Submitted:
19 June 2023
Posted:
19 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. The Molecular Properties of KChIPs
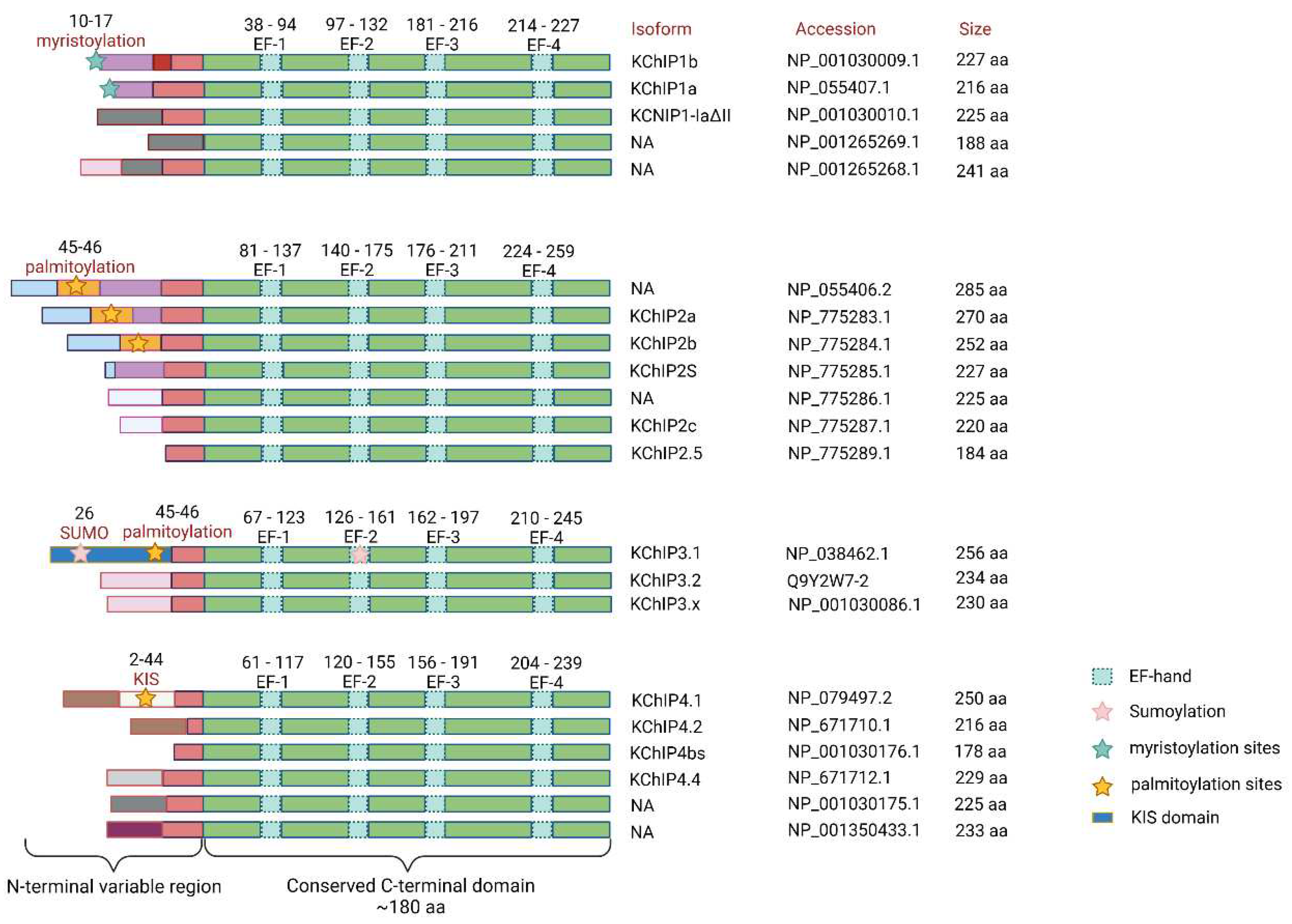
2.1. The Structure of KChIPs
2.2. Regulation of KChIPs expression
2.2.1. Transcriptional Level
2.2.2. Protein Level
3. Biological Function of KChIPs
3.1. KChIPs are Auxiliary Subunits of KV4 Channels
3.1.1. The Interaction of KChIPs with KV4 Channels
3.1.2. KChIPs Modulate the Gating Properties of KV4 Channels
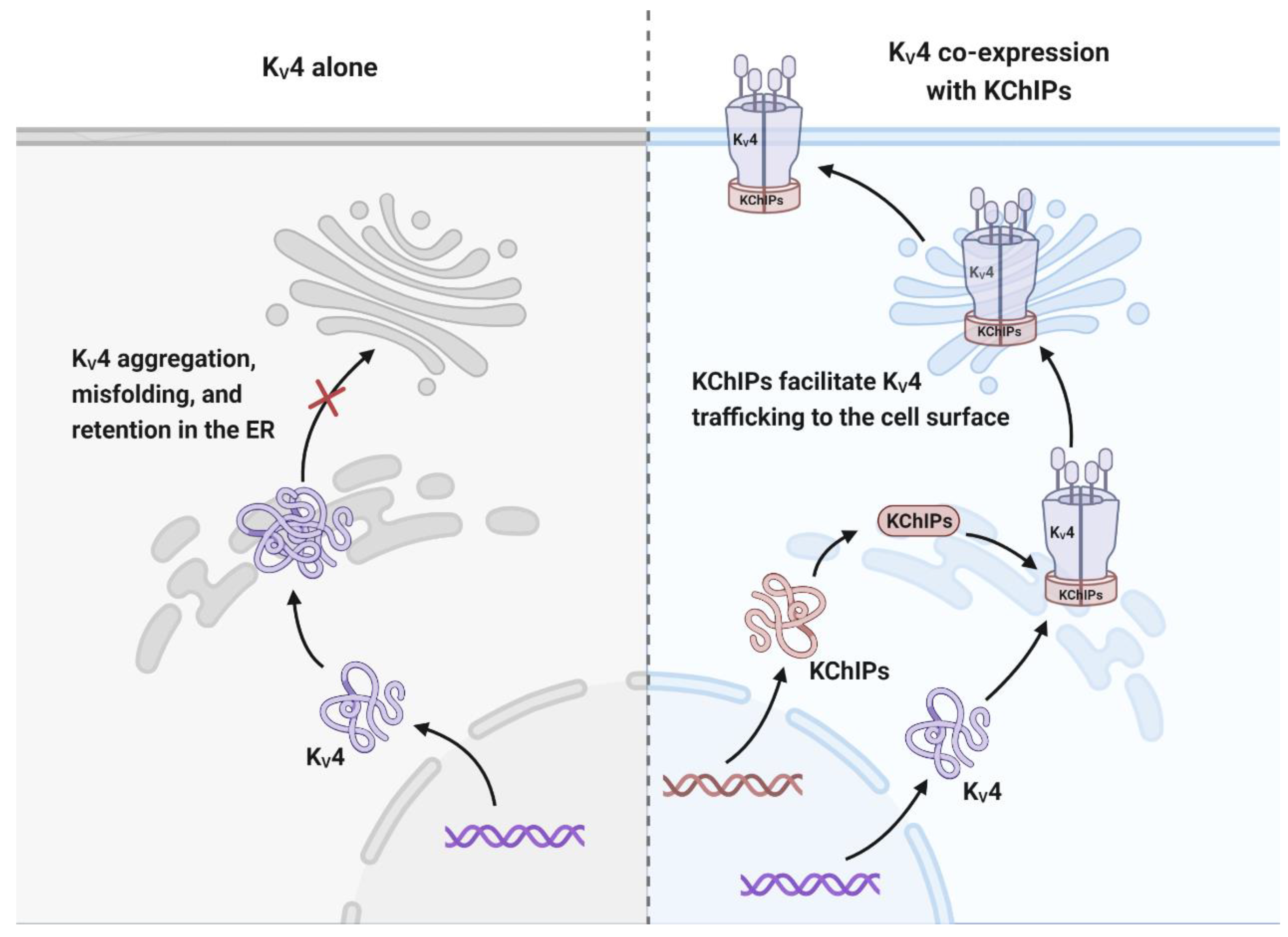
3.1.3. KChIPs Modulate the Trafficking of KV4 Channels
3.1.4. KChIP Ligands Affect KChIP Regulation on KV4
3.2. Role of KChIPs in Regulating Other Ion Channels
3.2.1. KV1.5
3.2.2. CaV1.2
3.2.3. NaV1.5
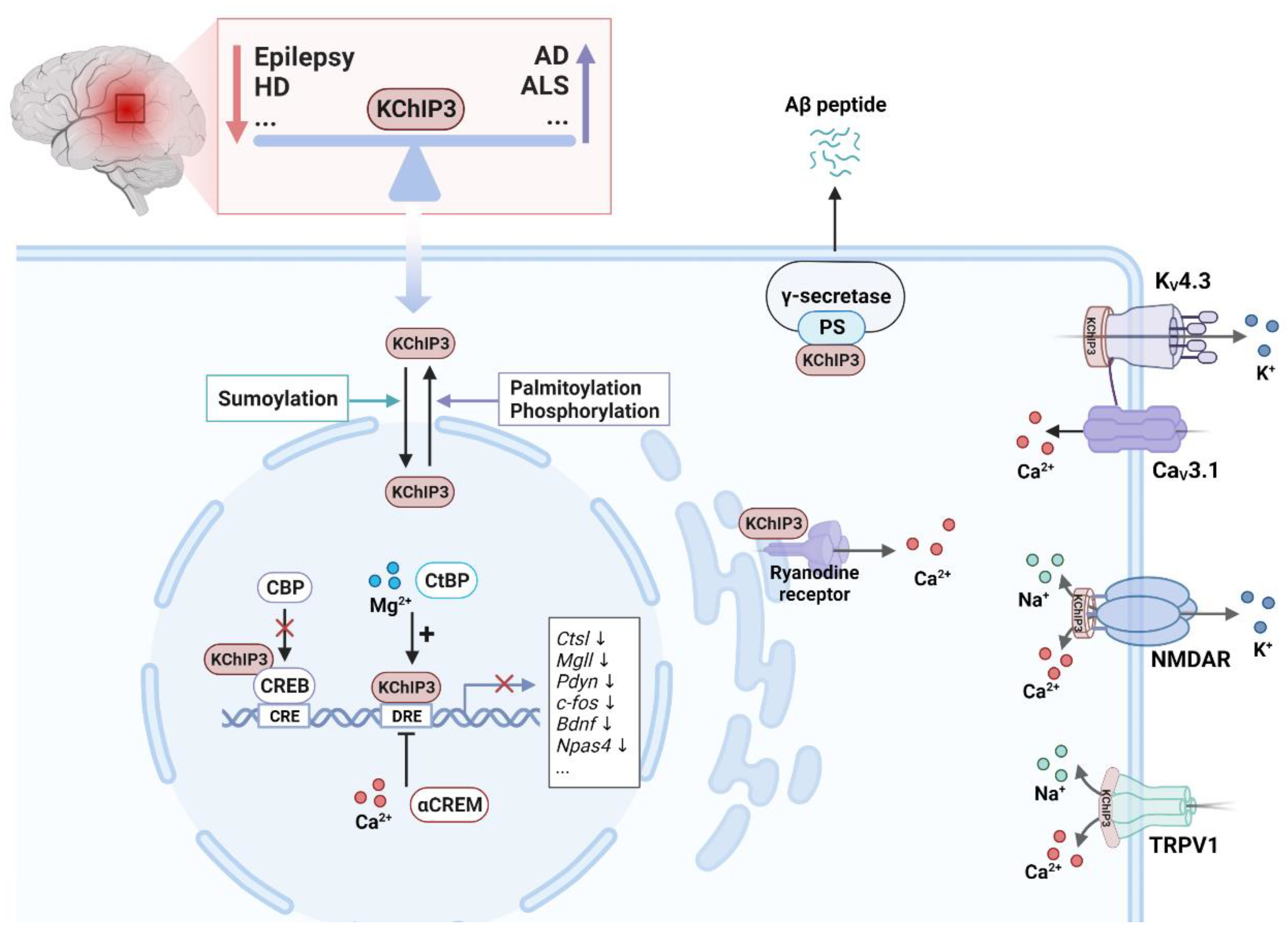
3.3. KChIPs are Ca2+-Dependent Transcriptional Factors
4. KChIPs and Diseases
4.1. Neurological Diseases
4.1.1. Epilepsy
4.1.2. Pain
4.1.3. Memory Dysfunction
4.1.4. Alzheimer's Disease
4.1.5. Other Neurodegenerative Disorders
4.2. Cardiovascular Diseases
4.2.1. Arrhythmias
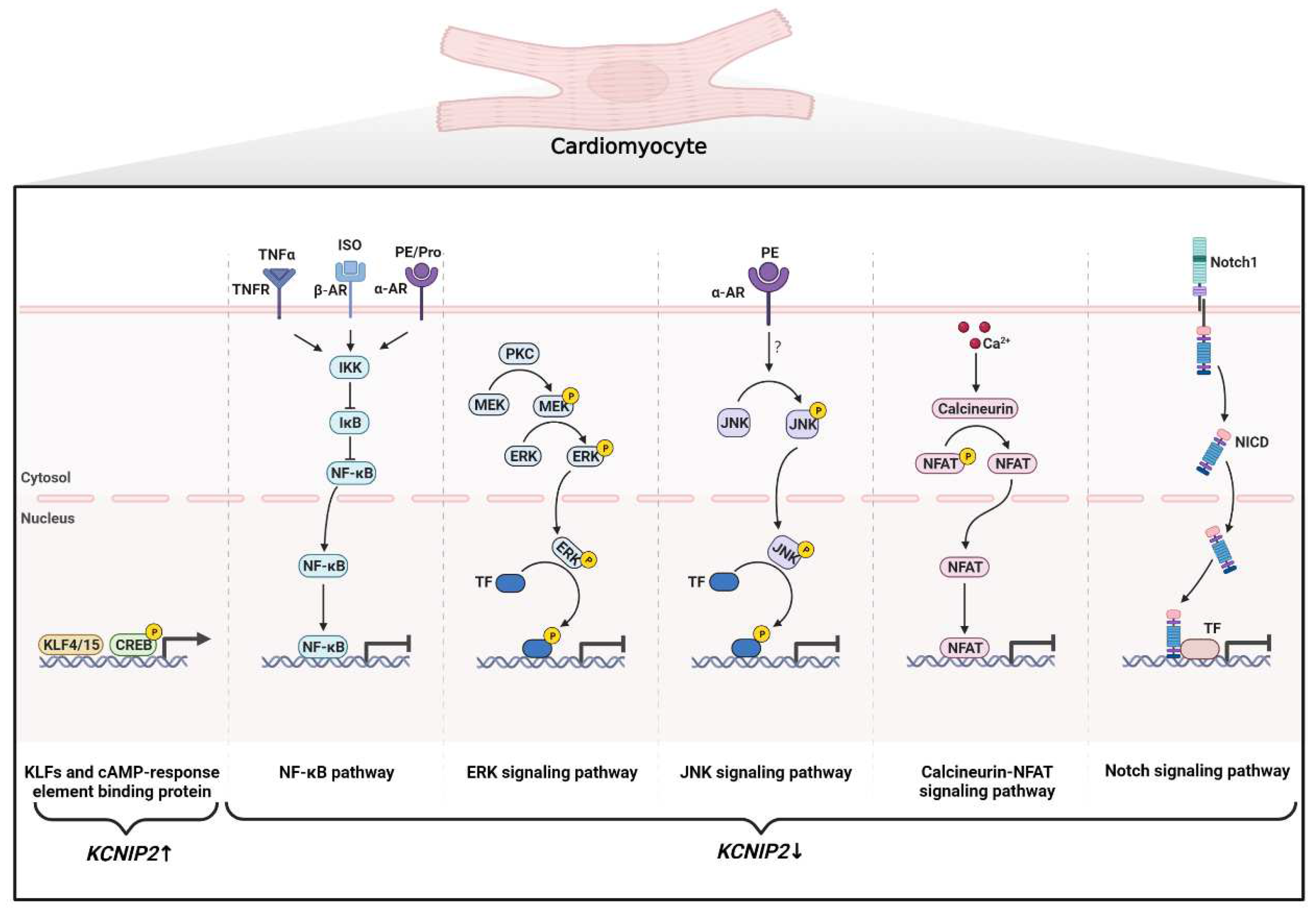
4.2.2. Cardiac Remodeling
4.2.3. Heart Failure
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosati, B.; Pan, Z.; Lypen, S.; Wang, H.S.; Cohen, I.; Dixon, J.E.; McKinnon, D. Regulation of KChIP2 potassium channel beta subunit gene expression underlies the gradient of transient outward current in canine and human ventricle. The Journal of physiology 2001, 533, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Maniak, P.J.; Ingbar, D.H.; O'Grady, S.M. Adult alveolar epithelial cells express multiple subtypes of voltage-gated K+ channels that are located in apical membrane. American journal of physiology. Cell physiology 2003, 284, C1614–1624. [Google Scholar] [CrossRef]
- Bonne, A.; Vreede, L.; Kuiper, R.P.; Bodmer, D.; Jansen, C.; Eleveld, M.; van Erp, F.; Arkesteijn, G.; Hoogerbrugge, N.; van Ravenswaaij, C.; et al. Mapping of constitutional translocation breakpoints in renal cell cancer patients: identification of KCNIP4 as a candidate gene. Cancer genetics and cytogenetics 2007, 179, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Amberg, G.C.; Koh, S.D.; Hatton, W.J.; Murray, K.J.; Monaghan, K.; Horowitz, B.; Sanders, K.M. Contribution of Kv4 channels toward the A-type potassium current in murine colonic myocytes. The Journal of physiology 2002, 544, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Beckett, E.A.; McCloskey, C.; O'Kane, N.; Sanders, K.M.; Koh, S.D. Effects of female steroid hormones on A-type K+ currents in murine colon. The Journal of physiology 2006, 573, 453–468. [Google Scholar] [CrossRef]
- Zhang, Y.; MacLean, J.N.; An, W.F.; Lanning, C.C.; Harris-Warrick, R.M. KChIP1 and frequenin modify shal-evoked potassium currents in pyloric neurons in the lobster stomatogastric ganglion. Journal of neurophysiology 2003, 89, 1902–1909. [Google Scholar] [CrossRef]
- Buxbaum, J.D.; Choi, E.K.; Luo, Y.; Lilliehook, C.; Crowley, A.C.; Merriam, D.E.; Wasco, W. Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nature medicine 1998, 4, 1177–1181. [Google Scholar] [CrossRef]
- Carrión, A.M.; Link, W.A.; Ledo, F.; Mellström, B.; Naranjo, J.R. DREAM is a Ca2+-regulated transcriptional repressor. Nature 1999, 398, 80–84. [Google Scholar] [CrossRef]
- An, W.F.; Bowlby, M.R.; Betty, M.; Cao, J.; Ling, H.P.; Mendoza, G.; Hinson, J.W.; Mattsson, K.I.; Strassle, B.W.; Trimmer, J.S.; et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature 2000, 403, 553–556. [Google Scholar] [CrossRef]
- Morohashi, Y.; Hatano, N.; Ohya, S.; Takikawa, R.; Watabiki, T.; Takasugi, N.; Imaizumi, Y.; Tomita, T.; Iwatsubo, T. Molecular cloning and characterization of CALP/KChIP4, a novel EF-hand protein interacting with presenilin 2 and voltage-gated potassium channel subunit Kv4. The Journal of biological chemistry 2002, 277, 14965–14975. [Google Scholar] [CrossRef]
- Cho, J. Downstream Regulatory Element Antagonist Modulator (DREAM), a target for anti-thrombotic agents. Pharmacological research 2017, 117, 283–287. [Google Scholar] [CrossRef]
- Cercós, P.; Peraza, D.A.; Benito-Bueno, A.; Socuéllamos, P.G.; Aziz-Nignan, A.; Arrechaga-Estévez, D.; Beato, E.; Peña-Acevedo, E.; Albert, A.; González-Vera, J.A.; et al. Pharmacological Approaches for the Modulation of the Potassium Channel K(V)4.x and KChIPs. International journal of molecular sciences 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Néant, I.; Haiech, J.; Kilhoffer, M.C.; Aulestia, F.J.; Moreau, M.; Leclerc, C. Ca(2+)-Dependent Transcriptional Repressors KCNIP and Regulation of Prognosis Genes in Glioblastoma. Frontiers in molecular neuroscience 2018, 11, 472. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.Q.; Liang, P.; Zhou, J.; Lu, Y.; Lei, L.; Bian, X.; Wang, K. Auxiliary KChIP4a suppresses A-type K+ current through endoplasmic reticulum (ER) retention and promoting closed-state inactivation of Kv4 channels. The Journal of biological chemistry 2013, 288, 14727–14741. [Google Scholar] [CrossRef]
- Jerng, H.H.; Pfaffinger, P.J. Modulatory mechanisms and multiple functions of somatodendritic A-type K (+) channel auxiliary subunits. Frontiers in cellular neuroscience 2014, 8, 82. [Google Scholar] [CrossRef]
- Bähring, R. Kv channel-interacting proteins as neuronal and non-neuronal calcium sensors. Channels (Austin, Tex.) 2018, 12, 187–200. [Google Scholar] [CrossRef]
- O'Callaghan, D.W.; Hasdemir, B.; Leighton, M.; Burgoyne, R.D. Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2+ sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K+ channels. Journal of cell science 2003, 116, 4833–4845. [Google Scholar] [CrossRef]
- Takimoto, K.; Yang, E.K.; Conforti, L. Palmitoylation of KChIP splicing variants is required for efficient cell surface expression of Kv4.3 channels. The Journal of biological chemistry 2002, 277, 26904–26911. [Google Scholar] [CrossRef] [PubMed]
- Murthy, A.; Workman, S.W.; Jiang, M.; Hu, J.; Sifa, I.; Bernas, T.; Tang, W.; Deschenes, I.; Tseng, G.N. Dynamic palmitoylation regulates trafficking of K channel interacting protein 2 (KChIP2) across multiple subcellular compartments in cardiac myocytes. Journal of molecular and cellular cardiology 2019, 135, 1–9. [Google Scholar] [CrossRef]
- Palczewska, M.; Casafont, I.; Ghimire, K.; Rojas, A.M.; Valencia, A.; Lafarga, M.; Mellström, B.; Naranjo, J.R. Sumoylation regulates nuclear localization of repressor DREAM. Biochimica et biophysica acta 2011, 1813, 1050–1058. [Google Scholar] [CrossRef]
- Choi, E.K.; Miller, J.S.; Zaidi, N.F.; Salih, E.; Buxbaum, J.D.; Wasco, W. Phosphorylation of calsenilin at Ser63 regulates its cleavage by caspase-3. Molecular and cellular neurosciences 2003, 23, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gomez, A.; Mellström, B.; Tornero, D.; Morato, E.; Savignac, M.; Holguín, H.; Aurrekoetxea, K.; González, P.; González-García, C.; Ceña, V.; et al. G protein-coupled receptor kinase 2-mediated phosphorylation of downstream regulatory element antagonist modulator regulates membrane trafficking of Kv4.2 potassium channel. The Journal of biological chemistry 2007, 282, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, M.H.; Cao, J.; Hernandez-Pineda, R.; Jacobson, M.D.; Carroll, K.I.; Sung, M.A.; Betty, M.; Ge, P.; Gilbride, K.J.; Brown, M.E.; et al. Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proceedings of the National Academy of Sciences of the United States of America 2002, 99, 1035–1040. [Google Scholar] [CrossRef]
- Jerng, H.H.; Pfaffinger, P.J. Multiple Kv channel-interacting proteins contain an N-terminal transmembrane domain that regulates Kv4 channel trafficking and gating. The Journal of biological chemistry 2008, 283, 36046–36059. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, J.; Zolles, G.; Kandias, N.G.; Neubauer, I.; Kalbacher, H.; Covarrubias, M.; Fakler, B.; Bentrop, D. NMR analysis of KChIP4a reveals structural basis for control of surface expression of Kv4 channel complexes. The Journal of biological chemistry 2008, 283, 18937–18946. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Wang, H.; Chen, H.; Cui, Y.; Gu, L.; Chai, J.; Wang, K. Structural Insights into KChIP4a Modulation of Kv4.3 Inactivation. The Journal of biological chemistry 2009, 284, 4960–4967. [Google Scholar] [CrossRef]
- Liu, W.; Wang, G.; Zhang, C.; Ding, W.; Cheng, W.; Luo, Y.; Wei, C.; Liu, J. MG53, A Novel Regulator of KChIP2 and I(to,f), Plays a Critical Role in Electrophysiological Remodeling in Cardiac Hypertrophy. Circulation 2019, 139, 2142–2156. [Google Scholar] [CrossRef]
- Wagner, S.; Hacker, E.; Grandi, E.; Weber, S.L.; Dybkova, N.; Sossalla, S.; Sowa, T.; Fabritz, L.; Kirchhof, P.; Bers, D.M.; et al. Ca/calmodulin kinase II differentially modulates potassium currents. Circulation. Arrhythmia and electrophysiology 2009, 2, 285–294. [Google Scholar] [CrossRef]
- Rossow, C.F.; Dilly, K.W.; Santana, L.F. Differential calcineurin/NFATc3 activity contributes to the Ito transmural gradient in the mouse heart. Circulation research 2006, 98, 1306–1313. [Google Scholar] [CrossRef]
- Jia, Y.; Takimoto, K. Mitogen-activated protein kinases control cardiac KChIP2 gene expression. Circulation research 2006, 98, 386–393. [Google Scholar] [CrossRef]
- Huang, H.; Tang, Y.; Wu, G.; Mei, Y.; Liu, W.; Liu, X.; Wan, N.; Liu, Y.; Huang, C. ALK7 protects against pathological cardiac hypertrophy in mice. Cardiovascular research 2015, 108, 50–61. [Google Scholar] [CrossRef]
- Ying, S.; Cao, H.; Hu, H.; Wang, X.; Tang, Y.; Huang, C. Alk7 Depleted Mice Exhibit Prolonged Cardiac Repolarization and Are Predisposed to Ventricular Arrhythmia. PloS one 2016, 11, e0149205. [Google Scholar] [CrossRef] [PubMed]
- Borghetti, G.; Eisenberg, C.A.; Signore, S.; Sorrentino, A.; Kaur, K.; Andrade-Vicenty, A.; Edwards, J.G.; Nerkar, M.; Qanud, K.; Sun, D.; et al. Notch signaling modulates the electrical behavior of cardiomyocytes. American journal of physiology. Heart and circulatory physiology 2018, 314, H68–h81. [Google Scholar] [CrossRef]
- Kaur, K.; Zarzoso, M.; Ponce-Balbuena, D.; Guerrero-Serna, G.; Hou, L.; Musa, H.; Jalife, J. TGF-β1, released by myofibroblasts, differentially regulates transcription and function of sodium and potassium channels in adult rat ventricular myocytes. PloS one 2013, 8, e55391. [Google Scholar] [CrossRef] [PubMed]
- Panama, B.K.; Latour-Villamil, D.; Farman, G.P.; Zhao, D.; Bolz, S.S.; Kirshenbaum, L.A.; Backx, P.H. Nuclear factor kappaB downregulates the transient outward potassium current I(to,f) through control of KChIP2 expression. Circulation research 2011, 108, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Panama, B.K.; Korogyi, A.S.; Aschar-Sobbi, R.; Oh, Y.; Gray, C.B.; Gang, H.; Brown, J.H.; Kirshenbaum, L.A.; Backx, P.H. Reductions in the Cardiac Transient Outward K+ Current Ito Caused by Chronic β-Adrenergic Receptor Stimulation Are Partly Rescued by Inhibition of Nuclear Factor κB. The Journal of biological chemistry 2016, 291, 4156–4165. [Google Scholar] [CrossRef]
- Xie, Y.; Mai, J.T.; Wang, F.; Lin, Y.Q.; Yuan, W.L.; Luo, N.S.; Fang, M.C.; Wang, J.F.; Chen, Y.X. Effects of C-reactive protein on K(+) channel interaction protein 2 in cardiomyocytes. American journal of translational research 2015, 7, 922–931. [Google Scholar] [CrossRef]
- Ozgen, N.; Lau, D.H.; Shlapakova, I.N.; Sherman, W.; Feinmark, S.J.; Danilo, P., Jr.; Rosen, M.R. Determinants of CREB degradation and KChIP2 gene transcription in cardiac memory. Heart rhythm 2010, 7, 964–970. [Google Scholar] [CrossRef]
- Chowdhury, S.K.; Liu, W.; Zi, M.; Li, Y.; Wang, S.; Tsui, H.; Prehar, S.; Castro, S.; Zhang, H.; Ji, Y.; et al. Stress-Activated Kinase Mitogen-Activated Kinase Kinase-7 Governs Epigenetics of Cardiac Repolarization for Arrhythmia Prevention. Circulation 2017, 135, 683–699. [Google Scholar] [CrossRef]
- Jeyaraj, D.; Haldar, S.M.; Wan, X.; McCauley, M.D.; Ripperger, J.A.; Hu, K.; Lu, Y.; Eapen, B.L.; Sharma, N.; Ficker, E.; et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature 2012, 483, 96–99. [Google Scholar] [CrossRef]
- Menegola, M.; Trimmer, J.S. Unanticipated region- and cell-specific downregulation of individual KChIP auxiliary subunit isotypes in Kv4.2 knock-out mouse brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 2006, 26, 12137–12142. [Google Scholar] [CrossRef] [PubMed]
- Norris, A.J.; Foeger, N.C.; Nerbonne, J.M. Interdependent roles for accessory KChIP2, KChIP3, and KChIP4 subunits in the generation of Kv4-encoded IA channels in cortical pyramidal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 2010, 30, 13644–13655. [Google Scholar] [CrossRef] [PubMed]
- Nerbonne, J.M.; Gerber, B.R.; Norris, A.; Burkhalter, A. Electrical remodelling maintains firing properties in cortical pyramidal neurons lacking KCND2-encoded A-type K+ currents. The Journal of physiology 2008, 586, 1565–1579. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Maffie, J.K.; Lin, L.; Petralia, R.S.; Rudy, B.; Hoffman, D.A. DPP6 establishes the A-type K(+) current gradient critical for the regulation of dendritic excitability in CA1 hippocampal neurons. Neuron 2011, 71, 1102–1115. [Google Scholar] [CrossRef]
- Kuo, Y.L.; Cheng, J.K.; Hou, W.H.; Chang, Y.C.; Du, P.H.; Jian, J.J.; Rau, R.H.; Yang, J.H.; Lien, C.C.; Tsaur, M.L. K(+) Channel Modulatory Subunits KChIP and DPP Participate in Kv4-Mediated Mechanical Pain Control. The Journal of neuroscience : the official journal of the Society for Neuroscience 2017, 37, 4391–4404. [Google Scholar] [CrossRef]
- Mellström, B.; Sahún, I.; Ruiz-Nuño, A.; Murtra, P.; Gomez-Villafuertes, R.; Savignac, M.; Oliveros, J.C.; Gonzalez, P.; Kastanauskaite, A.; Knafo, S.; et al. DREAM controls the on/off switch of specific activity-dependent transcription pathways. Molecular and cellular biology 2014, 34, 877–887. [Google Scholar] [CrossRef]
- Kise, Y.; Kasuya, G.; Okamoto, H.H.; Yamanouchi, D.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Nakajo, K.; Nureki, O. Structural basis of gating modulation of Kv4 channel complexes. Nature 2021, 599, 158–164. [Google Scholar] [CrossRef]
- Jerng, H.H.; Pfaffinger, P.J.; Covarrubias, M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Molecular and cellular neurosciences 2004, 27, 343–369. [Google Scholar] [CrossRef]
- Patel, S.P.; Campbell, D.L. Transient outward potassium current, 'Ito', phenotypes in the mammalian left ventricle: underlying molecular, cellular and biophysical mechanisms. The Journal of physiology 2005, 569, 7–39. [Google Scholar] [CrossRef]
- Kim, L.A.; Furst, J.; Gutierrez, D.; Butler, M.H.; Xu, S.; Goldstein, S.A.; Grigorieff, N. Three-dimensional structure of I(to); Kv4.2-KChIP2 ion channels by electron microscopy at 21 Angstrom resolution. Neuron 2004, 41, 513–519. [Google Scholar] [CrossRef]
- Wang, H.; Yan, Y.; Liu, Q.; Huang, Y.; Shen, Y.; Chen, L.; Chen, Y.; Yang, Q.; Hao, Q.; Wang, K.; et al. Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits. Nature neuroscience 2007, 10, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Catte, A.; Ferbel, L.; Bhattacharjee, N.; Jan Akhunzada, M.; D'Agostino, T.; Brancato, G. In silico investigation of the interaction between the voltage-gated potassium channel Kv4.3 and its auxiliary protein KChIP1. Physical chemistry chemical physics : PCCP 2019, 21, 25290–25301. [Google Scholar] [CrossRef] [PubMed]
- Fineberg, J.D.; Szanto, T.G.; Panyi, G.; Covarrubias, M. Closed-state inactivation involving an internal gate in Kv4.1 channels modulates pore blockade by intracellular quaternary ammonium ions. Scientific reports 2016, 6, 31131. [Google Scholar] [CrossRef] [PubMed]
- Bähring, R.; Dannenberg, J.; Peters, H.C.; Leicher, T.; Pongs, O.; Isbrandt, D. Conserved Kv4 N-terminal domain critical for effects of Kv channel-interacting protein 2.2 on channel expression and gating. The Journal of biological chemistry 2001, 276, 23888–23894. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.P.; Campbell, D.L.; Strauss, H.C. Elucidating KChIP effects on Kv4.3 inactivation and recovery kinetics with a minimal KChIP2 isoform. The Journal of physiology 2002, 545, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Zhao, H.; Dai, Y.; Wang, Y.; Lo, Y.H.; Jan, L.Y.; Lee, C.H. Activation and closed-state inactivation mechanisms of the human voltage-gated K(V)4 channel complexes. Molecular cell 2022, 82, 2427–2442. [Google Scholar] [CrossRef]
- Shibata, R.; Misonou, H.; Campomanes, C.R.; Anderson, A.E.; Schrader, L.A.; Doliveira, L.C.; Carroll, K.I.; Sweatt, J.D.; Rhodes, K.J.; Trimmer, J.S. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. The Journal of biological chemistry 2003, 278, 36445–36454. [Google Scholar] [CrossRef]
- Foeger, N.C.; Marionneau, C.; Nerbonne, J.M. Co-assembly of Kv4 {alpha} subunits with K+ channel-interacting protein 2 stabilizes protein expression and promotes surface retention of channel complexes. The Journal of biological chemistry 2010, 285, 33413–33422. [Google Scholar] [CrossRef]
- Li, Y.; Duan, H.; Yi, J.; Wang, G.; Cheng, W.; Feng, L.; Liu, J. Kv4.2 phosphorylation by PKA drives Kv4.2-KChIP2 dissociation, leading to Kv4.2 out of lipid rafts and internalization. American journal of physiology. Cell physiology 2022, 323, C190–c201. [Google Scholar] [CrossRef]
- Holmqvist, M.H.; Cao, J.; Knoppers, M.H.; Jurman, M.E.; Distefano, P.S.; Rhodes, K.J.; Xie, Y.; An, W.F. Kinetic modulation of Kv4-mediated A-current by arachidonic acid is dependent on potassium channel interacting proteins. The Journal of neuroscience : the official journal of the Society for Neuroscience 2001, 21, 4154–4161. [Google Scholar] [CrossRef]
- Bowlby, M.R.; Chanda, P.; Edris, W.; Hinson, J.; Jow, F.; Katz, A.H.; Kennedy, J.; Krishnamurthy, G.; Pitts, K.; Ryan, K.; et al. Identification and characterization of small molecule modulators of KChIP/Kv4 function. Bioorganic & medicinal chemistry 2005, 13, 6112–6119. [Google Scholar] [CrossRef]
- Naranjo, J.R.; Zhang, H.; Villar, D.; González, P.; Dopazo, X.M.; Morón-Oset, J.; Higueras, E.; Oliveros, J.C.; Arrabal, M.D.; Prieto, A.; et al. Activating transcription factor 6 derepression mediates neuroprotection in Huntington disease. The Journal of clinical investigation 2016, 126, 627–638. [Google Scholar] [CrossRef]
- Lopez-Hurtado, A.; Peraza, D.A.; Cercos, P.; Lagartera, L.; Gonzalez, P.; Dopazo, X.M.; Herranz, R.; Gonzalez, T.; Martin-Martinez, M.; Mellström, B.; et al. Targeting the neuronal calcium sensor DREAM with small-molecules for Huntington's disease treatment. Scientific reports 2019, 9, 7260. [Google Scholar] [CrossRef] [PubMed]
- Peraza, D.A.; Cercós, P.; Miaja, P.; Merinero, Y.G.; Lagartera, L.; Socuéllamos, P.G.; Izquierdo García, C.; Sánchez, S.A.; López-Hurtado, A.; Martín-Martínez, M.; et al. Identification of IQM-266, a Novel DREAM Ligand That Modulates K(V)4 Currents. Frontiers in molecular neuroscience 2019, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- de Benito-Bueno, A.; Socuellamos, P.G.; Merinero, Y.G.; Cercos, P.; Izquierdo, C.; Daniel-Mozo, M.; Marín-Olivero, I.; Perez-Lara, A.; Gonzalez-Vera, J.A.; Orte, A.; et al. Modulation of K(V)4.3-KChIP2 Channels by IQM-266: Role of DPP6 and KCNE2. International journal of molecular sciences 2022, 23. [Google Scholar] [CrossRef]
- Witzel, K.; Fischer, P.; Bähring, R. Hippocampal A-type current and Kv4.2 channel modulation by the sulfonylurea compound NS5806. Neuropharmacology 2012, 63, 1389–1403. [Google Scholar] [CrossRef]
- Gonzalez, W.G.; Pham, K.; Miksovska, J. Modulation of the voltage-gated potassium channel (Kv4.3) and the auxiliary protein (KChIP3) interactions by the current activator NS5806. The Journal of biological chemistry 2014, 289, 32201–32213. [Google Scholar] [CrossRef]
- Calloe, K.; Soltysinska, E.; Jespersen, T.; Lundby, A.; Antzelevitch, C.; Olesen, S.P.; Cordeiro, J.M. Differential effects of the transient outward K(+) current activator NS5806 in the canine left ventricle. Journal of molecular and cellular cardiology 2010, 48, 191–200. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Wang, C.; Wang, Y.; Zou, R.; Shi, C.; Guan, B.; Gamper, N.; Xu, Y. Auxiliary subunits control biophysical properties and response to compound NS5806 of the Kv4 potassium channel complex. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2020, 34, 807–821. [Google Scholar] [CrossRef]
- Cheng, H.; Cannell, M.B.; Hancox, J.C. Differential responses of rabbit ventricular and atrial transient outward current (I(to)) to the I(to) modulator NS5806. Physiological reports 2017, 5. [Google Scholar] [CrossRef]
- Ravens, U.; Wettwer, E. Ultra-rapid delayed rectifier channels: molecular basis and therapeutic implications. Cardiovascular research 2011, 89, 776–785. [Google Scholar] [CrossRef]
- Li, H.; Guo, W.; Mellor, R.L.; Nerbonne, J.M. KChIP2 modulates the cell surface expression of Kv 1.5-encoded K(+) channels. Journal of molecular and cellular cardiology 2005, 39, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.B.; Sosunov, E.A.; Anyukhovsky, E.P.; Ozgen, N.; Boyden, P.A.; Rosen, M.R. Deleting the accessory subunit KChIP2 results in loss of I(to,f) and increased I(K,slow) that maintains normal action potential configuration. Heart rhythm 2009, 6, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, F.; Flockerzi, V.; Kahl, S.; Wegener, J.W. L-type CaV1.2 calcium channels: from in vitro findings to in vivo function. Physiological reviews 2014, 94, 303–326. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.B.; Wang, C.; Ozgen, N.; Wang, H.G.; Rosen, M.R.; Pitt, G.S. Accessory subunit KChIP2 modulates the cardiac L-type calcium current. Circulation research 2009, 104, 1382–1389. [Google Scholar] [CrossRef]
- Thomsen, M.B.; Foster, E.; Nguyen, K.H.; Sosunov, E.A. Transcriptional and electrophysiological consequences of KChIP2-mediated regulation of CaV1.2. Channels (Austin, Tex.) 2009, 3, 308–310. [Google Scholar] [CrossRef]
- Ronkainen, J.J.; Hänninen, S.L.; Korhonen, T.; Koivumäki, J.T.; Skoumal, R.; Rautio, S.; Ronkainen, V.P.; Tavi, P. Ca2+-calmodulin-dependent protein kinase II represses cardiac transcription of the L-type calcium channel alpha(1C)-subunit gene (Cacna1c) by DREAM translocation. The Journal of physiology 2011, 589, 2669–2686. [Google Scholar] [CrossRef] [PubMed]
- Clatot, J.; Neyroud, N.; Cox, R.; Souil, C.; Huang, J.; Guicheney, P.; Antzelevitch, C. Inter-Regulation of K(v)4.3 and Voltage-Gated Sodium Channels Underlies Predisposition to Cardiac and Neuronal Channelopathies. International journal of molecular sciences 2020, 21. [Google Scholar] [CrossRef]
- Deschênes, I.; Armoundas, A.A.; Jones, S.P.; Tomaselli, G.F. Post-transcriptional gene silencing of KChIP2 and Navbeta1 in neonatal rat cardiac myocytes reveals a functional association between Na and Ito currents. Journal of molecular and cellular cardiology 2008, 45, 336–346. [Google Scholar] [CrossRef]
- Osawa, M.; Dace, A.; Tong, K.I.; Valiveti, A.; Ikura, M.; Ames, J.B. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. The Journal of biological chemistry 2005, 280, 18008–18014. [Google Scholar] [CrossRef]
- Lusin, J.D.; Vanarotti, M.; Li, C.; Valiveti, A.; Ames, J.B. NMR structure of DREAM: Implications for Ca(2+)-dependent DNA binding and protein dimerization. Biochemistry 2008, 47, 2252–2264. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.F.; Kuplast, K.G.; Washicosky, K.J.; Kajiwara, Y.; Buxbaum, J.D.; Wasco, W. Calsenilin interacts with transcriptional co-repressor C-terminal binding protein(s). Journal of neurochemistry 2006, 98, 1290–1301. [Google Scholar] [CrossRef] [PubMed]
- Ledo, F.; Carrión, A.M.; Link, W.A.; Mellström, B.; Naranjo, J.R. DREAM-alphaCREM interaction via leucine-charged domains derepresses downstream regulatory element-dependent transcription. Molecular and cellular biology 2000, 20, 9120–9126. [Google Scholar] [CrossRef] [PubMed]
- Ledo, F.; Kremer, L.; Mellström, B.; Naranjo, J.R. Ca2+-dependent block of CREB-CBP transcription by repressor DREAM. The EMBO journal 2002, 21, 4583–4592. [Google Scholar] [CrossRef]
- Rivas, M.; Mellström, B.; Naranjo, J.R.; Santisteban, P. Transcriptional repressor DREAM interacts with thyroid transcription factor-1 and regulates thyroglobulin gene expression. The Journal of biological chemistry 2004, 279, 33114–33122. [Google Scholar] [CrossRef]
- Scsucova, S.; Palacios, D.; Savignac, M.; Mellström, B.; Naranjo, J.R.; Aranda, A. The repressor DREAM acts as a transcriptional activator on Vitamin D and retinoic acid response elements. Nucleic acids research 2005, 33, 2269–2279. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Pitcher, G.M.; Laviolette, S.R.; Whishaw, I.Q.; Tong, K.I.; Kockeritz, L.K.; Wada, T.; Joza, N.A.; Crackower, M.; Goncalves, J.; et al. DREAM is a critical transcriptional repressor for pain modulation. Cell 2002, 108, 31–43. [Google Scholar] [CrossRef]
- Sanz, C.; Mellstrom, B.; Link, W.A.; Naranjo, J.R.; Fernandez-Luna, J.L. Interleukin 3-dependent activation of DREAM is involved in transcriptional silencing of the apoptotic Hrk gene in hematopoietic progenitor cells. The EMBO journal 2001, 20, 2286–2292. [Google Scholar] [CrossRef]
- Rivera-Arconada, I.; Benedet, T.; Roza, C.; Torres, B.; Barrio, J.; Krzyzanowska, A.; Avendaño, C.; Mellström, B.; Lopez-Garcia, J.A.; Naranjo, J.R. DREAM regulates BDNF-dependent spinal sensitization. Molecular pain 2010, 6, 95. [Google Scholar] [CrossRef]
- Savignac, M.; Pintado, B.; Gutierrez-Adan, A.; Palczewska, M.; Mellström, B.; Naranjo, J.R. Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. The EMBO journal 2005, 24, 3555–3564. [Google Scholar] [CrossRef] [PubMed]
- Savignac, M.; Mellström, B.; Bébin, A.G.; Oliveros, J.C.; Delpy, L.; Pinaud, E.; Naranjo, J.R. Increased B cell proliferation and reduced Ig production in DREAM transgenic mice. Journal of immunology (Baltimore, Md. : 1950) 2010, 185, 7527–7536. [Google Scholar] [CrossRef] [PubMed]
- Tiruppathi, C.; Soni, D.; Wang, D.M.; Xue, J.; Singh, V.; Thippegowda, P.B.; Cheppudira, B.P.; Mishra, R.K.; Debroy, A.; Qian, Z.; et al. The transcription factor DREAM represses the deubiquitinase A20 and mediates inflammation. Nature immunology 2014, 15, 239–247. [Google Scholar] [CrossRef] [PubMed]
- D'Andrea, B.; Di Palma, T.; Mascia, A.; Motti, M.L.; Viglietto, G.; Nitsch, L.; Zannini, M. The transcriptional repressor DREAM is involved in thyroid gene expression. Experimental cell research 2005, 305, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Link, W.A.; Ledo, F.; Torres, B.; Palczewska, M.; Madsen, T.M.; Savignac, M.; Albar, J.P.; Mellström, B.; Naranjo, J.R. Day-night changes in downstream regulatory element antagonist modulator/potassium channel interacting protein activity contribute to circadian gene expression in pineal gland. The Journal of neuroscience : the official journal of the Society for Neuroscience 2004, 24, 5346–5355. [Google Scholar] [CrossRef]
- Leclerc, G.M.; Boockfor, F.R. Calcium influx and DREAM protein are required for GnRH gene expression pulse activity. Molecular and cellular endocrinology 2007, 267, 70–79. [Google Scholar] [CrossRef]
- Cebolla, B.; Fernández-Pérez, A.; Perea, G.; Araque, A.; Vallejo, M. DREAM mediates cAMP-dependent, Ca2+-induced stimulation of GFAP gene expression and regulates cortical astrogliogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience 2008, 28, 6703–6713. [Google Scholar] [CrossRef]
- Gomez-Villafuertes, R.; Torres, B.; Barrio, J.; Savignac, M.; Gabellini, N.; Rizzato, F.; Pintado, B.; Gutierrez-Adan, A.; Mellström, B.; Carafoli, E.; et al. Downstream regulatory element antagonist modulator regulates Ca2+ homeostasis and viability in cerebellar neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 2005, 25, 10822–10830. [Google Scholar] [CrossRef]
- Dierssen, M.; Fedrizzi, L.; Gomez-Villafuertes, R.; de Lagran, M.M.; Gutierrez-Adan, A.; Sahún, I.; Pintado, B.; Oliveros, J.C.; Dopazo, X.M.; Gonzalez, P.; et al. Reduced Mid1 Expression and Delayed Neuromotor Development in daDREAM Transgenic Mice. Frontiers in molecular neuroscience 2012, 5, 58. [Google Scholar] [CrossRef]
- Calì, T.; Fedrizzi, L.; Ottolini, D.; Gomez-Villafuertes, R.; Mellström, B.; Naranjo, J.R.; Carafoli, E.; Brini, M. Ca2+-activated nucleotidase 1, a novel target gene for the transcriptional repressor DREAM (downstream regulatory element antagonist modulator), is involved in protein folding and degradation. The Journal of biological chemistry 2012, 287, 18478–18491. [Google Scholar] [CrossRef]
- Fontán-Lozano, A.; Capilla-Gonzalez, V.; Aguilera, Y.; Mellado, N.; Carrión, A.M.; Soria, B.; Hmadcha, A. Impact of transient down-regulation of DREAM in human embryonic stem cell pluripotency: The role of DREAM in the maintenance of hESCs. Stem cell research 2016, 16, 568–578. [Google Scholar] [CrossRef]
- Baczyk, D.; Kibschull, M.; Mellstrom, B.; Levytska, K.; Rivas, M.; Drewlo, S.; Lye, S.J.; Naranjo, J.R.; Kingdom, J.C. DREAM mediated regulation of GCM1 in the human placental trophoblast. PloS one 2013, 8, e51837. [Google Scholar] [CrossRef]
- Jacobson, D.A.; Cho, J.; Landa, L.R., Jr.; Tamarina, N.A.; Roe, M.W.; Buxbaum, J.D.; Philipson, L.H. Downstream regulatory element antagonistic modulator regulates islet prodynorphin expression. American journal of physiology. Endocrinology and metabolism 2006, 291, E587–595. [Google Scholar] [CrossRef]
- Li, J.; Kumari, T.; Barazia, A.; Jha, V.; Jeong, S.Y.; Olson, A.; Kim, M.; Lee, B.K.; Manickam, V.; Song, Z.; et al. Neutrophil DREAM promotes neutrophil recruitment in vascular inflammation. The Journal of experimental medicine 2022, 219. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, I.E.; Berkovic, S.; Capovilla, G.; Connolly, M.B.; French, J.; Guilhoto, L.; Hirsch, E.; Jain, S.; Mathern, G.W.; Moshé, S.L.; et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017, 58, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Maizuliana, H.; Usui, N.; Terada, K.; Kondo, A.; Inoue, Y. Clinical, semiological, electroencephalographic, and neuropsychological features of "pure" neocortical temporal lobe epilepsy. Epileptic disorders : international epilepsy journal with videotape 2020, 22, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Maillard, L.; Vignal, J.P.; Gavaret, M.; Guye, M.; Biraben, A.; McGonigal, A.; Chauvel, P.; Bartolomei, F. Semiologic and electrophysiologic correlations in temporal lobe seizure subtypes. Epilepsia 2004, 45, 1590–1599. [Google Scholar] [CrossRef] [PubMed]
- Villa, C.; Combi, R. Potassium Channels and Human Epileptic Phenotypes: An Updated Overview. Frontiers in cellular neuroscience 2016, 10, 81. [Google Scholar] [CrossRef]
- Hong, Y.M.; Jo, D.G.; Lee, M.C.; Kim, S.Y.; Jung, Y.K. Reduced expression of calsenilin/DREAM/KChIP3 in the brains of kainic acid-induced seizure and epilepsy patients. Neuroscience letters 2003, 340, 33–36. [Google Scholar] [CrossRef]
- Wang, H.G.; He, X.P.; Li, Q.; Madison, R.D.; Moore, S.D.; McNamara, J.O.; Pitt, G.S. The auxiliary subunit KChIP2 is an essential regulator of homeostatic excitability. The Journal of biological chemistry 2013, 288, 13258–13268. [Google Scholar] [CrossRef]
- Monaghan, M.M.; Menegola, M.; Vacher, H.; Rhodes, K.J.; Trimmer, J.S. Altered expression and localization of hippocampal A-type potassium channel subunits in the pilocarpine-induced model of temporal lobe epilepsy. Neuroscience 2008, 156, 550–562. [Google Scholar] [CrossRef]
- Kuner, R. Central mechanisms of pathological pain. Nature medicine 2010, 16, 1258–1266. [Google Scholar] [CrossRef]
- Costigan, M.; Woolf, C.J. No DREAM, No pain. Closing the spinal gate. Cell 2002, 108, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, Z.; Yu, P.; Li, J.; Bai, N.; He, Z.; Yang, S.; Guo, Q. [Construction of shRNA lentivirus vector on rat DREAM gene and its analgesic effect on CCI rats]. Zhong nan da xue xue bao. Yi xue ban = Journal of Central South University. Medical sciences 2009, 34, 723–730. [Google Scholar] [PubMed]
- Benedet, T.; Gonzalez, P.; Oliveros, J.C.; Dopazo, J.M.; Ghimire, K.; Palczewska, M.; Mellstrom, B.; Naranjo, J.R. Transcriptional repressor DREAM regulates trigeminal noxious perception. Journal of neurochemistry 2017, 141, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Tian, N.X.; Xu, Y.; Yang, J.Y.; Li, L.; Sun, X.H.; Wang, Y.; Zhang, Y. KChIP3 N-Terminal 31-50 Fragment Mediates Its Association with TRPV1 and Alleviates Inflammatory Hyperalgesia in Rats. The Journal of neuroscience : the official journal of the Society for Neuroscience 2018, 38, 1756–1773. [Google Scholar] [CrossRef] [PubMed]
- Matsuyoshi, H.; Takimoto, K.; Yunoki, T.; Erickson, V.L.; Tyagi, P.; Hirao, Y.; Wanaka, A.; Yoshimura, N. Distinct cellular distributions of Kv4 pore-forming and auxiliary subunits in rat dorsal root ganglion neurons. Life sciences 2012, 91, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Tseng, P.Y.; Hoon, M.A. Molecular Genetics of Kappa Opioids in Pain and Itch Sensations. Handbook of experimental pharmacology 2022, 271, 255–274. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, P.; Liang, P.; Liu, T.; Liu, X.; Liu, X.Y.; Zhang, B.; Han, T.; Zhu, Y.B.; Yin, D.M.; et al. The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. The Journal of neuroscience : the official journal of the Society for Neuroscience 2010, 30, 7575–7586. [Google Scholar] [CrossRef]
- Cheng, C.F.; Wang, W.C.; Huang, C.Y.; Du, P.H.; Yang, J.H.; Tsaur, M.L. Coexpression of auxiliary subunits KChIP and DPPL in potassium channel Kv4-positive nociceptors and pain-modulating spinal interneurons. The Journal of comparative neurology 2016, 524, 846–873. [Google Scholar] [CrossRef]
- Anderson, D.; Mehaffey, W.H.; Iftinca, M.; Rehak, R.; Engbers, J.D.; Hameed, S.; Zamponi, G.W.; Turner, R.W. Regulation of neuronal activity by Cav3-Kv4 channel signaling complexes. Nature neuroscience 2010, 13, 333–337. [Google Scholar] [CrossRef]
- Socuéllamos, P.G.; Olivos-Oré, L.A.; Barahona, M.V.; Cercós, P.; Pérez Pascual, M.; Arribas-Blázquez, M.; Naranjo, J.R.; Valenzuela, C.; Gutiérrez-Rodríguez, M.; Artalejo, A.R. IQM-PC332, a Novel DREAM Ligand with Antinociceptive Effect on Peripheral Nerve Injury-Induced Pain. International journal of molecular sciences 2022, 23. [Google Scholar] [CrossRef]
- Guo, Y.P.; Zhi, Y.R.; Liu, T.T.; Wang, Y.; Zhang, Y. Global Gene Knockout of Kcnip3 Enhances Pain Sensitivity and Exacerbates Negative Emotions in Rats. Frontiers in molecular neuroscience 2019, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zuo, Y. Synaptic modifications in learning and memory - A dendritic spine story. Seminars in cell & developmental biology 2022, 125, 84–90. [Google Scholar] [CrossRef]
- Bin Ibrahim, M.Z.; Benoy, A.; Sajikumar, S. Long-term plasticity in the hippocampus: maintaining within and 'tagging' between synapses. The FEBS journal 2022, 289, 2176–2201. [Google Scholar] [CrossRef]
- Lisman, J.; Buzsáki, G.; Eichenbaum, H.; Nadel, L.; Ranganath, C.; Redish, A.D. Viewpoints: how the hippocampus contributes to memory, navigation and cognition. Nature neuroscience 2017, 20, 1434–1447. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.C.; McDermott, C.M.; Tunur, T.; Rands, V.; Stelly, C.; Karhson, D.; Bowlby, M.R.; An, W.F.; Sweatt, J.D.; Schrader, L.A. The role of calsenilin/DREAM/KChIP3 in contextual fear conditioning. Learning & memory (Cold Spring Harbor, N.Y.) 2009, 16, 167–177. [Google Scholar] [CrossRef]
- Fontán-Lozano, A.; Romero-Granados, R.; del-Pozo-Martín, Y.; Suárez-Pereira, I.; Delgado-García, J.M.; Penninger, J.M.; Carrión, A.M. Lack of DREAM protein enhances learning and memory and slows brain aging. Current biology : CB 2009, 19, 54–60. [Google Scholar] [CrossRef]
- Chen, Q.Y.; Li, X.H.; Zhuo, M. NMDA receptors and synaptic plasticity in the anterior cingulate cortex. Neuropharmacology 2021, 197, 108749. [Google Scholar] [CrossRef]
- Dore, K.; Malinow, R. Elevated PSD-95 Blocks Ion-flux Independent LTD: A Potential New Role for PSD-95 in Synaptic Plasticity. Neuroscience 2021, 456, 43–49. [Google Scholar] [CrossRef]
- Wu, L.J.; Mellström, B.; Wang, H.; Ren, M.; Domingo, S.; Kim, S.S.; Li, X.Y.; Chen, T.; Naranjo, J.R.; Zhuo, M. DREAM (downstream regulatory element antagonist modulator) contributes to synaptic depression and contextual fear memory. Molecular brain 2010, 3, 3. [Google Scholar] [CrossRef]
- Lugo, J.N.; Brewster, A.L.; Spencer, C.M.; Anderson, A.E. Kv4.2 knockout mice have hippocampal-dependent learning and memory deficits. Learning & memory (Cold Spring Harbor, N.Y.) 2012, 19, 182–189. [Google Scholar] [CrossRef]
- Soria Lopez, J.A.; González, H.M.; Léger, G.C. Alzheimer's disease. Handbook of clinical neurology 2019, 167, 231–255. [Google Scholar] [CrossRef]
- Robakis, N.K. Cell signaling abnormalities may drive neurodegeneration in familial Alzheimer disease. Neurochemical research 2014, 39, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Pham, K.; Miksovska, J. Molecular insight of DREAM and presenilin 1 C-terminal fragment interactions. FEBS letters 2016, 590, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Levy-Lahad, E.; Wasco, W.; Poorkaj, P.; Romano, D.M.; Oshima, J.; Pettingell, W.H.; Yu, C.E.; Jondro, P.D.; Schmidt, S.D.; Wang, K.; et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science (New York, N.Y.) 1995, 269, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.G.; Lee, J.Y.; Hong, Y.M.; Song, S.; Mook-Jung, I.; Koh, J.Y.; Jung, Y.K. Induction of pro-apoptotic calsenilin/DREAM/KChIP3 in Alzheimer's disease and cultured neurons after amyloid-beta exposure. Journal of neurochemistry 2004, 88, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.F.; Berezovska, O.; Choi, E.K.; Miller, J.S.; Chan, H.; Lilliehook, C.; Hyman, B.T.; Buxbaum, J.D.; Wasco, W. Biochemical and immunocytochemical characterization of calsenilin in mouse brain. Neuroscience 2002, 114, 247–263. [Google Scholar] [CrossRef]
- Jo, D.G.; Chang, J.W.; Hong, H.S.; Mook-Jung, I.; Jung, Y.K. Contribution of presenilin/gamma-secretase to calsenilin-mediated apoptosis. Biochemical and biophysical research communications 2003, 305, 62–66. [Google Scholar] [CrossRef]
- Jo, D.G.; Jang, J.; Kim, B.J.; Lundkvist, J.; Jung, Y.K. Overexpression of calsenilin enhances gamma-secretase activity. Neuroscience letters 2005, 378, 59–64. [Google Scholar] [CrossRef]
- Naranjo, R.; González, P.; Lopez-Hurtado, A.; Dopazo, X.M.; Mellström, B.; Naranjo, J.R. Inhibition of the Neuronal Calcium Sensor DREAM Modulates Presenilin-2 Endoproteolysis. Frontiers in molecular neuroscience 2018, 11, 449. [Google Scholar] [CrossRef]
- Cascella, R.; Cecchi, C. Calcium Dyshomeostasis in Alzheimer's Disease Pathogenesis. International journal of molecular sciences 2021, 22. [Google Scholar] [CrossRef]
- Ferreiro, E.; Resende, R.; Costa, R.; Oliveira, C.R.; Pereira, C.M. An endoplasmic-reticulum-specific apoptotic pathway is involved in prion and amyloid-beta peptides neurotoxicity. Neurobiology of disease 2006, 23, 669–678. [Google Scholar] [CrossRef]
- Lilliehook, C.; Chan, S.; Choi, E.K.; Zaidi, N.F.; Wasco, W.; Mattson, M.P.; Buxbaum, J.D. Calsenilin enhances apoptosis by altering endoplasmic reticulum calcium signaling. Molecular and cellular neurosciences 2002, 19, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Fedrizzi, L.; Lim, D.; Carafoli, E.; Brini, M. Interplay of the Ca2+-binding protein DREAM with presenilin in neuronal Ca2+ signaling. The Journal of biological chemistry 2008, 283, 27494–27503. [Google Scholar] [CrossRef] [PubMed]
- Grillo, M.A.; Grillo, S.L.; Gerdes, B.C.; Kraus, J.G.; Koulen, P. Control of Neuronal Ryanodine Receptor-Mediated Calcium Signaling by Calsenilin. Molecular neurobiology 2019, 56, 525–534. [Google Scholar] [CrossRef]
- López-Hurtado, A.; Burgos, D.F.; González, P.; Dopazo, X.M.; González, V.; Rábano, A.; Mellström, B.; Naranjo, J.R. Inhibition of DREAM-ATF6 interaction delays onset of cognition deficit in a mouse model of Huntington's disease. Molecular brain 2018, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Larrodé, P.; Calvo, A.C.; Moreno-Martínez, L.; de la Torre, M.; Moreno-García, L.; Molina, N.; Castiella, T.; Iñiguez, C.; Pascual, L.F.; Mena, F.J.M.; et al. DREAM-Dependent Activation of Astrocytes in Amyotrophic Lateral Sclerosis. Molecular neurobiology 2018, 55, 1–12. [Google Scholar] [CrossRef]
- Singh, D. Astrocytic and microglial cells as the modulators of neuroinflammation in Alzheimer's disease. Journal of neuroinflammation 2022, 19, 206. [Google Scholar] [CrossRef]
- Tse, G. Mechanisms of cardiac arrhythmias. Journal of arrhythmia 2016, 32, 75–81. [Google Scholar] [CrossRef]
- Kant, R.; Hu, Z.; Malhotra, J.K.; Krogh-Madsen, T.; Christini, D.J.; Heerdt, P.M.; Abbott, G.W. NHE isoform switching and KChIP2 upregulation in aging porcine atria. PloS one 2013, 8, e82951. [Google Scholar] [CrossRef]
- Monnerat-Cahli, G.; Alonso, H.; Gallego, M.; Alarcón, M.L.; Bassani, R.A.; Casis, O.; Medei, E. Toll-like receptor 4 activation promotes cardiac arrhythmias by decreasing the transient outward potassium current (Ito) through an IRF3-dependent and MyD88-independent pathway. Journal of molecular and cellular cardiology 2014, 76, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Liu, T.; Li, Y.; Qin, M.; Tang, Y.; Shen, J.Y.; Liang, J.; Yang, B.; Huang, C. Chronic N-methyl-D-aspartate receptor activation induces cardiac electrical remodeling and increases susceptibility to ventricular arrhythmias. Pacing and clinical electrophysiology : PACE 2014, 37, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.T.; Hsieh, C.S.; Chang, S.N.; Chuang, E.Y.; Ueng, K.C.; Tsai, C.F.; Lin, T.H.; Wu, C.K.; Lee, J.K.; Lin, L.Y.; et al. Genome-wide screening identifies a KCNIP1 copy number variant as a genetic predictor for atrial fibrillation. Nature communications 2016, 7, 10190. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.C.; Cheng, C.F.; Clark, R.B.; Lin, J.J.; Lin, J.L.; Hoshijima, M.; Nguyêñ-Trân, V.T.; Gu, Y.; Ikeda, Y.; Chu, P.H.; et al. A defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of I(to) and confers susceptibility to ventricular tachycardia. Cell 2001, 107, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Foeger, N.C.; Wang, W.; Mellor, R.L.; Nerbonne, J.M. Stabilization of Kv4 protein by the accessory K(+) channel interacting protein 2 (KChIP2) subunit is required for the generation of native myocardial fast transient outward K(+) currents. The Journal of physiology 2013, 591, 4149–4166. [Google Scholar] [CrossRef]
- Nassal, D.M.; Wan, X.; Liu, H.; Maleski, D.; Ramirez-Navarro, A.; Moravec, C.S.; Ficker, E.; Laurita, K.R.; Deschênes, I. KChIP2 is a core transcriptional regulator of cardiac excitability. eLife 2017, 6. [Google Scholar] [CrossRef]
- Cohn, J.N.; Ferrari, R.; Sharpe, N. Cardiac remodeling--concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. Behalf of an International Forum on Cardiac Remodeling. Journal of the American College of Cardiology 2000, 35, 569–582. [Google Scholar] [CrossRef]
- Liu, X.S.; Jiang, M.; Zhang, M.; Tang, D.; Clemo, H.F.; Higgins, R.S.; Tseng, G.N. Electrical remodeling in a canine model of ischemic cardiomyopathy. American journal of physiology. Heart and circulatory physiology 2007, 292, H560–571. [Google Scholar] [CrossRef]
- Wakisaka, Y.; Niwano, S.; Niwano, H.; Saito, J.; Yoshida, T.; Hirasawa, S.; Kawada, H.; Izumi, T. Structural and electrical ventricular remodeling in rat acute myocarditis and subsequent heart failure. Cardiovascular research 2004, 63, 689–699. [Google Scholar] [CrossRef]
- Oh, S.; Kim, K.B.; Ahn, H.; Cho, H.J.; Choi, Y.S. Remodeling of ion channel expression in patients with chronic atrial fibrillation and mitral valvular heart disease. The Korean journal of internal medicine 2010, 25, 377–385. [Google Scholar] [CrossRef]
- Kawada, H.; Niwano, S.; Niwano, H.; Yumoto, Y.; Wakisaka, Y.; Yuge, M.; Kawahara, K.; Izumi, T. Tumor necrosis factor-alpha downregulates the voltage gated outward K+ current in cultured neonatal rat cardiomyocytes: a possible cause of electrical remodeling in diseased hearts. Circulation journal : official journal of the Japanese Circulation Society 2006, 70, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.Z.; Jiang, H.; Li, L.; Xie, P.; Li, J.Y.; Lu, Z.B.; He, B. Semaphorin 3A attenuates electrical remodeling at infarct border zones in rats after myocardial infarction. The Tohoku journal of experimental medicine 2011, 225, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Kobayashi, T.; Kuno, A.; Miki, T.; Tanno, M.; Kouzu, H.; Itoh, T.; Ishikawa, S.; Kojima, T.; Miura, T.; et al. Type 2 diabetes induces subendocardium predominant reduction in transient outward K+ current with downregulation of Kv4.2 and KChIP2. American journal of physiology. Heart and circulatory physiology 2014, 306, H1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Näbauer, M.; Kääb, S. Potassium channel down-regulation in heart failure. Cardiovascular research 1998, 37, 324–334. [Google Scholar] [CrossRef]
- Jin, H.; Hadri, L.; Palomeque, J.; Morel, C.; Karakikes, I.; Kaprielian, R.; Hajjar, R.; Lebeche, D. KChIP2 attenuates cardiac hypertrophy through regulation of Ito and intracellular calcium signaling. Journal of molecular and cellular cardiology 2010, 48, 1169–1179. [Google Scholar] [CrossRef]
- Chiale, P.A.; Etcheverry, D.; Pastori, J.D.; Fernandez, P.A.; Garro, H.A.; González, M.D.; Elizari, M.V. The multiple electrocardiographic manifestations of ventricular repolarization memory. Current cardiology reviews 2014, 10, 190–201. [Google Scholar] [CrossRef]
- Yu, H.; McKinnon, D.; Dixon, J.E.; Gao, J.; Wymore, R.; Cohen, I.S.; Danilo, P., Jr.; Shvilkin, A.; Anyukhovsky, E.P.; Sosunov, E.A.; et al. Transient outward current, Ito1, is altered in cardiac memory. Circulation 1999, 99, 1898–1905. [Google Scholar] [CrossRef]
- Radicke, S.; Cotella, D.; Graf, E.M.; Banse, U.; Jost, N.; Varró, A.; Tseng, G.N.; Ravens, U.; Wettwer, E. Functional modulation of the transient outward current Ito by KCNE beta-subunits and regional distribution in human non-failing and failing hearts. Cardiovascular research 2006, 71, 695–703. [Google Scholar] [CrossRef]
- Choudhury, S.; Schnell, M.; Bühler, T.; Reinke, Y.; Lüdemann, J.; Nießner, F.; Brinkmeier, H.; Herda, L.R.; Staudt, A.; Kroemer, H.K.; et al. Antibodies against potassium channel interacting protein 2 induce necrosis in isolated rat cardiomyocytes. Journal of cellular biochemistry 2014, 115, 678–689. [Google Scholar] [CrossRef]
- Odagiri, F.; Inoue, H.; Sugihara, M.; Suzuki, T.; Murayama, T.; Shioya, T.; Konishi, M.; Nakazato, Y.; Daida, H.; Sakurai, T.; et al. Effects of candesartan on electrical remodeling in the hearts of inherited dilated cardiomyopathy model mice. PloS one 2014, 9, e101838. [Google Scholar] [CrossRef]
- Nassal, D.M.; Wan, X.; Liu, H.; Deschênes, I. Myocardial KChIP2 Expression in Guinea Pig Resolves an Expanded Electrophysiologic Role. PloS one 2016, 11, e0146561. [Google Scholar] [CrossRef]
- Nassal, D.M.; Wan, X.; Liu, H.; Laurita, K.R.; Deschênes, I. KChIP2 regulates the cardiac Ca2+ transient and myocyte contractility by targeting ryanodine receptor activity. PloS one 2017, 12, e0175221. [Google Scholar] [CrossRef]
- Speerschneider, T.; Grubb, S.; Metoska, A.; Olesen, S.P.; Calloe, K.; Thomsen, M.B. Development of heart failure is independent of K+ channel-interacting protein 2 expression. The Journal of physiology 2013, 591, 5923–5937. [Google Scholar] [CrossRef] [PubMed]
- Grubb, S.; Speerschneider, T.; Occhipinti, D.; Fiset, C.; Olesen, S.P.; Thomsen, M.B.; Calloe, K. Loss of K+ currents in heart failure is accentuated in KChIP2 deficient mice. Journal of cardiovascular electrophysiology 2014, 25, 896–904. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




