Brown, R. CpG Island Methylator Phenotype (CIMP) in Cancer: Causes and Implications. Cancer Letters 2008, 268, 177–186. [Google Scholar] [CrossRef]
- Krishnamurti, U.; Silverman, J.F. HER2 in Breast Cancer: A Review and Update. Advances in Anatomic Pathology 2014, 21, 100–107. [Google Scholar] [CrossRef]
- Billingsley, C.C.; Cohn, D.E.; Mutch, D.G.; Stephens, J.A.; Suarez, A.A.; Goodfellow, P.J. Polymerase ɛ ( POLE ) Mutations in Endometrial Cancer: Clinical Outcomes and Implications for Lynch Syndrome Testing: POLE Mutations and Endometrial Cancer. Cancer 2015, 121, 386–394. [Google Scholar] [CrossRef]
- Stanland, L.J.; Luftig, M.A. The Role of EBV-Induced Hypermethylation in Gastric Cancer Tumorigenesis. Viruses 2020, 12, 1222. [Google Scholar] [CrossRef]
- Dai, C.; Geng, R.; Wang, C.; Wong, A.; Qing, M.; Hu, J.; Sun, Y.; Lo, A.W.I.; Li, J. Concordance of Immune Checkpoints within Tumor Immune Contexture and Their Prognostic Significance in Gastric Cancer. Molecular Oncology 2016, 10, 1551–1558. [Google Scholar] [CrossRef]
- Panda, A.; Mehnert, J.M.; Hirshfield, K.M.; Riedlinger, G.; Damare, S.; Saunders, T.; Kane, M.; Sokol, L.; Stein, M.N.; Poplin, E.; et al. Immune Activation and Benefit From Avelumab in EBV-Positive Gastric Cancer. JNCI: Journal of the National Cancer Institute 2018, 110, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Hudler, P. Genetic Aspects of Gastric Cancer Instability. The Scientific World Journal 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schöniger, S.; Rüschoff, J. Mismatch Repair Deficiency and Microsatellite Instability. Encyclopedia 2022, 2, 1559–1576. [Google Scholar] [CrossRef]
- Gomes, D.S.; Porto, S.S.; Rocha, R.M.; Gobbi, H. Usefulness and Limitations of E-Cadherin and β-Catenin in the Classification of Breast Carcinomas in Situ with Mixed Pattern. Diagn Pathol 2013, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Buechel, D.; Sugiyama, N.; Rubinstein, N.; Saxena, M.; Kalathur, R.K.R.; Lüönd, F.; Vafaizadeh, V.; Valenta, T.; Hausmann, G.; Cantù, C.; et al. Parsing β-Catenin’s Cell Adhesion and Wnt Signaling Functions in Malignant Mammary Tumor Progression. Proc. Natl. Acad. Sci. U.S.A. 2021, 118, e2020227118. [Google Scholar] [CrossRef] [PubMed]
- Gottardi, C.J.; Gumbiner, B.M. Adhesion Signaling: How β-Catenin Interacts with Its Partners. Current Biology 2001, 11, R792–R794. [Google Scholar] [CrossRef] [PubMed]
- Canas-Marques, R.; Schnitt, S.J. E-Cadherin Immunohistochemistry in Breast Pathology: Uses and Pitfalls. Histopathology 2016, 68, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Mastracci, T.L.; Tjan, S.; Bane, A.L.; O’Malley, F.P.; Andrulis, I.L. E-Cadherin Alterations in Atypical Lobular Hyperplasia and Lobular Carcinoma in Situ of the Breast. Modern Pathology 2005, 18, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Guilford, P.; Hopkins, J.; Harraway, J.; McLeod, M.; McLeod, N.; Harawira, P.; Taite, H.; Scoular, R.; Miller, A.; Reeve, A.E. E-Cadherin Germline Mutations in Familial Gastric Cancer. Nature 1998, 392, 402–405. [Google Scholar] [CrossRef]
- Blair, V.R.; McLeod, M.; Carneiro, F.; Coit, D.G.; D’Addario, J.L.; Van Dieren, J.M.; Harris, K.L.; Hoogerbrugge, N.; Oliveira, C.; Van Der Post, R.S.; et al. Hereditary Diffuse Gastric Cancer: Updated Clinical Practice Guidelines. The Lancet Oncology 2020, 21, e386–e397. [Google Scholar] [CrossRef]
- Han, A.; Xiong, M.; Li, Z.; Liang, Y. [E-cadherin associated protein expression and its significance in invasive lobular carcinoma and invasive ductal carcinoma of breast]. Zhonghua Bing Li Xue Za Zhi 2001, 30, 27–30. [Google Scholar]
- Morrogh, M.; Andrade, V.P.; Giri, D.; Sakr, R.A.; Paik, W.; Qin, L.X.; Arroyo, C.D.; Brogi, E.; Morrow, M.; King, T.A. Cadherin–Catenin Complex Dissociation in Lobular Neoplasia of the Breast. Breast Cancer Res Treat 2012, 132, 641–652. [Google Scholar] [CrossRef]
- Cheng, X.-X.; Wang, Z.-C.; Chen, X.-Y.; Sun, Y.; Kong, Q.-Y.; Liu, J.; Gao, X.; Guan, H.-W.; Li, H. Frequent Loss of Membranous E-Cadherin in Gastric Cancers: A Cross-Talk with Wnt in Determining the Fate of β-Catenin. Clin Exp Metastasis 2005, 22, 85–93. [Google Scholar] [CrossRef]
- Huiping, C.; Kristjansdottir, S.; Jonasson, J.G.; Magnusson, J.; Egilsson, V.; Ingvarsson, S. Alterations of E-Cadherin and β-Catenin in Gastric Cancer. BMC Cancer 2001, 1, 16. [Google Scholar] [CrossRef]
- Van Laarhoven, H.W.M.; Derks, S. Claudin-18.2 Targeting by Zolbetuximab: Results of SPOTLIGHT in Perspective. The Lancet 2023, 401, 1630–1631. [Google Scholar] [CrossRef]
- Shah, M.A.; Ajani, J.A.; Al-Batran, S.-E.; Bang, Y.-J.; Catenacci, D.V.T.; Enzinger, P.C.; Ilson, D.H.; Kim, S.S.; Lordick, F.; Shitara, K.; et al. Zolbetuximab + CAPOX versus CAPOX in First-Line Treatment of Claudin18.2 + /HER2 – Advanced/Metastatic Gastric or Gastroesophageal Junction Adenocarcinoma: GLOW Phase 3 Study. JCO 2022, 40, TPS365–TPS365. [Google Scholar] [CrossRef]
- Sahin, U.; Türeci, Ö.; Manikhas, G.; Lordick, F.; Rusyn, A.; Vynnychenko, I.; Dudov, A.; Bazin, I.; Bondarenko, I.; Melichar, B.; et al. FAST: A Randomised Phase II Study of Zolbetuximab (IMAB362) plus EOX versus EOX Alone for First-Line Treatment of Advanced CLDN18.2-Positive Gastric and Gastro-Oesophageal Adenocarcinoma. Annals of Oncology 2021, 32, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Park, S.; Kim, H.; Kang, S.Y.; Ahn, S.; Kim, K.-M. Gastric Cancer: Mechanisms, Biomarkers, and Therapeutic Approaches. Biomedicines 2022, 10, 543. [Google Scholar] [CrossRef] [PubMed]
- Lima, Á.; Sousa, H.; Medeiros, R.; Nobre, A.; Machado, M. PD-L1 Expression in EBV Associated Gastric Cancer: A Systematic Review and Meta-Analysis. Discov Onc 2022, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Ooki, A.; Yamaguchi, K. The Dawn of Precision Medicine in Diffuse-Type Gastric Cancer. Ther Adv Med Oncol 2022, 14, 17588359221083048. [Google Scholar] [CrossRef] [PubMed]
- Fukamachi, H.; Kim, S.-K.; Koh, J.; Lee, H.S.; Sasaki, Y.; Yamashita, K.; Nishikawaji, T.; Shimada, S.; Akiyama, Y.; Byeon, S.; et al. A Subset of Diffuse-Type Gastric Cancer Is Susceptible to MTOR Inhibitors and Checkpoint Inhibitors. J Exp Clin Cancer Res 2019, 38, 127. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Choi, M.G.; Kim, K.; Kim, K.-M.; Kim, S.T.; Park, S.H.; Cristescu, R.; Peter, S.; Lee, J. High PD-L1 Expression in Gastric Cancer (GC) Patients and Correlation with Molecular Features. Pathology - Research and Practice 2020, 216, 152881. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Chen, M.; Guo, D.; Zhu, H.; Zhang, W.; Pan, J.; Zhong, X.; Li, X.; Qian, H.; Wang, X. PD-L1 and Gastric Cancer Prognosis: A Systematic Review and Meta-Analysis. PLoS One 2017, 12, e0182692. [Google Scholar] [CrossRef]
- Muro, K.; Chung, H.C.; Shankaran, V.; Geva, R.; Catenacci, D.; Gupta, S.; Eder, J.P.; Golan, T.; Le, D.T.; Burtness, B.; et al. Pembrolizumab for Patients with PD-L1-Positive Advanced Gastric Cancer (KEYNOTE-012): A Multicentre, Open-Label, Phase 1b Trial. The Lancet Oncology 2016, 17, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Janjigian, Y.Y.; Shitara, K.; Moehler, M.; Garrido, M.; Salman, P.; Shen, L.; Wyrwicz, L.; Yamaguchi, K.; Skoczylas, T.; Campos Bragagnoli, A.; et al. First-Line Nivolumab plus Chemotherapy versus Chemotherapy Alone for Advanced Gastric, Gastro-Oesophageal Junction, and Oesophageal Adenocarcinoma (CheckMate 649): A Randomised, Open-Label, Phase 3 Trial. The Lancet 2021, 398, 27–40. [Google Scholar] [CrossRef] [PubMed]
| Test | Type | Vendor, Catalogue number |
Clone | On-slide control | Conc. or RTU | Retrieval Solution and Time (min) | Antibody Incubation Time (min) and T | Platform | |
|---|---|---|---|---|---|---|---|---|---|
| EBER | ISH | Ventana, 800-2842 | - | Cell lines (Ventana) | As per package insert | ||||
| MLH-1 | IHC | Leica, PA0988 |
ES05 | Appendix | RTU | AR1, 30 | 15min RT |
Bond III |
|
| MSH-2 | IHC | Leica, PA0989 |
79H11 | Appendix | RTU | AR2, 30 | 15min RT |
Bond III | |
| E-cadherin | IHC | Leica, PA0387 |
36B5 | Skin | RTU | AR2, 20 | 15min RT |
Bond III | |
| Beta catenin | IHC | Leica, PA0083 |
17C2 | Skin | RTU | AR1, 20 | 15min RT |
Bond III | |
| p53 | IHC | Leica, P53-DO7-L-CE | DO-7 | Colon AC | 1:200 | AR2, 30 | 15min RT |
Bond III | |
| Her2 | IHC | Ventana, 790-4493 | 4B5 | Cell lines (Histiocyte) | RTU | CC1, 36 | 16min 36C |
Benchmark Ultra |
|
| DDISH | ISH | Ventana, 800-6043 | - | Cell lines (Histiocyte) | As per package insert | ||||
| PD-L1 (22C3) | IHC | Agilent, SK006 | 22C3 | Cell lines (Histiocyte) | RTU | TRS low pH, 20 | 30min RT |
Dako Autostainer Link 48 | |
| Claudin18.2 | IHC | Ventana, 790-7027 | 43-14A | Stomach | RTU | CC1, 64 | 16min 36C |
Benchmark |
|
| Molecular type | Prevalence | Laurén’s type |
|---|---|---|
| GC EBV(+) | 6% | Intestinal: 80% |
| Diffuse: 20% | ||
| GC dMMR | 20% | Intestinal: 88% |
| Diffuse: 13% | ||
| GC EMT | 14% | Intestinal: 9% |
| Diffuse: 91% | ||
| GC p53m | 23% | Intestinal: 94% |
| Diffuse: 6% | ||
| GC p53wt | 29% | Intestinal: 87% |
| Diffuse: 13% | ||
| GC NOS | 8% | Intestinal: 50% |
| Diffuse: 50% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




